ABSTRACT
We recently demonstrated that TP_0326 is a bona fide rare outer membrane protein (OMP) in Treponema pallidum and that it possesses characteristic BamA bipartite topology. Herein, we used immunofluorescence analysis (IFA) to show that only the β-barrel domain of TP_0326 contains surface-exposed epitopes in intact T. pallidum. Using the solved structure of Neisseria gonorrhoeae BamA, we generated a homology model of full-length TP_0326. Although the model predicts a typical BamA fold, the β-barrel harbors features not described in other BamAs. Structural modeling predicted that a dome comprised of three large extracellular loops, loop 4 (L4), L6, and L7, covers the barrel's extracellular opening. L4, the dome's major surface-accessible loop, contains mainly charged residues, while L7 is largely neutral and contains a polyserine tract in a two-tiered conformation. L6 projects into the β-barrel but lacks the VRGF/Y motif that anchors L6 within other BamAs. IFA and opsonophagocytosis assay revealed that L4 is surface exposed and an opsonic target. Consistent with B cell epitope predictions, immunoblotting and enzyme-linked immunosorbent assay (ELISA) confirmed that L4 is an immunodominant loop in T. pallidum-infected rabbits and humans with secondary syphilis. Antibody capture experiments using Escherichia coli expressing OM-localized TP_0326 as a T. pallidum surrogate further established the surface accessibility of L4. Lastly, we found that a naturally occurring substitution (Leu593 → Gln593) in the L4 sequences of T. pallidum strains affects antibody binding in sera from syphilitic patients. Ours is the first study to employ a “structure-to-pathogenesis” approach to map the surface topology of a T. pallidum OMP within the context of syphilitic infection.
IMPORTANCE Previously, we reported that TP_0326 is a bona fide rare outer membrane protein (OMP) in Treponema pallidum and that it possesses the bipartite topology characteristic of a BamA ortholog. Using a homology model as a guide, we found that TP_0326 displays unique features which presumably relate to its function(s) in the biogenesis of T. pallidum's unorthodox OM. The model also enabled us to identify an immunodominant epitope in a large extracellular loop that is both an opsonic target and subject to immune pressure in a human population. Ours is the first study to follow a structure-to-pathogenesis approach to map the surface topology of a T. pallidum rare OMP within the context of syphilitic infection.
INTRODUCTION
Within the outer membranes (OMs) of Gram-negative bacteria is a unique class of integral membrane proteins that fold into a β-barrel structure consisting of 8 to 26 antiparallel amphipathic β strands; typically, extensive hydrogen bonding between the first and last strands closes and stabilizes the barrel, often creating a central channel (1, 2). β-Barrel outer membrane proteins (OMPs) have two principal functions: (i) insertion/transport of proteins into or across the OM and (ii) formation of aqueous pores for the passive or selective uptake of nutrients required for cellular homeostasis (1, 2). OMPs are synthesized in the cytoplasm and transported across the inner membrane by the Sec translocon coincidently with removal of their signal peptides (3). Following export, periplasmic chaperones ferry the unfolded OMPs to the β-barrel assembly machinery (BAM), the molecular complex that catalyzes insertion and folding of nascent β-barrels into the OM (4–6). BamA, a member of the Omp85 superfamily (5, 7), is the central and essential component of the BAM. BamA consists of an OM-embedded C-terminal β-barrel and a periplasmic N-terminal region containing one or more polypeptide transport-associated (POTRA) domains (4, 7, 8). In addition to providing a scaffold for the other subunits of the BAM complex, the POTRA domains thread nascent OMPs toward the BamA β-barrel in a process called “β-augmentation” (9–11). Recently, three crystal structures from Neisseria gonorrhoeae, Haemophilus ducreyi (12), and Escherichia coli (13, 14) have provided insights into the mechanism by which the BamA β-barrel functions during OM biogenesis. After entering the channel, the OMP is thought to integrate into the bilayer through a lateral opening between the first and last β strands of the barrel, while its hydrophilic loops access the bacterial surface via the foreshortened β strands in the barrel wall above the opening (14, 15). In the solved BamA structures, the extracellular loops form a dome over the β-barrel, presumably functioning as a lid that guides OMP substrates within the channel toward the lateral gate (14, 15). While most recent studies have focused on BamA's role in OM biogenesis, it also is well established for a number of Gram-negative bacteria that immunization with BamA can induce a protective immune response (16–19).
Syphilis is a multistage, sexually transmitted illness caused by Treponema pallidum, an extracellular, highly invasive bacterium that establishes persistent infection in a substantial proportion of untreated individuals (20). The mechanisms underlying syphilis pathogenesis are poorly understood, in large part because its etiologic agent cannot be cultivated in vitro and, therefore, is not amenable to genetic manipulation (21). Although T. pallidum has both outer and cytoplasmic membranes, the composition and physical properties of its cell envelope differ considerably from those of Gram-negative bacteria (22, 23). In addition to lacking lipopolysaccharide (22), the OM of T. pallidum is a fluid and fragile bilayer with a much lower density of membrane-spanning proteins, often referred to as rare OMPs based upon their visualization as low-density intramembranous particles by freeze fracture electron microscopy (24, 25). The collective paucity of surface-exposed pathogen-associated molecular patterns (PAMPs) and OM-spanning proteins is believed to be the ultrastructural basis for the syphilis spirochete's impressive capacity to evade innate and adaptive host defenses, a parasitic strategy designated “stealth pathogenicity” (26). Not surprisingly, molecular characterization of the spirochete's OMP repertoire has long been regarded as critical for unraveling the many enigmas of syphilis as well as being the key to the development of an effective vaccine (22, 27). Rare OMPs presumably fulfill virulence-related and physiological functions in T. pallidum, an extreme auxotroph (28), analogous to those of their extensively characterized, nonorthologous Gram-negative counterparts (29). Moreover, epitopes on the spirochetal surface represent potential targets for immune responses that contribute to containment of the pathogen during natural infection and/or confer protection following artificial immunization (26, 27). It is also conceivable that sequence diversity in rare OMPs among circulating T. pallidum strains (30, 31) is a major contributory factor to the dynamics of disease transmission and reinfection in at-risk populations (32, 33).
Identification of rare OMPs is prerequisite for elucidating at the molecular level the interactions between the syphilis spirochete and its obligate human host (28). The lack of sequence relatedness between these entities and OMPs of Gram-negative bacteria has impeded efforts by investigators to achieve this critical objective, even with the availability of the spirochete's genomic sequence (28). To circumvent this problem, we devised a consensus computational framework that employs a battery of cellular localization and topological prediction tools to generate ranked clusters of candidate rare OMPs (29). Among our highest ranked candidates was TP_0326, discovered in 2000 as TP_92 by Cameron et al. (19) using a differential screening strategy to identify opsonic targets in T. pallidum. Recently, we confirmed the status of TP_0326 as a bona fide rare OMP (34) and, using recombinant constructs, established that it possesses the bipartite topology characteristic of members of the Omp85 superfamily (7). In the present study, we demonstrated by indirect immunofluorescence analysis (IFA) of T. pallidum that only the β-barrel of native TP_0326 contains surface-exposed epitopes. We then went on to generate a homology model of TP_0326 using the first solved crystal structure of a full-length BamA protein, that of Neisseria gonorrhoeae (12). While TP_0326 adopts a typical BamA structural fold, it also displays unique features which presumably relate to its function(s) in the biogenesis of T. pallidum's unorthodox OM and/or its constitutive residence at the host-pathogen interface throughout the disease process. Importantly, we identified an immunodominant large extracellular loop that is both an opsonic target and subject to immune pressure in a human population. Ours is the first study to follow a “structure-to-pathogenesis” approach to map the surface topology of a T. pallidum rare OMP within the context of syphilitic infection.
MATERIALS AND METHODS
Propagation and harvesting of T. pallidum.
Animal protocols described in this work strictly follow the recommendations of the Guide for the Care and Use of Laboratory Animals (8th edition) of the National Institutes of Health (35) and were approved by the University of Connecticut Health Center Animal Care Committee under the auspices of Animal Welfare Assurance A347-01. The Nichols-Farmington strain of T. pallidum subsp. pallidum was propagated by intratesticular inoculation of adult New Zealand White rabbits with 1 × 108 treponemes per testis; the treponemes were harvested approximately 12 days later and processed as described previously (29, 36).
Homology modeling and prediction of B cell epitopes.
The homology model of TP_0326 was generated by comparative modeling using the ModWeb server (https://modbase.compbio.ucsf.edu/modweb/) and the solved structure of full-length BamA (PDB accession no. 4K3B) of Neisseria gonorrhoeae (12) as the preferred template. The homology model was structurally analyzed by PyMOL and the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera/); structural figures were generated using PyMOL. Linear and discontinuous B cell epitopes were predicted using the ElliPro antibody (Ab) epitope server (37) and the DiscoTope-2.0 server (38), respectively.
Sequence alignment and calculation of conservation score.
Sequence alignment between BamAs from different strains of T. pallidum was generated with the programs Clustal W (39) and ESPript (version 3.0) (40). Variability scores for each position of TP_0326 were calculated using the Shannon entropy analysis of the protein variability server (PVS) (41).
Cloning and mutagenesis.
Recombinant constructs and oligonucleotide primers used in this study are presented in Fig. S1 and Table S1 in the supplemental material, respectively. DNA encoding full-length TP_0326 without its signal sequence (amino acid residues 22 through 837) was PCR amplified from T. pallidum genomic DNA. The resulting amplicon was cloned into the NheI (5′-end) and HindIII (3′-end) restriction sites of the expression vector pET23b (Novagen, San Diego, CA) in frame with the C-terminal polyhistidine tag. Using the homology model as a guide (see Fig. 2), selected regions of the TP_0326 β-barrel (loop 3β6 [L3β6] [residues 519 to 548], L4 [residues 568 to 602], L6 [residues 682 to 725], the L6 construct used for antibody generation [L6Ab; residues 687 to 725], and L7 [residues 749 to 787]) were PCR amplified from the pET23b plasmid harboring full-length TP_0326. The amplicon for L3β6 was cloned into the BamHI (5′-end) and HindIII (3′-end) sites of pET41a (Novagen) in frame with the N-terminal glutathione S-transferase (GST) tag and the C-terminal polyhistidine tag. The amplicons for L4, L6, and L7 were cloned into the BamHI (5′-end) and HindIII (3′-end) restriction sites of pET29a (Novagen) in frame with the N-terminal S tag and the C-terminal polyhistidine tag. To generate 326pel (mature TP_0326 expressed with an N-terminal E. coli PelB signal sequence), the region encoding full-length TP_0326 without its signal sequence was PCR amplified from T. pallidum genomic DNA and cloned into the BamHI (5′-end) and HindIII (3′-end) restriction sites of pET26b (Novagen, San Diego, CA) downstream and in frame with an N-terminal PelB signal sequence and C-terminal polyhistidine tag. To generate a POTRA arm, the portion of TP_0326 encoding all five of the predicted POTRA domains (residues 22 through 433) was PCR amplified from the pET23b plasmid harboring full-length TP_0326 and cloned into the NheI (5′-end) and HindIII (3′-end) sites of pET23b in frame with the C-terminal polyhistidine tag. The β-barrel region of TP_0326 (residues 434 through 837) was PCR amplified from the codon-optimized version of TP_0326 (synthesized by GeneScript) and cloned into the NheI (5′-end) and HindIII (3′-end) sites of pET23b in frame with the C-terminal polyhistidine. The skp gene was PCR amplified from E. coli DNA and cloned into the NheI (5′-end) and XhoI (3′-end) restriction sites of pET23b (Novagen, San Diego, CA) in frame with the C-terminal polyhistidine. The L4 open reading frame in pET29a was mutated using a site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). Nucleotide sequencing was performed by Genewiz, Inc. (South Plainfield, NJ).
