Abstract
Expression of the genome requires RNA polymerase II (RNAPII) to transcribe across many natural and unnatural barriers, and this transcription across barriers is facilitated by protein complexes called elongation factors (EFs). Genetic studies in Saccharomyces cerevisiae yeast suggest that multiple EFs collaborate to assist RNAPII in completing the transcription of genes, but the molecular mechanisms of how they cooperate to promote elongation are not well understood. The Ccr4-Not complex participates in multiple steps of mRNA metabolism and has recently been shown to be an EF. Here we describe how Ccr4-Not and TFIIS cooperate to stimulate elongation. We find that Ccr4-Not and TFIIS mutations show synthetically enhanced phenotypes, and biochemical analyses indicate that Ccr4-Not and TFIIS work synergistically to reactivate arrested RNAPII. Ccr4-Not increases the recruitment of TFIIS into elongation complexes and enhances the cleavage of the displaced transcript in backtracked RNAPII. This is mediated by an interaction between Ccr4-Not and the N terminus of TFIIS. In addition to revealing insights into how these two elongation factors cooperate to promote RNAPII elongation, our study extends the growing body of evidence suggesting that the N terminus of TFIIS acts as a docking/interacting site that allows it to synergize with other EFs to promote RNAPII transcription.
INTRODUCTION
Transcription of genes by RNA polymerase II (RNAPII) is a well-orchestrated process that involves steps of initiation, elongation, and termination. Following promoter clearance, RNAPII enters the phase of productive elongation that is achieved by a Brownian ratchet mechanism, in which the RNAPII oscillates between a pretranslocated and a posttranslocated state. After nucleotide addition to the 3′ end of the RNA, the incoming nucleoside triphosphate (NTP) locks RNAPII in a posttranslocated form, readying it for the next cycle (1–4). However, productive elongation is not a product of efficient addition of nucleotides by RNAPII alone. During transcription elongation, RNAPII encounters several blocks, including sequence-specific pause sites, nucleotide limitations, DNA lesions, negative elongation factors, and DNA-bound proteins, which cause RNAPII to pause, arrest, or terminate transcription (5, 6). A myriad of elongation factors helps rescue paused/arrested polymerases and stimulate transcription (7, 8). Each elongation factor acts via a different mechanism, and often, one works in combination with others.
One of the best-characterized elongation factors known to rescue backtracked RNAPII is TFIIS. TFIIS promotes transcription elongation by stimulating the nucleolytic activity of RNAPII and realigning the 3′ end of the transcript in the active site of arrested RNAPII (for reviews, see references 9 and 10). New evidence suggests that the cleavage-promoting activity of TFIIS is not the only way in which it stimulates elongation (11–13). Biochemical and biophysical studies have only recently begun to uncover the mechanisms by which TFIIS functions with TFIIF, ELL, DSIF/NELF, and the Paf1c complex to stimulate elongation (11, 12, 14–18). The presence of multiple factors working to promote elongation strongly suggests redundancy in the functions of these factors but, more importantly, that these factors may work cooperatively during transcription. The roles of transcription factors in stimulating elongation in vivo and interactions among them have traditionally been identified by genetic analyses of Saccharomyces cerevisiae mutants and studies showing defects in transcription. However, fewer detailed biochemical studies have tried to understand how these factors act directly on RNAPII and the mechanisms by which they stimulate elongation.
The Ccr4-Not complex has been well studied for its functions in mRNA metabolism, especially for its role in regulating mRNA decay and transcription initiation (19–21). Though the possibility that it has the ability to affect the elongation stage of transcription has been suggested by genetic studies (23), only recently have biochemical analysis confirmed that it plays a direct role in stimulating transcription elongation (22, 24). The Ccr4-Not complex binds to elongation complexes (ECs) by directly interacting with both the Rpb4/Rpb7 module of RNAPII and the emerging transcript (25). Once it is bound to arrested ECs, it reactivates backtracked RNAPII, which apparently does not involve transcript cleavage. Evidence for this mode of action is that Ccr4-Not cannot stimulate elongation if arrest is achieved by the incorporation of a chain-terminating nucleotide, O-methyl-GTP (O-me-GTP), onto the 3′ end of the transcript (24). Rescue of this type of arrest requires transcript cleavage. In this regard, it functions through a mechanism different from that of the well-characterized elongation factor TFIIS (9). In vivo studies indicate that deleting DST1, the gene encoding TFIIS, enhances the phenotypes of a ccr4Δ mutation (22). This result suggests that the two genes genetically interact, but the cause of the enhanced phenotypes is not clear. Since Ccr4-Not and TFIIS play multiple roles in gene expression, including preinitiation complex formation, the cause of the enhanced phenotypes is not necessarily rooted in altered elongation.
In our current study, we have characterized the mechanism by which Ccr4-Not and TFIIS work cooperatively to regulate transcription elongation by RNAPII. We show that Ccr4-Not directly interacts with TFIIS and recruits it into arrested ECs, resulting in the enhanced cleavage of the displaced transcript in backtracked RNAPII and enhanced transcription. Interestingly, the interaction with Ccr4-Not requires the N terminus of TFIIS. We provide direct evidence of how these two elongation factors work cooperatively to regulate transcription and propose a function for a poorly understood region of TFIIS.
MATERIALS AND METHODS
Yeast strains and GLAM assay.
