ABSTRACT
Iron acquisition at the outer membrane (OM) of Gram-negative bacteria is powered by the proton motive force (PMF) of the cytoplasmic membrane (CM), harnessed by the CM-embedded complex of ExbB, ExbD, and TonB. Its stoichiometry, ensemble structural features, and mechanism of action are unknown. By panning combinatorial phage libraries, periplasmic regions of dimerization between ExbD and TonB were predicted. Using overexpression of full-length His6-tagged exbB-exbD and S-tagged tonB, we purified detergent-solubilized complexes of ExbB-ExbD-TonB from Escherichia coli. Protein-detergent complexes of ∼230 kDa with a hydrodynamic radius of ∼6.0 nm were similar to previously purified ExbB4-ExbD2 complexes. Significantly, they differed in electronegativity by native agarose gel electrophoresis. The stoichiometry was determined to be ExbB4-ExbD1-TonB1. Single-particle electron microscopy agrees with this stoichiometry. Two-dimensional averaging supported the phage display predictions, showing two forms of ExbD-TonB periplasmic heterodimerization: extensive and distal. Three-dimensional (3D) particle classification showed three representative conformations of ExbB4-ExbD1-TonB1. Based on our structural data, we propose a model in which ExbD shuttles a proton across the CM via an ExbB interprotein rearrangement. Proton translocation would be coupled to ExbD-mediated collapse of extended TonB in complex with ligand-loaded receptors in the OM, followed by repositioning of TonB through extensive dimerization with ExbD. Here we present the first report for purification of the ExbB-ExbD-TonB complex, molar ratios within the complex (4:1:1), and structural biology that provides insights into 3D organization.
IMPORTANCE Receptors in the OM of Gram-negative bacteria allow entry of iron-bound siderophores that are necessary for pathogenicity. Numerous iron-acquisition strategies rely upon a ubiquitous and unique protein for energization: TonB. Complexed with ExbB and ExbD, the Ton system links the PMF to OM transport. Blocking iron uptake by targeting a vital nanomachine holds promise in therapeutics. Despite much research, the stoichiometry, structural arrangement, and molecular mechanism of the CM-embedded ExbB-ExbD-TonB complex remain unreported. Here we demonstrate in vitro evidence of ExbB4-ExbD1-TonB1 complexes. Using 3D EM, we reconstructed the complex in three conformational states that show variable ExbD-TonB heterodimerization. Our structural observations form the basis of a model for TonB-mediated iron acquisition.
INTRODUCTION
Iron, universally found at the center of metabolic processes in almost all organisms (1), may be acquired by Gram-negative bacteria in chelated form as iron-bound siderophores. The Fur protein regulates the expression of proteins involved in iron homeostasis (2, 3), such that iron-limiting conditions lead to induction of genes necessary for its import from the environment (4). High-affinity receptors in the outer membrane (OM) of Gram-negative bacteria recognize different siderophores (5). Each receptor protein forms a 22-stranded transmembrane β-barrel that contains an N-terminal occlusion termed the plug domain (6). This highly solvated globular domain is composed of a four-stranded β-sheet inclined 45° to the membrane normal. At its periplasmic face, the plug contains a semiconserved motif, the Ton box (7, 8). More than 98 OM receptors across multiple species demonstrate complexation with TonB, hence the term TonB-dependent transporters (TBDTs) (7, 9). Upon substrate binding at the cell surface, structural signals are propagated to the periplasmic side of the OM. In the case of the ferrichrome receptor FhuA of Escherichia coli, binding induces unwinding of a short α-helix, resulting in a disordered Ton box in the periplasm (10–12). Substrate passage into the periplasm may occur by conformational rearrangement of the plug domain following mechanical interactions with the Ton box (10, 13). These interactions require input of energy that is generated at the cytoplasmic membrane (CM) in the form of the proton motive force (PMF) (14, 15), harnessed and coupled to OM transport by the CM-embedded complex of ExbB, ExbD, and TonB (16). The Ton system is an attractive target for development of therapeutics due to its central position in powering active nutrient import in all Gram-negative bacteria.
In E. coli, ExbB (26.3 kDa) and ExbD (15.5 kDa) are encoded by the same operon (17), reflecting their linked functionality. With three transmembrane helices (18), ExbB has more residues facing the cytoplasm than exposed to the periplasm. ExbD is also membrane bound with a single transmembrane domain; 22 residues are predicted to face the cytoplasm, and ∼98 residues face the periplasm (19). With few cytoplasmic residues and a single transmembrane domain, TonB (26.1 kDa) is located mostly in the periplasm (20). The N terminus of TonB requires ExbB for function (21) through its interactions with the first ExbB transmembrane domain (18). Indeed, ExbB has been demonstrated to stabilize both ExbD and TonB (16). Apparently, no ExbB residue directly participates in proton translocation across the CM (18). Rather, ExbB has a structural “scaffolding” role, assembling into dimers and tetramers (22) and potentially propagating signals between the cytoplasm and the periplasm (18).
In contrast, ExbD contains the sole residue, Asp25, in the Ton complex that responds to the PMF (18, 23). Nuclear magnetic resonance (NMR) revealed a solution structure of an isolated periplasmic fragment from ExbD (24) as a folded globular domain accounting for residues 64 to 133, surrounded by flexible and extended residues (43 to 63 and 134 to 141). Only weak interactions were measured between this soluble construct and peptides representing periplasmic segments of TonB. However, considering the nonphysiological conditions (pH 3 and low salt) and a propensity for protein aggregation, it is not surprising that this study did not reflect in vivo evidence of ExbD-TonB interactions (15, 23, 25–27). Postle and coworkers identified periplasmic regions involved in ExbD and TonB interactions by disulfide cross-linking (26, 28). In the absence of TonB, the same ExbD regions tend to promote their homodimerization (26). This in vivo evidence described an extended and dynamically flexible ExbD periplasmic domain. The biological relevance of the NMR structure (24) has therefore been questioned (26, 28).
The TonB C-terminal domain forms high-affinity interactions with TBDTs (29) and has been cocrystallized with FhuA (30) and with the cobalamin receptor BtuB (31). However, no definitive structural information for TonB's N-terminal transmembrane domain or for its central linker region (residues 33 to 150) (20) exists. A stretch of residues (70 to 102) rich in proline was found to adopt an extended and rigid conformation by NMR (32), and yet it was dispensable for TonB-dependent transport in E. coli (33). This suggests that the remainder of the central linker region would assume a highly extended conformation, as described by Köhler et al. (34). Indeed, the TonB C-terminal domain in the cocrystal structures (30, 31) accounts for approximately one-third of total residues (∼151 to 239), yet it occupies only ∼2.5 nm of the ∼21-nm periplasmic thickness (35, 36). Overall, TonB's flexible and dynamic periplasmic domain (28, 37) renders it an active protein in the periplasm. For example, certain regions of TonB have been demonstrated to mediate contacts with the periplasmic binding protein FhuD (38). There is currently no ensemble analysis of the role played by TonB's periplasmic domain in its work cycle or how its interactions with ExbD lead to mechanical input at the Ton box.
Although the monomeric TonB C-terminal domain crystallized in complex with TBDTs (30, 31), publications document in vivo (39, 40) and in vitro (29, 41) evidence for TonB dimerization. Despite various outcomes using several TonB truncations in vitro, studies investigating full-length TonB have yet to be reported. The absence of the TonB transmembrane domain has been implicated in the nonnatural crystal structures of its periplasmic residues (42). Moreover, a functional TonB transmembrane domain was shown to play a role in its energy-transducing work cycle (43). The transmembrane domains of all Ton system proteins may participate in energy transduction and mechanical interactions between each other and TBDTs. Current models are limited in explaining biochemical observations.
The stoichiometry of TonB-containing complexes also remains unknown. Higgs et al. measured seven ExbB and two ExbD proteins for every TonB protein in E. coli (44). This ratio may not reflect the actual stoichiometry of the complex, as TonB is known to be labile in vivo (21, 45). Indeed, strong TonB overexpression led to degradation products, but moderate overexpression relative to ExbB and ExbD was tolerated (44), providing ExbB-ExbD complexes that are poised for synthesis and incorporation of TonB. The measured cellular ratio, therefore, could represent a mixture of three-protein complexes (ExbB-ExbD-TonB) and two-protein complexes (ExbB-ExbD).
Our recent in vitro investigations characterized copurifying proteins of stoichiometry ExbB4-ExbD2 (46). By single-particle electron microscopy (EM), we noted occasional periplasmic homodimerization by ExbD that correlated with two different arrangements of ExbB's cytoplasmic domains: a tetrameric arrangement and a dimer of dimers. Despite the lack of a functional assay, our results are consistent with in vivo data (18, 22, 26). Our biophysical analyses were expanded by the use of amphipols (47). In both detergent and amphipols, we observed identical structural conformations of the ExbB4-ExbD2 complex. Since amphipols are known to retain activity of some membrane proteins (48, 49), these data validated our choice of detergent for preserving in vivo structural conformations.
Here, we present the first report for copurification of ExbB, ExbD, and TonB as complexes having a 4:1:1 ratio, hence the designation ExbB4-ExbD1-TonB1. Consistent with our predictions from phage display, structural investigations by single-particle EM revealed two forms of periplasmic heterodimerization between ExbD and TonB. A model of transenvelope energy transduction by the Ton system is derived from structural observations that show conformational plasticity of the intact complex.
MATERIALS AND METHODS
Cloning, expression, and purification of periplasmic domains: ExbD43–141 and TonB33–239.