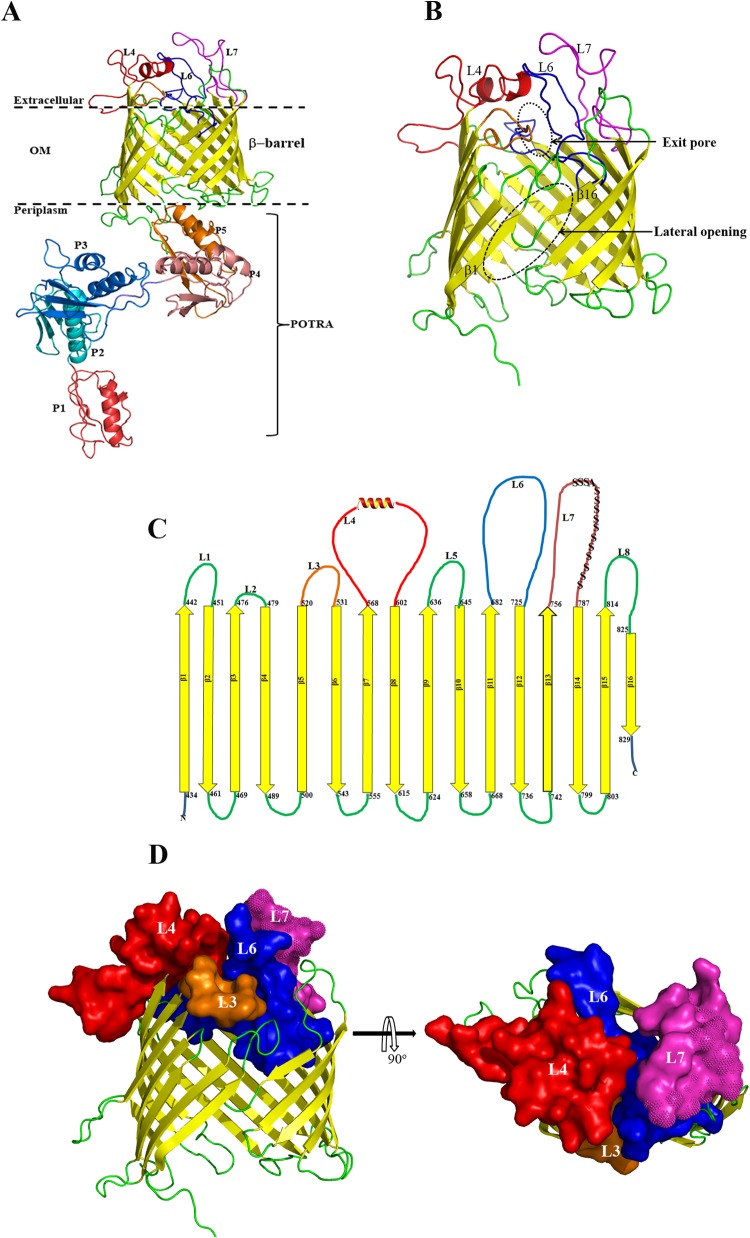
FIG 2.
(A) Homology model of full-length TP_0326 BamA based on the crystal structure of Neisseria gonorrhoeae BamA (12) showing the 16-stranded β-barrel and the five-domain periplasmic POTRA arm (P1 to P5). The three large predicted extracellular loops L4, L6, and L7 are shown in red, blue, and magenta, respectively. (B) Enlargement of β-barrel domain depicted as a ribbon model. The lateral opening and exit pore in the homology model are outlined by the dashed oval. (C) Predicted two-dimensional topology of the TP_0326 β-barrel. The amino acids predicted to demarcate the extracellular and periplasmic boundaries of each β strand are indicated. The polyserine tract in L7 is indicated by S's. (D) Side and top views of the β-barrel. Surface representations of L3, L4, L6, and L7 are shown using the same colors as in panels A and C. The polyserine tract in L7 is indicated with spikes.
Protein expression and purification.
All constructs were expressed in Overexpress C41(DE3) (Lucigen Corporation, Middleton, WI). For batch purification of full-length TP_0326, β-barrel, and L4, 500 ml of Luria broth (LB) medium was inoculated with 10 ml of overnight culture grown at 37°C; isopropyl β-d-1-thiogalactopyranoside (IPTG; final concentration, 0.1 mM) was added when the culture reached an optical density at 600 nm of between 0.2 and 0.3. Cells were grown for an additional 4 h and then harvested by centrifugation at 6,000 × g for 15 min at 4°C. The pellets were resuspended with 20 ml of 50 mM Tris (pH 7.5), 100 μg of lysozyme, and 100 μl of protease inhibitor cocktail (PIC; Sigma-Aldrich, St. Louis, MO) and stored at −20°C. After thawing, the bacterial suspension was lysed by sonication. After sonication, the pellet was recovered by centrifugation at 20,000 × g for 30 min at 4°C and then incubated in solubilization buffer (100 mM NaH2PO4 [pH 8.0], 10 mM Tris, 8 M urea) for 30 min at 4°C; the remaining insoluble material was removed by centrifugation at 20,000 × g for 30 min at 4°C. The supernatant was added to a Ni-nitrilotriacetic acid (NTA)–agarose matrix (Qiagen, Valencia, CA) that had been equilibrated in solubilization buffer and incubated with shaking at room temperature (RT) for 30 min. The matrix was washed with wash buffer (100 mM NaH2PO4 [pH 6.3], 10 mM Tris, 8 M urea) and subsequently eluted with elution buffer (100 mM NaH2PO4 [pH 4.5], 10 mM Tris, 8 M urea). For batch purification of the POTRA arm, L6, L6Ab, and L7, 500 ml of LB medium was inoculated with 50 ml of overnight culture grown at 37°C. At an optical density at 600 nm of 0.6, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM. The cells were grown for an additional 3 h and then harvested by centrifugation at 6,000 × g for 15 min at 4°C. The pellets were resuspended with 20 ml of 50 mM Tris (pH 7.5), 10% glycerol, 100 μg of lysozyme (Sigma-Aldrich), and 100 μl of PIC (Sigma-Aldrich). After the cells were thawed on ice, the bacterial suspension was lysed by sonication. Following sonication, NaCl and β-mercaptoethanol were added to the lysates to give final concentrations of 0.5 M and 0.1%, respectively. The supernatants were then cleared of cellular debris by centrifugation at 18,000 × g for 20 min at 4°C and applied to a Superflow Ni-NTA (Qiagen, Valencia, CA) immobilized metal-affinity chromatography (IMAC) column, previously equilibrated with 25 mM Tris (pH 7.5), 0.5 M NaCl, 10% glycerol, and 0.1% β-mercaptoethanol (buffer A). The protein was eluted with buffer A supplemented with 300 mM imidazole. In the case of L3β6, pooled fractions from the Ni-NTA column were mixed with preswollen glutathione-agarose beads (Sigma-Aldrich) and incubated overnight at 4°C. The following day, the glutathione-agarose with bound protein was loaded onto an empty column and washed extensively with 50 mM Tris (pH 7.5), 100 mM NaCl, and 10% glycerol. To excise the N-terminal GST tag, 100 U of thrombin (Sigma-Aldrich) was added to the resin, followed by incubation for 12 to 14 h at 4°C on a rocker. Cleaved L3 in the flowthrough was loaded onto a cation exchange column packed with SP Sepharose (Bio-Rad, Hercules, CA), washed extensively with 25 mM Tris (pH 7.5)–10% glycerol, and eluted with a gradient of 0 M to 1.0 M NaCl.
Immunologic reagents.
Rat polyclonal antiserum directed against the POTRA arm (29, 34) and FlaA (TP_0249) (36) were described previously. Antisera directed against the entire β-barrel, L3β6, L4, L6Ab, L7, and E. coli Skp were generated in 6-week-old female Sprague-Dawley rats as described previously (42). Polyclonal antibodies recognizing the POTRA arm and L4 were generated in rabbits by Rockland, Inc., according to their established protocol. The sensitivities of the various rat and rabbit TP_0326 antisera were assessed by immunoblot analysis using recombinant full-length TP_0326 and β-barrel proteins and T. pallidum lysates (see Fig. S2A to C in the supplemental material). The reactivity of L3β6 antisera was assessed by enzyme-linked immunosorbent assay (ELISA) against an L3 peptide (GLPHPYTSREQ; synthesized by GenScript) (see Fig. S2D in the supplemental material). Immune rabbit serum (IRS) was generated as described previously (36). Normal human serum (NHS) was purchased from Fisher Scientific Company (catalog no. MT35060CI). Deidentified, banked sera from HIV-negative patients with secondary syphilis were from two distinct geographic sources: (i) Dallas, TX (approved for use by the Institutional Review Board of the University of Connecticut Health Center [UCHC]), and (ii) Cali, Colombia, collected after we obtained informed consent under protocols approved by the human subject boards at the Connecticut Children's Medical Center (CCMC), UCHC, and the CIDEIM (43, 44).
Immunofluorescence analysis of T. pallidum encapsulated in gel microdroplets.
