Abstract
Ex vivo high-resolution magic-angle spinning (HRMAS) provides metabolic information with higher sensitivity and spectral resolution than in vivo magnetic resonance spectroscopy (MRS). Therefore, we used both techniques to better characterize the metabolic pattern of the infarct and the neural progenitor cells (NPCs) in the ipsilateral subventricular zone (SVZi). Ischemic stroke rats were divided into three groups: G0 (non-stroke controls, n=6), G1 (day 1 after stroke, n=6), and G7 (days 6 to 8 after stroke, n=12). All the rats underwent MRS. Three rats per group were analyzed by HRMAS. The remaining rats were used for immunohistochemical studies. In the infarct, both techniques detected significant metabolic changes. The most relevant change was in mobile lipids (2.80 ppm) in the G7 group (a 5.53- and a 3.95-fold increase by MRS and HRMAS, respectively). In the SVZi, MRS did not detect any significant metabolic change. However, HRMAS detected a 2.70-fold increase in lactate and a 0.68-fold decrease in N-acetylaspartate in the G1 group. None of the metabolites correlated with the 1.37-fold increase in NPCs detected by immunohistochemistry in the G7 group. In conclusion, HRMAS improves the metabolic characterization of the brain in experimental ischemic stroke. However, none of the metabolites qualifies as a surrogate biomarker of NPCs.
Keywords: HRMAS, MRS, neurogenesis, spectroscopy, stroke
Introduction
Ischemic stroke changes the metabolic pattern in the territory irrigated by the occluded artery (infarct area) and in other regions such as the ipsilateral subventricular zone (SVZi) where neural progenitor cells (NPCs) proliferation is enhanced in the mammalian1 and human2 brains.
In vivo magnetic resonance spectroscopy (MRS) is used commonly to assess the metabolic pattern either in the clinical or preclinical studies of stroke. For example, an increase in lactate (Lac; 1.30 ppm) related to anerobic metabolism and a decrease in N-acetylaspartate (NAA; 2.02 ppm) related to neuronal dysfunction are found consistently in the infarct area.3, 4, 5
Regarding NPCs proliferation, a resonance at 1.28 ppm was described as an in vivo MRS biomarker of NPCs in the hippocampus of rats and humans.6 However, many authors disagree with this interpretation7, 8, 9, 10, 11 as this resonance has been found in different cell types and during different developmental stages.12
The 1.28 ppm resonance corresponds to mobile lipids (MLs). Mobile lipid resonances arise from fatty acyl chains of neutral lipids from cytosolic droplets in the cells.13 Different functional groups of MLs give rise to resonances in the proton spectra: ML1 (methyl of fatty acids, 0.90 ppm) and ML2 (methylene of fatty acids, 1.28 ppm), which are related to cell proliferation14 and ML3 (bis-alyllic protons of polyunsaturated fatty acids, 2.80 ppm), which is related to cell apoptosis.15, 16
Therefore, although a specific metabolic pattern of NPCs may be detectable by in vivo MRS, there are two main limitations. First, a very low number of NPCs might not be detectable. Second, in vivo MRS has limited spectral resolution. For example, it is unable to differentiate between Lac and ML2 resonances that overlap at short echo time, without additional post-processing of the spectral pattern.3, 4, 17, 18
To overcome these limitations, we used an experimental stroke model with a big infarct volume to increase the number of NPCs, because maximum proliferation occurs around day 7 after stroke. We analyzed the subventricular zones (SVZs) instead of the hippocampus because it is the major NPCs reservoir in the brain.19 We used ex vivo high-resolution magic-angle spinning (HRMAS) spectroscopy to achieve more spectral resolution than with in vivo MRS.
High-resolution magic-angle spinning allows the analysis of tissue samples. By spinning the sample at the magic angle (54.7º), line broadening effects because of dipolar interactions are removed resulting in high-resolution quality spectra.20
Nonetheless, in ex vivo HRMAS, postmortem metabolic changes could affect the final metabolic pattern. These changes in animal models can be avoided using focused microwave irradiation (FMW) for euthanasia. This device delivers pulsed microwaves that rapidly elevate brain temperature to 80 to 90 °C leading to an irreversible inactivation of enzymes causing metabolism arrest.21 However, to our knowledge, there are no studies using this methodology in experimental stroke.
The aim of this study was to perform in vivo MRS and complementary ex vivo HRMAS with prior FMW irradiation to demonstrate that this method is useful in stroke experimental studies to better characterize the metabolic changes in the infarct area and the SVZi.
Materials and Methods
Animal Preparation
Animal use was approved by the Animal Care Committee of the Universitat Autònoma de Barcelona and was conducted in compliance with the Spanish legislation (Departament de Medi Ambient i Habitatge (DMAH) registry numbers 5620 and 5596) and in accordance with the Directives of the European Union. A total of 24 male Sprague–Dawley rats (Charles River Laboratories, L'Arbresle, France) of approximately 10 weeks of age weighing 301±33 g were used. Eighteen rats were subjected to transient middle cerebral artery (MCA) occlusion. For the generation of the stroke model, rats were anesthetized in an induction box at 4% isofluorane (IsoVet, Braun VetCare, Spain) in O2. During surgery, general anesthesia was maintained with 2% isofluorane (IsoVet) in O2 via a face mask. The temperature was monitored by a rectal thermometer (TES-1307 Datalogging K/J Thermometer, TES Electrical Electronic Corporation, Taiwan) and maintained at 37±0.5 °C with a heating pad. The MCA was occluded with a 4/0 nylon intraluminal filament (Ethicon blue nylon 4/0, Sangüesa, Spain) ended with a rounded tip.22 The filament was inserted to occlude the entrance of the right MCA during 90 minutes. After this period, the filament was withdrawn. The surgery was performed under a microscope (Leica MZ9.5, Leica Microsystems, Wetzlar, Germany) equipped with an halogen reflector lamp (KL1500 LCD, Leica Microsystems).
When the surgery was finished, analgesia (Metacam, Boheringer Inghelm GmbH, Inghelm, Germany, 1.0 mg/kg body weight) was administered subcutaneously. After recovery from anesthesia, the rats were housed at 25±1 °C and were kept on a 12/12 hours light/darkness cycle with ad libitum access to food and water. Only two rats were discarded because of mortality during the reperfusion period.
The study was designed to achieve a high number of cells in the SVZi around day 7 after stroke. The animals were divided in groups according to different time points after stroke: non-stroke controls (G0) (n=6), day 1 after stroke (G1) (n=6), and days 6 to 8 after stroke (G7) (n=12).
In Vivo Magnetic Resonance Studies
In vivo magnetic resonance (MR) studies were carried out at the joint nuclear magnetic resonance facility of the Universitat Autònoma de Barcelona and Centro de Investigación Biomédica en Red—Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN) (Cerdanyola del Vallès, Spain) in a 7 Tesla horizontal magnet (BioSpec 70/30, Bruker BioSpin, Ettlingen Germany) equipped with actively shielded gradients (B-GA12 gradient coil inserted into a B-GA20S gradient system). For signal reception, a rat brain-phased array coil was used actively decoupled from a 72 mm inner diameter volume resonator. During exploration, the rats were anesthetized with 1% to 2% isofluorane (IsoVet), and breathing and temperature were constantly monitored (SA Instruments, New York, NY, USA).
