Abstract
Fluorine-18 labelled N,N-diethyl-2-(2-[4-(2-fluoroethoxy)phenyl]-5,7-dimethylpyrazolo[1,5-α]pyrimidine-3-yl)acetamide ([18F]DPA-714) binds to the 18-kDa translocator protein (TSPO) with high affinity. The aim of this initial methodological study was to develop a plasma input tracer kinetic model for quantification of [18F]DPA-714 binding in healthy subjects and Alzheimer's disease (AD) patients, and to provide a preliminary assessment whether there is a disease-related signal. Ten AD patients and six healthy subjects underwent a dynamic positron emission tomography (PET) study along with arterial sampling and a scan protocol of 150 minutes after administration of 250±10 MBq [18F]DPA-714. The model that provided the best fits to tissue time activity curves (TACs) was selected based on Akaike Information Criterion and F-test. The reversible two tissue compartment plasma input model with blood volume parameter was the preferred model for quantification of [18F]DPA-714 kinetics, irrespective of scan duration, volume of interest, and underlying volume of distribution (VT). Simplified reference tissue model (SRTM)-derived binding potential (BPND) using cerebellar gray matter as reference tissue correlated well with plasma input-based distribution volume ratio (DVR). These data suggest that [18F]DPA-714 cannot be used for separating individual AD patients from heathy subjects, but further studies including TSPO binding status are needed to substantiate these findings.
Keywords: [18F]DPA-714, Alzheimer's disease, neuroinflammation, PET quantification, TSPO
Introduction
The 18-kDa translocator protein (TSPO), formerly known as peripheral benzodiazepine receptor (PBR) is involved in several cellular functions such as response to apoptosis, regulation of cell proliferation, and modulation of mitochondrial respiration.1, 2 Activation of microglia, the main event leading to neuroinflammation, is a triggered immune response to any pathologic stimulus to the brain and leads to increased mitochondrial TSPO expression in these cells.3 Under physiologic conditions, TSPO expression is low and it is restricted mainly to ependymal cells, cells of the olfactory bulb, and the choroid plexus as well as resting glial cells, including microglia.4, 5 Increased TSPO expression in activated microglia has been associated with a number of diseases, including early stages of Alzheimer's disease (AD).6 The latter has also been shown in in vivo PET studies using the TSPO tracers (R)-[11C]PK11195 (ref. 7, 8, 9) and [11C]PBR28 (ref. 10). In vivo molecular imaging of TSPO may be useful for the assessment of early stages of AD by monitoring microglia activation.
The most widely used radiotracer for the detection of neuroinflammation today, (R)-[11C]PK11195, binds to TSPO with nanomolar affinity. Unfortunately, it has high nonspecific binding in the brain.3 High nonspecific binding complicates accurate quantification of the specific signal. Hence, several other TSPO ligands, such as [11C]PBR28, [18F]PBR06, and [18F]PBR111, with high affinity and improved signal-to-noise ratio, are under investigation.11, 12
Fluorine-18 labelled N,N-diethyl-2-(2-[4-(2-fluoroethoxy)phenyl]-5,7-dimethylpyrazolo[1,5-α]pyrimidine-3-yl)acetamide ([18F]DPA-714) is a TSPO ligand thought to be superior to (R)-[11C]PK11195 in terms of specificity and brain uptake.13 In addition, it is labelled with fluorine-18, allowing for distribution to imaging centers without an on-site cyclotron. Comparative studies of [18F]DPA-714 and (R)-[11C]PK11195 in a rat model of acute neuroinflammation have suggested that [18F]DPA-714 could be a good alternative for (R)-[11C]PK11195, as it showed better bioavailability to brain tissue together with a low level of nonspecific binding and increased binding potential.14 This, in turn, could result in more accurate quantification and easier visual interpretation. So far, some preliminary studies have been performed using [18F]DPA-714,15, 16 but a complete quantitative assessment of its in vivo binding characteristics in healthy subjects and AD patients has not been performed yet. Therefore, the primary aim of this initial methodological study was to develop a plasma input tracer kinetic model for quantifying [18F]DPA-714 binding in healthy subjects and AD patients. The secondary aim was to evaluate the diagnostic potential of [18F]DPA-714 for discriminating AD patients from healthy subjects.
Materials and methods
Subjects
The study was conducted in line with the Helsinki Declaration and approved by the Finnish Medicines Agency (FIMEA) and the Netherlands Central Committee on Research Involving Human Subjects (CCMO), the local Ethics Committee of the Southwest Hospital District of Finland and of the VU University Medical Center (VUmc). The study was registered at www.ClinicalTrials.gov (NCT01009359). All subjects gave written informed consent for participation in the study.