Freshly harvested T. pallidum cells were encapsulated in low-melting-point agarose (Sigma-Aldrich) microdroplets as previously described (29, 45). Encapsulated organisms were probed in a two-step process. In the first step, rat antiserum directed against β-barrel, the POTRA arm, L3β6, L4, or FlaA (36) (each diluted 1:100) was added to the bead suspensions (0.2 to 0.3 ml) in the presence or absence of 0.01% (vol/vol) Triton X-100 and incubated for 2 h. In the second step, beads were incubated with gentle mixing in a 34°C water bath for 2 h. The beads then were washed three times with CMRL medium (Connaught Medical Research Laboratories) by low-speed centrifugation (100 × g) and resuspended in CMRL medium followed by incubation for 1 h at 34°C with 1 μg/ml of goat anti-rat antibody–Alexa Fluor 488 conjugates (Invitrogen, Grand Island, NY). The beads then were washed three times with CMRL medium and observed with a epifluorescence Olympus BX-41 microscope using a 100× (1.4-numerical-aperture [NA]) oil immersion objective equipped with a Retiga Exi charge-coupled-device (CCD) camera (Q Imaging, Tucson, AZ) and fluorescein isothiocyanate (FITC) and rhodamine omega filter sets. For each sample, three slides were prepared and approximately 100 organisms were scored for labeling.
SDS-PAGE and immunoblotting.
Samples were resolved by Any kD Mini-Protean TGX gels (Bio-Rad, Hercules, CA) and transferred to nitrocellulose membranes (0.45-μm pore size; GE Healthcare) at 25 V for 25 min using a semidry apparatus (Bio-Rad, Hercules, CA). All sera, including IRS, normal rabbit serum (NRS), pooled human secondary syphilitic sera (HSS), and NHS, were preadsorbed with crude lysates of E. coli C41(DE3) cells. Membranes were blocked for 1 h with phosphate-buffered saline (PBS), 5% nonfat dry milk, 5% fetal bovine serum (FBS), and 0.1% Tween 20 and probed overnight at 4°C with antisera directed against the POTRA arm (1:1,000), β-barrel (1:1,000), L3 (1:1,000), L4 (1:1,000), L7 (1:500), NRS (1:500), and IRS (1:500). After being washed with PBS and 0.05% Tween 20 (PBST), the membranes were incubated for 1 h at 4°C with conjugated goat anti-rat or anti-mouse antibodies (Southern Biotech, Birmingham, AL) at dilutions of 1:30,000. In the case of human samples (NHS and HSS), membranes were blocked overnight with PBS, 5% nonfat dry milk, 5% FBS, and 0.1% Tween 20 and probed for 3 h at room temperature with NHS and HSS at dilutions of 1:250. After being washed with PBST, the membranes were incubated for 1 h at 4°C with a horseradish peroxidase (HRP)-conjugated goat IgG anti-human antibody (Pierce, Rockford, IL) at a dilution of 1:30,000. Following washes with PBST, the immunoblots were developed using the SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific, Waltham, MA).
ELISAs.
Ninety-six-well Maxi-Sorb immunoplates (Nunc, Naperville, IL) were coated with 100 μl of 10-μg/ml proteins in PBS and incubated overnight at 4°C. Plates were washed with PBST and blocked with 0.25% bovine serum albumin (BSA) in PBS. All rabbit and human sera from rabbit and humans were preadsorbed with crude lysates of E. coli C41(DE3) cells to remove Abs directed against E. coli. One hundred microliters of serum diluted to a final concentration of 1:1,000 in PBS with 0.25% BSA was added to each well and incubated at 4°C overnight. The plates were washed five times with PBST, followed by the addition to each well of 100 μl of HRP-conjugated goat IgG anti-rabbit or goat IgG anti-human antibody diluted 1:10,000 in PBS with 0.25% BSA. After 1 h of incubation at RT, the plates were washed five times with PBST, followed by the addition of the substrate {1 mM ABTS [(NH4)2(2, 2′-azino-bis-3-ethylbenzothiazoline-6-sulfonic acid)] in 50 mM phosphate citrate buffer, pH 4.2} and hydrogen peroxide (0.1% vol/vol). Absorbance measurements (415-nm wavelength) were taken after 30 min. After subtraction of background (no protein) values, means ± standard errors of the means (SEMs) of results from triplicate wells were calculated.
Fractionation of E. coli expressing 326pel.
E. coli C41(DE3) cells were transformed with plasmid pET26b harboring 326pel. Five hundred milliliters of LB medium was inoculated with a single colony picked from a transformed LB plate incubated overnight at 37°C; IPTG (final concentration, 0.3 mM) was added when the culture reached an optical density at 600 nm of between 0.2 and 0.3. Cells were grown for an additional 3 to 4 h and then harvested by centrifugation at 8,000 × g for 15 min at 4°C. The pellet was dissolved in 2× Laemmli sample buffer with 8 M urea (SB+U) prior to SDS-PAGE without boiling and immunoblotted with antiserum generated against the POTRA arm. Subcellular fractionation of E. coli cells expressing 326pel was based on previously published protocols (46).
Immunofluorescence analysis of E. coli cells expressing 326pel.
E. coli cells expressing 326pel were grown to mid-logarithmic phase, and IPTG (final concentration, 0.3 mM) was added to induce expression. E. coli cells expressing 326pel were grown for an additional 3 to 4 h, and 1 ml of induced cells was pelleted, washed, and resuspended in PBS. E. coli cells expressing 326pel were fixed in 4% paraformaldehyde for 30 min at RT, followed by several washes with PBS. To permeabilize cells prior to immunolabeling them, cells were resuspended in buffer P (PBS with 0.1% Triton X-100 and 10 mM EDTA) with 100 μg/ml lysozyme for 30 min at RT. Samples containing whole or permeabilized cells were resuspended in PBS or buffer P, respectively, containing 2% bovine serum albumin prior to overnight incubation at 4°C with rat antisera generated against the POTRA arm, β-barrel, L3, L4, and E. coli Skp, each at a dilution of 1:500, followed by incubation with Alexa Fluor 488-conjugated goat anti-rat IgG or Alexa Fluor 594-conjugated goat anti-rat IgG (1:300) for 1 h. After being washed, cells were resuspended in PBS and 20 μl of resuspended cells was mixed with 20 μl of Vectashield with DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories, Burlingame, CA) and placed on glass slides. Images were obtained using an Olympus microscope as described above. Images were processed and analyzed using ImageJ (http://rsb.info.nih.gov/ij/).
Antibody capture using E. coli cells expressing 326pel.
E. coli C41(DE3) cells transformed with plasmid pET26b harboring 326pel were grown to mid-logarithmic phase, and IPTG (final concentration, 0.3 mM) was added to induce expression. Cells were grown for an additional 3 to 4 h, and 10 ml of induced cells was pelleted, washed, and resuspended in PBS. IRS was preadsorbed with crude lysates of C41(DE3) cells to remove antibodies directed against E. coli. IRS was incubated overnight with induced cells expressing 326pel. The E. coli lysate incubated with IRS was washed two times with PBST. Bound antibodies were eluted by incubating the membrane with 1 ml of 100 mM glycine (pH 2.5) for 2 min and promptly added to tubes containing 100 μl of 2 M Tris-HCl (pH 8.5). The eluted antibodies were dialyzed against PBS (pH 7.4).
Opsonophagocytosis assays.
Opsonophagocytosis assays were performed as described previously (42, 47). Live treponemes were preincubated with 10% heat-inactivated sera for 2 h at 37°C in CMRL medium; at the end of preincubation, the motility of treponemes was confirmed by dark-field microscopy. Rabbit peritoneal macrophages then were incubated for 4 h at 37°C in a reduced-oxygen atmosphere (3% O2 and 5% CO2) at a multiplicity of infection (MOI) of 10:1. After incubation with macrophages, supernatants were removed to count the remaining T. pallidum organisms. Internalization of treponemes by macrophages was determined by IFA. Slides were fixed with 4% (vol/vol) paraformaldehyde and 0.1% Triton X-100 for 10 min at RT, blocked with CMRL medium supplemented with 10% FCS for 30 min at RT, and incubated with rat FlaA antiserum (1:75) for 1 h at RT. The slides then were washed with PBST and incubated with Alexa Fluor 488-conjugated goat anti-rat antiserum (1:75) (Invitrogen) for 30 min at RT. The slides were given one final wash and mounted in Vectashield containing DAPI. Fluorescent images were acquired on an epifluorescence Olympus BX-41 microscope using a 40× (1.4-NA) oil immersion objective equipped with a Retiga Exi CCD camera (Q Imaging, Tucson, AZ) and the following Omega filter sets: DAPI, FITC, and rhodamine. The percentages of rabbit macrophages containing degraded fluorescent spirochetes were systematically quantified for each of the conditions studied.
PCR amplification of TP_0326 β-barrel sequences from skin biopsy specimens obtained from two patients with secondary syphilis.
After obtaining informed consent, we took two punch biopsy specimens from the affected skin of two different patients with untreated secondary syphilis (designated Cali-77 and Cali-84) seen at our Cali, Colombia, study site (43, 44) according to protocols approved by the human subject boards at CCMC, UCHC, and CIDEIM (43, 44). DNA was extracted using the QIAamp DNA minikit (Qiagen) according to procedures recommended by the manufacturer, eluted in 100 μl of elution buffer at 70°C, and stored at −80°C. The concentration of DNA was determined spectrophotometrically by examining absorbance at 260/280 nm. The β-barrel region was amplified using the primers listed in Table S1 in the supplemental material, cloned into the pCR2.1-TOPO vector (Invitrogen), and sequenced.
Statistical analysis.
Statistical analysis was performed using Prism 4.0 (GraphPad Software, San Diego, CA). In the gel microdroplet assays and ELISAs, data are presented as grouped columns and were analyzed by using an unpaired Student t test.
Nucleotide sequence accession numbers.
The GenBank accession numbers for Cali-77 and Cali-84 are KP713715 and KP713716, respectively.
RESULTS
Bipartite architecture of TP_0326/BamA in T. pallidum.