Diffusion tensor imaging and three-dimensional time-of-flight angiography were performed in rats during the occlusion period, to ensure adequate ischemia in the infarct area.
Diffusion tensor imaging was carried out using a spin-echo four-shot echo-planar readout sequence approximately at 30 minutes after occlusion, when changes in diffusion are already visible.23 Diffusion weighted images were acquired along 12 diffusion directions and two diffusion weightings (b=0 seconds/mm2 and b=1,000 seconds/mm2). The acquisition parameters were: repetition time/effective echo time (TR/TEeff)=3,100/30 ms, diffusion gradient duration=4 milliseconds, diffusion gradient separation=16 milliseconds, matrix size=128 × 128 (250 × 250 μm/pixel); field of view=32 × 32 mm, number of slices=12, total acquisition time (TAT)=3 minutes and 30 seconds. Maps of the apparent diffusion coefficient were derived using Paravision 5.0 (Bruker BioSpin).
Three-dimensional time-of-flight angiography was acquired using a Fast Low-Angle Shot sequence. The acquisition parameters were: field of view=32 × 32 × 32 mm, matrix size=128 × 128 × 128 (250 × 250 × 250 μm/pixel), repetition time/echo time (TR/TE)=15/2.5 milliseconds, flip angle=20°, number of acquisitions=1, TAT=3 minutes and 4 seconds. The reconstruction of angiography was performed using maximum intensity projection in the axial plane with Paravision 5.0 software (Bruker BioSpin).
T2-weighted image was performed in all the rats at day 1 after stroke and at the final time point in each group. The analysis at day 1 after stroke was performed to assure that the infarct volume was similar in all the rats included in the study. Only rats with both striatal and cortical infarct were included. Approximately 25% of the rats were discarded for having small infarct volume detected by T2-weighted image. Afterwards, rats were randomized to the G1 or the G7 group. The investigators were not masked to the group allocation.
T2-weighted image was acquired using a rapid acquisition with a relaxation enhancement sequence. The acquisition parameters were: orientation=coronal plane, echo train length=8, field of view=32 × 32 mm, matrix size=256 × 256 (125 × 125 μm/pixel), number of slices=30, slice thickness=0.5 mm, interslice distance=0.1 mm, TR/TEeff=4,560/60 milliseconds, number of acquisitions=4, TAT=7 minutes and 17 seconds.
T2-weighted images were used to calculate infarct volume. The hyperintense area corresponding to the infarcted tissue was traced manually using the region of interest (ROI) tool of ParaVision 5.0 (Bruker BioSpin) at the MR workstation. The infarct volume was calculated according to equation (1):
Equation (1): Infarct volume calculation from T2-weighted images. IV refers to infarct volume (in mm3), ROI refers to region of interest (in number of pixels), ST refers to slice thickness (in mm), GD refers to gap distance (in mm) and PA refers to pixel area (in mm2).
Single voxel in vivo proton MRS was acquired using point-resolved spectroscopy localization and variable pulse power and optimized relaxation delay water suppression. Previously acquired T2-weighted image was used for voxel positioning. The rectangular volumes of interest (2 × 2 × 4.5 mm, 18 μl) were positioned in four regions of the brain (the infarct area, the SVZi, the contralateral subventricular zone (SVZc), and the contralateral area (symmetric to the infarct area)).
The acquisition parameters were: TR/TE=1,800/12 milliseconds, spectral width=13.34 ppm (4,006.41 Hz), number of acquisitions=256; TAT=7 minutes and 48 seconds.
Focused Microwave Irradiation
Immediately after in vivo MRS, before recovering from anesthesia, the rats (n=9) for ex vivo HRMAS studies were euthanized by FMW irradiation using the Muromachi Microwave Fixation System (Muromachi Kikai, Tokyo, Japan). This system was equipped with a rat holder adjusted to the animal's weight. Optimal irradiation parameters were set as 5 kW with 1.2 seconds of exposure time. After irradiation, the brains were removed and frozen in liquid nitrogen.
Ex Vivo High-Resolution Magic-Angle Spinning Studies
Frozen brains were allowed to thaw at room temperature (22±2 ºC). Then, tissue samples (15±5 mg) from the regions corresponding to the same volume of interest where in vivo MRS had been performed were dissected. The T2-weighted image with the in vivo MRS voxel superposed and anatomic cues of the brain (e.g., vicinity to ventricles and corpus callosum) were used as a guide. The dissected samples were placed in a zirconium rotor (Cortecnet, Voisins-le-Bretonneux, France) and deutered saline (0.15 M NaCl) (Euriso-top, Centre d'études de Saclay, Saclay, France) was added (10.0±5.0 μl) to provide a lock frequency to the sample. The zirconium rotor (4 mm in diameter, 50 μl in capacity) was mounted as previously described.24 The proton HRMAS experiments were carried out in a Bruker Avance III 400 spectrometer operating at 9.4 Tesla equipped with a 1H/13C/31P HRMAS probe (Bruker BioSpin). A spinning rate of 3,000 Hz at the magic angle (54.7°) was used. The probe temperature was set to 309.1 K, which corresponded to the 37 °C temperature in the sample with a Bruker Cooling Unit for temperature control. Two different types of spectra were acquired using the following sequences: (A) pulse-and-acquire with 2-second water presaturation during relaxation delay, (B) spin-echo with 2-second water presaturation, TR/TE=1,000/136 milliseconds. The spin-echo sequence was used essentially to determine Lac content without the influence of ML2. The acquisition parameters were: spectral width=10 ppm (4,000 Hz), number of acquisitions=256, TAT=15 minutes and 34 seconds.
Immunohistochemical Studies
The rats (n=18) for immunohistochemical studies were euthanized by transcardial perfusion with 0.9% saline (Sigma Aldrich, St Louis, MO, USA) and 4% paraformaldehyde (Panreac, Barcelona, Spain) once in vivo MRS was finished. After post-fixation with 4% paraformaldehyde, the brains were washed in 0.1 M phosphate buffer, pH 7.4, and six series for coronal free-floating sections of 25 μm each were cut with vibratome (VT 1000 M, Leica Microsystems) in the region of the lateral ventricles to evaluate the SVZs. Another six series were cut to evaluate the MCA areas. To determine proliferation in the SVZs, sections were incubated in blocking solution for 1 hour at room temperature (22±2 ºC) and incubated with the primary antibody (Ki67) (1:100, Novocastra, New England, UK). Then, sections were washed and incubated with biotinylated antibody anti-mouse, later with the ABC Elite complex (Vector, Burlingame, CA, USA) treated with diamobenzidine (DAB 0.05% Sigma Aldrich). To determine apoptosis in the MCA areas, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) was performed according to the manufacturer's protocol using the Apoptag Fluorescein In Situ Apoptosis Detection Kit (Merck Millipore, Darmstadt, Germany). Also sections were stained with 4',6-diamidino-2-phenylindole dilactate (Sigma Aldrich) which is marker for locating the nucleus. Sections were analyzed using fluorescence microscopy (Leica Microsystems). A Hamamatsu CCD camera (Hamamatsu Photonics K.K, Hamamatsu, Japan) interfaced with a computer was used to capture images.