The S. cerevisiae strains used in this work are described in Table 1. Double mutants were isolated by mating and dissection of diploid strains. Cells were typically grown at 30°C in YP medium (1% yeast extract, 2% peptone) containing 2% dextrose (YPD). Deletion of genes by homologous recombination was carried out using knockout cassettes produced by PCR (26). Mutants were confirmed by using primers directed to the open reading frames of the genes. Spot testing on solid media was performed by growing cells to stationary phase in YPD and then diluting them in distilled water to optical densities at 600 nm (OD600s) of 1.0, 0.1, and 0.01. Cells were spotted onto synthetic complete (SC) medium with or without 2.5, 5, and 10 μg/ml mycophenolic acid (MPA; Sigma-Aldrich) and grown at 30°C. The plates were scanned daily for up to 4 days. The gene-length accumulation of mRNA (GLAM) assay (27) was performed with strains containing the GAL1 promoter inserted upstream of the YLR454W open reading frame, as described in previous publications (28, 29). Cells were grown to late mid-log phase at 30°C in YP medium supplemented with 2% galactose and 2% raffinose. RNA was extracted and subjected to Northern blotting as described previously (30). Probes directed to the open reading frames of GAL1 and YLR454W were used to detect the accumulation of the short and long transcripts, respectively. The ratio of long/short transcripts was calculated for each strain and compared to the ratio for the wild-type strain.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| JR1390 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 NOT4-TAP::HIS3 | This study |
| JR1408 | MATa NOT4-TAP::HIS3 dst1Δ::URA3 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 24 |
| JR1523 | MATa CAF40-TAP::HIS3 dst1Δ::URA3 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 25 |
| JR1533 | MATa/α his3Δ1/his3Δ0 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 CCR4/cct4::NATmX GAL1-YLR454::KanMX/YLR454dst1Δ::URA3/DST1 | This study |
| JR1534 | MATa/α his3Δ1/his3Δ0 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 NOT4/not4Δ::NATmX GAL1-YLR454::KanMX/YLR454dst1Δ::URA3/DST1 | This study |
| JR1535 | MATa/α his3Δ1/his3Δ0 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 CAF40/caf40Δ::NATmX GAL1-YLR454::KanMX/YLR454 dst1Δ::URA3/DST1 | This study |
| JR1536 | MATa/α his3Δ1/his3Δ0 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 DHH1/dhh1Δ::HIS3 GAL1-YLR454::KanMX/YLR454 dst1Δ::URA3/DST1 | This study |
| JR1537 | MATa/α his3Δ1/his3Δ0 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 NOT5/not5Δ::HIS3 GAL1-YLR454::KanMX/YLR454 dst1Δ::URA3/DST1 | This study |
| JR1538 | MATa his3 leu2Δ0 lys2Δ0 ura3Δ0 GAL1-YLR454::KanMX | This study |
| JR1540 | MATa his3 leu2Δ0 lys2Δ0 ura3Δ0 GAL1-YLR454::KanMX dst1Δ::URA3 | This study |
| JR1542 | MATa his3 leu2Δ0 lys2Δ0 ura3Δ0 GAL1-YLR454::KanMX dhh1Δ::HIS3 | This study |
| JR1544 | MATa his3 leu2Δ0 lys2Δ0 ura3Δ0 GAL1-YLR454::KanMX dhh1Δ::HIS3 dst1Δ::URA3 | This study |
| JR1550 | MATa his3 leu2Δ0 lys2Δ0 ura3Δ0 GAL1-YLR454::KanMX not5Δ::HIS3 | This study |
| JR1554 | MATa his3 leu2Δ0 lys2Δ0 ura3Δ0 GAL1-YLR454::KanMX not5Δ::HIS3 dst1Δ::URA3 | This study |
| JR1559 | MATa his3 leu2Δ0 ura3Δ0 GAL1-YLR454::KanMX ccr4Δ::NATMx | This study |
| JR1562 | MATa his3 leu2Δ0 ura3Δ0 GAL1-YLR454::KanMX ccr4Δ::NATMx dst1::URA3 | This study |
| JR1567 | MATa his3 leu2Δ0 lys2Δ0 ura3Δ0 GAL1-YLR454::KanMX caf40Δ::NATMx | This study |
| JR1570 | MATa his3 leu2Δ0 ura3Δ0 GAL1-YLR454::KanMX caf40Δ::NATMx dst1::URA3 | This study |
| JR1581 | MATa his3 leu2Δ0 lys2Δ0 ura3Δ0 GAL1-YLR454::KanMX not4Δ::NATMx | This study |
| JR1584 | MATα his3 leu2Δ0 lys2Δ0 ura3Δ0 GAL1-YLR454::KanMX not4Δ::NATMx dst1::URA3 | This study |
Purification of Ccr4-Not complex.
The Ccr4-Not complex was purified from 16-liter cultures of yeast containing a tandem affinity purification (TAP)-tagged version of CAF40 and a deletion of DST1, as described in previous publications (24, 25). The purified complex was dialyzed in dialysis buffer (20 mM HEPES, pH 7.6, 150 mM KCl, 10% glycerol), followed by concentration using a microfiltration device. The concentration of the Ccr4-Not complex was estimated by comparing the intensity of the bands to that of bands for known amounts of bovine serum albumin (BSA) on a silver-stained SDS-polyacrylamide gel. RNAPII was purified through a TAP-tagged version of Rpb4 and immunoaffinity chromatography on 8WG16 beads (31). The amount of RNAPII was estimated by the Bradford assay. Aliquots of protein were flash frozen in liquid nitrogen and stored at −80°C.
Cloning, expression, and purification of TFIIS.