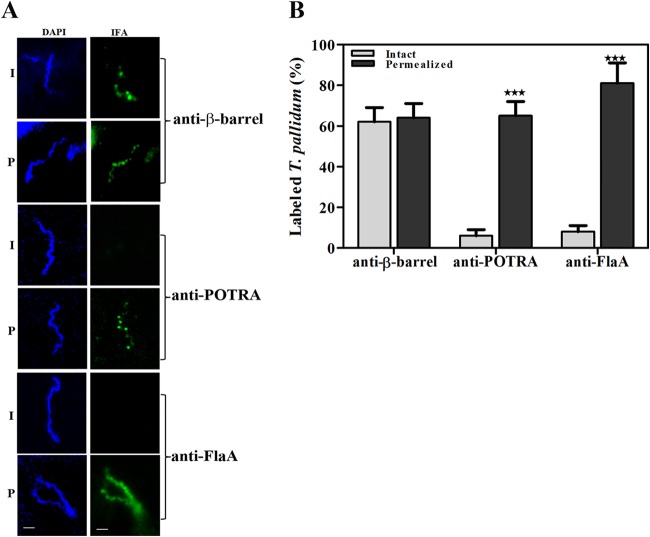
Previously, we used biophysical methodologies to establish that recombinant TP_0326 possesses the characteristic bipartite topology (i.e., the 5-domain POTRA arm and β-barrel) of a BamA ortholog (4, 34). We also demonstrated by proteinase K accessibility that native TP_0326 is surface exposed in T. pallidum (34). We began the present study by using our gel microdroplet immunofluorescence assay (29, 45) to determine the cellular locations of the β-barrel and the POTRA arm in live treponemes. This method is advantageous because it not only preserves the integrity of the spirochete's fragile OM during immunolabeling but also allows for its controlled removal by detergent solubilization in order to expose periplasmic antigens. Because T. pallidum expresses BamA at a very low copy number (∼100 molecules per cell) (34), at the outset, we confirmed that our rat β-barrel and POTRA antisera could recognize their target antigens at nanogram levels with recombinant proteins as well as in T. pallidum lysates (see Fig. S2 in the supplemental material). Representative images and graphed data from three independent experiments are shown in Fig. 1A and B, respectively. More than 60% (62% ± 7%) of the treponemes were labeled by antibodies against the β-barrel in the absence of detergent, far greater than the low percentage (6% ± 3%) of treponemes labeled with antibodies against the POTRA arm under the same conditions. The result for the POTRA antiserum is consistent with the background percentage of OM-damaged organisms typically observed in this assay (29, 36) and herein (8% ± 3%) based upon labeling with antibodies against the periplasmic protein FlaA. Importantly, the percentage of treponemes labeled by the POTRA antiserum increased dramatically in the presence of detergent (65% ± 7%), as was observed for FlaA (81% ± 10%). Also noteworthy was the punctate labeling observed with both β-barrel and POTRA antisera as opposed to the uniform labeling by the antiserum to the flagellar sheath protein FlaA. As a whole, these experiments establish definitively that only the β-barrel domain of TP_0326 contains surface-exposed epitopes.
FIG 1.
Bipartite topology of TP_0326 (BamA) in T. pallidum. (A) Following encapsulation in gel microdroplets, intact (I) T. pallidum or organisms permeabilized (P) with 0.05% Triton X-100 were probed with rat antisera directed against the β-barrel or POTRA regions of TP_0326 or FlaA. Antibody binding was detected with Alexa Fluor 488-conjugated goat anti-rat antibody (green). Scale bars = 2.5 μm. (B) Mean percentages of labeling ± SEMs from three independent gel microdroplet experiments. Statistical significance (I versus P) was assigned according to the following scheme: ***, P < 0.0001.
A homology model identifies putative surface-exposed regions in TP_0326.
Having confirmed the general membrane topology of native BamA in T. pallidum, we next turned our attention to the broader questions of its roles in OM biogenesis and disease pathogenesis. We reasoned that a structural model would be a powerful tool for advancing both lines of experimentation. Accordingly, we used ModWeb to generate a three-dimensional (3D) homology model for full-length TP_0326 based upon the recently solved crystal structure of N. gonorrhoeae BamA in the lateral open conformation (root mean square difference [RMSD] = 0.76) (12); the model is presented in Fig. 2. Of note, the 3D model for the β-barrel (Fig. 2B) readily superimposes on the solved structure of the E. coli BamA β-barrel (RMSD = 1.9) (14). Along with the five domains of the periplasmic POTRA arm, each containing a canonical β-α-α-β-β motif (8, 34), TP_0326 is predicted to contain a 16-stranded β-barrel in which the β1 and β16 strands can separate, presumably to enable insertion of transmembrane segments of OMP precursors into the OM bilayer (15). This supposition is supported by the finding that the shorter β1 and β16 strands of TP_0326 contain six residues similar to those shown by paired cysteine mutagenesis to be in proximity at the site of lateral opening in E. coli BamA (15) (Fig. S3). In Gram-negative BamAs, the fourth residue from the C terminus is a glycine thought to promote an inward kink in β16, further weakening its interactions with β1 (14). Although this residue is absent in TP_0326, T. pallidum BamA does contain a 4-residue extension at its C terminus that likely serves a similar purpose. The model predicts that the TP_0326 β-barrel possesses three large extracellular loops, L4, L6, and L7 (Fig. 2C and D), the latter containing a large polyserine tract unique to T. pallidum BamA (19, 34). L4 and L7 abut each other on the surface of the protein, largely overlying L6, which projects downward into the barrel. Together, these three loops comprise another structural characteristic of BamA, a dome that occludes the barrel's extracellular opening. L4 forms a major portion of the dome and contains a small α-helix that runs nearly parallel to the plane of the membrane, another highly conserved structural feature of BamAs (12). L7 sits obliquely atop the side of the barrel opposite the site of lateral opening, adopting a two-tiered conformation in which the polyserine tract largely covers the remainder of the loop. Electrostatics analysis (Fig. S4A) reveals that charged residues on the dome surface are asymmetrically distributed and located mainly in L4; the surface-exposed residues of L7 are largely neutral due to the presence of the polyserine tract. The electrostatics of the barrel interior is mostly positive (Fig. S4B). In solved structures for E. coli and N. gonorrhoeae BamAs, L6 attaches to the interior of the barrel wall via salt bridges between an Arg in a VRGF/Y motif within the loop and conserved Glu and Asp residues in β12 and β13 (14). Surprisingly, L6 of TP_0326 lacks this motif and is predicted to be stabilized instead by salt bridges between Glu692 (L6) and Arg648 (β11) and between Lys695 (L6) and Glu479 (L3) (Fig. S4C).
Lastly, we used the homology model to localize variability in TP_0326 sequences. We included, in addition to the Nichols, Dal-1, Chicago, SS14, and Mexico A strains, β-barrel sequences amplified from two skin biopsy specimens obtained from non-HIV-infected secondary syphilis patients (Cali-77 and Cali-84) enrolled in our Cali, Colombia, study site (see Fig. S5A and B in the supplemental material for the alignments of the POTRA arms and β-barrels, respectively). Conservation values for each residue were then generated and superimposed onto the model. Remarkably, the heterogeneity resides only on the extracellular face of the β-barrel domain (Fig. 3), results that support the model's robustness.
FIG 3.
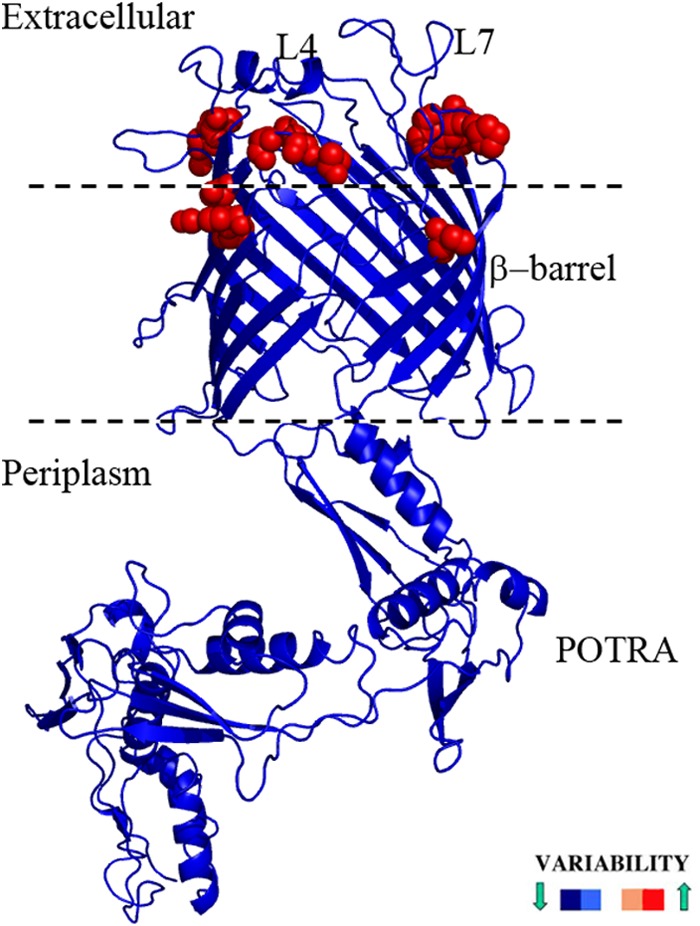
Conserved and variable regions of TP_0326, defined by sequence alignments between different strains of T. pallidum, superimposed on the homology model and colored according to variability scores. Variable amino acids are shown as red spheres.
L4 is surface exposed and an opsonic target.
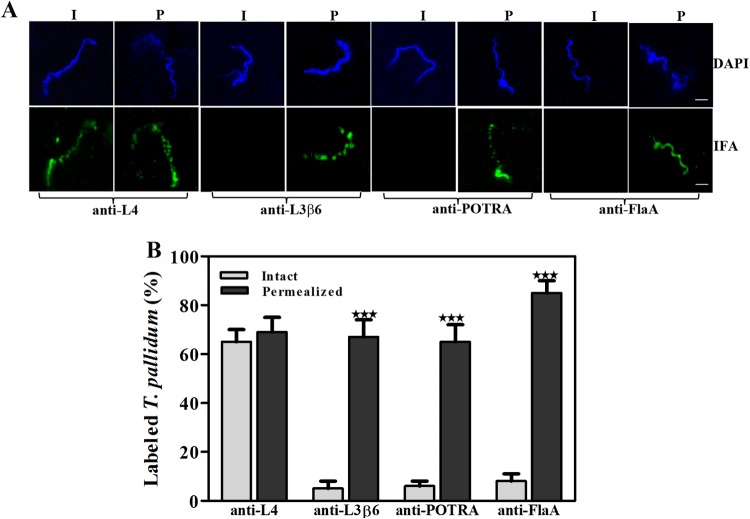
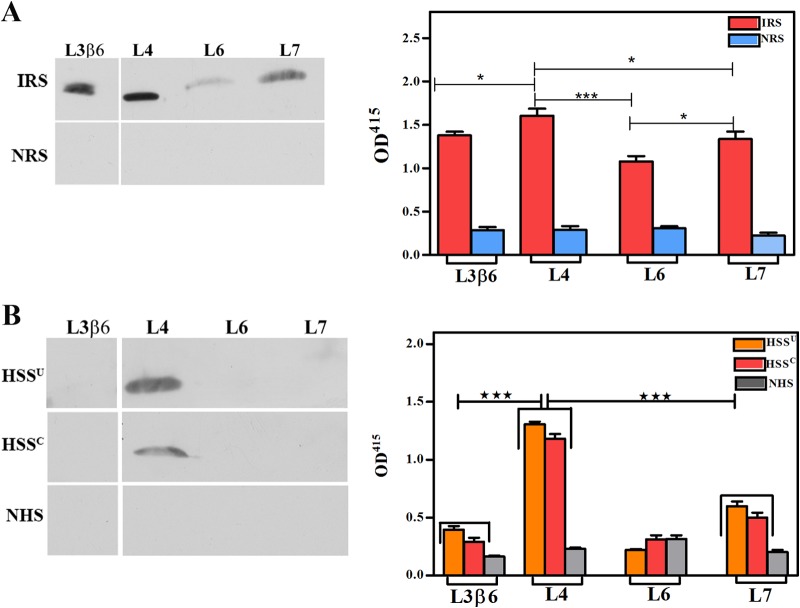
Studies next were conducted to assess the homology model's prediction that L4 and L7 are the principal surface-exposed loops in TP_0326. Whereas L4 proved to be highly immunogenic, we were unable to generate L6 and L7 antisera with the requisite sensitivity for surface localization experiments despite multiple immunization attempts (see Fig. S2 in the supplemental material). To enhance the interpretation of our localization experiments, we also generated antiserum against a region of the β-barrel predicted not to be surface exposed—the β6 transmembrane strand plus adjoining small loop L3 (Fig. 2C). The resulting construct, designated L3β6, induced a strong antibody response, which included L3 (Fig. S2D).