Spectral Analysis
Spectra were processed using Topspin 1.3 software (Bruker Daltonik Gmbh, Rheinstetten, Germany). Fourier transformation was applied with previous line broadening (4 Hz for in vivo MRS spectra and 0.5 Hz for ex vivo HRMAS spectra). Manual zero- and first-order phase corrections were applied and the chemical shifts were referenced to total creatine (TCr).
All the spectra were normalized to unit length (UL2).24 This methodology was used as it is useful to compare data in vivo and ex vivo without previous knowledge of metabolites contributing to the resonance variation because of stroke or the SVZ. This is especially useful in the ML determination as fitting lipid resonances in HRMAS spectra may be complex. This simple methodology based on UL2 normalized peak heights can provide robust information about relative changes.
Ex vivo HRMAS spectral data were normalized to UL2 according to equation (2) as previously described25 for the region between 0.5 and 4.5 ppm This region was digitized with 13,113 complex data points.
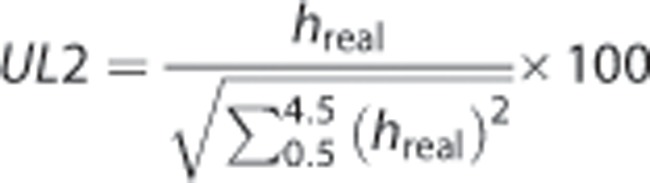 |
Equation (2): Unit length normalized (UL2) spectra. hreal refer to the peak heights at each data point in the real part of the acquired spectra between 0.5 and 4.5 ppm.
In vivo MRS spectral data were normalized to unit length to the contralateral area (UL2CA) according to equation (3) for the region between 0 and 4.5 ppm. This region was digitized with 1,331 complex data points.
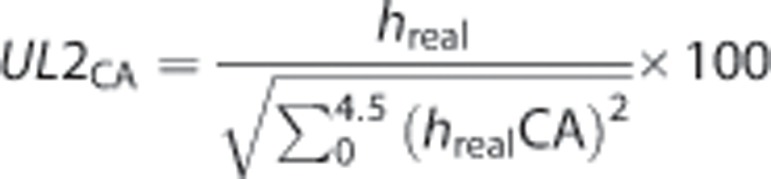 |
Equation (3): Unit length normalized to the contralateral area (UL2CA) spectra. hreal refers to the peak heights at each data point in the real part of the acquired spectra and hrealCA refers to the peak heights between 0 and 4.5 ppm from the contralateral area spectra of each animal.
For calculating the PCr/Cr ratio in the ex vivo HRMAS spectra, area integration was performed using TopSpin v. 1.3. (Bruker BioSpin GmbH, Rheinstetten, Germany) for PCr (3.95 ppm) and Cr (3.93 ppm). The integration range was 3.92 to 3.94 ppm for the singlet of Cr and 3.94 to 3.96 for the singlet of PCr.
The metabolites analyzed in the in vivo MRS spectra were total choline (TCho, 3.21 ppm), total creatine (TCr, 3.03 ppm), mobile lipids 3 (ML3, 2.80 ppm), NAA (2.02 ppm), lactate+mobile lipids 2 (Lac+ML2, 1.30 ppm), and mobile lipids 1 (ML1, 0.90 ppm). Tentative assignments were performed according to literature.4, 21
The metabolites analyzed in the ex vivo HRMAS spectra were phosphocreatine (PCr, 3.95 ppm), creatine (Cr, 3.93 ppm) N-CH2 region, TCho (3.25 ppm), TCr (3.03 ppm), mobile lipids 3 apparent doublet (ML3, 2.78 to 2.82 ppm), NAA (2.02 ppm), lactate doublet (Lac, 1.31 to 1.33 ppm), mobile lipids 2 (ML2, 1.28 ppm), and mobile lipids 1 (ML1, 0.90 ppm). Tentative assignments were performed according to literature.4, 21, 25, 26
Data Analysis
Unit length normalized metabolite peak heights of in vivo and ex vivo spectra were used for analysis. The results were given as mean relative changes (fold-change) for MCA areas (infarct area compared with contralateral area) or SVZs (SVZi compared with SVZc). Mean spectra representation was obtained with home-written R software scripts. For immunohistochemical results, the number of positively stained cells per mm2 was recorded.
All the data were evaluated for compliance with a normal distribution using Shapiro–Wilk test. Then the data were analyzed by the unpaired Student's t-test (normal distribution) or Mann–Whitney U-test (non-normal distribution) to evaluate the significant changes using SPSS 15.0 software (SPSS, Chicago, IL, USA). The significance level was set at P<0.05.
Results
Postmortem Metabolism Arrest
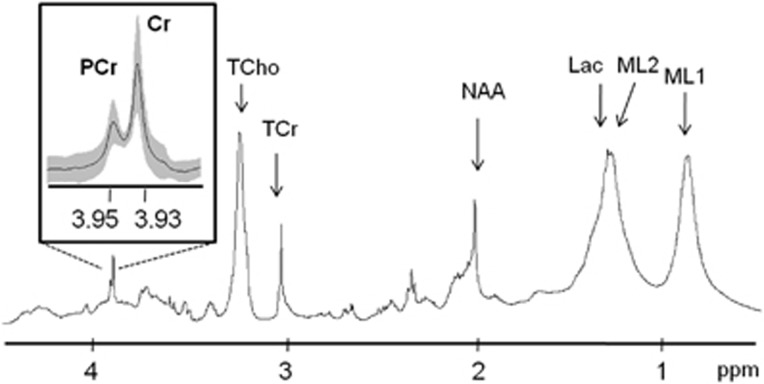
Postmortem metabolism arrest by FMW irradiation was evaluated by analyzing the PCr/Cr ratio (3.95/3.93 ppm) in ex vivo HRMAS pulse-and-acquire spectra (Figure 1). This ratio was determined in the intact tissue samples of both hemispheres in the G0 group (n=6) and in the contralateral area in the G1 group (n=3) and the G7 group (n=3). The PCr/Cr mean ratio (n=12) was 0.69±0.13.
Figure 1.
Ex vivo HRMAS pulse-and-acquire mean spectra (n=12) of intact tissue areas at 37 ºC. The inset shows an expanded view of the phosphocreatine (PCr)–creatine (Cr) resonances. Mean spectra are in black and standard deviation is in gray shadow in the inset only. The other assigned metabolites correspond to: total choline (TCho), total creatine (TCr), N-acetylaspartate (NAA), lactate (Lac) and mobile lipids (ML1, ML2). HRMAS, high-resolution magic-angle spinning.