Ten AD patients (age 71.8±9.9 years, mini-mental state examination (MMSE) 24.1±3.0, disease duration 2.5±1.6 years) and six healthy subjects (age 64.5±5.5, MMSE 28.8±0.8) were included. Fifteen out of the sixteen subjects were scanned at Turku University Hospital and only one AD patient at VUmc. To avoid possible interinstitutional and/or interscanner variability, the latter patient was excluded from further analysis. The age difference between the remaining nine AD patients and healthy subjects became significant (P=0.045). The AD patients were considered only when they fulfilled National Institute of Neurological and Communicative Disorders and Stroke - Alzheimer's Disease and Related Disorders Association and Diagnostic and Statistical Manual of Mental Disorders, 4th Edition criteria. Tests included neurologic and neuropsychologic examinations, blood and urine analyses (hematology, clotting status, dip stick, serum chemistry, virology/serology, and urine pregnancy test), and magnetic resonance imaging (MRI) of the brain to identify structural lesions. Patients underwent an MMSE for assessment of severity of dementia. Healthy subjects needed to have normal performance on physical and neuropsychologic examinations, normal laboratory screening results, and MRI findings in accordance with their age. Additionally, neither AD patients nor healthy subjects were allowed to show signs of systemic autoimmune or inflammatory disease. Participants with current medication acting on the central nervous system (including anti-inflammatory medication) were also excluded to avoid interference with radioligand binding. Demographic data of all subjects are listed in Table 1.
Table 1. Details of subjects included.
| Sex | Dose (MBq) | Weight (kg) | Length (cm) | MMSE | Age (years) | Duration of disease (years) | ||
|---|---|---|---|---|---|---|---|---|
| A1 | AD | M | 261 | 85 | 173 | 22 | 73 | 5.5 |
| A2 | AD | M | 235 | 77 | 170 | 26 | 80 | 1.5 |
| A3 | AD | F | 248 | 57 | 160 | 24 | 74 | 5 |
| A4a | AD | M | 250 | 71 | 173 | 26 | 81 | 2 |
| A5 | AD | F | 265 | 83 | 190 | 29 | 63 | 1 |
| A6 | AD | F | 252 | 70 | 176 | 26 | 82 | 2.5 |
| A7a | AD | M | 256 | 58 | 158 | 22 | 82 | 1.5 |
| A8 | AD | M | 239 | 70 | 172 | 26 | 60 | 1 |
| A9 | AD | F | 243 | 49 | 154 | 20 | 68 | 2.5 |
| N1 | C | F | 265 | 76 | 171 | 29 | 73 | |
| N2 | C | M | 234 | 66 | 168 | 29 | 64 | |
| N3 | C | F | 243 | 57 | 161 | 30 | 69 | |
| N4 | C | F | 252 | 59 | 168 | 28 | 60 | |
| N5 | C | F | 261 | 72 | 166 | 28 | 59 | |
| N6 | C | F | 247 | 52 | 155 | 29 | 62 |
Abbreviations: AD, Alzheimer's disease; MMSE, mini-mental state examination.
No arterial input function available.
Synthesis of [18F]DPA-714
[18F]DPA-714 radiosynthesis was performed using in-house built automatic devices. [18F]DPA-714 was produced with a specific activity of 1,000±330 GBq/μmole, a radioactivity concentration of 240±120 MBq/mL, and a radiochemical purity of at least 99.5%. A detailed description of the synthesis of [18F]DPA-714 can be found in Supplementary Material 1.
Positron Emission Tomography Scanning Protocol and Image Reconstruction
Positron emission tomography scans were acquired on an ECAT EXACT HR+ scanner (CTI/Siemens, Knoxville, TN, USA) after administration of 250±10 MBq [18F]DPA-714 (tracer mass dose⩽9.7 μg) through an intravenous bolus injection over 30 seconds. On the basis of anticipated kinetics of [18F]DPA-714, two dynamic emission scans were acquired, the first from time of injection to 90 minutes, and the second from 120 minutes to 150 minutes after injection. After the first scan, subjects were given a 20-minute break, which was followed by a 10-minute period to prepare for the next emission scan. During the break, patients were lying comfortably to ensure that there were no significant physiologic changes. Nondiagnostic low-dose 3-D CT scans preceded both PET acquisitions and were used to correct the subsequent emission scans for tissue attenuation. Taking into account appropriate corrections for attenuation, scatter, and randoms, PET data were reconstructed using the FORE reprojection method with a Hanning filter (cutoff 0.5), a matrix size of 256 × 256 × 47, a field of view of 15.7 cm, and a final voxel size of 1.17 × 1.17 × 3.27 mm3. Both emission scans were reconstructed into a single dynamic data set of 36 frames (6 × 5, 3 × 10, 3 × 20, 4 × 60, 6 × 180, 11 × 360, 3 × 600 seconds), in which the first 33 frames were from the first scan and the last 3 from the second.