Chromosomal DNA encoding residues 43 to 141 of ExbD (ExbD43–141) from E. coli was cloned by PCR into a modified pET15b expression vector, resulting in the addition of 23 residues (including the His6 tag) to its N terminus and two residues to its C terminus. ExbD43–141 was expressed in E. coli BL21(DE3)/pLysS grown in Luria-Bertani (LB; Fisher) medium supplemented with 50 μg/ml ampicillin (Sigma) and 34 μg/ml chloramphenicol (Sigma). When cultures reached an optical density at 600 nm (OD600) of 0.5 to 0.6, expression was induced by addition of 0.4 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG; Bio Vectra) followed by shaking for 2 h at 37°C. Cell pellets were collected by centrifugation at 7,000 × g and stored at −20°C. Pellets were resuspended in binding buffer (20 mM Tris [pH 8.0], 150 mM NaCl) supplemented with 0.4 mM phenylmethylsulfonyl fluoride (Sigma), 25 μg/ml DNase (Roche), 25 μg/ml RNase (Roche), 5 mM MgCl2, and one tablet of EDTA-free cOmplete Protease Inhibitor cocktail (Roche). Cells were lysed on ice by two passes through an EmulsiFlex-C5 (Avestin) at 17,500 kPa, and the lysate was centrifuged at 34,000 × g for 1 h at 4°C; supernatant was retained. After clarification by filtration (0.45-μm pore size), supernatant was loaded onto a 10-ml nickel-nitrilotriacetic (Ni+-NTA) Profinity immobilized metal ion affinity chromatography (IMAC; Bio-Rad) column preequilibrated with binding buffer. All purifications were performed using a Biologic DuoFlow chromatography system (Bio-Rad). Nonspecifically bound material was removed by extensive washing (20 mM Tris [pH 8.0], 150 mM NaCl, 18 mM imidazole). His-tagged ExbD43–141 was eluted from the resin by an imidazole gradient to 300 mM and extensively dialyzed against 20 mM Tris-HCl (pH 8.0), 150 mM NaCl for storage at 4°C.
Chromosomal DNA encoding residues 33 to 239 of E. coli TonB (TonB33–239) was cloned by PCR into a modified pET28 expression vector, resulting in the addition of 20 residues to its N terminus, including an N-terminal His6 tag. The TonB33–239 construct (TonB33–239) was expressed in E. coli BL21(DE3)/pLysS and purified by Ni+-NTA Superflow resin (Qiagen) and SP-Sepharose (Amersham Biosciences) cation exchange as we previously described (50).
Cloning, expression, and purification of ExbB-ExbD-TonB.
The His6-tagged exbB-exbD (exbB-exbDHis6) coding sequence was PCR amplified from pExbBD (46) and inserted between restriction sites NcoI and EcoRI of the dual-gene expression vector pETDuet-1 (Novagen). Full-length tonB was PCR amplified from E. coli DH5α cells and inserted between restriction sites BglII and KpnI in the second multiple cloning site of pETDuet-1. This strategy resulted in the addition of 26 C-terminal residues to TonB, 15 of which specify the S-tag (51, 52). This plasmid was transformed into E. coli BL21(DE3)/pLysS, and cells were grown with shaking at 37°C in LB broth supplemented with 50 μg/ml carbenicillin and 34 μg/ml chloramphenicol until the OD600 reached 0.8. Following induction with 0.5 mM IPTG, cultures were left to grow for 3.5 h at 25°C with shaking. Cells were collected by low-speed centrifugation (7,000 × g) for 20 min at 4°C and frozen for storage at −20°C. Thawed bacterial pellets were suspended in lysis buffer (50 mM Tris [pH 7.5], 200 mM NaCl, 5 mM MgCl2, 100 μg/ml DNase, 100 μg/ml RNase, 10 μg/ml lysozyme, and one tablet of EDTA-free cOmplete Protease Inhibitor cocktail). The resuspended mixture was lysed by two passes through an EmulsiFlex-C5. Following low-speed centrifugation (4,300 × g) to remove unlysed cells and other debris, samples were ultracentrifuged for 1 h at 4°C and 250,000 × g to isolate membrane fractions. Membrane pellets were resuspended in 50 mM Tris (pH 7.5), 200 mM NaCl, 20 mM imidazole, and 1% (wt/vol) n-dodecyl-β-d-maltopyranoside (DDM; Anatrace) with EDTA-free cOmplete Protease Inhibitor cocktail for 2 h at 4°C. DDM-solubilized lysate, clarified by centrifugation at 34,500 × g, was applied to a 5-ml Ni+-NTA Profinity IMAC column preequilibrated with 50 mM Tris pH 7.5, 200 mM NaCl, 20 mM imidazole and 0.02% (wt/vol) DDM. Following elution of nonbinding lysate, the column was washed with 50 mM Tris (pH 7.5), 500 mM NaCl, and 0.02% (wt/vol) DDM and then with 50 mM Tris (pH 7.5), 200 mM NaCl, 50 mM imidazole, and 0.02% (wt/vol) DDM. Specifically bound material was eluted with 25 mM Tris (pH 7.5), 150 mM NaCl, 200 mM imidazole, and 0.02% (wt/vol) DDM.
Complexes of ExbB4-ExbD2 in DDM were purified as described previously (46) using a 5-ml Ni+-NTA Profinity IMAC column followed by preparative size exclusion chromatography (SEC) on a 120-ml (16 by 600 mm) Superdex pg column (GE Healthcare). Samples were eluted at 0.5 ml/min in 25 mM Tris (pH 7.5), 150 mM NaCl, and 0.02% (wt/vol) DDM. When necessary, protein-detergent complexes (PDCs) were concentrated in centrifugal filter devices (Thermo Scientific) having a nominal molecular mass cutoff (MMCO) of 150 kDa. Following a 10-min centrifugation at 2,000 × g, the filtrate and retentate were assayed for protein.
Assays for protein purity, concentration, and immunodetection.
Purified proteins were analyzed by SDS-PAGE under denaturing and reducing conditions by silver staining or by colloidal Coomassie blue G-250 staining (Thermo Scientific). For S-tag detection, separated proteins were transferred from polyacrylamide gels to an Immobilon-P polyvinylidene fluoride (Millipore) membrane and then blocked for 30 min with 2% bovine serum albumin (BSA) in Tris-buffered saline (TBS), consisting of 50 mM Tris (pH 8) and 150 mM NaCl. The membrane was washed extensively with TBS–0.1% (vol/vol) Tween 20 before and after incubation with mouse anti-S-tag IgG2b antibody (Novagen) and goat anti-mouse alkaline-phosphatase-conjugated antibody (Jackson ImmunoResearch). Immunoblots were developed with a nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolyl phosphate solution (Roche).
Concentrations of ExbD43–141 and TonB33–239 were determined using the bicinchoninic acid (Thermo Scientific) and Bradford (Thermo Scientific) assays, respectively. Concentrations of ExbB-ExbD and ExbB-ExbD-TonB were measured with a NanoDrop 1000 (Thermo Scientific) using approximate extinction coefficients derived from potential complex stoichiometries and the Beer-Lambert law. Extinction coefficients were calculated from primary sequence using ProtParam from ExPASy (http://web.expasy.org/protparam).
Phage panning and analysis of affinity-selected peptides.
Phage panning against TonB33–239 was performed as we previously described (53). Purified ExbD43–141 was diluted to 100 μg/ml in 0.1 M NaHCO3 (pH 8.6), and 150 μl of protein was adsorbed to a polystyrene microtiter plate (Thermo Scientific). Plates coated with immobilized ExbD43–141 were incubated for 16 h at 4°C followed by blocking (2 h at 37°C) with 5 mg/ml BSA in 0.1 M NaHCO3 (pH 8.6) containing 0.02% (wt/vol) NaN3 (Fisher). A Ph.D.-12 phage library or Ph.D.-C7C phage library (1.5 × 1011 PFU each; New England BioLabs) was then added. Phage panning, clone isolation, and DNA sequencing were performed under conditions identical to those used for TonB33–239 (53). Pairwise alignments of affinity-selected peptides from ExbD43–141 and from TonB33–239 phage panning were performed against the sequences of E. coli K-12 TonB and ExbD using the Match program from the Receptor Ligand Contacts (RELIC) server (54). This program uses a default scoring window of five; the threshold score requires at minimum three identities and one similarity between selected peptides and alignment sequence based on a modified block amino acid substitution matrix (BLOSUM62). Identities are given a score of +4, whereas similarities are given a score of +1. Alignment mismatches are given a score of either 0 or −1. For a window size of 5 residues, the threshold for Match is a score of 13. Improvements to the pairwise alignments were made using the program MatchScan (P. D. Pawelek, unpublished data), which calculates alignment scores for a range of scoring windows. For MatchScan, the minimum scoring threshold is increased or decreased by 1 for each residue added or subtracted, respectively, to the default scoring window.
Protein identification by mass spectrometry.
Following SDS-PAGE and Coomassie blue G-250 staining, protein bands were excised, destained, and vacuum centrifuged. The samples were resolubilized in 2% acetonitrile–1% formic acid, followed by reduction and alkylation. Trypsin-digested samples were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on an LTQ Orbitrap Velos (Thermo Scientific). Alcohol dehydrogenase was injected between samples as a quality control and blank to monitor carryover. Identified peptides were analyzed using Mascot (version 2.3.02; Matrix Science), searching the NCBInr Escherichia coli database (357,495 entries) assuming trypsin digestion. Search parameters included a fragment ion mass tolerance of 0.6 Da, parent ion tolerance of 15 ppm, cysteine carbamidomethyl fixed modification, and methionine oxidation variable modification. Results were validated using Scaffold (version Scaffold_3.6.2; Proteome Software Inc.).
Analysis of size and molecular mass of protein-detergent complexes.
The mass of the IMAC-purified ExbB-ExbD-TonB complex was measured by blue-native PAGE (BN-PAGE). Complexes were separated on 4 to 16% gradient acrylamide gels containing 0.05% (wt/vol) DDM. Samples were run at 4°C with cathode buffer (25 mM Tris and 192 mM glycine [pH 8.8]) containing 0.02% (wt/vol) Coomassie blue G-250 at 100 V. After 45 min, the Coomassie blue-containing cathode buffer was replaced with clear cathode buffer, and electrophoresis was resumed for 3 h at 200 V. PDC masses were determined by comparison with comigrating protein standards (NativeMark; Life Technologies).
IMAC-eluted material was analyzed by analytical SEC using a 24-ml (10- by 300-mm) Superdex 200 GL column (GE Healthcare). Samples (20 to 40 μg) were injected onto the column equilibrated in SEC running buffer and eluted at 0.5 ml/min. The elution volume of known standards (Bio-Rad) was used to determine the hydrodynamic radii of eluting complexes.
SEC-MALLS.