Analysis of Middle Cerebral Artery Areas
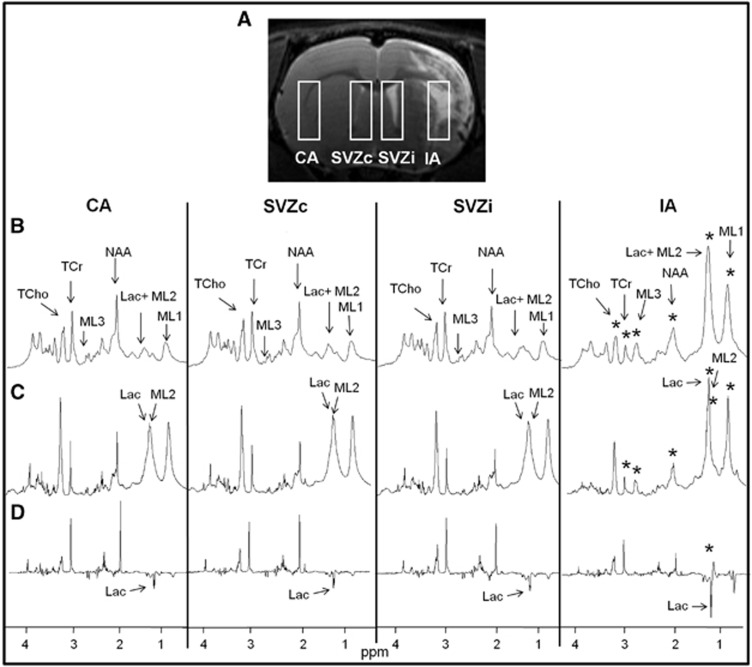
The mean infarct volume calculated from T2-weighted images at day 1 after stroke for the G1 and the G7 groups was 255±80 mm3. Changes in the metabolic pattern from in vivo MRS and ex vivo HRMAS spectra between the infarct area and the contralateral area were analyzed at different time points. There were no significant metabolic changes between the MCA areas in the G0 control group. In contrast, there were significant metabolic changes between the MCA areas in the G1 and the G7 groups. Mean spectra of the MCA areas in the G7 group (in which more representative metabolic changes took place) are shown in Figure 2 and a list of the metabolic changes for all the groups is shown in Table 1.
Figure 2.
Analysis of the four brain regions: contralateral area (CA), contralateral subventricular zone (SVZc), ipsilateral subventricular zone (SVZi), and infarct area (IA) in the G7 group. (A) Coronal T2-weighted image from a rat brain in the G7 group with rectangular single voxels superimposed, (B) in vivo MRS mean spectra (n=12), (C) ex vivo HRMAS pulse-and-acquire mean spectra (n=3), (D) ex vivo HRMAS spin-echo mean spectra (n=3). Statistically significant changes between IA and CA are indicated with stars (*P<0.05). No significant changes were detected between the SVZi and the SVZc in the G7 group. Assigned metabolites correspond to: total choline (TCho), total creatine (TCr), N-acetylaspartate (NAA), lactate (Lac), and mobile lipids (ML1, ML2, ML3). HRMAS, high-resolution magic-angle spinning.
Table 1. Metabolic fold changes (mean±s.d.) from middle cerebral artery areas and subventricular zones.
| Group | Metabolite | ppm | Technique |
MCA areas (IA/CA)a |
SVZs (SVZi/SVZc)a |
||
|---|---|---|---|---|---|---|---|
| Fold-change | P-value | Fold-change | P-value | ||||
| G0 | TCho | 3.21 | MRS | 1.09±0.23 | 0.51 | 0.98±0.27 | 0.71 |
| 3.25 | HRMAS | 0.95±0.25 | 0.56 | 1.18±0.19 | 0.23 | ||
| TCr | 3.03 | MRS | 1.03±0.20 | 0.72 | 0.93±0.16 | 0.57 | |
| HRMAS | 1.12±0.03 | 0.48 | 1.23±0.17 | 0.09 | |||
| ML3 | 2.80 | MRS | 1.15±0.46 | 0.77 | 0.92±0.39 | 0.67 | |
| 2.78–2.82b | HRMAS | 1.10±0.44 | 0.74 | 1.04±0.36 | 0.99 | ||
| NAA | 2.02 | MRS | 1.01±0.20 | 0.94 | 0.92±0.09 | 0.49 | |
| HRMAS | 1.11±0.10 | 0.65 | 1.18±0.12 | 0.12 | |||
| Lac + ML2 | 1.30 | MRS | 1.07±0.28 | 0.68 | 0.95±0.23 | 0.67 | |
| Lac | 1.31–1.33b | HRMAS-SEc | 1.49±0.93 | 0.99 | 1.06±0.24 | 0.80 | |
| ML2 | 1.28 | HRMAS | 0.95±0.07 | 0.21 | 0.98±0.03 | 0.41 | |
| ML1 | 0.90 | MRS | 1.08±0.31 | 0.66 | 0.97±0.07 | 0.67 | |
| HRMAS | 0.98±0.09 | 0.93 | 1.03±0.08 | 0.65 | |||
| G1 | TCho | 3.21 | MRS | 0.55±0.10 | <0.01* | 0.94±0.07 | 0.13 |
| 3.25 | HRMAS | 0.78±0.04 | 0.18 | 0.91±0.06 | 0.40 | ||
| TCr | 3.03 | MRS | 0.46±0.13 | <0.01* | 1.10±0.19 | 0.82 | |
| HRMAS | 0.68±0.19 | 0.19 | 0.79±0.08 | 0.28 | |||
| ML3 | 2.80 | MRS | 2.02±1.88 | 0.31 | 1.15±0.76 | 0.91 | |
| 2.78–2.82b | HRMAS | 0.85±0.07 | 0.07 | 1.06±0.57 | 0.86 | ||
| NAA | 2.02 | MRS | 0.36±0.07 | <0.01* | 1.07±0.20 | 0.51 | |
| HRMAS | 0.49±0.15 | 0.05* | 0.68±0.06 | 0.03* | |||
| Lac+ML2 | 1.30 | MRS | 2.26±0.59 | <0.01* | 1.23±0.36 | 0.58 | |
| Lac | 1.31–1.33b | HRMAS-SEc | 5.83±0.75 | <0.01* | 2.70±0.75 | 0.01* | |
| ML2 | 1.28 | HRMAS | 1.19±0.26 | 0.26 | 1.06±0.04 | 0.43 | |
| ML1 | 0.90 | MRS | 0.91±0.21 | 0.34 | 1.02±0.14 | 0.79 | |
| HRMAS | 1.12±0.22 | 0.30 | 1.01±0.02 | 0.55 | |||
| G7 | TCho | 3.21 | MRS | 0.66±0.24 | <0.01* | 0.91±0.19 | 0.24 |
| 3.25 | HRMAS | 0.52±0.11 | 0.10 | 1.03±0.23 | 0.95 | ||
| TCr | 3.03 | MRS | 0.34±0.12 | <0.01* | 1.02±0.20 | 0.82 | |
| HRMAS | 0.35±0.08 | 0.03* | 1.02±0.15 | 0.95 | |||
| ML3 | 2.80 | MRS | 5.53±3.69 | <0.01* | 1.09±0.65 | 0.60 | |
| 2.78–2.82b | HRMAS | 3.95±0.80 | <0.01* | 1.13±0.26 | 0.95 | ||
| NAA | 2.02 | MRS | 0.45±0.15 | <0.01* | 0.94±0.20 | 0.21 | |
| HRMAS | 0.46±0.04 | <0.01* | 0.95±0.12 | 0.70 | |||
| Lac+ML2 | 1.30 | MRS | 5.36±2.78 | <0.01* | 1.06±0.35 | 0.63 | |
| Lac | 1.31–1.33b | HRMAS-SEc | 2.88±1.06 | 0.04* | 1.09±0.31 | 0.92 | |
| ML2 | 1.28 | HRMAS | 1.55±0.26 | <0.01* | 0.99±0.12 | 0.79 | |
| ML1 | 0.90 | MRS | 2.64±1.40 | <0.01* | 1.05±0.14 | 0.35 | |
| HRMAS | 1.20±0.17 | 0.04* | 1.01±0.09 | 0.88 | |||
Abbreviations: HRMAS, high-resolution magic-angle spinning; Lac, lactate; ML, mobile lipid; MRS, magnetic resonance spectroscopy; NAA, N-acetylaspartate; TCho, total choline; TCr, total creatine.