Continuous arterial sampling was performed using an automated blood sampling system (ABSS, Mariefred, Allogg, Sweden). In addition, manual blood samples were collected at seven different time points for estimation of whole blood and plasma concentrations, and for measurement of labelled metabolite fractions. A detailed description of the method to determine metabolites and intact [18F]DPA-714 in plasma can be found in Supplementary Material 2. Finally, blood time activity curves (TACs) obtained were corrected for plasma to whole blood ratios, metabolites, dispersion, and time delay, yielding a metabolite corrected plasma input function.
Data Analysis
Each subject underwent a T1-weighted MRI scan on a Philips Intera 1.5 Tesla scanner (Philips Medical Systems, Best, The Netherlands), reconstructed into a matrix of 256 × 256 × 158 with a voxel size of 1.00 × 1.00 × 1.00 mm3. In case of head motion, automatic frame realignment of the dynamic PET images was performed using the Vinci Software.17 Next, an average PET image (average of frames 5 to 22, i.e., 0.3 minute to 24 minutes) was used to coregister the T1-weighted MRI image with the PET data. Delineation of volumes of interest (VOIs) on the coregistered MRI images was performed using a probabilistic VOI template implemented in PVElab software.18, 19 PVElab uses SPM8 segmentation for automatic gray, white and cerebral spinal fluid segmentation. Delineated gray-matter VOIs were projected automatically onto the dynamic PET images yielding regional tissue TACs.
To find the optimal tracer kinetic model for analyzing [18F]DPA-714 data, standard single tissue (1T2k), irreversible (2T3k) and reversible (2T4k) two tissue compartment models, all with and without additional blood volume (VB) parameter, were evaluated. Regional TACs were fitted to these models using standard nonlinear regression.
In addition to plasma input models, performance of the simplified reference tissue model (SRTM)20 was evaluated using gray matter of the cerebellum as a reference tissue. Analyses were performed for several intervals (0 to 48, 0 to 60, 0 to 90, and 0 to 150 minutes post injection) to assess the impact of scan duration. The SRTM results were compared with those from plasma input analyses. Models and their abbreviations are listed in Supplementary Table 1. Comparisons were performed by considering 90 minutes as the basic data set. The 150-minute data set was not considered as a gold standard, since in a few subjects correction for head motion between the two emission scans was problematic.
For two of the nine AD patients no arterial blood data were available and they were excluded from plasma input analysis. Their data were, however, still included in the SRTM analysis. Finally, a preliminary and exploratory comparison of pharmacokinetic results with demographic data (Table 1) and/or disease status (AD/Healthy) was performed.
Statistical Analysis
To determine the best plasma input model, both Akaike Information Criterion21 and F-test were used. Accuracy and precision of several outcome measures for varying scan durations were evaluated by comparing them with the corresponding values derived from the 90-minute data sets. The validity of SRTM was assessed based on the correlation between SRTM-derived nondisplaceable binding potential (BPND) and plasma input-derived distribution volume ratio (DVR). Regression analyses of various pharmacokinetic parameter values with age and MMSE were performed for both AD and healthy subjects.
Results
Demographic Data
Main demographic data for the subjects included in this study are given in Table 1. There was a small, but significant (P=0.045) difference in age between healthy subjects and AD patients. Average disease duration of the AD patients was 2.5±1.7 years, and they had significantly lower MMSE scores than healthy subjects (P=0.0025). To illustrate [18F]DPA-714 uptake and distribution in the brain, Supplementary Figure 1 shows representative plasma input parametric volume of distribution (VT) images generated using Logan graphical analysis22 for both an AD patient and a healthy subject.
Plasma Input Analysis
There were no significant differences in injected dose and specific activity between healthy subjects and AD patients. The fraction of unchanged [18F]DPA-714 as function of time is shown in Supplementary Figure 2 for both AD patients and healthy subjects, indicating relatively slow metabolism with still >50% parent [18F]DPA-714 at 120 minutes. At all-time points, these fractions were not significantly different between subject groups (P=0.79).