Protein complexes in DDM were analyzed by SEC coupled to multiangle laser light scattering (MALLS) in-line detectors. Following 150-kDa MMCO concentration, IMAC-purified PDCs were applied to a 24-ml Superdex 200 column operated by an Alliance high-pressure liquid chromatographer (HPLC; Waters Corporation) at 0.5 ml/min. OD280, refractive index, and laser light scattering were measured using the 2489 UV/Vis (Waters Corporation), OptiLab rEX (Wyatt Technology), and miniDAWN TREOS (Wyatt Technology) detectors, respectively. Data were processed using the “protein conjugate” analysis module of the Astra software package version 5.3.4.18 (Wyatt Technology). The protein and detergent dn/dc values used were 0.187 and 0.143 ml/g (55–57), respectively.
Native agarose gel electrophoresis.
Purified protein samples (5 μg) were loaded into a horizontal 0.8% (wt/vol) agarose gel immersed in nondenaturing buffer (25 mM Tris, 19.2 mM glycine [pH 7.5], 0.02% [wt/vol] DDM). Migration was carried out at 50 V for 1 h. Samples migrated according to their apparent isoelectric point (pI) (58). All theoretical pI analyses were calculated using the translated sequences of our constructs that include tags on the ExPASy program ProtParam (http://web.expasy.org/protparam). The pIs of protein standards lysozyme (59) and BSA (60) were taken to be 11.0 and 5.1, respectively.
Electron microscopy and single-particle analysis.
The IMAC-purified ExbB-ExbD-TonB complex (1 ng/μl) was adsorbed for 1 min on negatively glow-discharged carbon-coated EM copper grids, followed by negative staining with freshly prepared 1.5% (wt/vol) uranyl formate (pH 5) for 90 s. Grids were imaged at 200 kV at the Facility for Electron Microscopy Research, McGill University, with an FEI Tecnai G2 F20 equipped with a Gatan Ultrascan 4k by 4k digital charge-coupled device (CCD) camera. Digital transmission electron microscopy (TEM) micrographs were recorded at a nominal magnification of approximately ×67,000, corresponding to a 2.2-Å pixel size at the specimen level. A total of 83 micrographs were imaged untilted and tilted −60° using the UCSF Tomo package (61) for the random conical tilt (RCT) reconstruction method. The untilted micrographs collected for the RCT method were combined with another 92 untilted micrographs that were imaged manually. All contrast transfer function parameters were determined using CTFFIND3 (62). For the RCT reconstructions, approximately 9,500 tilt pairs were selected manually in Xmipp 3.1 (63). All particles were windowed into 128- by 128-pixel images. Boxed images of particles were normalized and subjected to two-dimensional (2D) classification using the CL2D algorithm of the Xmipp package (64). Using the calculated in-plane rotations and triangulated tilt angles, three-dimensional (3D) initial models were reconstructed using the corresponding images of the tilted RCT particles (65).
Particles from the additional 92 untilted micrographs were selected semiautomatically in EMAN2 (66). Combined untilted particles (∼38,000) were phase flipped, aligned, and submitted to the Sparx program “iterative stable alignment and clustering” (ISAC) for 10 generations (67).
The program RELION was used for 3D classification and refinement (68). All steps implemented phase flipping, as suggested by the programmers for negative-stain data. Symmetry was never imposed during calculations. The particles (∼33,000) belonging to RELION 2D classes resembling validated ISAC classes were accepted for 3D classification and refinement. For 3D classification, we chose three RCT models that represent the principal conformations most frequently observed in 2D class averaging. Initial models were low-pass filtered to 50 Å. This resulted in three subsets of particles belonging to the same conformational class. Each subset was then used to refine its 50-Å low-pass-filtered RCT model. The resultant refined models were used for a second round of 3D classification of all particles followed by refinement with the new subsets. All 3D visualizations were rendered in UCSF Chimera (69) (version 1.9; build 39798). Measurements of 3D volume applied a partial specific volume of 1.21 Å3/Da (70). Gold standard resolution was calculated within RELION refinement using the 0.143 Fourier shell correlation cutoff (68, 71).
The three 3D-EM volumes have been deposited in EMDataBank under accession numbers EMD-2859, EMD-2934, and EMD-2935.
RESULTS
Phage display predicts periplasmic interactions between TonB and ExbD.
Periplasmic interactions between TonB and ExbD were predicted using phage display. This molecular genetics strategy subjects an immobilized protein to a combinatorial bacteriophage M13 library engineered to express random sequences of 12 (Ph.D.-12) or seven (Ph.D.-C7C) amino acids on its surface-exposed pIII coat protein. Phages that bind strongly to the immobilized protein are subjected to DNA sequencing, thus identifying the affinity-selected peptides. The peptides are then computationally aligned to the primary sequence of potential binding partners; interaction sites are predicted from localized alignment clusters.
A soluble construct of ExbD (ExbD43–141) was expressed and purified. By SDS-PAGE, His6-tagged ExbD43–141 exhibited near homogeneity and migrated to a molecular mass of ∼13.7 kDa, in concordance with the expected molecular mass for this construct (see Fig. S1A in the supplemental material). Soluble TonB33–239 was isolated to greater than 95% purity and migrated to its higher-than-anticipated molecular mass, ∼35 kDa (see Fig. S1B in the supplemental material). Although the predicted molecular mass of our construct is 24.9 kDa, aberrant migration of TonB on SDS-PAGE gels has long been reported (41, 72). The purified periplasmic domain of ExbD (ExbD43–141) was subjected to phage panning against both peptide libraries. Ph.D.-C7C panning and Ph.D.-12 panning returned 131 and 110 unique peptides, respectively. Affinity-selected peptides against TonB33–239 were obtained from our previous study (53).
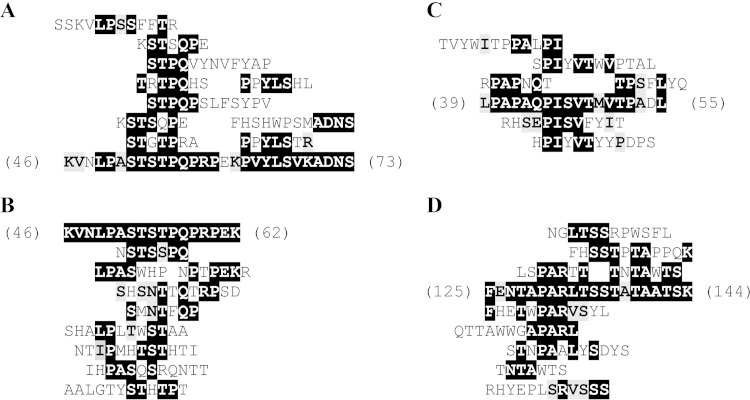
Unique ExbD43–141 affinity-selected peptides (11 from Ph.D.-C7C and 10 from Ph.D.-12 libraries [see Table S1 in the supplemental material]) were aligned to the sequence of ExbD to identify potential regions of homodimerization. These peptides clustered between residues 46 to 73 of ExbD (Fig. 1A), the periplasmic region immediately following the transmembrane domain. To identify potential regions of heterodimerization, we aligned TonB33–239 affinity-selected peptides to the sequence of ExbD (9 peptides from the Ph.D.-C7C library and 10 from the Ph.D.-12 library [see Table S2 in the supplemental material]). These clustered to the same region on ExbD: residues 46 to 62 (Fig. 1B). All of these ExbD residues were found in the region previously described as extended and flexible in the NMR solution structure of a similar construct (24). Similarly, to identify residues on TonB that may interact with ExbD, ExbD43–141 affinity-selected peptides (see Table S3 in the supplemental material) clustered at two regions on TonB. Peptides (4 from Ph.D.-C7C and 9 from Ph.D.-12 libraries) matched TonB periplasmic residues 39 to 55, a region immediately following the transmembrane domain (Fig. 1C). Another 19 peptides (7 from Ph.D.-C7C and 12 from Ph.D.-12 libraries) matched residues 125 to 144 of TonB's periplasmic domain (Fig. 1D). The latter set of residues map between the proline-rich region and the C-terminal domain that has previously been crystallized in complex with outer membrane receptors (30, 31). Under our stringent conditions, no TonB affinity-selected peptides clustered to the sequence of TonB itself.
FIG 1.
(A and B) Alignment of ExbD43–141 and TonB33–239 affinity-selected Ph.D.-C7C and Ph.D.-12 peptides to the ExbD sequence. (A) The top 10 scoring ExbD43–141 affinity-selected peptides aligned to residues 46 to 73 of the periplasmic domain of ExbD. See also Table S1 in the supplemental material for peptide match scores and window sizes. (B) The top nine scoring TonB33–239 affinity-selected peptides aligned to residues 46 to 62 of the periplasmic domain of ExbD. See also Table S2 in the supplemental material for peptide match scores and window sizes. (C and D) Alignment of ExbD43–141 affinity-selected Ph.D.-C7C and Ph.D.-12 peptides to TonB sequence. (C) The top six scoring peptides aligned to residues 38 to 55 of the periplasmic domain of TonB. (D) The top nine scoring peptides aligned to residues 125 to 144 of the central region of periplasmic TonB. See also Table S3 in the supplemental material for peptide match scores and window sizes. Only top-scoring peptides are shown for clarity; see the corresponding tables in the supplemental material for all peptides.
ExbB and TonB copurify in a complex with ExbD.
Having predictions of periplasmic interactions between TonB and ExbD and wishing to expand on our recent copurification of ExbB with ExbD (46), we pursued copurification of the three full-length proteins. The exbB-exbDHis6 operon from pExbBD was subcloned into a pETduet vector along with the tonB gene, a cloning strategy that resulted in the addition of 11 residues and the S-tag to the C terminus of TonB. The DDM-solubilized membrane fraction was applied onto an IMAC column followed by elution of strongly bound proteins. The eluate was analyzed by SDS-PAGE and colloidal Coomassie blue staining, identifying three bands migrating to 37, 26, and 17 kDa (Fig. 2A). Immunodetection using anti-S-tag antibodies identified the 37-kDa band as the S-tagged TonB (Fig. 2B). The identity of all proteins, including ExbB at 26 kDa and ExbD at 17 kDa, was confirmed using LC-MS/MS (see Fig. S2 in the supplemental material). Again, TonB showed aberrant migration on SDS-PAGE despite a calculated molecular mass of 28.8 kDa.
FIG 2.