Using the gel microdroplet technique (see Fig. 4A and B, respectively, for representative images and summary graphs), the percentage of intact treponemes labeled with the L4 antiserum (65% ± 5%) was virtually identical to that labeled by antibodies against the β-barrel (Fig. 1B). In contrast, L3β6 antibodies failed to label intact treponemes above background levels. Labeling with this antiserum increased dramatically in the presence of detergent (65% ± 7%), as observed for FlaA (81% ± 10%) and the POTRA arm (67% ± 7%). Moreover, as with the β-barrel and POTRA antisera (Fig. 1A), L4 and L3β6 antibodies produced a punctate labeling pattern clearly distinguishable from the continuous labeling obtained with antibodies against the flagellar sheath.
FIG 4.
Surface localization of L4 in T. pallidum. (A) Following encapsulation in gel microdroplets, intact (I) T. pallidum or organisms permeabilized (P) with 0.05% Triton X-100 were probed with rat antisera directed against L4, the POTRA arm of TP_0326, or FlaA. Antibody binding was detected with Alexa Fluor 488-conjugated goat anti-rat antibodies (green). Scale bar = 2.5 μm. (B) Mean labeling values ± SEMs from three independent experiments are shown. Statistical significance (I versus P) was assigned according to the following scheme: ***, P < 0.0001.
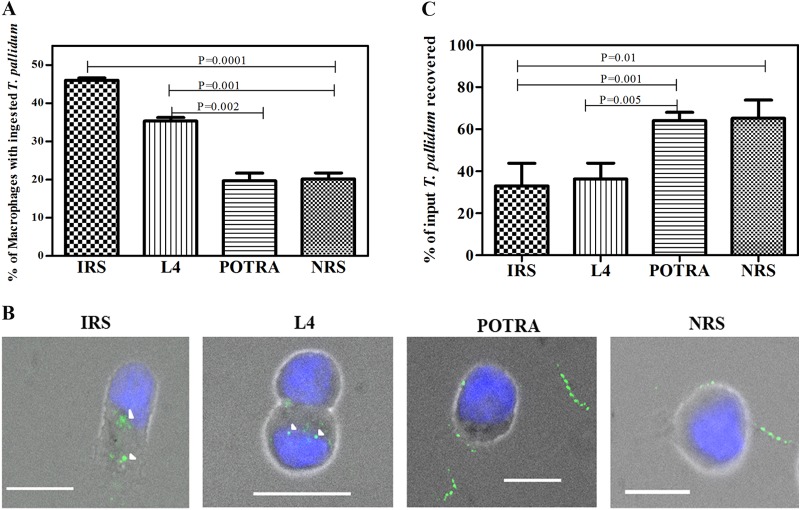
To complement the surface immunolabeling results, we next assessed the ability of rabbit L4 antiserum to promote internalization of motile treponemes by rabbit peritoneal macrophages. Compared to NRS and rabbit POTRA antiserum, L4 antibodies significantly increased the uptake of organisms, although to a lesser extent than did IRS (Fig. 5A and B). As a qualitative measure of the degree of surface antibody binding by the treponemal populations incubated with the various antisera, in each experiment we also enumerated spirochetes in supernatants collected at the conclusion of the 4-h incubation period. Supernatants containing spirochetes incubated with IRS and L4 antiserum contained significantly lower percentages of organisms (using input values as 100%) than supernatants containing spirochetes incubated with NRS or the POTRA antiserum (Fig. 5C), consistent with the internalization results.
FIG 5.
L4 is an opsonic target in T. pallidum. (A) Percentages of rabbit peritoneal macrophages containing internalized treponemes following a 4-h incubation period with the indicated sera. (B) Representative micrographs. Each image is a composite of bright-field nuclei stained with DAPI and Alexa Fluor 488-labeled T. pallidum. Arrowheads indicate degraded T. pallidum in phagolysosomes. Scale bars, 10 μm. (C) Percentages of treponemes in supernatants recovered at the conclusion of the 4-h incubation period. The results shown in panels A and C are means ± SEMs for three independent experiments; P values of <0.05 were considered to be significant.
L4 is an immunodominant extracellular loop.
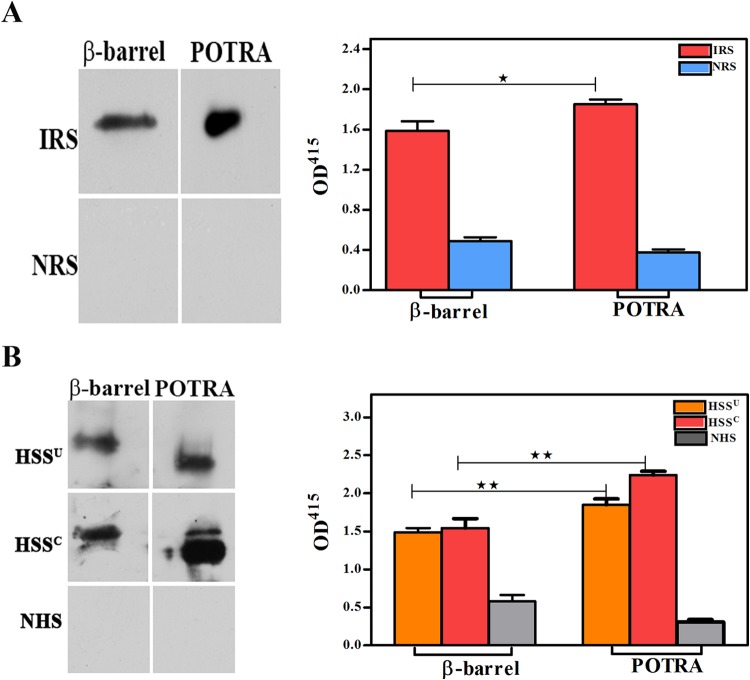
We next sought to determine which surface-exposed features of TP_0326 elicit a humoral response during syphilitic infection. We began by performing an in silico analysis of full-length TP_0326 using the ElliPro (37) and DispcoTope-2.0 (38) servers to identify, respectively, linear and conformational B cell epitopes. These algorithms predicted that the POTRA arm contains only linear B cell epitopes, predominantly in the first of its five POTRA domains, but that the β-barrel contains both linear and conformational epitopes, mainly within L4 (Table 1; see also Fig. S6 in the supplemental material). To test these predictions, we first assessed the reactivity of pooled IRS and two demographically distinct pools of human secondary syphilitic sera (from the United States and Colombia [HSSU and HSSC, respectively]) against the β-barrel and POTRA arm by immunoblotting and ELISA. Both the rabbit and patient sera reacted well with both domains, although in all cases the values were significantly greater for the POTRA arm than for the β-barrel (Fig. 6). Of note, we previously reported that HSS contains antibodies directed exclusively against the POTRA arm (34). We attribute this unanticipated discrepancy to our use in the present study of IgG-specific immunoconjugates (see Materials and Methods), whereas we formerly used an immunoconjugate pan-reactive for IgG, IgA, and IgM (34). To follow up these results, we examined the reactivities of the sera against L3β6, L4, L6, and L7. By immunoblotting and ELISA, the pooled IRS reacted strongly with L4, less so with L3β6 and L7, and weakly with L6 (Fig. 7A). Both HSS pools recognized only L4 by immunoblotting, while a weak response to L7 was detected by ELISA (Fig. 7B).
TABLE 1.
Predicted linear B cell epitopes in TP_0326
| Start position | End position | Amino acid sequence | Region | Scorea |
|---|---|---|---|---|
| 33 | 112 | ISAISFEGLEYIARGQLDTIFSQYKGQKWTYELYLEILQKVYDLEYFSEVSPKAVPTDPEYQYVMLQFTVKERPSVKGI | POTRA arm (P1) | 0.911 |
| 335 | 350 | EEHRDTAEKTLSFKIT | POTRA arm (P4) | 0.873 |
| 187 | 204 | VSRIQFKGNKAFTESVLK | POTRA arm (P2) | 0.813 |
| 581 | 601 | NFYDKDNNQPFDLTVKEQLNW | β-Barrel (L4) | 0.803 |
Cutoff score, ≥0.8.
FIG 6.
T. pallidum-immune rabbits and humans with secondary syphilis mount an antibody response against the β-barrel and the POTRA arm. Reactivities of pooled IRS (A) and pooled sera from the U.S. (HSSU) and Colombian (HSSC) HIV-negative patients with secondary syphilis (B) against the β-barrel and POTRA arms of TP_0326 assessed by immunoblotting (1 μg of protein) and ELISA (100 ng of protein). Results shown for ELISAs are means ± SEMs from three independent experiments. Statistical significance was assigned according to the following scheme: *, P < 0.05; **, P < 0.005. OD415, optical density at 415 nm.
FIG 7.
L4 is an immunodominant surface feature in TP_0326. Reactivities of pooled IRS (A) and pooled sera from U.S. (HSSU) and Colombian (HSSC) HIV-negative patients with secondary syphilis (B) against L3β6, L4, L6, and L7 assessed by immunoblotting (1 μg of protein) and ELISA (100 ng of protein). Results shown for ELISAs are means ± SEMs from three independent experiments. Statistical significance was assigned according to the following scheme: *, P < 0.05; ***, P < 0.0001.
Use of E. coli expressing outer membrane-localized TP_0326 to confirm surface accessibility of L4 antibodies elicited during syphilitic infection.