The expression vectors for wild-type TFIIS, TFIIS D290A and E291A mutants, and TFIIS mutants with deletions of residues 290 deletion (290Δ) and 291 (291Δ) were obtained from Jesper Svejstrup (32). The proteins were expressed in Escherichia coli BL21(DE3)pLysS cells. Cells were grown in 1 liter Luria-Bertani (LB) medium to an OD600 of 0.7 and induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Sigma Aldrich) at 30°C for 3 h. The cell pellets were resuspended in cell lysis buffer (20 mM HEPES, pH 7.5, 500 mM NaCl, 10 mM imidazole, 10 μM ZnCl2, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM benzamidine hydrochloride) and lysed by freezing and thawing on ice, followed by sonication. The resulting crude cell lysate was centrifuged at 16,000 rpm and 4°C for 30 min, and the clarified lysate was incubated with 1 ml of Talon beads (Clontech) for 1 h. The resin was loaded into a column and washed with 50 ml of wash buffer (20 mM HEPES, pH 7.5, 500 mM NaCl, 20 mM imidazole, 10 μM ZnCl2, 10% glycerol, 1 mM PMSF, 2 mM benzamidine hydrochloride). The proteins were eluted in elution buffer (20 mM HEPES, pH 7.5, 500 mM NaCl, 150 mM imidazole, 10 μM ZnCl2, 10% glycerol, 1 mM PMSF, 2 mM benzamidine hydrochloride). The peak protein-containing fractions were pooled, dialyzed in dialysis buffer (20 mM HEPES, pH 7.9, 100 mM KCl, 10% glycerol, 5 mM beta-mercaptoethanol, 10 μM ZnCl2), and stored at −80°C. TFIIS truncation mutants were prepared by PCR, amplifying regions corresponding to amino acids (aa) 1 to 130 (TFIIS mutant 1 [TFIISmut1]), 131 to 309 (TFIISmut2), and 1 to 265 (TFIISmut3), and cloned into the NdeI and BamHI restriction sites in pET15b. Primer sequences are available upon request. Plasmids were confirmed by DNA sequencing.
Formation of elongation complexes and transcription runoff assays.
DNA templates were prepared by PCR using a plasmid containing a 70-nucleotide (nt)-long G-less cassette followed by a G tract (GGGG) and 80 nucleotides (33). The forward primer used in the PCR mixture contained a BglII restriction site. The PCR products were digested with BglII, followed by dephosphorylation with Antarctic phosphatase (New England BioLabs). The 5′-phosphorylated oligonucleotide containing the sequence GATCAAAAAAAATTA was ligated to the template, and the DNA was purified by agarose gel electrophoresis. Radiolabeled ECs were prepared in a manner similar to that described earlier (24, 33) with some minor modifications. Approximately 100 ng of tailed DNA template was preincubated with ∼0.25 pmol of purified yeast RNAPII (the amount was estimated from the protein content) in a 15-μl volume of transcription buffer (50 mM HEPES, pH 7.6, 100 mM KCl, 1 mM MnCl2, 12% glycerol, 0.5 mM dithiothreitol [DTT], 0.5 mM UpG, 20 U of RNasin [Promega], 100 ng/ml BSA) at room temperature for 10 min. ECs were formed at 30°C for 20 min by adding 5 μl of NTP mix, yielding final concentrations of 0.1 mM ATP, 0.1 mM CTP, 5 μM UTP, 5 μM O-methyl-GTP, and 4 μCi of [α-32P]UTP. Purified Ccr4-Not complex or an equivalent amount of BSA was added, and the mixture was incubated for 5 min at room temperature. Wild-type or mutant TFIIS was added where indicated, and after a 5-min incubation, runoff was initiated by the addition of UTP and GTP to 50 and 100 μM, respectively. Samples were removed and added to tubes containing 40 μl stop buffer (20 mM EDTA, pH 8.0, 200 mM NaCl, 1% SDS, 0.5 mg/ml yeast total RNA, 1 mg/ml proteinase K). The RNA was purified by extraction with phenol-chloroform-isoamyl alcohol (25:24:1) and ethanol precipitation. The products were separated on 10% urea-polyacrylamide gels, and the gels were dried, exposed to phosphorimager screen, and scanned using a Typhoon system (Molecular Dynamics-GE Life Sciences). The gel files were analyzed using ImageJ software, and the percent runoff was calculated as the amount of runoff product divided by the sum of the amount of the runoff product and the amount of EC remaining.
Immobilized elongation complex assays.
ECs were prepared as described above, except that the template was prepared using a biotinylated reverse primer and the DNA was bound to streptavidin Dynabeads (Life Technologies). For the TFIIS cleavage assay, the ECs were washed three times with wash buffer (50 mM HEPES, pH 7.5, 100 mM KCl, 0.5 mM DTT, 12% glycerol, 0.02% NP-40, 100 ng/ml BSA, 20 U of RNasin) and resuspended in transcription buffer (50 mM HEPES, pH 7.6, 100 mM KCl, 1 mM MgCl2, 12% glycerol, 0.5 mM DTT, 20 U of RNasin, 100 ng/ml BSA) lacking nucleotides. The Ccr4-Not complex or BSA and TFIIS were added to the transcription reaction mixtures, and the mixtures were incubated for 5 min at room temperature. The beads were collected, and then the RNA was purified from the beads by phenol-chloroform-isoamyl alcohol (25:24:1) extraction and ethanol precipitation. Samples were resolved on 12% denaturing gels. For the TFIIS binding assays, ECs were formed on immobilized templates for 20 min as described above, and purified Ccr4-Not complex was added for 5 min, followed by the addition of TFIIS mutants in which the acidic residues were changed to alanines for another 5 min. BSA was added to keep the amount of protein added to the assay mixtures constant. ECs were collected by a magnet and washed twice with wash buffer (20 mM HEPES, pH 7.5, 100 mM KCl, 1 mM DTT, 0.02% NP-40, 100 ng BSA). The proteins were liberated from the beads in elution buffer (20 mM HEPES, pH 7.5, 100 mM KCl, 1 mM DTT, 6 mM MgCl2, 2 mM CaCl2, 0.2 U DNase I, 400 ng RNase A) for 20 min at 37°C. The supernatants were boiled in SDS-PAGE loading buffer, and the proteins were separated on 12% SDS-polyacrylamide gels. The proteins were then transferred to a nitrocellulose membrane (Whatman) and probed with anti-6His (Sigma-Aldrich), anti-Rpb1 (monoclonal antibody 8WG16; Covance), and Not4 (polyclonal) antibodies. A Cy5-labeled goat anti-mouse secondary antibody (GE Life Sciences) was used for detection, and the blots were scanned on a Typhoon system. The signals were quantified using ImageJ software. The level of TFIIS binding relative to that of a defined amount of input on the gel was quantified.