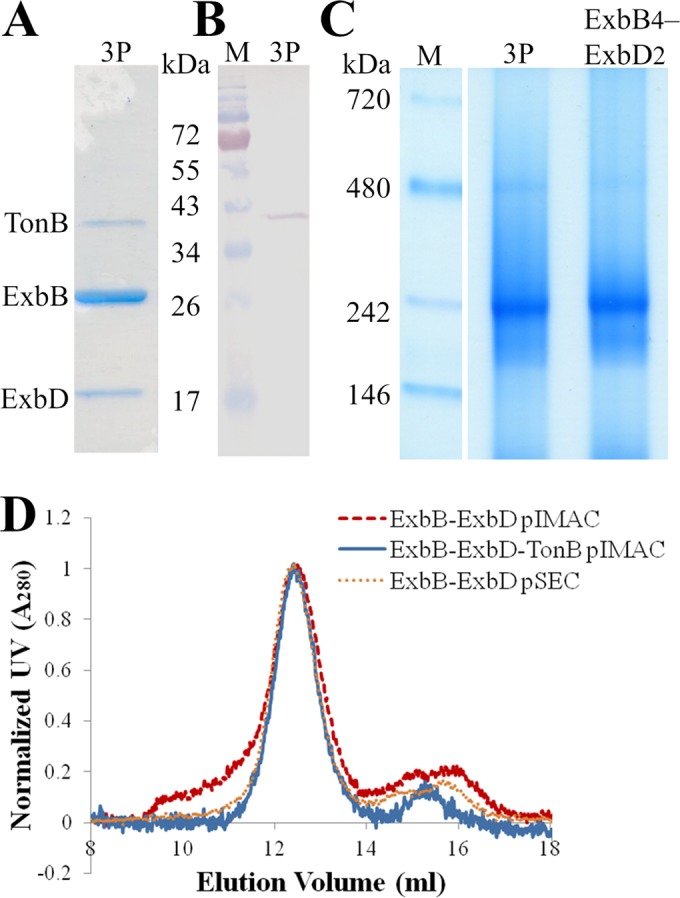
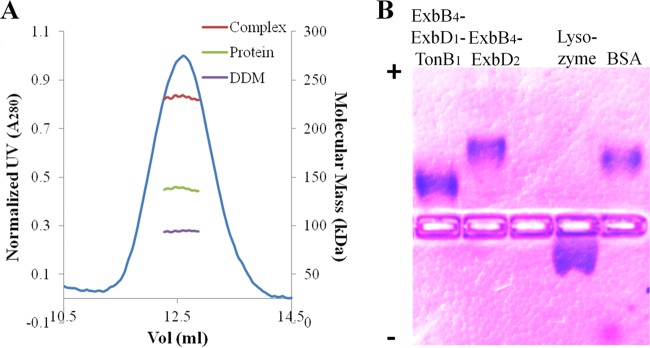
Purification and biophysical properties of the ExbB-ExbD-TonB complex. (A) SDS-PAGE analysis shows three coeluting proteins (3P) migrating to 37 (TonB), 26 (ExbB), and 17 (ExbD) kDa, stained with colloidal Coomassie blue. (B) Anti-S-tag immunodetection of IMAC-eluted proteins (3P) reveals S-tagged TonB. (C) BN-PAGE analysis showing that ExbB, ExbD, and TonB form a complex (3P) with a molecular mass similar to that of DDM-solubilized ExbB4-ExbD2: ∼240 kDa. (D) Analytical SEC comparing IMAC-eluted ExbB and ExbD, IMAC-eluted ExbB, ExbD, and TonB, and SEC-purified ExbB4-ExbD2. The peak at ∼12.4 ml corresponds to a hydrodynamic radius of ∼6.0 nm. Larger oligomers of ExbB-ExbD elute at 9.5 to 11 ml. M, molecular mass markers.
We previously showed (46) that the DDM-solubilized ExbB4-ExbD2 complex migrated to ∼240 kDa by BN-PAGE. Figure 2C shows that ExbB, ExbD, and TonB form a DDM-solubilized complex that migrates to a similar position by BN-PAGE. Whereas ExbB copurified with ExbD in several oligomeric states (Fig. 2D, red dashed line), the cooverexpression of TonB with ExbB and ExbD resulted in a single monodispersed complex (Fig. 2D, blue solid line). Such monodispersity was achieved for ExbB4-ExbD2 only after preparative SEC (Fig. 2D, orange dotted line). Comparison of the IMAC-purified three-protein complex to SEC-purified ExbB4-ExbD2 shows that the two share a hydrodynamic radius of ∼6.0 nm. Considering that the similar biophysical properties of the two PDCs most likely reflect comparable protein content, we turned to SEC-MALLS to identify the stoichiometry of the three proteins.
Proteins purify in complexes of ExbB4-ExbD1-TonB1 stoichiometry.
Stoichiometry of the monodisperse three-protein complex was deduced following the method of Slotboom et al. (73). Simultaneous measurements of light scattering, absorbance, and refractive index (three-detector method) during elution from gel filtration allow calculation of the absolute molecular mass and detergent contribution to a PDC. However, since the stoichiometry for the complex of ExbB, ExbD, and TonB remained unknown, a self-consistent method (74) was applied to determine the molar ratios. The band size and staining intensity of ExbB (Fig. 2A) reduced the stoichiometric candidates to those having ExbB in excess. Because the three-protein complex has a molecular mass (Fig. 2C) and a hydrodynamic radius (Fig. 2D) similar to those of DDM-solubilized ExbB4-ExbD2 (see Table S4 in the supplemental material), the total number of protein units within the complex was limited to six. Under these criteria, the most consistent stoichiometry was that of ExbB4-ExbD1-TonB1 (Fig. 3A). The absolute molecular mass of the PDC (∼230 kDa) is in agreement with that observed via BN-PAGE (Fig. 2C) and with estimates of molecular mass based on analytical SEC regression curves of standard proteins (data not shown). Furthermore, the measured detergent contribution by refractive index (∼40.5%) is similar to amounts that we assayed by 1D NMR for the ExbB4-ExbD2 complex (47).
FIG 3.
Determination of stoichiometry for the ExbB4-ExbD1-TonB1 complex. (A) Protein and detergent contributions of the DDM-solubilized ExbB-ExbD-TonB complex were measured by SEC-MALLS using the three-detector method. From candidate extinction coefficients, the best derived stoichiometry was ExbB4-ExbD1-TonB1. The resultant complex size was ∼230 kDa, ∼40.5% of which is DDM. (B) Native agarose gel electrophoresis (pH 7.5) separated proteins based on pI. Lysozyme (pI 11.0) migrated toward the cathode (indicated by a minus sign); BSA (pI 5.1) migrated toward the anode (indicated by a plus sign). The three-protein complex showed less migration than ExbB4-ExbD2, consistent with its predicted electronegativity. For clarity, the brightness of the image has been adjusted.
To further investigate whether our deduced stoichiometry could be supported by independent criteria, we looked at the resultant pIs of the candidate stoichiometries. Following removal of the predicted transmembrane domains of each protein, the potential stoichiometries displayed sufficiently different expected pIs (see Table S4 in the supplemental material). Using native agarose gel electrophoresis, the ExbB4-ExbD2 complex migrated with strong electronegativity, comparable to what was seen with the BSA control (Fig. 3B). In contrast, the deduced ExbB4-ExbD1-TonB1 stoichiometry was consistent with a reduced electronegative migration of the three-protein complex. Moreover, this experiment confirmed that there is no apparent mixture of three-protein and two-protein complexes. The three-protein complex migrated as a single species to a unique location without any recognizable band corresponding to the ExbB4-ExbD2 complex. Although the exact pIs of the three proteins are unknown, an analysis of their soluble domains can inform on the observed migrations. The soluble residues of ExbB are less electronegative (pI 6.85) than the soluble residues of ExbD (pI 5.78). DDM is nonionic, and we assume that it masks the surface charge of all transmembrane domains. Soluble residues of the ExbB4-ExbD2 complex are predicted to have a pI of 6.23, yet the complex migrates further, with an apparent pI of ∼5.5. This result indicates that ExbD's soluble residues may contribute strongly to the overall pI along with surface masking of basic residues, probably by extensive cytoplasmic contacts of ExbB (18, 46). The soluble residues of TonB show an equally strong, yet opposite, overall charge compared to ExbD. With a predicted pI of 9.57, these residues would largely neutralize ExbD's electronegativity. Assuming that basic residues remain masked in the ExbB soluble domains, modest migration of the three-protein complex toward the anode supports equal amounts of ExbD and TonB in the complex. Considering our phage display predictions of periplasmic interactions of ExbD with TonB and having purified the ExbB4-ExbD1-TonB1 complex, we turned to structural studies to visualize our predictions.
Electron microscopy shows heterodimerization between ExbD and TonB.
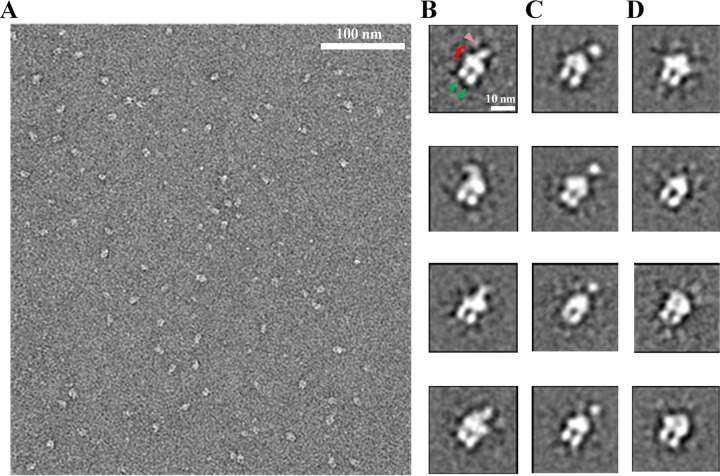
Monodisperse complexes of DDM-solubilized ExbB4-ExbD1-TonB1 were stained with uranyl formate and imaged by EM. Similar to the ExbB4-ExbD2 complex in DDM (46), uniform particles of ∼10-nm diameter were observed, apparently in various orientations (Fig. 4A). Single particles (∼38,000) were extracted, aligned, and submitted to 2D analysis using the iterative algorithm ISAC (see Fig. S3 in the supplemental material for the single particles) (67). Instead of forcing particles into a user-defined number of averages, ISAC validates only stable and reproducible class averages. Some validated class averages (Fig. 4B) showed features previously described for the ExbB4-ExbD2 particles (46). The major features are conserved, including the thickest density (red bracket), which had been identified as the DDM micelle surrounding the transmembrane domains. The two extensions descending from the micelle (green arrowheads) were inferred to be the ExbB cytoplasmic domains. The lone density on the periplasmic side of the micelle (pink arrowhead) represented homodimerized ExbD. Figure 4C shows a new conformation not observed with the ExbB4-ExbD2 complex. It shows a thick point of density above the micelle, yet not connected by observable density. Whereas most ExbB4-ExbD2 particles did not show observable periplasmic density, fewer ExbB4-ExbD1-TonB1 particles adopted this conformation (Fig. 4D). As individual, extended polypeptides cannot be observed in negative-staining EM, these class averages may represent complexes without dimerization in their periplasmic domains.