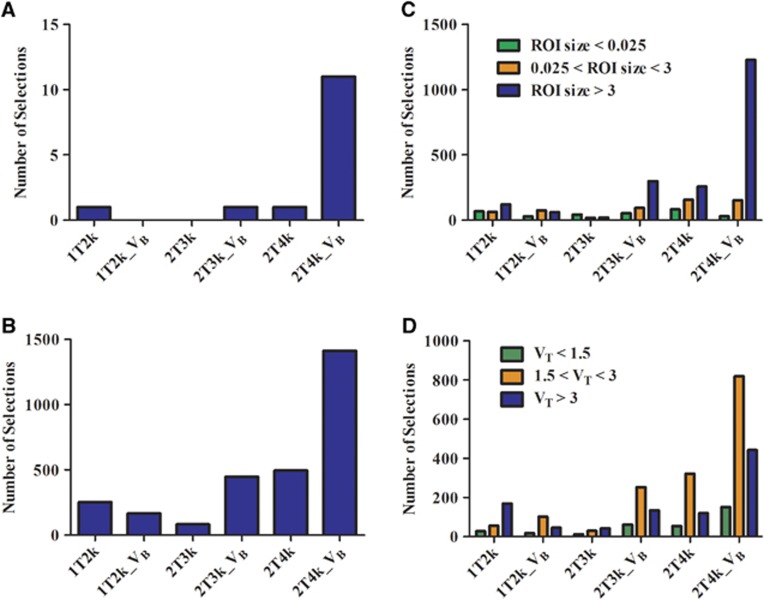
Figure 1A shows histograms of the preference for the various plasma input models according to the Akaike criterion per patient, per VOI, and per VOI of different sizes. It is clear from these histograms that, overall, the reversible two tissue compartment model with blood volume parameter best described observed kinetics of [18F]DPA-714. Supplementary Figure 3 illustrates several model fits for a typical region TAC. To assess effects of scan duration on model selection, similar comparisons were performed for 48-minute, 60-minute, and 150-minute data sets and no significant variation in model preference was observed. Dependence of model preferences on varying VT values was also evaluated and 2T4k_VB consistently was the most preferred model (Figure 1B).
Figure 1.
Histograms indicating number of selections (y axis) per model (x axis) according to the Akaike criterion for (A) subjects, (B) regions, (C) regions according to size and (D) varying volumes of distribution (VT).
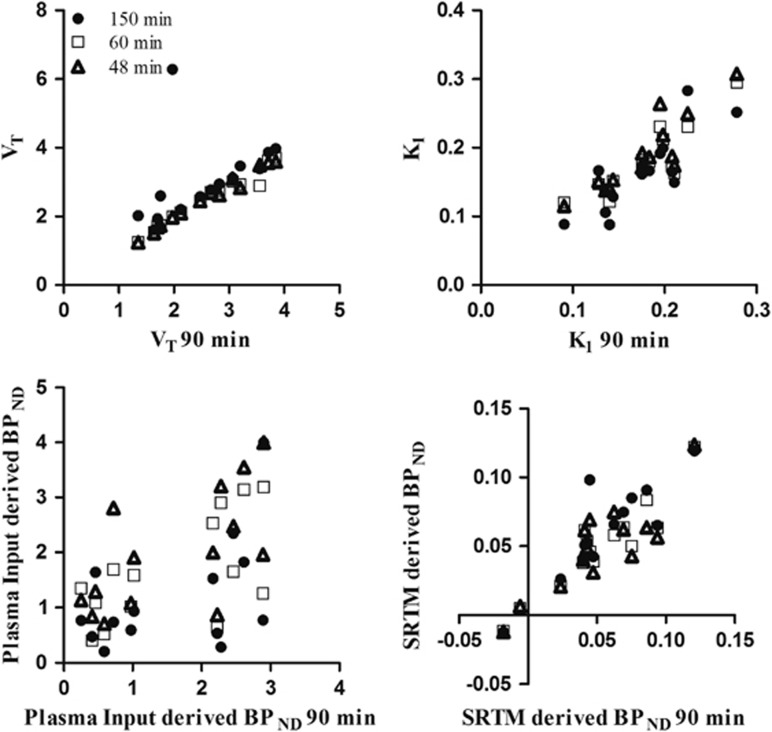
Finally, effects of scan duration on macroparameter values (VT, BPND) and K1 values were assessed by comparing parameter estimates for different study durations with those obtained for 90 minutes of data (Figure 2). Results of this comparison (correlation and slope) are listed in Table 2. Data from one healthy subject were excluded from the 150-minute comparison, as it was not possible to obtain a good fit due to poor coregistration of the two PET sessions.
Figure 2.
Kinetic parameter values (VT, K1, plasma input-derived nondisplaceable binding potential (BPND) and simplified reference tissue model (SRTM)-derived BPND) for different scan durations plotted against those obtained for 90 minutes of positron emission tomography (PET) data.
Table 2. Correlation coefficients (r 2) and slopes for linear regressions between parameter estimates obtained for different scan durations and those for 90 minutes.
| 150 minutes | 60 minutes | 48 minutes | ||||
|---|---|---|---|---|---|---|
| r2 | Slope | r2 | Slope | r2 | Slope | |
| VT | 0.9 | 0.84 | 0.96 | 0.89 | 0.98 | 0.94 |
| K1 | 0.67 | 0.96 | 0.78 | 0.9 | 0.79 | 1 |
| Plasma input-derived BPND | 0.25 | 0.51 | 0.39 | 0.59 | 0.42 | 0.69 |
| SRTM-derived BPND | 0.75 | 0.85 | 0.87 | 0.82 | 0.75 | 0.76 |
Abbreviations: BPND, nondisplaceable binding potential; SRTM, simplified reference tissue model.