TFIIS-pulldown assays.
Approximately 5 μg of purified wild-type and mutant TFIIS was immobilized on 20 μl Ni-nitrilotriacetic acid (NTA) beads (Qiagen). The immobilized beads were then incubated for 1 h with 1 μg of Ccr4-Not complex, collected, and washed twice with 200 μl each of wash buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10% glycerol, 0.01% NP-40). The bound proteins were eluted from the beads in 25 μl of elution buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 150 mM imidazole, 10% glycerol, 0.01% NP-40). The samples were boiled in SDS-PAGE loading buffer and subjected to Western blotting using polyclonal antibodies to Caf1 and Not1. A Cy5-labeled goat anti-rabbit secondary antibody (GE Life Sciences) was used for detection, and the blots were scanned on a Typhoon system.
RESULTS
Ccr4-Not and TFIIS genetically interact and function to promote elongation.
Loss of CCR4 results in enhanced phenotypes in combination with elongation factor mutants, including a dst1Δ mutant (TFIIS) (20, 34). Ccr4-Not is a modular complex composed of a Ccr4 group and a Not group (19, 21, 34). Genes in each of these groups display different genetic and physical interactions, and it cannot be taken for granted that deletion of other subunits, especially those in the Not group, would show synthetically enhanced phenotypes with a dst1Δ mutation. Therefore, we isolated mutants containing a deletion of DST1 and representative members of the Ccr4-Not complex. DHH1, CAF1, and CCR4 encode subunits of the nuclease module, NOT4 and NOT5 are nonessential subunits of the Not module, and CAF40 is a member of neither and its function is unknown (21, 34). Elongation factor mutants are sensitive to mycophenolic acid (MPA), which decreases intracellular GTP levels and leads to transcriptional stress (7, 35). Genetic interactions between Ccr4-Not genes and DST1 were assessed by growing single and double mutants at 30°C on synthetic complete (SC) medium or SC medium supplemented with 5 μg/ml MPA. Combining the dst1Δ allele with the ccr4Δ, not5Δ, not4Δ, or caf1Δ mutation resulted in reduced growth on SC medium and rich medium (Fig. 1A and data not shown). However, the double mutants displayed significantly enhanced sensitivity to MPA (Fig. 1A). Even the double caf40Δ dst1Δ mutant, which showed the less severe phenotypes, displayed an enhanced sensitivity to the drug when large amounts were used (data not shown). Thus, deletion of either the Ccr4 group or the Not group of genes causes synthetically enhanced phenotypes in combination with a loss of TFIIS activity.
FIG 1.
Ccr4-Not and DST1 mutations display synthetically enhanced phenotypes. (A) Single and double Ccr4-Not and dst1Δ mutants were spotted onto SC medium or SC medium containing 5 μg/ml MPA. The plates were incubated at 30°C for either 2 or 4 days, as indicated. (B) Schematic of the constructs used in the GLAM assay. (C) GLAM assay. Strains were grown to log phase in YP medium supplemented with galactose and raffinose. Northern blotting using probes directed to GAL1 (short) and YLR454W (long) was performed, and the ratio of the transcripts was calculated for each strain and compared to that for the wild type (WT), which was set equal to 1.0. Data are presented as the averages and standard deviations from 4 assays.
We next examined the effects of deleting Ccr4-Not subunits and TFIIS on transcription elongation. We used an assay that compares transcription across a long GAL1p-regulated gene (GAL1p-YLR454W) and the shorter endogenous GAL1 locus (Fig. 1B). Since the YLR454W gene is longer than GAL1, it is more sensitive to transcription elongation defects arising from mutations in transcription factors. Changes in transcription initiation would be controlled for since both genes are driven by the same promoter. This assay, referred to as the gene-length accumulation of mRNA (GLAM) assay, has been used extensively to detect elongation defects in yeast mutants (27–29). Deletion of DST1 did not result in a reduced GLAM ratio (Fig. 1C). It was previously noted that although TFIIS is a bona fide elongation factor, deleting DST1 does not cause a reduced GLAM ratio or changes in RNAPII transcription across genes (27, 36). Analysis of transcript abundance ratios showed that of the Ccr4-Not components tested, mutants with deletion of NOT4 and NOT5 individually displayed reduced GLAM ratios (Fig. 1C), suggesting an elongation defect. Deletion of CCR4 or CAF40 did not alter GLAM ratios significantly; however, the same mutants containing a deletion of DST1 had GLAM ratios lower than the GLAM ratio for either single mutant alone (Fig. 1C). Deleting DST1 in the not5Δ background only slightly reduced the GLAM ratio compared to that for the single mutants (Fig. 1C), but the ratio was already quite low in the single not5Δ mutant. Unfortunately, the not4Δ dst1Δ double mutant grew too poorly in galactose medium to be analyzed in this assay. Collectively, these genetic results support the hypothesis that Ccr4-Not and TFIIS cooperate to regulate transcription elongation in vivo.
Ccr4-Not enhances the ability of TFIIS to reactivate arrested RNAPII.