FIG 4.
2D image analysis of the ExbB4-ExbD1-TonB1 complex. (A) A typical TEM micrograph showing uniform particles of ∼10-nm diameter in various orientations. Following extraction, single particles were classified using ISAC. (B) Four class averages showing the DDM micelle (bracket), the cytoplasmic domains of ExbB (green arrowheads), and dimerized protein in the periplasmic domain (pink arrowhead). These averages show dimerization beginning at the micelle, whereas the periplasmic dimerization in panel C is at a point further from the micelle. (D) Four class averages with no observable density on the periplasmic side of the micelle. Single, extended polypeptides are not expected to be visualized by negative-staining EM.
Taken together, these 2D class averages reveal two distinct forms of heterodimerization between ExbD and TonB in their periplasmic domains. The first shows extensive dimerization beginning at the micelle (Fig. 4B), as was seen with ExbD homodimerization. However, unlike the observed flexibility of the ExbD periplasmic homodimer, the ExbD-TonB periplasmic heterodimer tends to remain in a position normal to the micelle. The “floating ball” conformation may represent a second form of heterodimerization (distal dimerization), where TonB and ExbD form periplasmic contacts further away from the micelle (Fig. 4C).
Regardless of the presence or absence of periplasmic dimerization, some side view class averages showed differences in the cytoplasmic arrangement of ExbB. While generally of similar lengths, one of the two thick cytoplasmic extensions tended to descend further away from the micelle than the second extension (Fig. 4B, rows 2 and 4; Fig. 4D, row 3). Overall, 2D image analyses of the ExbB4-ExbD1-TonB1 complex revealed homogeneous particles captured in multiple conformations. A 3D classification approach was taken to reconstruct certain snapshots of this dynamic complex.
3D reconstruction shows multiple states of the ExbB4-ExbD1-TonB1 complex.
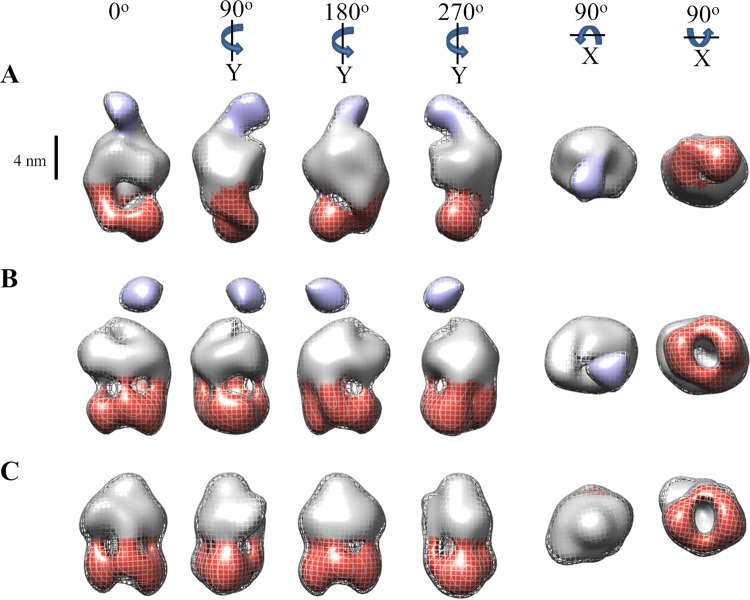
Initial 3D models were constructed with the RCT method. The resultant 3D RCT models closely resembled the 2D averages used in their determinations (see Fig. S4 in the supplemental material). 3D single-particle classification in RELION (68) using three representative RCT models abrogated their missing-cone effect (65). The improved (no longer flattened) references were used for a more precise 3D classification of single particles. Based on similarity in 3D, single particles were grouped into the extensive dimerization (∼9,400 particles), distal dimerization (∼12,200), and unobserved dimerization (∼10,900) classes. Approximately two-thirds of the ExbB4-ExbD1-TonB1 particles grouped to the two heterodimerization classes, as opposed to one-third of ExbB4-ExbD2 particles belonging to classes with periplasmic density (46). Each group of single particles was used to refine its low-pass-filtered reference model, leading to the final 3D volumes (Fig. 5). The final 3D volumes showing extensive, distal, and unobserved periplasmic dimerization have resolutions of 28, 23, and 23 Å, respectively (see Fig. S5 in the supplemental material), and are consistent with 2D averages (see Fig. S6 in the supplemental material). The resultant 3D volumes indicate consensus representations of the ExbB4-ExbD1-TonB1 complex in the extensive periplasmic heterodimerization conformation (Fig. 5A), the distal periplasmic heterodimerization conformation (Fig. 5B), and no observed dimerization in the periplasmic domain (Fig. 5C). The last two volumes further support our deduced stoichiometry by clearly showing four regions of density extending from the micelle into the cytoplasmic domain, forming a ringlike arrangement parallel to and below the micelle. The first volume, however, has two thicker regions of density descending unequally from the micelle, previously interpreted as a dimer of ExbB dimers (46).
FIG 5.
3D volumes representing the consensus conformational states of the ExbB4-ExbD1-TonB1 complex. Following 3D classification using RCT reference models, particles were grouped and used for refinement. The resultant volumes represent consensus structures of the ExbB4-ExbD1-TonB1 complex in three conformational states: extensive periplasmic dimerization (A), distal periplasmic dimerization (B), and no observed dimerization on the periplasmic side of the micelle (C). Volumes in panels B and C show four regions of density descending from the micelle, which can be attributed to a tetramer of ExbB. The distal portions of these cytoplasmic domains form a ringlike arrangement below and parallel to the micelle. The volume in panel A shows two thick regions of density descending unequally from the micelle, attributable to a dimer of ExbB dimers. All rotations are relative to 0°. The mesh contour represents volumes of ∼230 kDa, with the inner volume threshold decreased to display features. A scale bar (4 nm) delineates the potential CM boundary. Cytoplasmic and periplasmic domains are colored red and purple, respectively.
DISCUSSION
Current mechanistic models for the Ton system's energizing TBDTs derive from sometimes contradictory structural data for isolated domains of individual proteins. In vivo cross-linking studies identified important residues and their associated phenotypes but have not provided basic information on stoichiometry of the complex or ensemble views of conformational states. In this study, we used His-tagged ExbD to copurify interacting partners ExbB and TonB in complexes having estimated stoichiometry ExbB4-ExbD1-TonB1. We overexpressed the full-length exbB-exbDHis6 and S-tagged tonB genes using identical promoters on a single plasmid, proposed to approximate Fur-regulated transcriptional activity (17, 75). Using this strategy, stoichiometrically relevant complexes were anticipated to form in the CM of E. coli cells. Our attempts to purify TonB alone, either fused to stabilizing proteins or with terminal affinity tags or coexpressed as tonB and exbB-exbD on two separate plasmids, were unrewarded.
OM receptors greatly outnumber TonB molecules (44). They can even bind the TonB C-terminal domain in their apo form (29, 41, 76). The role of TonB, therefore, is continually to engage OM receptors and potentially other periplasmic proteins, as described in the “membrane surveillance” model (77). This physical wear most likely explains TonB's observed lability in vivo (16, 45, 78, 79). What might happen to the ExbB tetramer and a single ExbD during TonB turnover? Considering in vivo protein quantitation (44), we propose that a second molecule of ExbD occupies the place of TonB. This coordination could be mediated by multifunctional or essential housekeeping proteins that have been noted by in vivo cross-linking to ExbB (18, 22) and to ExbD (15, 27). Indeed, such ExbB4-ExbD2 complexes readily formed and purified in the absence of TonB overexpression (46). Alternatively, if the TonB work cycle is restricted to a single productive energy transduction potentially leading to its cleavage, then transient ExbD homodimerization may be necessary to “prime” one of the ExbD molecules with a proton in anticipation of TonB's reintegration into the complex.
TonB undergoes a complicated work cycle, in part as a dimer (29, 39, 40, 42), and yet acts as a monomer during Ton box interactions with TBDTs (29–31). EM observations (see below) support our biophysical data (Fig. 2C and D) of monomeric TonB with ExbD (Fig. 4B and C). These results are at variance with those of a study by Sauter et al., in which an in vivo two-hybrid assay demonstrated transmembrane-mediated TonB homodimerization irrespective of the presence of ExbB and ExbD (39). Our purification strategy would not have isolated complexes lacking ExbD. However, the findings presented here do not rule out periplasmic TonB homodimerization in vivo but establish ExbB4-ExbD1-TonB1 as the basic CM-embedded complex. Two molecules of TonB may undergo periplasmic homodimerization, potentially upon binding to peptidoglycan (29, 77) or to TBDTs (50), without interaction in their transmembrane domains.
We exploited phage panning and clustering of sequence alignments to discover potential regions of interaction between the periplasmic domains of ExbD and TonB. Our data identify a region of residues immediately following the ExbD transmembrane domain that is predicted to mediate ExbD homodimeric and ExbD-TonB heterodimeric interactions (Fig. 1A and B, respectively). This outcome accords with our structural observations of ExbD homodimerization beginning at the micelle in the ExbB4-ExbD2 complex (46). Although an extensive cysteine-scanning mutagenesis study of ExbD homodimerization did not investigate this region (26), a subsequent study of residue block deletions identified this region as playing a role in PMF-dependent conversion of heterodimeric ExbD-TonB to an energized form (27). Indeed, our data also predict the region immediately following TonB's transmembrane domain as interacting with the ExbD periplasmic domain (Fig. 1C). Such heterodimerization may extend from the region adjacent to the transmembrane domain to a point approximately one-third from the TonB C terminus (Fig. 1D). Taken together, our two predicted periplasmic regions of interaction may form the basis of the mutual protection from complete proteinase K degradation as detailed in previous studies (23, 43). Consistent with those findings, extensive heterodimerization according to our data would leave the final one-third of TonB's C-terminal residues susceptible to cleavage by proteinase K. This heterodimeric conformation (“stage II”) was found to be associated with correct periplasmic assembly of TonB before the input of PMF energy by ExbD (23).