*Statistically significant changes (P<0.05).
Fold-change values in middle cerebral artery (MCA) areas were obtained comparing the infarct area (IA) to the contralateral area (CA), in the subventricular zones (SVZs) were obtained comparing the ipsilateral subventricular zone (SVZi) to the contralateral subventricular zone (SVZc).
To calculate the fold change for Lac (1.31 and 1.33 ppm) and ML3 (2.78 and 2.82 ppm) doublets, the average value of the two UL2 normalized peak heights was used.
Fold-change values of the metabolites were calculated from in vivo MRS and ex vivo HRMAS pulse-and-acquire sequence. However, for lactate (Lac), the fold-change value from HRMAS spin-echo sequence (HRMAS-SE) is also given.
Most of the metabolic changes were in agreement using both techniques although for TCr (3.03 ppm) in the G1 group and TCho in the G1 and the G7 group, there were metabolic changes that were significant with in vivo MRS but not with ex vivo HRMAS.
One of the major changes between MCA areas was in Lac. In vivo Lac+ML2 (1.30 ppm) appeared to increase continuously over time: 2.26-fold increase (P<0.01) in the G1 group and 5.36-fold increase (P<0.01) in the G7 group. Ex vivo HRMAS spin-echo sequence detected a 5.83-fold increase (P<0.01) of Lac in the G1 group and a 2.88-fold increase (P=0.04) in the G7 group while a 1.55-fold increase in ML2 (P<0.01) was detected only in the G7 group with the pulse-and-acquire sequence.
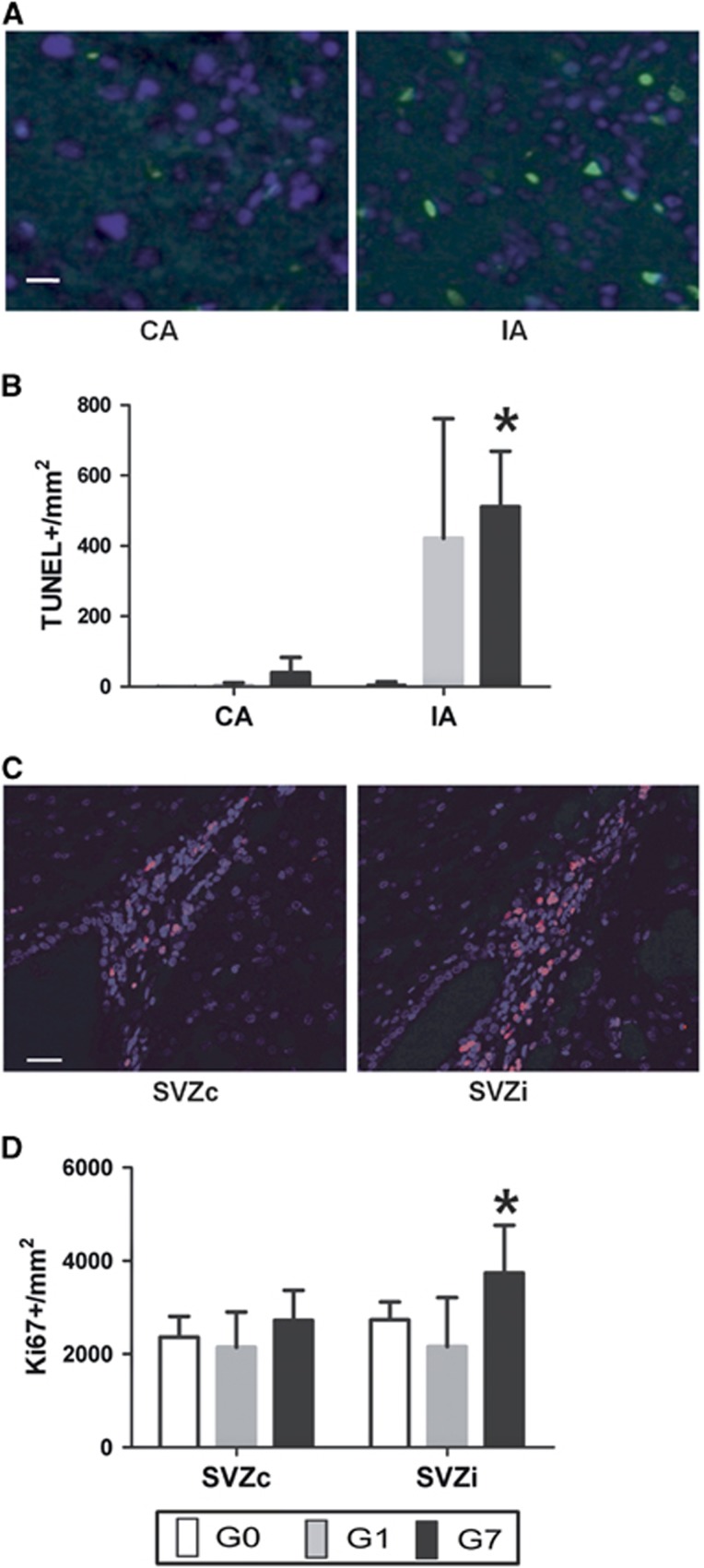
Another important metabolic change was in ML3 (2.80 ppm) in the G7 group, 5.53-fold increase (P<0.01) in vivo and a 3.95-fold increase (P<0.01) ex vivo between the MCA areas. Immunohistochemical data with TUNEL-labeling showed a significant change of a 13-fold increase in apoptotic cells (P<0.01) in the G7 group (Figures 3A and 3B).