Genetic analyses described here and elsewhere suggest that Ccr4-Not and TFIIS work together to promote elongation, but genetic interactions can be indirect. Thus, we turned to biochemistry to determine how these elongation factors cooperate to regulate elongation. Both Ccr4-Not and TFIIS can reactivate arrested RNAPII. However, Ccr4-Not cannot rescue RNAPII arrested by the incorporation of O-me-GTP into the 3′ end of the transcript, indicating that it functions differently than TFIIS (24). During our initial studies on Ccr4-Not, we found that a complex purified from a strain containing DST1 stimulated the rescue of RNAPII blocked by the incorporation of O-me-GTP into the transcript but that a complex purified from a dst1Δ strain could not (data not shown). We speculated that a trace amount of TFIIS was copurifying with Ccr4-Not and that two elongation factors cooperated to promote transcription. To characterize the interplay between these two factors more precisely, we examined the ability of Ccr4-Not and TFIIS to stimulate elongation when used in combination. We employed an in vitro run-on transcription assay where transcription was initiated on a tailed template using the UpG dinucleotide. A 70-nt transcript is produced from a G-less cassette, and arrest occurs at the first G in a G tract (33). While it is documented that some end-initiated templates produce RNA-DNA hybrids (R loops) and the transcript displaces the nontemplate strand (NTS) behind RNAPII (37, 38), the template used here is not prone to such artifacts. Transcripts produced from this template are resistant to digestion with RNase H, which cleaves RNA in RNA-DNA duplexes (33), but are sensitive to single-stranded RNase (J. B. Crickard and J. C. Reese, unpublished data). Further, permanganate footprinting of the NTS of ECs formed on this template indicates that the nontemplate strand reanneals with the template strand behind polymerase (Crickard and Reese, unpublished).
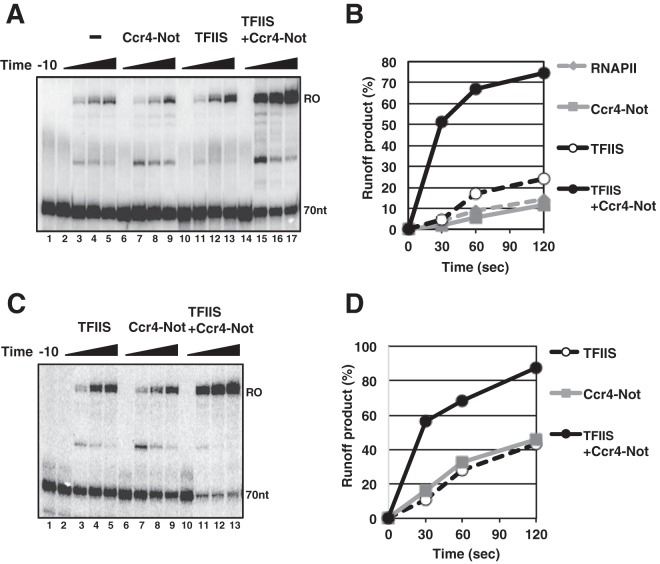
RNAPII ECs were formed in the presence of O-me-GTP, which prevents elongation and causes backtracking of RNAPII when the chain-terminating nucleotide is incorporated into the 3′ end of the transcript. As we reported previously (24), adding Ccr4-Not did not stimulate elongation from complexes arrested by O-me-GTP incorporation (Fig. 2A, lanes 6 to 9). We established nonactivating conditions where the amount of TFIIS did not cause an appreciable increase in the amount of runoff product (Fig. 2A, lanes 10 to 13). Interestingly, when Ccr4-Not was added to reaction mixtures containing the nonactivating amounts of TFIIS, a robust stimulation of elongation was observed (Fig. 2A, lanes 14 to 17). Results of quantification of the experiment are provided in Fig. 2B. Because Ccr4-Not cannot stimulate elongation when arrest is achieved by the incorporation of O-me-GTP into the transcript, the results suggest that Ccr4-Not is enhancing the activity of TFIIS. However, when assays were performed in the absence of O-me-GTP and arrest was achieved by omitting GTP, enhanced activation of transcription was also observed, suggesting that the synergy between Ccr4-Not and TFIIS is not a unique feature of chain terminator-induced arrest (Fig. 2C and D).
FIG 2.
Ccr4-Not and TFIIS synergize to rescue arrested RNAPII. (A) Runoff transcription assay in the presence of TFIIS and Ccr4-Not. Arrested RNAPII elongation complexes (70-nt ECs) were formed in the presence of O-me-GTP, as described in the Materials and Methods section, for 10 min (−10, lane 1). Afterwards, 50 fmol of TFIIS or 0.5 μg of Ccr4-Not or both were added. After 10 min, GTP and UTP were added to produce runoff (RO) transcripts. Reactions were stopped at 0 s (lanes 2, 6, 10, and 14), 30 s (lanes 3, 7, 11, and 15), 60 s (lanes 4, 8, 12, and 16), and 120 s (lanes 5, 9, 13, and 17) after the addition of the nucleotides. The transcripts were purified and analyzed on a 10% urea-polyacrylamide gel. (B) The percentage of runoff products from the gel in panel A was calculated and plotted over time. (C) Stalled elongation complexes were formed as described in the legend to panel A, except that the O-me-GTP transcription terminator was not added to the reaction mixture. Approximately 50 fmol of purified TFIIS (lanes 2 to 5 and 10 to 13) or 0.5 μg of the Ccr4-Not complex (lanes 6 to 9 and 10 to 13) was added to the transcription reaction mixtures and incubated for 10 min. The transcription reaction was then resumed with the addition of GTP and UTP. Reactions were stopped at 0, 30, 60, and 120 s, and transcripts were purified and analyzed on a 10% denaturing gel. (D) The percentage of runoff products was calculated and plotted as a function of time.
The results of the experiments described above suggest that Ccr4-Not allows TFIIS to stimulate elongation when the concentrations of TFIIS are limiting. This was examined more closely by carrying out transcription assays while titrating in increasing amounts of TFIIS in the presence or absence of a fixed amount of Ccr4-Not. While TFIIS only poorly stimulated elongation of arrested RNAPII complexes at the amounts tested, a greatly enhanced stimulation was observed in the presence of Ccr4-Not (Fig. 3A and B). This suggests that Ccr4-Not and TFIIS work synergistically to stimulate transcription elongation and that Ccr4-Not enhances the effective concentration of TFIIS.
FIG 3.

Ccr4-Not reduces the concentration of TFIIS required to rescue arrested RNAPII. (A) ECs were formed as described in the legend to Fig. 2A, and then TFIIS was titrated in (0, 25, 50, 100, and 250 fmol) with (lanes 6 to 10) or without (lanes 1 to 5) 0.5 μg Ccr4-Not complex. GTP and UTP were added to allow runoff for 120 s. (B) The percentage of runoff products was calculated from the gel in panel A and plotted as a function of the amount of TFIIS.