Although we could not establish which region of ExbD may mediate the above-mentioned heterodimerization with TonB, others suggest that ExbD Leu132 is involved (23, 80). Whereas ExbD Ala92 was shown (15) to interact with TonB Ala150 (adjacent to our predicted region), it most likely contributes a different form of heterodimerization. A follow-up study by the same authors found that ExbD residues 92, 97, and 109 specifically and strongly cross-linked with TonB residues 150 to 212 (28). While at first these results may seem inconsistent with our phage display data, Ollis et al. did not mutate the TonB residues identified here. Furthermore, their strong cross-linking depended on a PMF-competent form of ExbD; we used soluble periplasmic domains of each protein. It appears as though the latter set of ExbD residues are involved in the correct positioning of TonB C-terminal residues for interaction with TBDTs (“stage III”), a conformation associated with proteinase K sensitivity of all of TonB species (23, 28).
Structural biology using EM has the benefit of allowing the observation of protein complexes simultaneously in their different physical states (81). 3D-EM structure determination is optimal for large (>500-kDa), rigid complexes that display symmetry (82, 83). The DDM-solubilized ExbB4-ExbD1-TonB1 complex, relatively small for EM, shows both structural heterogeneity and asymmetry. Furthermore, 2D and 3D classification is complicated by variations in the negative stain itself (84). Nevertheless, with both single particles and 2D ISAC classes, we recognized different positions of ExbB's cytoplasmic domain and different periplasmic states of ExbD and TonB (Fig. 4). Furthermore, reliable 3D models were reconstructed with the RCT method (65) from 2D classes (see Fig. S4 in the supplemental material).
Considering that uranyl formate staining reveals domains of at least 10 kDa (84), it is not surprising that single particles failed to show a highly extended lone polypeptide, as described for TonB's central region (34). As we previously showed for ExbD (46), structural observations on the periplasmic side of the micelle are attributable to protein-protein interactions that have enough mass to be discernible by negative stain. In the present study, we identify two forms of periplasmic density: extensive dimerization (Fig. 4B) and distal dimerization (Fig. 4C). Although we cannot exclude the possibility that the “floating ball” is in fact the structured TonB C-terminal domain as seen in complex with TBDTs (30, 31), this would be unlikely due to the absence of a ligand-loaded TBDT and the previously described dynamic nature of the TonB C-terminal domain (28, 37). The extensive ExbD-TonB heterodimerization is consistent with our phage display predictions (Fig. 1B, C, and D) involving residues immediately following the two transmembrane domains up to TonB residue ∼144. The distal ExbD-TonB heterodimerization may result from protein-protein interactions promoted by, but not limited to, TonB residues 150 to 212 and ExbD residues 92, 97, and 109 (28). We used 3D reconstruction to gain more information about the two heterodimeric states and their relationship to ExbB4-ExbD1-TonB1 that lacked periplasmic dimerization (Fig. 4D).
While we noted less flexibility of the ExbD-TonB heterodimer than the ExbD homodimer (46) relative to the micelle, the varying positions of the heterodimeric densities constituted limiting structural heterogeneity. In addition to particles having no observable periplasmic density (Fig. 4D) and differences in the ExbB cytoplasmic domain positions, 3D classification and refinement proved difficult. The refined 3D volumes (Fig. 5) resemble their RCT reference models and serve as consensus structures for the three conformations chosen for reconstruction, rather than static conformations of ExbB4-ExbD1-TonB1. The distal-dimerization (Fig. 5B) and nondimerized (Fig. 5C) classes are similar to the ExbD-undefined class of ExbB4-ExbD2 (46) in terms of ExbB's arrangement of the cytoplasmic domain. The extensive-dimerization class (Fig. 5A), however, shares a comparable arrangement of ExbB's cytoplasmic domain with the ExbD-defined models of ExbB4-ExbD2.
Combining in vivo and in vitro data from the literature with our structural observations of the ExbB4-ExbD1-TonB1 complex, we propose a model for function of the Ton system. Baker and Postle have recently demonstrated that ExbD residue Asp25 is the sole residue capable of responding to the PMF (18). Whereas the authors suspect that unidentified proteins may lend proton translocating transmembrane domains to create a proton relay pathway, a simpler explanation may lie in the nature of the interaction between TonB and ligand-loaded TBDTs. Steered molecular dynamics simulations of the TonB C-terminal domain in complex with BtuB described a downward pulling (into the periplasm) by TonB (85). While the plug domain's β-sheet “unzipped” (10), TonB's C-terminal domain and interstrand contact with the Ton box remained intact. A downward pulling motion was also advocated by Köhler et al. when they found that TonB residues 56 to 126 adopt a left-handed polyproline-II helix conformation that would have more than enough length to span the periplasm (34). Such extended secondary structures need not be composed of a majority of, or even any, proline residues (86), thereby explaining why the E. coli TonB mutant lacking residues 66 to 100 [TonBΔ(66–100)] remained functionally active (33) and why some TonB proteins of other Gram-negative bacteria have reduced proline content or shorter linker regions (20). Polyproline-II helices can mutarotate to polyproline-I helices, resulting in a length contraction of 40% (34, 87) and thus providing the requisite downward pulling force. Based on our observations of single particles and 2D classes showing one set of ExbB dimers further away from the micelle, we hypothesize that ExbB interprotein rearrangements lead to a pulling down (into the cytoplasm) of ExbD and hence induce a contractile motion with TonB (Fig. 6).
FIG 6.
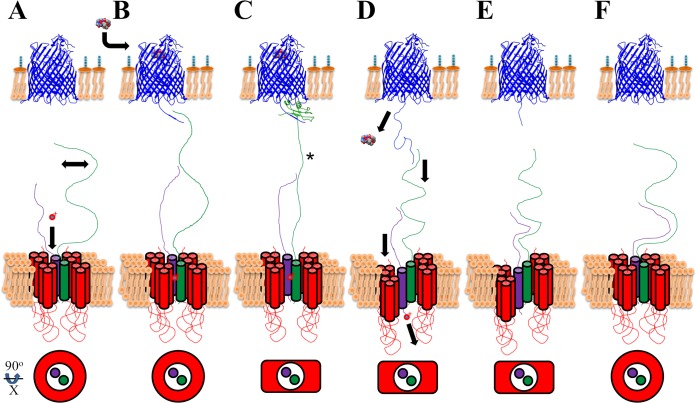
Proposed model of transenvelope energy transduction, whereby proton translocation across the CM couples ligand import at the OM. This conceptual model shows proteins of the cellular envelope: FhuA (PDB 2GRX) in blue, ExbB in red, ExbD in purple, and TonB in green. (A) The Ton system work cycle begins by “charging” ExbD's Asp25 with a proton (represented by red o+). TonB, in its extended conformation, scans (double-headed arrow) the OM for a disordered Ton box. ExbB is arranged in its tetrameric, ringlike form around the transmembrane domains of TonB and ExbD. (B) Energy transduction-competent ExbD binds TonB distally. The binding of ligand (ferricrocin crystal structure) triggers disordering of the Ton box. Recognition of ligand-loaded TBDTs (C) propagates a signal to the CM (*), inducing an interprotein rearrangement of ExbB. (D) Relative to one ExbB dimer, the other ExbB dimer transiently mediates cellular entry of ExbD, thus releasing its proton into the cytoplasm. Pulling down of ExbD would induce a contraction of TonB, thereby unzipping the TBDT β-sheet plug domain, resulting in periplasmic entry of its ligand. (E) The work cycle resets through ExbD-mediated repositioning of TonB (extensive dimerization) to its extended conformation (F), during which ExbB assumes its natural, ringlike arrangement.
How can a single residue translocate a proton across the CM? The simplest mechanism might be facilitated translocation. As diagrammed in Fig. 6A, a proton would bind Asp25 of ExbD without ExbD-TonB periplasmic dimerization (represented by Fig. 5C). This corresponds to the dimer of ExbB dimers as a ringlike, tetrameric arrangement in the cytoplasm, while TonB scans the periplasmic side of the OM for extended Ton boxes, indicating TBDT-bound ligand. Independent of contacts between TonB and the Ton box, a protonated ExbD leads to distal dimerization (Fig. 6B) with TonB, potentially at TonB residues 150 to 212 (28) and as presented in Fig. 5B. Although depicted by a simple contact, this interaction would undoubtedly involve the creation of a quaternary protein structure, sufficient for observable density by EM (Fig. 4C). This conformation would be the PMF-dependent, proteinase K-sensitive conformation described previously (23). Once TonB assumes a conformation identified in crystal structures (30, 31), a signal must be propagated to the CM (Fig. 6C, asterisk). Potentially, the “stiffening” of TonB may be sensed between TonB's transmembrane domain and the first ExbB transmembrane domain (16, 18, 88). Rearrangement of ExbB would follow (Fig. 6C and D), with subsequent shuttling of the proton into the cytoplasm via temporary cellular entry of ExbD, inducing a contraction of TonB (34), unzipping of the β-sheet plug domain, and entry of the TonB-dependent ligand (13, 85). Ton box release and repositioning of TonB by ExbD (Fig. 6E) would reset the work cycle (28). ExbD would make extensive dimerization contacts (Fig. 1 and 5A), followed by the dimer of ExbB dimers returning to the original arrangement (Fig. 6F), resembling the PMF-independent proteinase K-resistant conformation (23). Our model is an oversimplification based on structural observations; we cannot define proton energetics or the exact nature of ExbB-mediated proton shuttling by ExbD. Previously described TonB-FhuD interactions as a ternary complex with FhuA (38) are congruent with the proposition presented here. Further experimentation is needed to confirm our model. As direct electron detectors and computational software greatly improve (89–91), high-resolution cryo-EM is well suited to refining our structural observations.