Validation of Simplified Reference Tissue Model
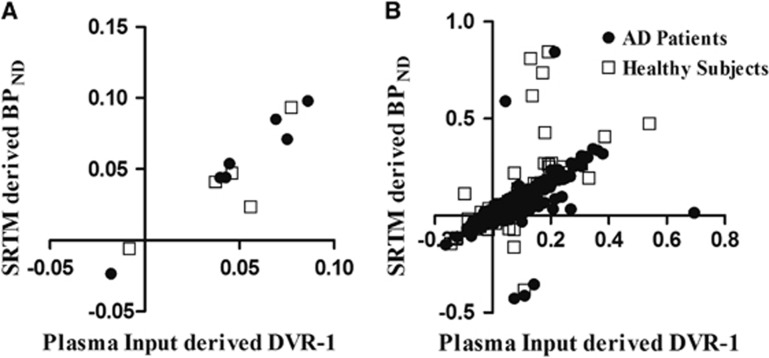
Figure 2 shows SRTM-derived BPND values for various scan intervals against those of the 90-minute data set. Figure 3 shows the relationship between SRTM-derived BPND and plasma input-derived DVR-1. Correlations and slopes of this comparison are listed in Table 2.
Figure 3.
Validation of simplified reference tissue model (SRTM) using cerebellum as a reference region: comparison of plasma input modelling-based distribution volume ratio (DVR)-1 with SRTM-based nondisplaceable binding potential (BPND) for (a) the total gray matter volume of interest (VOI) and (b) all gray-matter VOIs. LOI is the line of identity.
Preliminary Assessment of Potential Group Differences
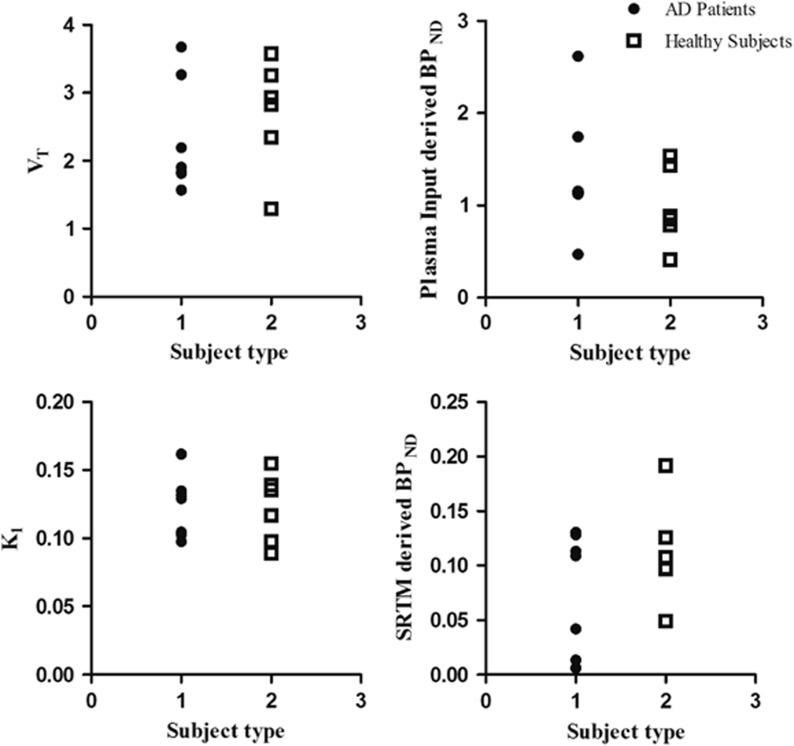
A preliminary evaluation of potential differences in whole brain gray-matter parameter values between healthy subjects and AD patients is shown in Figure 4. For none of the macroparameters evaluated (plasma input-derived BPND, VT, and K1/k2 and SRTM-derived BPND) could a significant difference be seen between both subject groups (Table 3) at either whole brain or regional levels.
Figure 4.
Total gray matter VT, nondisplaceable binding potential (BPND) and K1 for both healthy subjects and Alzheimer's disease (AD) patients.