Ccr4-Not enhances the ability of TFIIS to reactivate arrested RNAPII by stimulating transcript cleavage.
TFIIS reactivates backtracked ECs by stimulating nucleolytic cleavage of the displaced transcript, thus reregistering the 3′ end with the active site of polymerase (9). However, multiple studies indicate that TFIIS can also rescue RNAPII elongation in a cleavage-independent manner. This may involve TFIIS loosening the interaction of backtracked RNA with the funnel of polymerase, realigning nucleic acids in the active center, or affecting the location of the trigger loop (13, 39, 40). Therefore, we next determined if Ccr4-Not enhances the TFIIS-dependent cleavage of the transcript by performing transcript cleavage assays on ECs immobilized on streptavidin beads (Fig. 4A). Following formation of arrested complexes in the presence of O-me-GTP, ECs were recovered by magnetic collection and washed. TFIIS and/or the Ccr4-Not complex was added, and the lengths of the transcripts associated with the EC were analyzed on gels. Addition of saturating amounts of Ccr4-Not did not lead to an appreciable increase in the cleavage of the 70-nt transcript bound within the EC (Fig. 4B, lane 1 versus lane 2), and a small amount of shorter products was observed in the gel. The origins of these products are not clear, but they may result from the degradation of the transcript from the 5′ end by a trace amount of nuclease contaminating the Ccr4-Not preparations. Adding a 2-fold excess of TFIIS over RNAPII led to a shortening of the 70-nt transcript (Fig. 4B, lane 1 versus lane 3). This result is in good agreement with findings in published work indicating that TFIIS shortens transcripts in backtracked RNAPII by 7 to 9 bases and is consistent with the length of RNA observed in the funnel of backtracked polymerase (3, 40–42). Shorter products were also observed and most likely resulted from the progressive shortening of the transcript caused by reiterative rounds of backtracking of RNAPII and cleavage by TFIIS (42–44). When a substoichiometric amount of TFIIS was added (0.3:1), no cleavage was detected (Fig. 4B, lane 4). However, when the same amount of TFIIS was added in the presence of Ccr4-Not, a significant amount of transcript cleavage was observed (Fig. 4B; compare lanes 4 and 5). Thus, Ccr4-Not can promote the TFIIS-dependent cleavage of the transcript.
FIG 4.
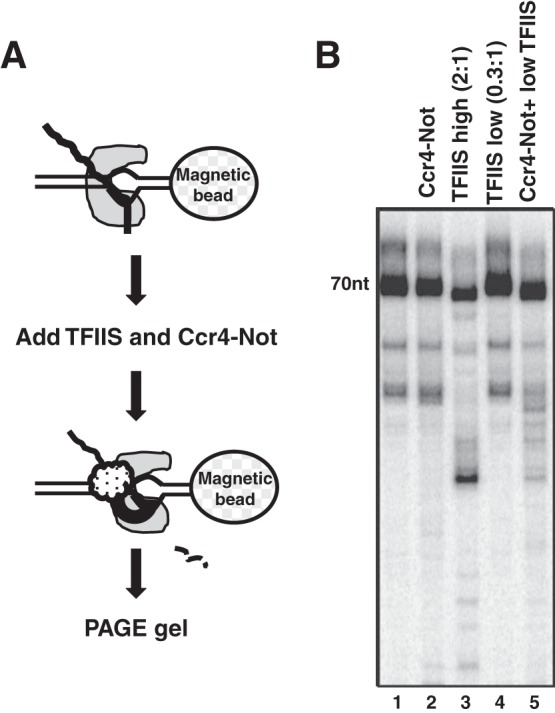
Ccr4-Not enhances TFIIS-stimulated transcript cleavage. (A) Schematic of the cleavage assay. ECs were assembled onto a biotinylated template immobilized onto streptavidin magnetic beads. Nucleotides were washed out, and then TFIIS or Ccr4-Not or both were added for 5 min. TFIIS was added in excess of RNAPII (2:1) or in substoichiometric amounts (0.3:1), as indicated in panel B. Beads were recovered by magnetic separation and washed, and the transcripts associated with the beads were purified and analyzed on urea-polyacrylamide gels. (B) Phosphorimage of the gel. The location of the 70-nt transcript is indicated.
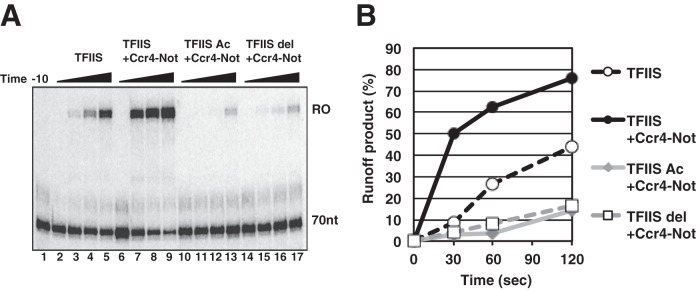
Next we determined if the transcript cleavage-promoting activity of TFIIS is required for it to function with Ccr4-Not. The loop of domain III inserts into the active site of RNAPII to stimulate transcript cleavage (39, 45). This activity of TFIIS is dependent on two conserved acidic residues in the loop of domain III (D290 and E291), and it has been shown that substituting these residues for alanines or deleting them blocks the TFIIS-dependent cleavage of the transcript and the rescue of backtracked RNAPII (32, 45) (data not shown). Transcription reactions were repeated in the presence of Ccr4-Not and substoichiometric amounts of wild-type TFIIS, TFIIS with point mutations (D290A and E291A), or TFIIS with deletion of the conserved acidic residues (290Δ and 291Δ). As expected, the mutant TFIIS proteins did not synergize with Ccr4-Not to rescue arrested RNAPII complexes (Fig. 5A and B). Therefore, the cleavage-promoting activity of TFIIS is required for the synergy between Ccr4-Not and TFIIS in rescuing arrested RNAPII.