Using predictive molecular genetics, protein purification, and structural biology, we identified the stoichiometry of the Ton complex, predicted and observed periplasmic interactions, and combined our structural data into a model for function. Considering that iron acquisition is a virulence factor (1, 92), an advanced understanding of the Ton system is imperative. Recently, bacteriostatic compounds targeting the Ton system were discovered (93). A combination of mutational in vivo work and the structural biology presented here holds potential to elucidate the mechanism of action of these newly described compounds.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by operating grants to J.W.C. from the Canadian Institutes of Health Research (CIHR reference number 200709MOP-178048-BMA-CFAA-11449). The Groupe d'Étude des Protéines Membranaires (GÉPROM), supported by the Fonds de Recherche du Québec-Santé (FRQ-S), awarded a Projet Novateur to J.W.C. A.S. was awarded fellowships from the CREATE program, Cellular Dynamics of Macromolecular Complexes, Natural Sciences and Engineering Research Council (NSERC) of Canada; from GÉPROM; and from the F. C. Harrison and the Rozanis Funds, Department of Microbiology and Immunology, McGill University.
Canada Foundation for Innovation provided infrastructure for the Facility for Electron Microscope Research, McGill University (www.mcgill.ca/femr). Mass spectrometry services were provided by Denis Faubert, Proteomics Discovery Platform at the Institut de Recherches Cliniques de Montréal (www.ircm.qc.ca/plateaux/proteomique). This work was facilitated by computing resources from CLUMEQ, under Compute/Calcul Canada.
We appreciate laboratory support from Nathalie Croteau and critical suggestions on the manuscript by B. Cousineau, S. Gruenheid, J. Kollman, H. LeMoual, G. Marczynski, I. Rouiller, and J. A. Kashul.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00069-15.
REFERENCES
- 1.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porcheron G, Garénaux A, Proulx J, Sabri M, Dozois CM. 2013. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front Cell Infect Microbiol 3:90. doi: 10.3389/fcimb.2013.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Escolar L, Pérez-Martín J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181:6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hantke K. 2001. Iron and metal regulation in bacteria. Curr Opin Microbiol 4:172–177. doi: 10.1016/S1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 5.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 7.Noinaj N, Guillier M, Barnard Travis J, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schramm E, Mende J, Braun V, Kamp RM. 1987. Nucleotide sequence of the colicin B activity gene cba: consensus pentapeptide among TonB-dependent colicins and receptors. J Bacteriol 169:3350–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krewulak KD, Vogel HJ. 2011. TonB or not TonB: is that the question? Biochem Cell Biol 89:87–97. doi: 10.1139/O10-141. [DOI] [PubMed] [Google Scholar]
- 10.Chimento DP, Kadner RJ, Wiener MC. 2005. Comparative structural analysis of TonB-dependent outer membrane transporters: implications for the transport cycle. Proteins 59:240–251. doi: 10.1002/prot.20416. [DOI] [PubMed] [Google Scholar]
- 11.Merianos HJ, Cadieux N, Lin CH, Kadner RJ, Cafiso DS. 2000. Substrate-induced exposure of an energy-coupling motif of a membrane transporter. Nat Struct Biol 7:205–209. doi: 10.1038/73309. [DOI] [PubMed] [Google Scholar]
- 12.Fanucci GE, Coggshall KA, Cadieux N, Kim M, Kadner RJ, Cafiso DS. 2003. Substrate-induced conformational changes of the periplasmic N-terminus of an outer-membrane transporter by site-directed spin labeling. Biochemistry 42:1391–1400. doi: 10.1021/bi027120z. [DOI] [PubMed] [Google Scholar]
- 13.Faraldo-Gómez JD, Smith GR, Sansom MSP. 2003. Molecular dynamics simulations of the bacterial outer membrane protein FhuA: a comparative study of the ferrichrome-free and bound states. Biophys J 85:1406–1420. doi: 10.1016/S0006-3495(03)74573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hancock RW, Braun V. 1976. Nature of the energy requirement for the irreversible adsorption of bacteriophages T1 and φ80 to Escherichia coli. J Bacteriol 125:409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ollis AA, Manning M, Held KG, Postle K. 2009. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol Microbiol 73:466–481. doi: 10.1111/j.1365-2958.2009.06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer E, Gunter K, Braun V. 1989. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J Bacteriol 171:5127–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eick-Helmerich K, Braun V. 1989. Import of biopolymers into Escherichia coli: nucleotide sequences of the exbB and exbD genes are homologous to those of the tolQ and tolR genes, respectively. J Bacteriol 171:5117–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker KR, Postle K. 2013. Mutations in Escherichia coli ExbB transmembrane domains identify scaffolding and signal transduction functions and exclude participation in a proton pathway. J Bacteriol 195:2898–2911. doi: 10.1128/JB.00017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kampfenkel K, Braun V. 1992. Membrane topology of the Escherichia coli ExbD protein. J Bacteriol 174:5485–5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu BH, Peacock RS, Vogel H. 2007. Bioinformatic analysis of the TonB protein family. Biometals 20:467–483. doi: 10.1007/s10534-006-9049-4. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson M, Hannavy K, Higgins CF. 1993. A sequence-specific function for the N-terminal signal-like sequence of the TonB protein. Mol Microbiol 8:379–388. doi: 10.1111/j.1365-2958.1993.tb01581.x. [DOI] [PubMed] [Google Scholar]
- 22.Jana B, Manning M, Postle K. 2011. Mutations in the ExbB cytoplasmic carboxy terminus prevent energy-dependent interaction between the TonB and ExbD periplasmic domains. J Bacteriol 193:5649–5657. doi: 10.1128/JB.05674-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ollis AA, Postle K. 2012. ExbD mutants define initial stages in TonB energization. J Mol Biol 415:237–247. doi: 10.1016/j.jmb.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Herrero A, Peacock RS, Howard SP, Vogel HJ. 2007. The solution structure of the periplasmic domain of the TonB system ExbD protein reveals an unexpected structural homology with siderophore-binding proteins. Mol Microbiol 66:872–889. doi: 10.1111/j.1365-2958.2007.05957.x. [DOI] [PubMed] [Google Scholar]
- 25.Higgs PI, Myers PS, Postle K. 1998. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J Bacteriol 180:6031–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ollis AA, Postle K. 2011. The same periplasmic ExbD residues mediate in vivo interactions between ExbD homodimers and ExbD-TonB heterodimers. J Bacteriol 193:6852–6863. doi: 10.1128/JB.06190-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ollis AA, Kumar A, Postle K. 2012. The ExbD periplasmic domain contains distinct functional regions for two stages in TonB energization. J Bacteriol 194:3069–3077. doi: 10.1128/JB.00015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ollis AA, Postle K. 2012. Identification of functionally important TonB-ExbD periplasmic domain interactions in vivo. J Bacteriol 194:3078–3087. doi: 10.1128/JB.00018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freed DM, Lukasik SM, Sikora A, Mokdad A, Cafiso DS. 2013. Monomeric TonB and the Ton Box are required for the formation of a high-affinity transporter-TonB complex. Biochemistry 52:2638–2648. doi: 10.1021/bi3016108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. 2006. Structure of TonB in complex with FhuA, E coli outer membrane receptor. Science 312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 31.Shultis DD, Purdy MD, Banchs CN, Wiener MC. 2006. Outer membrane active transport: structure of the BtuB:TonB complex. Science 312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 32.Evans JS, Levine BA, Trayer IP, Dorman CJ, Higgins CF. 1986. Sequence-imposed structural constraints in the TonB protein of E. coli. FEBS Lett 208:211–216. doi: 10.1016/0014-5793(86)81020-1. [DOI] [PubMed] [Google Scholar]
- 33.Larsen RA, Wood GE, Postle K. 1993. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol Microbiol 10:943–953. doi: 10.1111/j.1365-2958.1993.tb00966.x. [DOI] [PubMed] [Google Scholar]
- 34.Köhler SD, Weber A, Howard SP, Welte W, Drescher M. 2010. The proline-rich domain of TonB possesses an extended polyproline II-like conformation of sufficient length to span the periplasm of Gram-negative bacteria. Protein Sci 19:625–630. doi: 10.1002/pro.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. 2014. Structure of the AcrAB-TolC multidrug efflux pump. Nature 509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matias VRF, Al-Amoudi A, Dubochet J, Beveridge TJ. 2003. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J Bacteriol 185:6112–6118. doi: 10.1128/JB.185.20.6112-6118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh J, Postle K. 2004. Evidence for dynamic clustering of carboxy-terminal aromatic amino acids in TonB-dependent energy transduction. Mol Microbiol 51:203–213. doi: 10.1046/j.1365-2958.2003.03816.x. [DOI] [PubMed] [Google Scholar]
- 38.Carter DM, Miousse IR, Gagnon J-N, Martinez E, Clements A, Lee J, Hancock MA, Gagnon H, Pawelek PD, Coulton JW. 2006. Interactions between TonB from Escherichia coli and the periplasmic protein FhuD. J Biol Chem 281:35413–35424. doi: 10.1074/jbc.M607611200. [DOI] [PubMed] [Google Scholar]
- 39.Sauter A, Howard SP, Braun V. 2003. In vivo evidence for TonB dimerization. J Bacteriol 185:5747–5754. doi: 10.1128/JB.185.19.5747-5754.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosh J, Postle K. 2005. Disulphide trapping of an in vivo energy-dependent conformation of Escherichia coli TonB protein. Mol Microbiol 55:276–288. doi: 10.1111/j.1365-2958.2004.04384.x. [DOI] [PubMed] [Google Scholar]
- 41.Moeck GS, Letellier L. 2001. Characterization of in vitro interactions between a truncated TonB protein from Escherichia coli and the outer membrane receptors FhuA and FepA. J Bacteriol 183:2755–2764. doi: 10.1128/JB.183.9.2755-2764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Postle K, Kastead KA, Gresock MG, Ghosh J, Swayne CD. 2010. The TonB dimeric crystal structures do not exist in vivo. mBio 1(5):e00307-10. doi: 10.1128/mBio.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsen RA, Thomas MG, Postle K. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol Microbiol 31:1809–1824. doi: 10.1046/j.1365-2958.1999.01317.x. [DOI] [PubMed] [Google Scholar]