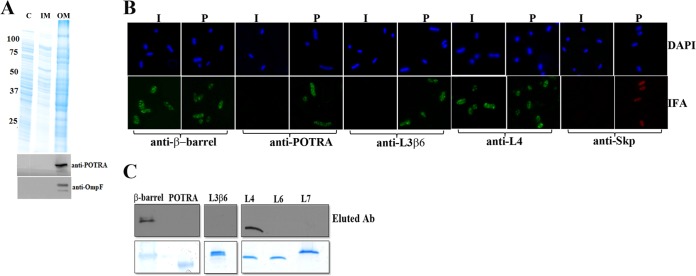
A key question arising from the previous results is whether the L4 antibodies in syphilitic sera bind to epitopes on the spirochetal surface. Adsorption experiments might address this issue but cannot be performed readily with T. pallidum given the spirochete's noncultivability and, especially, the fragility of its OM. We reasoned, however, that it might be possible to answer this question using E. coli expressing OM-localized TP_0326 as a T. pallidum surrogate. Toward this end, we cloned into E. coli C41 a pET26b plasmid construct (326pel) (see Fig. S7A and B in the supplemental material) encoding a PelB signal sequence followed by the mature (i.e., processed) portion of TP_0326. As shown in Fig. 8A, following subcellular fractionation of IPTG-induced E. coli cells expressing 326pel, TP_0326 was recovered exclusively in the OM fraction. To assess the surface expression and membrane orientation of 326pel, we performed IFA of intact and permeabilized E. coli cells expressing 326pel; OM integrity was established by probing cells in parallel with antibodies against the E. coli periplasmic chaperone Skp (48). Representative images of results obtained with IPTG-induced cells are shown in Fig. 8B. Labeling of intact cells was observed with antibodies to the β-barrel but not to the POTRA arm. Antibodies against the POTRA arm did, however, label permeabilized E. coli expressing 326pel, thereby confirming that 326pel not only was OM localized but also had adopted a native, bipartite topology. Importantly, intact cells also were labeled with antibodies to L4 but not to L3β6, further supporting the use of E. coli cells expressing 326pel as a T. pallidum surrogate. No labeling of uninduced E. coli cells expressing 326pel was observed with antibodies to the β-barrel, POTRA arm, or L4 (data not shown).
FIG 8.
TP_0326 with a PelB signal sequence localizes to the E. coli outer membrane and displays bipartite topology. (A) Fractionation of E. coli 326pel was performed as described by Thein et al. (46). Samples from different subcellular fractions were subjected to immunoblot analysis with monospecific rat antiserum against the POTRA arm of TP_0326. The membrane then was stripped and reprobed with antiserum against E. coli OmpF. C, IM, and OM, cytoplasmic, inner membrane, and outer membrane fractions, respectively. Molecular mass standards (in kilodaltons) are indicated on the left. (B) IFA of intact (I) and permeabilized (P) E. coli expressing 326pel probed with preabsorbed rat antisera directed against the β-barrel, POTRA arm, L3β6, or L4 of TP_0326 or E. coli Skp. Antibody binding was detected with either Alexa Fluor 488 (green)- or Alexa Fluor 596 (red)-conjugated goat anti-rat antibodies. (C) Antibodies captured by adsorption with E. coli 326pel were immunoblotted against the designated regions of TP_0326.
Having established that 326pel is properly oriented in the E. coli OM, we next confirmed that pooled IRS adsorbed with E. coli C41(DE3) reacted specifically with the protein in induced but not uninduced 326pel-expressing E. coli lysates (see Fig. S7C in the supplemental material). We then incubated the adsorbed IRS with intact E. coli expressing 326pel and immunoblotted the eluted antibodies against the recombinant β-barrel and POTRA arm. The eluted antibodies reacted only with the β-barrel (Fig. 8C), providing further evidence for the bipartite membrane topology of 326pel. When the eluted antibodies were immunoblotted against recombinant L3β6, L4, L6, and L7, reactivity was observed only against L4 (Fig. 8C).
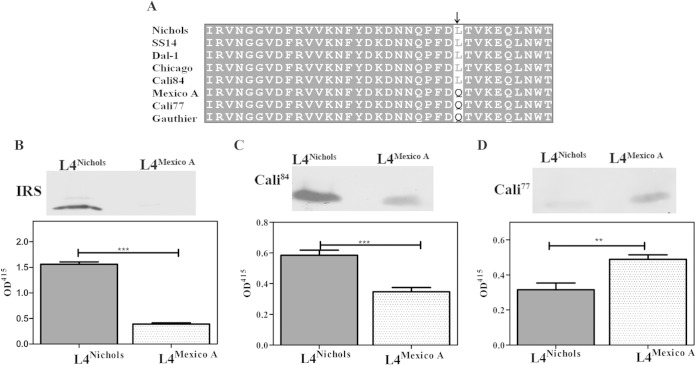
A nonconservative amino acid substitution in L4 markedly affects binding by antibodies in syphilitic sera.
As noted earlier (Table 1), ElliPro identified a linear B cell epitope in L4 spanning residues 581 to 601. From the alignments of the TP_0326 β-barrel sequences (Fig. 9A; see also Fig. S5B in the supplemental material), we noted that the generally conserved Leu593 residue in L4 in the Nichols strain is replaced by Gln593 in the Mexico A strain (30) and in the Mexico A-like β-barrel sequence amplified from Cali patient 77; the L4 sequence from Cali patient 84 is that of Nichols. We next examined the effect of this substitution on the reactivities of L4 antibodies in syphilitic sera. The immunoblots and ELISAs in Fig. 9B demonstrate that a Leu593 → Gln593 substitution virtually abrogated the reactivity of IRS from animals infected with T. pallidum Nichols. As shown in Fig. 9C and D, each patient's serum reacted more strongly with the homologous sequence, with the differences in ELISA optical density values reaching statistical significance.
FIG 9.
Leu593 in L4 is essential for binding of antibodies in the sera of secondary syphilis patients. (A) Multiple-sequence alignment of the L4 regions of BamAs from geographically diverse T. pallidum subsp. pallidum strains, sequences amplified from skin biopsy specimens from two secondary syphilis patients (Cali-77 and Cali-84) enrolled at our Cali, Colombia, study site (43, 44), and the Gauthier strain of the T. pallidum subsp. pertenue. All strains of T. pallidum subsp. pertenue in the database have same the L4 sequence. The arrow indicates residue 593. Immunoblotting and ELISA reactivities of pooled IRS from rabbits infected with the T. pallidum Nichols strain (B) and sera from Cali-84 (C) and Cali-77 (D) against L4Nichols and L4Mexico A. The ELISA results are means ± SEMs from three independent experiments. Statistical significance was assigned according to the following scheme: **, P < 0.005; ***, P < 0.0001.
DISCUSSION
From the time of their discovery, it was presumed that the low-density particles visualized in freeze fracture electron micrographs of the T. pallidum OM represent proteins containing membrane-spanning domains (24, 25). Over the years, efforts to molecularly characterize these entities have been impeded by their low abundance, the spirochete's recalcitrance to in vitro cultivation, the fragility of the T. pallidum OM, and the paucity of genes encoding proteins related to well-characterized bacterial OMPs in the spirochete's genome (23, 49). The conceptual breakthrough in our quest for T. pallidum rare OMPs was the use of a structural bioinformatics approach to mine the spirochete's genome for proteins predicted to adopt a β-barrel conformation; TP_0326/BamA, the only OMP in T. pallidum related to OMPs in Gram-negative bacteria, emerged as a top candidate in the putative T. pallidum OM proteome (29). The finding of BamA (29) and subsequently a BAM complex (34) implied the existence of other β-barrel OMPs which serve as substrates for the OM biogenesis machinery, a supposition confirmed by biophysical characterization and surface localization of the T. pallidum repeat C (TprC) subfamily of proteins (42, 62). Because TP_0326 resides constitutively at the host-pathogen interface, one might anticipate that its involvement in the disease process extends beyond its role in OM biogenesis. Indeed, these aspects of BamA are likely to be two sides of the same evolutionary coin. The homology model of T. pallidum BamA has furnished a structural template with which we can begin to dissect the molecular underpinnings of stealth pathogenicity along with the host's efforts to contain it by targeting the surface-exposed epitopes of this rare OMP.
The BAM pathway begins with the interception of newly exported OMP precursors within the periplasmic space by chaperones, which then transfer them to the POTRA arm (3). The POTRA arm, assisted by ancillary factors, then threads the precursors to the BamA barrel for insertion into the OM (4–6). Notably, with the exception of BamA, the components of the BAM complex are poorly conserved across prokaryotic taxa (50). This phylogenetic divergence suggests that bacterial species tailor the OM assembly process as they adapt to the environmental pressures that shape their OMP repertoires. T. pallidum appears to well exemplify this phenomenon of parallel evolution of OM constituents and assembly pathway. In contrast to E. coli, in which SurA is the major periplasmic chaperone and Skp plays a subsidiary role (48), T. pallidum harbors only Skp (28). Although the POTRA arm of TP_0326 contains five canonical structural domains, recent Small-angle X-ray scattering (SAXS) analysis reveals that it is much less flexible than those of E. coli and other pathogenic spirochetes (A. Luthra, A. Anand, M. R. Kenedy, D. R. Akins, and J. D. Radolf, unpublished data), findings further indicative of atypical chaperone interactions and functionality within the periplasmic space. The large size of the T. pallidum BAM complex (∼400 kDa), its modularity, and the absence of orthologs for known BAM subunits (34) strongly suggest that the mechanisms whereby nascent OMPs are handed off by the POTRA arm in T. pallidum differ substantially from those in Gram-negative prototypes. The sequence alignment between TP_0326 and structurally solved BamAs (see Fig. S3 in the supplemental material), as well as homology modeling, extends this concept of structure-function divergence in the T. pallidum OM assembly pathway to the TP_0326 β-barrel, which shares only ∼15% identity with its N. gonorrhoeae and E. coli counterparts despite adopting a typical BamA fold. While TP_0326 possesses the requisite dome structure to block egress of OMP precursors to the extracellular space, its surface electrostatics and the loop-loop interactions that create it differ considerably from those of the solved BamA prototypes (12, 14). Recent mutagenesis studies with E. coli have established that passage of OMP precursors through the barrel depends upon tethering of L6 within the channel (12, 14). The predicted positioning of L6 in TP_0326 indicates that it fulfills a comparable in-channel guidance function, while the lack of an essential VRGF/Y tethering motif suggests that it does so in a variant manner. Lastly, given that many of the putative OMPs in the T. pallidum OM proteome have basic isoelectric points, the positive electrostatics inside the barrel seems noteworthy, as it would tend to restrict trafficking through the channel, directly impacting the OM's total protein content as well as its composition.