FIG 5.
Synergy requires the cleavage-promoting activity of TFIIS. (A) A runoff assay was performed as described in the legend to Fig. 2A. Fifty femtomoles of wild-type TFIIS (lanes 2 to 5 and 6 to 9), TFIIS with point mutations (D290A and E291A; TFIIS Ac), or TFIIS with deletion of residues 290 and 291 (290Δ and 291Δ; TFIIS del) and 0.5 μg of Ccr4-Not complex were added as indicated. Runoff was initiated by adding GTP and UTP. Reactions were stopped at 0 s (lanes 2, 6, 10, and 14), 30 s (lanes 3, 7, 11, and 15), 60 s (lanes 4, 8, 12, and 16), and 120 s (lanes 5, 9, 13, and 17) after the addition of the nucleotides, and transcripts were analyzed on a 10% urea denaturing gel. (B) The percentage of runoff products was calculated and plotted over time.
Ccr4-Not interacts with TFIIS and enhances its recruitment into elongation complexes.
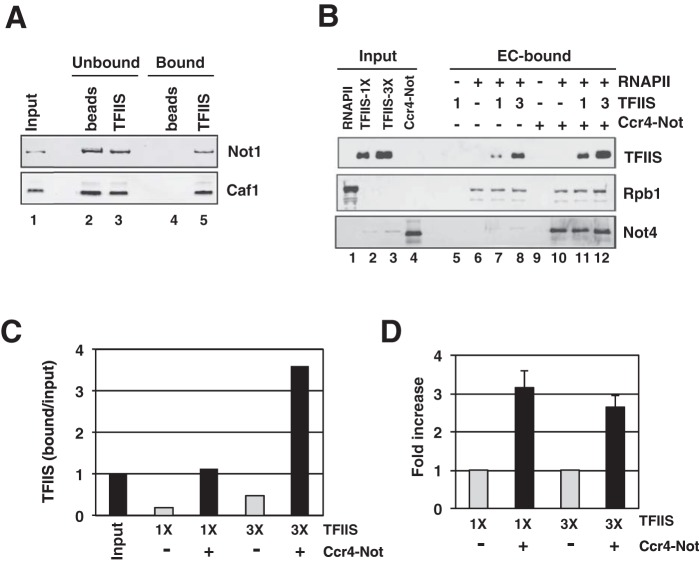
Transcription assays indicate that the concentration of TFIIS required to promote elongation is lowered in the presence of Ccr4-Not (Fig. 3A). One explanation for this result is that Ccr4-Not binds TFIIS and recruits it into the EC. A physical interaction would be consistent with the copurification of small amounts of TFIIS with the Ccr4-Not complex (see above), but this could also be explained if TFIIS interacted through another protein present in extracts. To test for a direct interaction between the two, we performed pulldown assays using recombinant TFIIS immobilized onto Ni-NTA beads via an N-terminal hexahistidine (6His) tag and purified Ccr4-Not. Ccr4-Not was then detected in the bound and supernatant fractions by immunoblotting. The results in Fig. 6A show that immobilized TFIIS, but not control Ni-NTA beads, retained Ccr4-Not. Thus, TFIIS binds directly to Ccr4-Not.
FIG 6.
Ccr4-Not binds TFIIS and enhances the recruitment of TFIIS into elongation complexes. (A) Pulldown assay with purified Ccr4-Not complex. Approximately 5 μg of purified TFIIS protein was immobilized on Ni-NTA beads and incubated for 1 h with 1 μg of Ccr4-Not. Half of the total of each fraction was loaded onto an SDS-polyacrylamide gel and analyzed by Western blotting using Not1 and Caf1 polyclonal antibodies. Naked beads were used as a control in the experiment. Protein bands were detected using a Cy5-conjugated secondary antibody and fluorescent imagining. Purified Ccr4-Not (200 ng of protein) was loaded in the input lane of the gel. (B) Binding of TFIIS to immobilized ECs. Elongation complexes were formed on a biotinylated DNA template in the presence of O-me-GTP using 500 fmol of yeast RNAPII for 20 min. Purified Ccr4-Not (1.2 μg) was added to the reaction mixture, followed by the addition of 600 (1×) or 1,800 (3×) fmol of TFIIS for 5 min. BSA was added to make up for the differences in the amount of the protein in the binding reaction. Beads were magnetically collected and washed thoroughly, and the proteins were released by nuclease digestion. Proteins were separated by SDS-PAGE and analyzed by Western blotting using anti-6His (TFIIS), anti-Not4, and anti-Rpb1 monoclonal antibodies. Protein bands were detected using a Cy5-conjugated secondary antibody. The bands in lanes 2 and 3 of the Not4 blot likely results from the cross-reaction of the antibody with an E. coli protein in the TFIIS preparations. There were unloaded lanes between the input and bound samples; these are not numbered. (C) Quantification of the binding of TFIIS to ECs from the gels shown in panel B. Signals in the bound lanes were compared to those obtained with a fixed amount of input on the gel (B, lane 2), which was set equal to a value of 1. (D) Average recruitment of TFIIS in ECs. The amount of TFIIS bound to ECs in the absence of Ccr4-Not for each experiment was set equal to 1.0, and the average fold increase in the presence of Ccr4-Not was calculated from three experiments. The error bars represent standard deviations. As in panels B and C, 1× and 3× designate the amount of TFIIS used in the binding assays.