Table 3. Results of kinetic analysis based on the 2T4k_VB plasma input model for both healthy subjects and AD patients.
| Brain region |
Healthy subjects |
AD patients |
||||
|---|---|---|---|---|---|---|
| VT | K1 | k3/k4 | VT | K1 | k3/k4 | |
| Frontal cortex | 2.80±0.80 | 0.18±0.05 | 1.73±1.02 | 2.40±0.82 | 0.17±0.04 | 1.55±1.05 |
| Temporal cortex | 2.67±0.77 | 0.16±0.04 | 1.76±0.99 | 2.36±0.82 | 0.16±0.03 | 1.43±0.74 |
| Parietal cortex | 2.68±0.69 | 0.19±0.05 | 1.66±1.14 | 2.22±0.74 | 0.17±0.04 | 1.20±0.75 |
| Occipital cortex | 2.61±0.68 | 0.18±0.04 | 1.22±0.65 | 2.28±0.79 | 0.17±0.03 | 1.08±0.64 |
| Insula | 3.01±0.97 | 0.17±0.03 | 2.00±0.96 | 2.55±0.95 | 0.18±0.04 | 2.02±0.57 |
| Hippocampus | 3.04±0.87 | 0.16±0.04 | 1.74±1.07 | 2.71±1.00 | 0.15±0.03 | 1.37±0.55 |
| Cerebellum | 2.75±0.83 | 0.19±0.06 | 1.78±1.35 | 2.16±0.72 | 0.21±0.06 | 1.40±0.52 |
| Caudate nucleus | 2.34±0.74 | 0.18±0.05 | 1.89±1.24 | 2.02±0.61 | 0.16±0.03 | 1.10±0.54 |
| Putamen | 3.02±0.97 | 0.21±0.05 | 2.04±1.03 | 2.60±0.95 | 0.21±0.05 | 0.98±0.58 |
| Thalamus | 3.29±1.11 | 0.20±0.05 | 2.29±0.90 | 2.75±0.89 | 0.21±0.03 | 2.00±0.94 |
| Gray matter | 2.67±0.78 | 0.18±0.04 | 1.66±1.16 | 2.29±0.80 | 0.17±0.04 | 1.35±0.79 |
| White matter | 2.71±0.81 | 0.12±0.03 | 0.98±0.42 | 2.32±0.81 | 0.12±0.02 | 1.34±0.67 |
Abbreviation: AD, Alzheimer's disease.
Discussion
Many studies9, 10, 23 have shown that TSPO is a target for imaging neuroinflammation in AD. In preclinical studies a variety of ligands24 with high binding affinity to TSPO have been identified. One of these ligands is [18F]DPA-714, which binds to TSPO with higher affinity than [11C]PK11195,13 the only established tracer for TSPO so far. Preclinical results of [18F]DPA-714 were promising,14, 25 indicating that it might be a good alternative for [11C]PK11195. The purpose of the present methodological study was first to determine the optimal plasma input model for quantitative analysis of [18F]DPA-714 data in humans and second to provide a preliminary evaluation whether there is a difference in kinetics of [18F]DPA-714 between AD patients and healthy subjects.
Akaike Information Criterion analysis showed that, in the majority of cases, kinetics of [18F]DPA-714 could best be described by a 2T4k_VB model, irrespective of subject group, region of interest, and region of interest size. Results of this Akaike Information Criterion analysis were similar for shorter (60 minutes) and longer (150 minutes) scan durations. Similar model preferences were obtained when using the F-test. In addition, fitted VT and K1 values were similar for all scan durations investigated. No trend was observed in plasma input-based BPND with varying scan duration, which could result from limitations in robust estimation of k3/k4. Based on reliability and stability of VT estimates, the scan duration of future [18F]DPA-714 studies may be reduced to 60 minutes.
Recently, it has been shown that, for several TSPO ligands, subjects may present as low, mixed, or high affinity binders.11 Therefore, in the present study, model preferences were studied for varying VT and similar comparisons were performed using plasma input-derived BPND. Comparable model preferences were obtained (general preference for two tissue compartmental model with reversible kinetics). Hence, it seems justified to assume that model preference of [18F]DPA-714 remains unchanged irrespective of the subject's binding status. Therefore, despite the fact that no information on binding status of subjects was available, it can be concluded that this does not affect the choice for the optimal plasma input model for quantification of [18F]DPA-714. Nevertheless, further studies are needed to substantiate this conclusion.
Quantitative assessment of the effect of scan duration on SRTM-derived BPND showed good correlations between the 90-minute data set and those of different duration. In addition, a linear trend was observed for the relationship between SRTM-derived BPND and DVR-1, suggesting no significant variation between SRTM-derived BPND values and plasma input-based DVRs. In other words, SRTM may be used to assess nondisplaceable binding potential. Nevertheless, use of cerebellum or any other region or method to obtain a reference tissue input function remains to be assessed. One way to obtain a suitable reference tissue input function would be to use methods such as supervised cluster analysis.26 This method, however, will require predefined kinetic classes derived from larger data sets of AD patients and healthy subjects.