The concept of stealth pathogenicity was based on the well-recognized ability of T. pallidum to disseminate rapidly and widely during early syphilis, even in the face of a robust cellular and humoral immune response. It was bolstered by the discovery of the spirochete's unorthodox OM ultrastructure and poor surface antigenicity in vitro (26). Host defenses do, however, eventually contain the pathogen, though months to years may elapse before this watershed event occurs (20). A wealth of ex vivo studies of the rabbit model (51, 52) and, more recently, humans (44, 53) indicates that opsonic antibodies elicited during infection are important for controlling spirochete burdens, with OMPs assumed to be the relevant antigenic targets. TP_0326/BamA introduces a new concept to the syphilis field; understanding how antibodies promote clearance of spirochetes requires not only identification of rare OMPs but detailed knowledge of their architectures and surface topologies. That TP_0326 is immunogenic during infection and capable of eliciting opsonic antibodies is well recognized (19, 54, 55). However, much of the antibody response is directed against the periplasmic POTRA arm and, in effect, wasted. The homology model and B cell epitope predictions enabled us to pinpoint a site of vulnerability on the molecule's surface, information of considerable importance for future vaccine-related studies. Whether one can extrapolate from the opsonophagocytosis assays conducted with L4 antisera to natural infection remains an open question. It seems reasonable to conjecture, however, that L4 antibodies elicited during infection do contribute to bacterial clearance.
Analysis of the dome region also yielded insights into the countermeasures that the spirochete has evolved to limit the damage that might be inflicted against this sizable, surface-exposed region. Much of the dome's surface is poorly immunogenic, a property attributable to the polyserine tract in L7, a unique feature of the T. pallidum BamA protein that has long intrigued investigators (19, 34). Moreover, the protection afforded by the polyserine tract may extend beyond its poor immunogenicity. Polyserine tracts tend to be highly flexible; the adsorption experiments with IRS and E. coli expressing 326pel suggest that this property enables it to shield L7 epitopes outside the tract from antibody binding. Since L4 likely serves an essential function in TP_0326, its inherent immunogenicity appears to be unavoidable, rendering it, in essence, an Achilles' heel for the spirochete. However, the ability of L4 antibodies to promote killing may be subject to limitations. L4 is an extremely low-copy-number target, and the level of binding of high-titer L4 antibodies by intact treponemes is well below 100% by IFA. Previously, we noted the heterogeneity of surface antibody binding within spirochete populations and proposed that it helps explain the duality of secondary syphilis (i.e., the parallel occurrence of bacterial clearance and immune evasion) (44). We contend that this phenomenon is reproduced ex vivo by the lack of internalization of sizable percentages of treponemes in opsonophagocytosis assays, as seen here and elsewhere (42, 44, 56), following incubation with IRS, patient sera, and antisera to surface-exposed regions of known OMPs. The punctate distribution of TP_0326 along the length of the spirochete, unaffected by removal of the OM, suggests that the mobility of the protein within the OM bilayer is restricted. Because efficient uptake of IgG-opsonized particles requires clustering of FcγR receptors (57, 58), limiting the mobility of TP_0326 represents another potential mechanism whereby the spirochete can resist killing by surface-bound L4 antibodies. In contrast to the natural human host, rabbits generate a much broader range of antibodies against the β-barrel, including all three large extracellular loops; however, the IFA and/or antibody capture experiments with E. coli expressing 326pel revealed that only the L4 antibodies can access their targets. Thus, even if one could circumvent the poor immunogenicity of L6 and L7 in humans, it seems doubtful that antibodies against these loops would contribute to a protective immune response.
Genomic analysis of T. pallidum strains has revealed a remarkably low degree of sequence diversity between T. pallidum strains (31). Nevertheless, sequence polymorphisms, typically amino acid substitutions resulting from single nucleotide changes, some of which are located in presumptive or proven OMPs, including members of the Tpr family, have been identified (31). Until our study, polymorphisms in a particular OMP had not been related to regions of the protein proven experimentally to be topologically and immunologically relevant. In 1953, the Mexico A strain of T. pallidum was isolated from a patient in that country with primary syphilis. Recently, Petrosova and coworkers sequenced the genome of the Mexico A strain and reported that its TP_0326 gene is a chimera of sequences from T. pallidum subsp. pallidum and T. pallidum subsp. pertenue, the agent of yaws (30). The sequence alignment in Fig. 9A shows that the T. pallidum subsp. pertenue L4 sequence (represented by the Gauthier strain) contains the same substitution (Gln593) as in the Mexico A strain. By chance, we amplified a Mexico A-like TP_0326 β-barrel sequence from T. pallidum DNA extracted from a secondary syphilis biopsy specimen obtained in our Cali, Colombia, site. We determined that one of the sequence polymorphisms in the Mexico A TP_0326 gene causes a nonconservative amino acid substitution in the L4 linear epitope that greatly diminishes its reactivity with sera from patients infected with strains containing the more conserved Nichols L4; the converse was also true. These results imply that anti-L4 antibodies of one type would have diminished opsonic potential against the other and, along with relevant polymorphisms in other OMPs, might collectively limit the degree of cross-protection in patients. This conjecture leads to scenarios whereby antigenically variant strains might be introduced into already-endemic populations and in which reinfection would facilitate circulation of divergent strains. An intriguing observation was that IRS from T. pallidum Nichols-infected rabbits showed virtually no reactivity with Mexico A L4, in contrast to the partial reactivity of serum from a patient with Nichols L4 antibodies. This result may reflect differences in the binding properties of human and rabbit antibodies produced against the major L4 epitope.
Investigations performed with refolded recombinant constructs and liposomes were instrumental in establishing the bipartite architecture of TP_0326 and confirming that its C-terminal domain forms an amphiphilic β-barrel (34), the secondary structural sine qua non of a prokaryotic OMP (1, 2). However, delineating surface topology with methodologies of this type is a different matter, since such experiments require both native conformation and proper orientation in a membrane environment. Expression of TP_0326 in E. coli in a form which localizes the β-barrel to the OM and the POTRA arm to the periplasm, therefore, represents a significant technical breakthrough. Substantial evidence presented herein argues that the OM-localized form of recombinant TP_0326 is a feasible heterologous platform for further topological mapping of the β-barrel region. This result is also notable because it demonstrates that the BAM of E. coli can recognize and efficiently fold an evolutionarily diverse BamA, including one, such as TP_0326, which lacks a canonical C-terminal recognition motif (34). Years ago, in our freeze fracture electron microscope analyses of the T. pallidum OM, we called attention to the spiral distribution pattern of intramembranous particles, their lack of mobility, and their location in areas of the OM away from the flagellar filaments, the regions of greatest separation between the outer and inner membranes (24, 59). At the time, we had no plausible explanation for this observation. Now, however, we can postulate that the BAM complex is static in T. pallidum due to interactions of the inflexible POTRA arm, which is long enough to reach the midzone of the periplasmic compartment, where the peptidoglycan sacculus resides (34, 60, 61; Luthra et al., unpublished data). A logical extension of this idea is that proteins insert into the OM via OM biogenesis zones established by the fixed location of BAM complexes. If so, this would represent a novel form of spatial organization that cannot be replicated in E. coli and must be studied in its native locale.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI-26756 (J.D.R.) and R03TW009172 (J.C.S. and A.R.C.), Colciencias (A.R.C.), Connecticut Children's Medical Center general research funds (J.D.R.), and Burr Curtis Strategic Research Funds (J.C.S.).
We are indebted to Susan K. Buchanan for her insights regarding BamA's structure. We also acknowledge the excellent technical assistance of Lady G. Ramirez and Carlos Valencia at CIDEIM. Lastly, we thank Nancy Saravia for her unflagging encouragement and support.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00086-15.
REFERENCES
- 1.Wimley WC. 2003. The versatile β-barrel membrane protein. Curr Opin Struct Biol 13:404–411. doi: 10.1016/S0959-440X(03)00099-X. [DOI] [PubMed] [Google Scholar]
- 2.Fairman JW, Noinaj N, Buchanan SK. 2011. The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr Opin Struct Biol 21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruiz N, Kahne D, Silhavy TJ. 2006. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol 4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 4.Ricci DP, Silhavy TJ. 2012. The Bam machine: a molecular cooper. Biochim Biophys Acta 1818:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misra R. 2012. Assembly of the β-barrel outer membrane proteins in Gram-negative bacteria, mitochondria, and chloroplasts. ISRN Mol Biol 2012:15. doi: 10.5402/2012/708203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selkrig J, Leyton DL, Webb CT, Lithgow T. 2014. Assembly of beta-barrel proteins into bacterial outer membranes. Biochim Biophys Acta 1843:1542–1550. doi: 10.1016/j.bbamcr.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Heinz E, Lithgow T. 2014. A comprehensive analysis of the Omp85/TpsB protein superfamily structural diversity, taxonomic occurrence, and evolution. Front Microbiol 5:370. doi: 10.3389/fmicb.2014.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. 2007. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 9.Bennion D, Charlson ES, Coon E, Misra R. 2010. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol Microbiol 77:1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. 2010. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure 18:1492–1501. doi: 10.1016/j.str.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knowles TJ, Jeeves M, Bobat S, Dancea F, McClelland D, Palmer T, Overduin M, Henderson IR. 2008. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol 68:1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- 12.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK. 2013. Structural insight into the biogenesis of beta-barrel membrane proteins. Nature 501:385–390. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albrecht R, Schutz M, Oberhettinger P, Faulstich M, Bermejo I, Rudel T, Diederichs K, Zeth K. 2014. Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr D Biol Crystallogr 70:1779–1789. doi: 10.1107/S1399004714007482. [DOI] [PubMed] [Google Scholar]