Figure 3.
Immunohistochemical analysis of the four brain regions: (A) Contralateral area (CA) and infarct area (IA) apoptotic cells determined by TUNEL (green dots) in a rat of the G7 group. Blue dots correspond to nucleus determined by DAPI; × 100 magnification; Scale bar, 7 μm. (B) Apoptotic cells count (TUNEL+/mm2). Error bars indicate the standard deviation. (C) Contralateral subventricular zone (SVZc) and ipsilateral subventricular zone (SVZi) NPCs determined by Ki67 (pink dots) in a rat of the G7 group. Blue dots correspond to nucleus determined by DAPI; × 20 magnification; Scale bar, 25 μm. (D) NPCs count (Ki67+/mm2). Statistically significant changes between MCA areas or SVZs are indicated with stars (*P<0.05). DAPI, 4',6-diamidino-2-phenylindole dilactate; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
Analysis in the Subventricular Zones
Immunohistochemical data with Ki67-labeling revealed a 1.37-fold increase (P<0.01) in NPCs in the SVZi compared with the SVZc in the G7 group (Figures 3C and 3D) while this increase was not yet detectable (0.99-fold decrease (P=0.99)) in the G1 group.
However, there were no significant in vivo MRS metabolic changes that correlated with the NPCs' increase in the G7 group (Figure 2, Table 1). Ex vivo HRMAS pulse-and-acquire sequence allowed the analysis of the ML2 (1.28 ppm) resonance specifically but no significant differences (0.99-fold change (P=0.79)) were detected in the G7 group.
Nonetheless, ex vivo HRMAS detected metabolic differences in the G1 group. Specifically, HRMAS spin-echo sequence detected a 2.70-fold increase (P<0.01) in Lac (1.31 and 1.33 ppm) and HRMAS pulse-and-acquire sequence detected a 0.68-fold decrease (P=0.03) in NAA (2.02 ppm).
Discussion
Our results indicate that ex vivo HRMAS spectroscopy with prior FMW irradiation can be applied to study an experimental model of stroke avoiding postmortem metabolic changes and obtaining higher spectral resolution than with in vivo MRS. Our results of the characterization of the metabolic pattern of NPCs support the hypothesis that neither ML2 nor the other analyzed metabolites can qualify as surrogate biomarkers of NPCs.
HRMAS offers signal-to-noise ratio (SNR) gain-related factors, which is important for low concentration metabolites and also for certain metabolites that are not visible or less visible in vivo. The metabolites where HRMAS gave complementary information were ML3 at 2.78 and 2.82 ppm (that can be better untangled from partially overlapping aspartate resonances than in the in vivo spectra) and lactate at 1.31 and 1.33 ppm (that can be clearly determined with the spin-echo sequence). Also, the mobilization properties of HRMAS can help to determine ‘invisible' lactate (that cannot be detected in vivo if it is bound to proteins)27 and the ML from the subcellular structures.26
However, to perform ex vivo HRMAS with similar conditions to in vivo MRS, euthanasia using FMW irradiation is necessary as it allows postmortem metabolism arrest. FMW irradiation may be comparable to low temperature freezing techniques that have been used in other ex vivo studies.28 However, freezing requires more time and may cause reactivation of enzymes when the tissue is thawed.21 An important advantage of FMW is that it produces irreversible enzyme inactivation, so ex vivo HRMAS studies can be performed at 37 °C allowing the detection of ML resonances (similarly to in vivo MRS), not detectable at lower temperatures (0 to 4 ºC).25, 26, 29
Adequate postmortem metabolism arrest in the ex vivo HRMAS spectra was defined by the PCr/Cr ratio. This ratio was used because one of the initial metabolic changes in an energy failure state is the conversion of PCr into Cr to phosphorylate ADP resulting in ATP production. According to this chemical reaction, the presence of high PCr in the HRMAS spectra indicates that any ATP demand in the cells is covered,30 so postmortem metabolism changes did not occur or were minimal.
Therefore, the metabolic pattern detected by ex vivo HRMAS is similar quantitatively and qualitatively to the in vivo MRS. With other euthanizing methods (e.g., transcardial perfusion, anesthesia overdose…) lower or inexistent PCr and also higher lactate would be expected making the comparison with in vivo MRS data more challenging.24
In the analysis between the MCA areas, there were TCr and TCho metabolic changes that were significant with in vivo MRS but not with ex vivo HRMAS. These differences could be explained because ex vivo HRMAS was performed in only three animals, whereas in vivo MRS was performed in all the animals of each group. Also, the differences in TCho could be explained because the maximum peak height origin is different in vivo and ex vivo. In vivo TCho corresponds mostly to phosphocholine+glicerophosphocholine (3.21 ppm)4 and ex vivo TCho acquired at 37 °C corresponds mostly to phosphatidilcholine (3.25 ppm).26
Ex vivo HRMAS studies between MCA areas allowed the detection of lactate and ML2 resonances separately. Lactate was essentially quantified using spin-echo sequence spectra, recording conditions which ‘filter' away most of the short T2 ML2 resonance. We could also perform this ‘filter' with in vivo MRS, but signal-to-noise ratio would have been much worse because of long echo time and low filling factor of the coil, as compared with the HRMAS probes.
We detected with HRMAS that the major increase in Lac was in the G1 group as expected because the Lac increase occurs at the onset of stroke.4 Our results also detected a significant increase in ML2 in the G7 group, as expected, as ML2 increases at least over 7 days after stroke because of an accumulation of neutral lipids (triglyceride) in cytosolic and extracellular droplets in apoptotic or necrotic cells.31
Moreover, we detected a significant increase in ML3 in the G7 group between MCA areas. This resonance can be detected both in vivo and ex vivo. There have been very few studies that describe this resonance in stroke.32, 33 However, in cancer studies, ML3 have been analyzed extensively and is considered evidence of apoptosis.15 Our immunohistochemical studies with TUNEL revealed that apoptosis in the infarct area was elevated in the G1 group at similar levels as in the G7 group, although a significant change in the number of apoptotic cells between the MCA areas was detected only in the G7 group.
The fact that a significant change in ML3 resonance was detected only in the G7 group may be related to an accumulation of apoptosis-caused effects through the entire post-stroke period. In this case, the lipid droplets would remain in the tissue even when cellular detritus after apoptosis (e.g., the ones providing TUNEL-labeling positivity) was removed.34 Consequently, ML3 may be a noninvasive surrogate biomarker of cumulative apoptosis in stroke.