Analyses at a regional level showed that differences in [18F]DPA-714 binding between AD patients and healthy subjects were extremely small. For none of the regions and for none of the models, a significant difference in either VT or BPND was observed between subject groups. It should be noted that the lack of a significant increase in [18F]DPA-714 binding in AD patients cannot be explained by the small, but significant, difference in age between AD patients and healthy subjects, because AD patients were slightly older than the healthy subjects and microglial activation tends to increase with healthy aging.27 Differences in specific binding between AD patients and healthy controls were not large enough to be detected at an individual level and it is likely that the overall number of subjects was too small to detect any differences at a group level. In addition, this study could have been affected by intersubject variability in binding status,11 although it is not known exactly to which extent [18F]DPA-714 uptake is affected. The major limitation for the present study is the lack of TSPO genotype information. In addition, accidental inclusion of non-AD patients is also possible, as no biomarker evidence was available. Clearly, further studies are needed to assess the effects of TSPO polymorphism on binding affinity of [18F]DPA-714 and the corresponding quantitative PET measures. Nevertheless, despite possible differences in binding status, the present study clearly showed that the reversible two tissue compartment plasma input model with blood volume parameter is the preferred model for quantification of [18F]DPA-714 data in both AD patients and healthy subjects.
Conclusions
The reversible two tissue compartment plasma input model with blood volume parameter is the preferred model for quantifying [18F]DPA-714 data. Scan durations can be reduced to 60 minutes without loss of accuracy. In addition, SRTM can also be used to assess nondisplaceable binding potential, although the optimal reference region needs to be determined in future studies. No significant differences in regional tracer binding were observed between AD patients and healthy controls, indicating that [18F]DPA-714 may not be suitable for early diagnosis of AD. However, further studies incorporating TSPO genotyping are needed to provide a definitive assessment of the potential role of [18F]DPA-714 in clinical studies.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Journal of Cerebral Blood Flow & Metabolism website (http://www.nature.com/jcbfm)
This study was sponsored by Bayer Healthcare AG, Berlin, Germany. This work was supported by the European Union's Seventh Framework Programme (FP7/2007-2013), grant agreement n° HEALTH-F2-2011-278850 (INMiND).
Supplementary Material
References
- Casellas P, Galiegue S, Basile AS. Peripheral benzodiazepine receptors and mitochondrial function. Neurochem Int. 2002;40:475–486. doi: 10.1016/s0197-0186(01)00118-8. [DOI] [PubMed] [Google Scholar]
- Galiegue S, Tinel N, Casellas P. The peripheral benzodiazepine receptor: a promising therapeutic drug target. Curr Med Chem. 2003;10:1563–1572. doi: 10.2174/0929867033457223. [DOI] [PubMed] [Google Scholar]
- Banati RB, Newcombe J, Gunn RN, Cagnin A, Turkheimer F, Heppner F, et al. The peripheral benzodiazepine binding site in the brain in multiple sclerosis: quantitative in vivo imaging of microglia as a measure of disease activity. Brain. 2000;123 (Pt 11:2321–2337. doi: 10.1093/brain/123.11.2321. [DOI] [PubMed] [Google Scholar]
- Bribes E, Carriere D, Goubet C, Galiegue S, Casellas P, Simony-Lafontaine J. Immunohistochemical assessment of the peripheral benzodiazepine receptor in human tissues. J Histochem Cytochem. 2004;52:19–28. doi: 10.1177/002215540405200103. [DOI] [PubMed] [Google Scholar]
- Scarf AM, Kassiou M. The translocator protein. J Nucl Med. 2011;52:677–680. doi: 10.2967/jnumed.110.086629. [DOI] [PubMed] [Google Scholar]
- Venneti S, Lopresti BJ, Wiley CA. The peripheral benzodiazepine receptor (Translocator protein 18kDa) in microglia: from pathology to imaging. Prog Neurobiol. 2006;80:308–322. doi: 10.1016/j.pneurobio.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokokura M, Mori N, Yagi S, Yoshikawa E, Kikuchi M, Yoshihara Y, et al. In vivo changes in microglial activation and amyloid deposits in brain regions with hypometabolism in Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2011;38:343–351. doi: 10.1007/s00259-010-1612-0. [DOI] [PubMed] [Google Scholar]
- Cagnin A, Brooks DJ, Kennedy AM, Gunn RN, Myers R, Turkheimer FE, et al. In-vivo measurement of activated microglia in dementia. Lancet. 2001;358:461–467. doi: 10.1016/S0140-6736(01)05625-2. [DOI] [PubMed] [Google Scholar]