- 44.Higgs PI, Larsen RA, Postle K. 2002. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol Microbiol 44:271–281. doi: 10.1046/j.1365-2958.2002.02880.x. [DOI] [PubMed] [Google Scholar]
- 45.Kadner RJ, McElhaney G. 1978. Outer membrane-dependent transport systems in Escherichia coli: turnover of TonB function. J Bacteriol 134:1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sverzhinsky A, Fabre L, Cottreau AL, Biot-Pelletier DMP, Khalil S, Bostina M, Rouiller I, Coulton JW. 2014. Coordinated rearrangements between cytoplasmic and periplasmic domains of the membrane protein complex ExbB-ExbD of Escherichia coli. Structure 22:791–797. doi: 10.1016/j.str.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 47.Sverzhinsky A, Qian S, Yang L, Allaire M, Moraes I, Ma D, Chung JW, Zoonens M, Popot J-L, Coulton JW. 2014. Amphipol-trapped ExbB-ExbD membrane protein complex from Escherichia coli: a biochemical and structural case study. J Membr Biol 247:1005–1018. doi: 10.1007/s00232-014-9678-4. [DOI] [PubMed] [Google Scholar]
- 48.Popot JL, Althoff T, Bagnard D, Banères JL, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, Crémel G, Dahmane T, de la Maza LM, Ebel C, Gabel F, Giusti F, Gohon Y, Goormaghtigh E, Guittet E, Kleindschmidt JH, Kuhlbrandt W, Bon CL, Martinez KL, Picard M, Pucci B, Sachs JN, Tribet C, van Heijenoort C, Wien F, Zito F, Zoonens M. 2011. Amphipols from A to Z. Annu Rev Biophys 40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]
- 49.Popot J-L. 2010. Amphipols, nanodiscs, and fluorinated surfactants: three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu Rev Biochem 79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- 50.Khursigara CM, De Crescenzo G, Pawelek PD, Coulton JW. 2004. Enhanced binding of TonB to a ligand-loaded outer membrane receptor: role of the oligomeric state of TonB in formation of a functional FhuA.TonB complex. J Biol Chem 279:7405–7412. doi: 10.1074/jbc.M311784200. [DOI] [PubMed] [Google Scholar]
- 51.Raines RT, McCormick M, Van Oosbree TR, Mierendorf RC. 2000. The S-tag fusion system for protein purification. Methods Enzymol 326:362–376. doi: 10.1016/S0076-6879(00)26065-5. [DOI] [PubMed] [Google Scholar]
- 52.Park J-H, Na S-Y, Lee H-H, Lee Y-J, Kim KL. 2001. Detection of pET-vector encoded, recombinant S-tagged proteins using the monoclonal antibody ATOM-2. Hybridoma 20:17–23. doi: 10.1089/027245701300060364. [DOI] [PubMed] [Google Scholar]
- 53.Carter DM, Gagnon J-N, Damlaj M, Mandava S, Makowski L, Rodi DJ, Pawelek PD, Coulton JW. 2006. Phage display reveals multiple contact sites between FhuA, an outer membrane receptor of Escherichia coli, and TonB. J Mol Biol 357:236–251. doi: 10.1016/j.jmb.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 54.Mandava S, Makowski L, Devarapalli S, Uzubell J, Rodi DJ. 2004. RELIC—a bioinformatics server for combinatorial peptide analysis and identification of protein-ligand interaction sites. Proteomics 4:1439–1460. doi: 10.1002/pmic.200300680. [DOI] [PubMed] [Google Scholar]
- 55.Roy A, Nury H, Wiseman B, Sarwan J, Jault J-M, Ebel C. 2013. Sedimentation velocity analytical ultracentrifugation in hydrogenated and deuterated solvents for the characterization of membrane proteins. Methods Mol Biol 1033:219–251. doi: 10.1007/978-1-62703-487-6_15. [DOI] [PubMed] [Google Scholar]
- 56.Salvay A, Ebel C. 2006. Analytical ultracentrifuge for the characterization of detergent in solution. Prog Colloid Polym Sci 131:74–82. doi: 10.1007/2882_006. [DOI] [Google Scholar]
- 57.Hayashi Y, Matsui H, Takagi T. 1989. Membrane protein molecular weight determined by low-angle laser light-scattering photometry coupled with high-performance gel chromatography. Methods Enzymol 172:514–528. doi: 10.1016/S0076-6879(89)72031-0. [DOI] [PubMed] [Google Scholar]
- 58.Kim R. 2011. Native agarose gel electrophoresis of multiprotein complexes. Cold Spring Harb Protoc 2011:884–887. doi: 10.1101/pdb.prot4558. [DOI] [PubMed] [Google Scholar]
- 59.Alderton G, Ward WH, Fevold HL. 1945. Isolation of lysozyme from egg white. J Biol Chem 157:43–58. [PubMed] [Google Scholar]
- 60.Jachimska B, Wasilewska M, Adamczyk Z. 2008. Characterization of globular protein solutions by dynamic light scattering, electrophoretic mobility, and viscosity measurements. Langmuir 24:6866–6872. doi: 10.1021/la800548p. [DOI] [PubMed] [Google Scholar]
- 61.Zheng SQ, Kollman JM, Braunfeld MB, Sedat JW, Agard DA. 2007. Automated acquisition of electron microscopic random conical tilt sets. J Struct Biol 157:148–155. doi: 10.1016/j.jsb.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142:334–347. doi: 10.1016/S1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 63.Scheres SHW, Nunez-Ramirez R, Sorzano COS, Carazo JM, Marabini R. 2008. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc 3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sorzano COS, Bilbao-Castro JR, Shkolnisky Y, Alcorlo M, Melero R, Caffarena-Fernández G, Li M, Xu G, Marabini R, Carazo JM. 2010. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol 171:197–206. doi: 10.1016/j.jsb.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Radermacher M, Wagenknecht T, Verschoor A, Frank J. 1987. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc 146(Part 2):113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 66.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 67.Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. 2012. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure 20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scheres SHW. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 70.Harpaz Y, Gerstein M, Chothia C. 1994. Volume changes on protein folding. Structure 2:641–649. doi: 10.1016/S0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 71.Scheres SHW, Chen S. 2012. Prevention of overfitting in cryo-EM structure determination. Nat Methods 9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Postle K, Reznikoff WS. 1979. Identification of the Escherichia coli tonB gene product in minicells containing tonB hybrid plasmids. J Mol Biol 131:619–636. doi: 10.1016/0022-2836(79)90011-1. [DOI] [PubMed] [Google Scholar]
- 73.Slotboom DJ, Duurkens RH, Olieman K, Erkens GB. 2008. Static light scattering to characterize membrane proteins in detergent solution. Methods 46:73–82. doi: 10.1016/j.ymeth.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 74.Wen J, Arakawa T, Philo JS. 1996. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal Biochem 240:155–166. doi: 10.1006/abio.1996.0345. [DOI] [PubMed] [Google Scholar]
- 75.Postle K. 1990. Aerobic regulation of the Escherichia coli tonB gene by changes in iron availability and the fur locus. J Bacteriol 172:2287–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choul-Li S, Adams H, Pattus F, Celia H. 2008. Visualization of interactions between siderophore transporters and the energizing protein TonB by native PAGE. Electrophoresis 29:1333–1338. doi: 10.1002/elps.200700612. [DOI] [PubMed] [Google Scholar]
- 77.Kaserer WA, Jiang X, Xiao Q, Scott DC, Bauler M, Copeland D, Newton SMC, Klebba PE. 2008. Insight from TonB hybrid proteins into the mechanism of iron transport through the outer membrane. J Bacteriol 190:4001–4016. doi: 10.1128/JB.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Skare JT, Postle K. 1991. Evidence for a TonB-dependent energy transduction complex in Escherichia coli. Mol Microbiol 5:2883–2890. doi: 10.1111/j.1365-2958.1991.tb01848.x. [DOI] [PubMed] [Google Scholar]
- 79.Ahmer BM, Thomas MG, Larsen RA, Postle K. 1995. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J Bacteriol 177:4742–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Braun V, Gaisser S, Herrmann C, Kampfenkel K, Killmann H, Traub I. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J Bacteriol 178:2836–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scheres SHW, Gao H, Valle M, Herman G, Eggermont J, Frank J, Carazo J-M. 2007. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods 4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 82.Glaeser RM, Hall RJ. 2011. Reaching the information limit in cryo-EM of biological macromolecules: experimental aspects. Biophys J 100:2331–2337. doi: 10.1016/j.bpj.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Orlova EV, Saibil HR. 2004. Structure determination of macromolecular assemblies by single-particle analysis of cryo-electron micrographs. Curr Opin Struct Biol 14:584–590. doi: 10.1016/j.sbi.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 84.Ohi M, Li Y, Cheng Y, Walz T. 2004. Negative staining and image classification—powerful tools in modern electron microscopy. Biol Proced Online 6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gumbart J, Wiener MC, Tajkhorshid E. 2007. Mechanics of force propagation in TonB-dependent outer membrane transport. Biophys J 93:496–504. doi: 10.1529/biophysj.107.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adzhubei AA, Sternberg MJE. 1993. Left-handed polyproline II helices commonly occur in globular proteins. J Mol Biol 229:472–493. doi: 10.1006/jmbi.1993.1047. [DOI] [PubMed] [Google Scholar]
- 87.Kakinoki S, Hirano Y, Oka M. 2005. On the stability of polyproline-I and II structures of proline oligopeptides. Polym Bull 53:109–115. doi: 10.1007/s00289-004-0317-6. [DOI] [Google Scholar]
- 88.Larsen RA, Deckert GE, Kastead KA, Devanathan S, Keller KL, Postle K. 2007. His20 provides the sole functionally significant side chain in the essential TonB transmembrane domain. J Bacteriol 189:2825–2833. doi: 10.1128/JB.01925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kühlbrandt W. 2014. The resolution revolution. Science 343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 90.Lu P, Bai X-c, Ma D, Xie T, Yan C, Sun L, Yang G, Zhao Y, Zhou R, Scheres SHW, Shi Y. 2014. Three-dimensional structure of human γ-secretase. Nature 512:166–170. doi: 10.1038/nature13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liao M, Erhu C, Julius D, Cheng T. 2013. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Torres AG, Redford P, Welch RA, Payne SM. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect Immun 69:6179–6185. doi: 10.1128/IAI.69.10.6179-6185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yep A, McQuade T, Kirchhoff P, Larsen M, Mobley HLT. 2014. Inhibitors of TonB function identified by a high-throughput screen for inhibitors of iron acquisition in uropathogenic Escherichia coli CFT073. mBio 5(2):e01089-13. doi: 10.1128/mBio.01089-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.