Immunohistochemical studies of the SVZs revealed that proliferation of NPCs was significantly enhanced in the SVZi in the G7 group as previously described.19 However, in our study, we could not detect any significant metabolic change between both SVZs either with in vivo MRS or ex vivo HRMAS in the G7 group. The analysis of the 1.28 ppm resonance (ML2) was performed using HRMAS pulse-and-acquire spectra but results were not significantly different between both SVZs. Therefore, our results are in agreement with other authors that demonstrated that the ML2 resonance, as mentioned in the introduction, is not a specific biomarker for NPCs.
Still, we could detect metabolic changes by ex vivo HRMAS in the SVZi in the G1 group (a Lac increase and a NAA decrease) but those were not correlated with a significant change in the NPCs count between SVZs. Consequently, they were not biomarkers of NPCs proliferation.
The changes in lactate could have an intracellular origin. Thus, tumors35 and also hematopoietic,36 embryonic and induced pluripotent stem cells37 accumulate lactate as a result of glycolytic energy production for increased cell proliferation. Still, Ki67 labeling for SVZi and SVZc in the G1 group would seem to discount this possibility, because similar lactate content should be expected in both SVZs, whereas higher lactate content in SVZi is detected. On the other hand, an increased lactate in the SVZi in the G1 group could originate in extracellular lactate diffusing away from the infarct area (high in lactate in the G1 group, see Table 1). This lactate content is clearly diminished in the G7 group as overall tissue perfusion may have improved.
Interestingly in NPCs cultures, increasing lactic acid concentration stimulates cell proliferation.38 Therefore, Lac from the infarct area may be a signal that activates NPCs proliferation after stroke.
Finally, changes in NAA in the G1 group could be intracellular, related with transient changes in the progenitor cells population in the SVZi. Oligodendrocyte-type 2 astrocytes (O-2A) accumulate NAA,39, 40 but not NPCs.10 Then, this could suggest that there is a transient decrease in the O-2A population in the SVZi, which returns to normal content in the G7 group. Also, an extra SVZ explanation is possible as dysfunctional mature neurons because of stroke are sampled along with the SVZi analyzed by HRMAS.
A limitation of our study is that a rat model with a big infarct volume may be suitable for the metabolic analysis of the infarct, but may have affected the metabolic results in the SVZi in the G1 group. Also, the different field strength used for the in vivo MRS (7 T) and the ex vivo HRMAS (9.4 T) could provide an additional advantage to obtain better sensitivity and resolution in HRMAS. Another limitation is the few animals that we analyzed by ex vivo HRMAS in each group (n=3). However, despite these few animals, significant HRMAS-detected metabolic changes were observed for MCA areas and SVZs.
In conclusion, ex vivo HRMAS with prior FMW irradiation improves the in vivo MRS characterization in an experimental model of stroke. We studied metabolic changes in the MCA areas and SVZs. ML3 could be an in vivo biomarker of cumulative apoptosis in the infarct area. However, none of the analyzed metabolites was related to an increase in NPCs in the SVZi that was detected by immunohistochemistry in the G7 group.
The authors declare no conflict of interest.
Footnotes
This study was partially funded by Ferrer International S.A, Redes Temáticas de Investigación Cooperativa RETICS-INVICTUS-RD12/014/0002, MARESCAN (SAF 2011-23870), Centro de Investigación Biomédica en Red – Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), [http://www.ciber-bbn.es/en]), an initiative of the Instituto de Salud Carlos III (Spain) co-funded by EU FEDER funds.
References
- Arvidsson A, Collin T, Kirik D, Kokaia Z, Lindvall O. Neuronal replacement from endogenous precursors in the adult brain after stroke. Nat Med. 2002;8:963–970. doi: 10.1038/nm747. [DOI] [PubMed] [Google Scholar]
- Marti-Fabregas J, Romaguera-Ros M, Gomez-Pinedo U, Martinez-Ramirez S, Jimenez-Xarrie E, Marin R, et al. Proliferation in the human ipsilateral subventricular zone after ischemic stroke Neurology 201074357–365.20054008 [Google Scholar]
- Lei H, Berthet C, Hirt L, Gruetter R. Evolution of the neurochemical profile after transient focal cerebral ischemia in the mouse brain. J Cereb Blood Flow Metab. 2009;29:811–819. doi: 10.1038/jcbfm.2009.8. [DOI] [PubMed] [Google Scholar]
- Berthet C, Lei H, Gruetter R, Hirt L. Early predictive biomarkers for lesion after transient cerebral ischemia. Stroke. 2011;42:799–805. doi: 10.1161/STROKEAHA.110.603647. [DOI] [PubMed] [Google Scholar]
- Alf MF, Lei H, Berthet C, Hirt L, Gruetter R, Mlynarik V. High-resolution spatial mapping of changes in the neurochemical profile after focal ischemia in mice. NMR Biomed. 2012;25:247–254. doi: 10.1002/nbm.1740. [DOI] [PubMed] [Google Scholar]
- Manganas LN, Zhang X, Li Y, Hazel RD, Smith SD, Wagshul ME, et al. Magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain. Science. 2007;318:980–985. doi: 10.1126/science.1147851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch JC, Maciejewski MW, Gryk MR.Comment on "Magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain" Science 2008321640author reply 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman SD.Comment on "Magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain" Science 2008321640author reply 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JF, Gearhart JD, Bulte JW.Comment on "Magnetic resonance spectroscopy identifies neural progenitor cells in the live human brain" Science 2008321640author reply 640. [DOI] [PubMed] [Google Scholar]
- Ramm P, Couillard-Despres S, Plotz S, Rivera FJ, Krampert M, Lehner B, et al. A nuclear magnetic resonance biomarker for neural progenitor cells: is it all neurogenesis. Stem Cells. 2009;27:420–423. doi: 10.1634/stemcells.2008-0816. [DOI] [PubMed] [Google Scholar]
- Loewenbruck KF, Fuchs B, Hermann A, Brandt M, Werner A, Kirsch M, et al. Proton MR spectroscopy of neural stem cells: does the proton-NMR peak at 1.28 ppm function as a biomarker for cell type or state. Rejuvenation Res. 2011;14:371–381. doi: 10.1089/rej.2010.1102. [DOI] [PubMed] [Google Scholar]