- Schuitemaker A, Kropholler MA, Boellaard R, van der Flier WM, Kloet RW, van der Doef TF, et al. Microglial activation in Alzheimer's disease: an (R)-[(1)(1)C]PK11195 positron emission tomography study. Neurobiol Aging. 2013;34:128–136. doi: 10.1016/j.neurobiolaging.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Kreisl WC, Lyoo CH, McGwier M, Snow J, Jenko KJ, Kimura N, et al. In vivo radioligand binding to translocator protein correlates with severity of Alzheimer's disease. Brain. 2013;136:2228–2238. doi: 10.1093/brain/awt145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DR, Gunn RN, Rabiner EA, Bennacef I, Fujita M, Kreisl WC, et al. Mixed-affinity binding in humans with 18-kDa translocator protein ligands. J Nucl Med. 2011;52:24–32. doi: 10.2967/jnumed.110.079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauveau F, Boutin H, Van CN, Dolle F, Tavitian B. Nuclear imaging of neuroinflammation: a comprehensive review of [11C]PK11195 challengers. Eur J Nucl Med Mol Imaging. 2008;35:2304–2319. doi: 10.1007/s00259-008-0908-9. [DOI] [PubMed] [Google Scholar]
- Peyronneau MA, Saba W, Goutal S, Damont A, Dolle F, Kassiou M, et al. Metabolism and quantification of [(18)F]DPA-714, a new TSPO positron emission tomography radioligand. Drug Metab Dispos. 2013;41:122–131. doi: 10.1124/dmd.112.046342. [DOI] [PubMed] [Google Scholar]
- Chauveau F, Van CN, Dolle F, Kuhnast B, Hinnen F, Damont A, et al. Comparative evaluation of the translocator protein radioligands 11C-DPA-713, 18F-DPA-714, and 11C-PK11195 in a rat model of acute neuroinflammation. J Nucl Med. 2009;50:468–476. doi: 10.2967/jnumed.108.058669. [DOI] [PubMed] [Google Scholar]
- Arlicot N, Vercouillie J, Ribeiro MJ, Tauber C, Venel Y, Baulieu JL, et al. Initial evaluation in healthy humans of [18F]DPA-714, a potential PET biomarker for neuroinflammation. Nucl Med Biol. 2012;39:570–578. doi: 10.1016/j.nucmedbio.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Corcia P, Tauber C, Vercoullie J, Arlicot N, Prunier C, Praline J, et al. Molecular imaging of microglial activation in amyotrophic lateral sclerosis. PLoS One. 2012;7:e52941. doi: 10.1371/journal.pone.0052941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmar S, Michel C, Treffert JT, Newport DF, Casey M, Knoss C, et al. HeinzelCluster: accelerated reconstruction for FORE and OSEM3D. Phys Med Biol. 2002;47:2651–2658. doi: 10.1088/0031-9155/47/15/307. [DOI] [PubMed] [Google Scholar]
- Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, et al. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp. 2003;19:224–247. doi: 10.1002/hbm.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarer C, Madsen K, Hasselbalch SG, Pinborg LH, Haugbol S, Frokjaer VG, et al. MR-based automatic delineation of volumes of interest in human brain PET images using probability maps. Neuroimage. 2005;24:969–979. doi: 10.1016/j.neuroimage.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Lammertsma AA, Hume SP. Simplified reference tissue model for PET receptor studies. Neuroimage. 1996;4:153–158. doi: 10.1006/nimg.1996.0066. [DOI] [PubMed] [Google Scholar]
- Akaike H.A new look at the statistical model identification19th ed. pp716–723.1974
- Logan J, Fowler JS, Volkow ND, Wolf AP, Dewey SL, Schlyer DJ, et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(-)-cocaine PET studies in human subjects. J Cereb Blood Flow Metab. 1990;10:740–747. doi: 10.1038/jcbfm.1990.127. [DOI] [PubMed] [Google Scholar]
- Suridjan I, Rusjan PM, Voineskos AN, Selvanathan T, Setiawan E, Strafella AP, et al. Neuroinflammation in healthy aging: a PET study using a novel Translocator Protein 18kDa (TSPO) radioligand, [(18)F]-FEPPA. Neuroimage. 2014;84:868–875. doi: 10.1016/j.neuroimage.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching AS, Kuhnast B, Damont A, Roeda D, Tavitian B, Dolle F. Current paradigm of the 18-kDa translocator protein (TSPO) as a molecular target for PET imaging in neuroinflammation and neurodegenerative diseases. Insights Imaging. 2012;3:111–119. doi: 10.1007/s13244-011-0128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harhausen D, Sudmann V, Khojasteh U, Muller J, Zille M, Graham K, et al. Specific imaging of inflammation with the 18 kDa translocator protein ligand DPA-714 in animal models of epilepsy and stroke. PLoS One. 2013;8:e69529. doi: 10.1371/journal.pone.0069529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaqub M, van Berckel BN, Schuitemaker A, Hinz R, Turkheimer FE, Tomasi G, et al. Optimization of supervised cluster analysis for extracting reference tissue input curves in (R)-[(11)C]PK11195 brain PET studies. J Cereb Blood Flow Metab. 2012;32:1600–1608. doi: 10.1038/jcbfm.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuitemaker A, van der Doef TF, Boellaard R, van der Flier WM, Yaqub M, Windhorst AD, et al. Microglial activation in healthy aging. Neurobiol Aging. 2012;33:1067–1072. doi: 10.1016/j.neurobiolaging.2010.09.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.