- 14.Ni D, Wang Y, Yang X, Zhou H, Hou X, Cao B, Lu Z, Zhao X, Yang K, Huang Y. 2014. Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. FASEB J 28:2677–2685. doi: 10.1096/fj.13-248450. [DOI] [PubMed] [Google Scholar]
- 15.Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK. 2014. Lateral opening and exit pore formation are required for BamA function. Structure 22:1055–1062. doi: 10.1016/j.str.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Thomas WR, Chong P, Loosmore SM, Klein MH. 1998. A 20-kilodalton N-terminal fragment of the D15 protein contains a protective epitope(s) against Haemophilus influenzae type a and type b. Infect Immun 66:3349–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruffolo CG, Adler B. 1996. Cloning, sequencing, expression, and protective capacity of the oma87 gene encoding the Pasteurella multocida 87-kilodalton outer membrane antigen. Infect Immun 64:3161–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su YC, Wan KL, Mohamed R, Nathan S. 2010. Immunization with the recombinant Burkholderia pseudomallei outer membrane protein Omp85 induces protective immunity in mice. Vaccine 28:5005–5011. doi: 10.1016/j.vaccine.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Cameron CE, Lukehart SA, Castro C, Molini B, Godornes C, Van Voorhis WC. 2000. Opsonic potential, protective capacity, and sequence conservation of the Treponema pallidum subspecies pallidum Tp92. J Infect Dis 181:1401–1413. doi: 10.1086/315399. [DOI] [PubMed] [Google Scholar]
- 20.Radolf JD, Tramont EC, Salazar JC. 2014. Syphilis (Treponema pallidum), p 2684–2709. In Bennett JE, Dolin R, Blaser MJ (ed), Mandell, Douglas and Bennett's principles and practice of infectious diseases, 8th ed Churchill Livingstone Elsevier, Philadelphia, PA. [Google Scholar]
- 21.Cox DL. 1994. Culture of Treponema pallidum. Methods Enzymol 236:390–405. doi: 10.1016/0076-6879(94)36029-4. [DOI] [PubMed] [Google Scholar]
- 22.Radolf JD, Robinson EJ, Bourell KW, Akins DR, Porcella SF, Weigel LM, Jones JD, Norgard MV. 1995. Characterization of outer membranes isolated from Treponema pallidum, the syphilis spirochete. Infect Immun 63:4244–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cameron CE. 2006. T. pallidum outer membrane and outer membrane proteins, p 237–266. In Radolf JD, Lukehart SA (ed), Pathogenic treponemes: cellular and molecular biology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 24.Radolf JD, Norgard MV, Schulz WW. 1989. Outer membrane ultrastructure explains the limited antigenicity of virulent Treponema pallidum. Proc Natl Acad Sci U S A 86:2051–2055. doi: 10.1073/pnas.86.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker EM, Zampighi GA, Blanco DR, Miller JN, Lovett MA. 1989. Demonstration of rare protein in the outer membrane of Treponema pallidum subsp. pallidum by freeze-fracture analysis. J Bacteriol 171:5005–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salazar JC, Hazlett KR, Radolf JD. 2002. The immune response to infection with Treponema pallidum, the stealth pathogen. Microbes Infect 4:1133–1140. doi: 10.1016/S1286-4579(02)01638-6. [DOI] [PubMed] [Google Scholar]
- 27.Cameron CE, Lukehart SA. 2014. Current status of syphilis vaccine development: need, challenges, prospects. Vaccine 32:1602–1609. doi: 10.1016/j.vaccine.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith HO, Venter JC. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 29.Cox DL, Luthra A, Dunham-Ems S, Desrosiers DC, Salazar JC, Caimano MJ, Radolf JD. 2010. Surface immunolabeling and consensus computational framework to identify candidate rare outer membrane proteins of Treponema pallidum. Infect Immun 78:5178–5194. doi: 10.1128/IAI.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrosova H, Zobanikova M, Cejkova D, Mikalova L, Pospisilova P, Strouhal M, Chen L, Qin X, Muzny DM, Weinstock GM, Smajs D. 2012. Whole genome sequence of Treponema pallidum subsp. pallidum, strain Mexico A, suggests recombination between yaws and syphilis strains. PLoS Negl Trop Dis 6:e1832. doi: 10.1371/journal.pntd.0001832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smajs D, Norris SJ, Weinstock GM. 2012. Genetic diversity in Treponema pallidum: implications for pathogenesis, evolution and molecular diagnostics of syphilis and yaws. Infect Genet Evol 12:191–202. doi: 10.1016/j.meegid.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grassly NC, Fraser C, Garnett GP. 2005. Host immunity and synchronized epidemics of syphilis across the United States. Nature 433:417–421. doi: 10.1038/nature03072. [DOI] [PubMed] [Google Scholar]
- 33.Garnett GP, Aral SO, Hoyle DV, Cates W Jr, Anderson RM. 1997. The natural history of syphilis. Implications for the transmission dynamics and control of infection. Sex Transm Dis 24:185–200. [DOI] [PubMed] [Google Scholar]
- 34.Desrosiers DC, Anand A, Luthra A, Dunham-Ems SM, LeDoyt M, Cummings MAD, Eshghi A, Cameron CE, Cruz AR, Salazar JC, Caimano MJ, Radolf JD. 2011. TP0326, a Treponema pallidum β-barrel assembly machinery A (BamA) ortholog and rare outer membrane protein. Mol Microbiol 80:1496–1515. doi: 10.1111/j.1365-2958.2011.07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed The National Academies Press, Washington, DC. [Google Scholar]
- 36.Hazlett KR, Sellati TJ, Nguyen TT, Cox DL, Clawson ML, Caimano MJ, Radolf JD. 2001. The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J Exp Med 193:1015–1026. doi: 10.1084/jem.193.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ponomarenko J, Bui HH, Li W, Fusseder N, Bourne PE, Sette A, Peters B. 2008. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics 9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kringelum JV, Lundegaard C, Lund O, Nielsen M. 2012. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol 8:e1002829. doi: 10.1371/journal.pcbi.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 40.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Boronat M, Diez-Rivero CM, Reinherz EL, Reche PA. 2008. PVS: a web server for protein sequence variability analysis tuned to facilitate conserved epitope discovery. Nucleic Acids Res 36:W35–W41. doi: 10.1093/nar/gkn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anand A, Luthra A, Dunham-Ems S, Caimano MJ, Karanian C, LeDoyt M, Cruz AR, Salazar JC, Radolf JD. 2012. TprC/D (Tp0117/131), a trimeric, pore-forming rare outer membrane protein of Treponema pallidum, has a bipartite domain structure. J Bacteriol 194:2321–2333. doi: 10.1128/JB.00101-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cruz AR, Pillay A, Zuluaga AV, Ramirez LG, Duque JE, Aristizabal GE, Fiel-Gan MD, Jaramillo R, Trujillo R, Valencia C, Jagodzinski L, Cox DL, Radolf JD, Salazar JC. 2010. Secondary syphilis in Cali, Colombia: new concepts in disease pathogenesis. PLoS Negl Trop Dis 4:e690. doi: 10.1371/journal.pntd.0000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cruz AR, Ramirez LG, Zuluaga AV, Pillay A, Abreu C, Valencia CA, La Vake C, Cervantes JL, Dunham-Ems S, Cartun R, Mavilio D, Radolf JD, Salazar JC. 2012. Immune evasion and recognition of the syphilis spirochete in blood and skin of secondary syphilis patients: two immunologically distinct compartments. PLoS Negl Trop Dis 6:e1717. doi: 10.1371/journal.pntd.0001717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cox DL, Akins DR, Porcella SF, Norgard MV, Radolf JD. 1995. Treponema pallidum in gel microdroplets: a novel strategy for investigation of treponemal molecular architecture. Mol Microbiol 15:1151–1164. doi: 10.1111/j.1365-2958.1995.tb02288.x. [DOI] [PubMed] [Google Scholar]
- 46.Thein M, Sauer G, Paramasivam N, Grin I, Linke D. 2010. Efficient subfractionation of Gram-negative bacteria for proteomics studies. J Proteome Res 9:6135–6147. doi: 10.1021/pr1002438. [DOI] [PubMed] [Google Scholar]
- 47.Lukehart SA, Miller JN. 1978. Demonstration of the in vitro phagocytosis of Treponema pallidum by rabbit peritoneal macrophages. J Immunol 121:2014–2024. [PubMed] [Google Scholar]
- 48.Sklar JG, Wu T, Kahne D, Silhavy TJ. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev 21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Radolf JD. 1995. Treponema pallidum and the quest for outer membrane proteins. Mol Microbiol 16:1067–1073. doi: 10.1111/j.1365-2958.1995.tb02332.x. [DOI] [PubMed] [Google Scholar]
- 50.Webb CT, Heinz E, Lithgow T. 2012. Evolution of the beta-barrel assembly machinery. Trends Microbiol 20:612–620. doi: 10.1016/j.tim.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Radolf JD, Lukehart SA. 2006. Immunology of syphilis, p 285–322. In Radolf JD, Lukehart SA (ed), Pathogenic treponemes: cellular and molecular biology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 52.Lafond RE, Lukehart SA. 2006. Biological basis for syphilis. Clin Microbiol Rev 19:29–49. doi: 10.1128/CMR.19.1.29-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore MW, Cruz AR, La Vake CJ, Marzo AL, Eggers CH, Salazar JC, Radolf JD. 2007. Phagocytosis of Borrelia burgdorferi and Treponema pallidum potentiates innate immune activation and induces gamma interferon production. Infect Immun 75:2046–2062. doi: 10.1128/IAI.01666-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Voorhis WC, Barrett LK, Lukehart SA, Schmidt B, Schriefer M, Cameron CE. 2003. Serodiagnosis of syphilis: antibodies to recombinant Tp0453, Tp92, and Gpd proteins are sensitive and specific indicators of infection by Treponema pallidum. J Clin Microbiol 41:3668–3674. doi: 10.1128/JCM.41.8.3668-3674.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brinkman MB, McKevitt M, McLoughlin M, Perez C, Howell J, Weinstock GM, Norris SJ, Palzkill T. 2006. Reactivity of antibodies from syphilis patients to a protein array representing the Treponema pallidum proteome. J Clin Microbiol 44:888–891. doi: 10.1128/JCM.44.3.888-891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cruz AR, Moore MW, La Vake CJ, Eggers CH, Salazar JC, Radolf JD. 2008. Phagocytosis of Borrelia burgdorferi, the Lyme disease spirochete, potentiates innate immune activation and induces apoptosis in human monocytes. Infect Immun 76:56–70. doi: 10.1128/IAI.01039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sobota A, Strzelecka-Kiliszek A, Gladkowska E, Yoshida K, Mrozinska K, Kwiatkowska K. 2005. Binding of IgG-opsonized particles to Fc gamma R is an active stage of phagocytosis that involves receptor clustering and phosphorylation. J Immunol 175:4450–4457. doi: 10.4049/jimmunol.175.7.4450. [DOI] [PubMed] [Google Scholar]
- 58.Greenberg S, Chang P, Wang DC, Xavier R, Seed B. 1996. Clustered syk tyrosine kinase domains trigger phagocytosis. Proc Natl Acad Sci U S A 93:1103–1107. doi: 10.1073/pnas.93.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bourell KW, Schulz W, Norgard MV, Radolf JD. 1994. Treponema pallidum rare outer membrane proteins: analysis of mobility by freeze-fracture electron microscopy. J Bacteriol 176:1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La Vake C, Gebhardt LL, Limberger RJ, Cox DL, Marko M, Radolf JD. 2009. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol 191:7566–7580. doi: 10.1128/JB.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J, Howell JK, Bradley SD, Zheng Y, Zhou ZH, Norris SJ. 2010. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J Mol Biol 403:546–561. doi: 10.1016/j.jmb.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anand A, LeDoyt M, Karanian C, Luthra A, Koszelak-Rosenblum M, Malkowski MG, Puthenveetil R, Vinogradova O, Radolf JD. 24 March 2015. Bipartite topology of Treponema pallidum repeat proteins C/D and I: outer membrane insertion and porin function require a C-terminal β-barrel domain. J Biol Chem doi: 10.1074/jbc.M114.629188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.