- Ramm Sander P, Hau P, Koch S, Schutze K, Bogdahn U, Kalbitzer HR, et al. Stem cell metabolic and spectroscopic profiling. Trends Biotechnol. 2013;31:204–213. doi: 10.1016/j.tibtech.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Hakumaki JM, Kauppinen RA. 1H NMR visible lipids in the life and death of cells. Trends Biochem Sci. 2000;25:357–362. doi: 10.1016/s0968-0004(00)01614-5. [DOI] [PubMed] [Google Scholar]
- Barba I, Cabanas ME, Arus C. The relationship between nuclear magnetic resonance-visible lipids, lipid droplets, and cell proliferation in cultured C6 cells. Cancer Res. 1999;59:1861–1868. [PubMed] [Google Scholar]
- Hakumaki JM, Poptani H, Sandmair AM, Yla-Herttuala S, Kauppinen RA. 1H MRS detects polyunsaturated fatty acid accumulation during gene therapy of glioma: implications for the in vivo detection of apoptosis. Nat Med. 1999;5:1323–1327. doi: 10.1038/15279. [DOI] [PubMed] [Google Scholar]
- Delikatny EJ, Chawla S, Leung DJ, Poptani H. MR-visible lipids and the tumor microenvironment. NMR Biomed. 2011;24:592–611. doi: 10.1002/nbm.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMRspectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- Stefan DDCF, Andrasescu A, Popa E, Lazariev A, Vescovo E, Strbak O, et al. Quantitation of magnetic resonance spectroscopy signals: the JMRUI software package. Meas Sci Technol. 2009;20:10. [Google Scholar]
- Zhang RL, Zhang ZG, Lu M, Wang Y, Yang JJ, Chopp M. Reduction of the cell cycle length by decreasing G1 phase and cell cycle reentry expand neuronal progenitor cells in the subventricular zone of adult rat after stroke. J Cereb Blood Flow Metab. 2006;26:857–863. doi: 10.1038/sj.jcbfm.9600237. [DOI] [PubMed] [Google Scholar]
- Beckonert O, Coen M, Keun HC, Wang Y, Ebbels TM, Holmes E, et al. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc. 5:1019–1032. doi: 10.1038/nprot.2010.45. [DOI] [PubMed] [Google Scholar]
- de Graaf RA, Chowdhury GM, Brown PB, Rothman DL, Behar KL. In situ 3D magnetic resonance metabolic imaging of microwave-irradiated rodent brain: a new tool for metabolomics research. J Neurochem. 2009;109:494–501. doi: 10.1111/j.1471-4159.2009.05967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longa EZ, Weinstein PR, Carlson S, Cummins R. Reversible middle cerebral artery occlusion without craniectomy in rats. Stroke. 1989;20:84–91. doi: 10.1161/01.str.20.1.84. [DOI] [PubMed] [Google Scholar]
- Neumann-Haefelin T, Kastrup A, de Crespigny A, Yenari MA, Ringer T, Sun GH, et al. Serial MRI after transient focal cerebral ischemia in rats: dynamics of tissue injury, blood-brain barrier damage, and edema formation Stroke 2000311965–1972.discussion 1972–1963. [DOI] [PubMed] [Google Scholar]
- Simões RVCA, Candiota AP, Julià-Sapé M, Arús C.In vivo magnetic resonance spectroscopic imaging and ex vivo quantitative neuropathology by high resolution magic angle spinning proton magnetic resonance spectroscopyMartínez-Murillo R, Martínez A (eds), Animal Models of Brain Tumors (Neuromehods)1st ednSpringer Science + Business Media: New York, NY, USA; 2013329–365. [Google Scholar]
- Davila M, Candiota AP, Pumarola M, Arus C. Minimization of spectral pattern changes during HRMAS experiments at 37 degrees celsius by prior focused microwave irradiation. Magn Reson Mater Phy. 2012;25:401–410. doi: 10.1007/s10334-012-0303-1. [DOI] [PubMed] [Google Scholar]
- Martin-Sitjar J, Delgado-Goni T, Cabanas ME, Tzen J, Arus C. Influence of the spinning rate in the HRMAS pattern of mobile lipids in c6 glioma cells and in artificial oil bodies. Magn Reson Mater Phy. 2012;25:487–496. doi: 10.1007/s10334-012-0327-6. [DOI] [PubMed] [Google Scholar]
- Bell JD, Brown JC, Kubal G, Sadler PJ. NMR-invisible lactate in blood plasma. FEBS Lett. 1988;235:81–86. doi: 10.1016/0014-5793(88)81238-9. [DOI] [PubMed] [Google Scholar]
- Yang M, Wang S, Hao F, Li Y, Tang H, Shi X. NMR analysis of the rat neurochemical changes induced by middle cerebral artery occlusion. Talanta. 2012;88:136–144. doi: 10.1016/j.talanta.2011.10.022. [DOI] [PubMed] [Google Scholar]
- Valverde-Saubi D, Candiota AP, Molins MA, Feliz M, Godino O, Davila M, et al. Short-term temperature effect on the HRMAS spectra of human brain tumor biopsies and their pattern recognition analysis. Magn Reson Mater Phy. 2010;23:203–215. doi: 10.1007/s10334-010-0218-7. [DOI] [PubMed] [Google Scholar]
- Hoehn M, Nicolay K, Franke C, van der Sanden B. Application of magnetic resonance to animal models of cerebral ischemia. J Magn Reson Imaging. 2001;14:491–509. doi: 10.1002/jmri.1213. [DOI] [PubMed] [Google Scholar]
- Harada K, Honmou O, Liu H, Bando M, Houkin K, Kocsis JD. Magnetic resonance lactate and lipid signals in rat brain after middle cerebral artery occlusion model. Brain Res. 2007;1134:206–213. doi: 10.1016/j.brainres.2006.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zijden JP, van Eijsden P, de Graaf RA, Dijkhuizen RM. 1H/13C MR spectroscopic imaging of regionally specific metabolic alterations after experimental stroke. Brain. 2008;131:2209–2219. doi: 10.1093/brain/awn139. [DOI] [PubMed] [Google Scholar]
- Saunders DE, Howe FA, van den Boogaart A, Griffiths JR, Brown MM. Discrimination of metabolite from lipid and macromolecule resonances in cerebral infarction in humans using short echo proton spectroscopy. J Magn Reson Imaging. 1997;7:1116–1121. doi: 10.1002/jmri.1880070626. [DOI] [PubMed] [Google Scholar]
- Griffin JL, Lehtimaki KK, Valonen PK, Grohn OH, Kettunen MI, Yla-Herttuala S, et al. Assignment of 1H nuclear magnetic resonance visible polyunsaturated fatty acids in BT4C gliomas undergoing ganciclovir-thymidine kinase gene therapy-induced programmed cell death. Cancer Res. 2003;63:3195–3201. [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CAt, Ramalho-Santos J, et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe KJ, Namba RM, Silverman TR, Bjugstad KB, Mahoney MJ. Impact of lactic acid on cell proliferation and free radical-induced cell death in monolayer cultures of neural precursor cells. Biotechnol Bioeng. 2009;103:1214–1. doi: 10.1002/bit.22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urenjak J, Williams SR, Gadian DG, Noble M. Specific expression of N-acetylaspartate in neurons, oligodendrocyte-type-2 astrocyte progenitors, and immature oligodendrocytes in vitro. J Neurochem. 1992;59:55–61. doi: 10.1111/j.1471-4159.1992.tb08875.x. [DOI] [PubMed] [Google Scholar]
- Bhakoo KK, Pearce D. In vitro expression of N-acetyl aspartate by oligodendrocytes: implications for proton magnetic resonance spectroscopy signal in vivo. J Neurochem. 2000;74:254–262. doi: 10.1046/j.1471-4159.2000.0740254.x. [DOI] [PubMed] [Google Scholar]