We next tested if Ccr4-Not increases the recruitment of TFIIS into arrested RNAPII elongation complexes. Arrested ECs were formed on biotinylated DNA templates, washed, and then incubated with increasing amounts of TFIIS with or without Ccr4-Not. The complexes were then washed, and the amounts of TFIIS and Rpb1 were detected by immunoblotting. Since adding wild-type TFIIS leads to the progressive shortening of the transcript due to the reiterative cycles of backtracking and transcript cleavage (Fig. 4B and data not shown), a cleavage-defective TFIIS mutant (D290A, E291S) was used. TFIIS did not bind to the template in the absence of RNAPII; however, TFIIS was retained when ECs were formed in the presence of RNAPII (Fig. 6B; compare lane 5 to lane 7). Importantly, adding Ccr4-Not led to a significant increase in the binding of TFIIS to ECs by approximately 2- to 3-fold compared to that when Ccr4-Not was omitted (Fig. 6B to D; compare lanes 7 and 11 and lanes 8 and 12 in Fig. 6B). Thus, Ccr4-Not increases the recruitment of TFIIS into ECs.
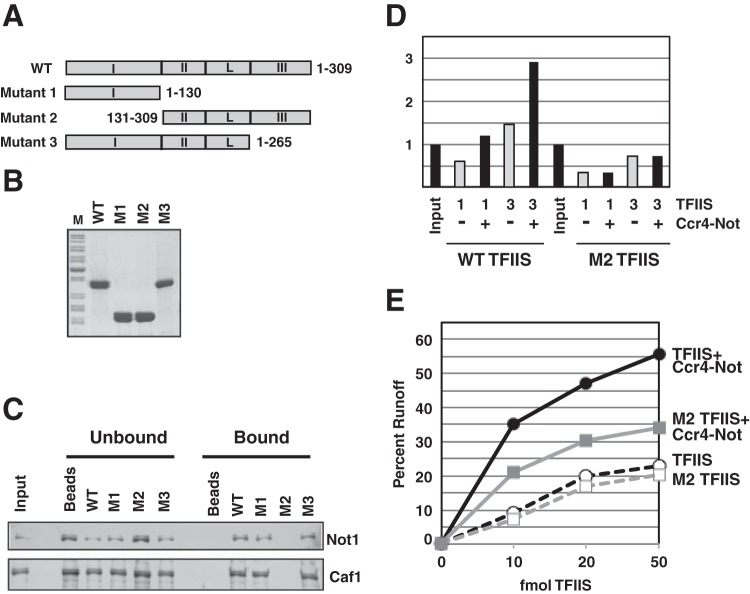
Many studies have mapped the domain structure of TFIIS (9, 46). The protein is divided into three domains and a linker region. Domain II (aa 131 to 240), the linker (aa 240 to 260), and domain III (aa 260 to 309) bind to RNAPII (46). High-resolution cocrystal structures of TFIIS with RNAPII have been solved and have revealed that domain III inserts into the secondary channel or funnel, placing two acidic residues into an active site to promote the cleavage of the displaced RNA transcript (1, 39, 40). The linker and domain II bind the core of RNAPII, with the latter contacting the jaw of Rpb1 (39). Domain I, the first 130 amino acids, was not visible in the structure, and biochemical and genetic studies indicate that it is dispensable for promoting transcript cleavage and rescuing backtracked RNAPII (9, 46). While dispensable for the biochemical activity of TFIIS, domain I interacts with several transcription factors (12, 47–49). In particular, a recent study showed that the N terminus is required for TFIIS recruitment into the preinitiation complex (50). In order to determine which domain of TFIIS interacts with Ccr4-Not, we constructed and purified truncated versions of TFIIS: mutant 1 (domain I), mutant 2 (domain II, linker, and domain III), and mutant 3 (domains I and II and linker) (Fig. 7A and B). Using a pulldown assay, we found that Ccr4-Not interacted with both mutants containing domain I (mutants 1 and 3) but failed to bind mutant 2, which lacked the N-terminal domain (Fig. 7C). This result indicates that domain I of TFIIS interacts with the Ccr4-Not complex.
FIG 7.
TFIIS domain I interacts with the Ccr4-Not complex. (A) Schematic representation of the domains of TFIIS and mutants used in the experiments. (B) A Coomassie blue-stained gel of immobilized TFIIS proteins used in the pulldown assays. The N terminus of TFIIS is very basic (pI 9.37) and runs very differently from its predicted molecular weight due to an abnormal charge-to-mass ratio. The aberrant migration of TFIIS mutants in SDS-polyacrylamide gels has been observed by others (see the figures in the supplemental material from reference 50). (C) Pulldown assays were performed as described in the legend to Fig. 6A, except that wild-type TFIIS, mutant 1 (M1; aa 1 to 130), mutant 2 (M2; aa 131 to 309), and mutant 3 (M3; aa 1 to 265) proteins were used. (D) TFIIS binding to ECs. Assays were performed and the results were quantified as described in the legends to Fig. 6B and C. (E) Runoff assays using wild-type TFIIS (TFIIS) and mutant 2 (M2 TFIIS) with and without Ccr4-Not.
To confirm that increased recruitment of TFIIS was dependent on its association with the Ccr4-Not complex, we repeated the TFIIS-EC binding assay using full-length TFIIS and mutant 2 (aa 131 to 309). Mutant 2 lacks domain I and does not bind Ccr4-Not but contains domains II and III, which bind RNAPII. While Ccr4-Not stimulated the recruitment of full-length TFIIS to RNAPII in ECs, deletion of domain I of TFIIS greatly reduced the Ccr4-Not-stimulated recruitment of TFIIS into ECs (Fig. 7D). These results suggest that Ccr4-Not directly binds the N terminus of TFIIS and this interaction is required for Ccr4-Not to recruit TFIIS into ECs.
To test if enhanced TFIIS recruitment was responsible for the cooperative action of Ccr4-Not and TFIIS on arrested RNAPII, we carried out run-on transcription assays using mutant 2. As shown in Fig. 2A, the addition of Ccr4-Not strongly enhanced the stimulation of elongation by substoichiometric amounts of wild-type TFIIS approximately 3-fold compared to that achieved with the addition of TFIIS alone, depending on the amount of TFIIS used (Fig. 7E). TFIIS mutant 2 (aa 131 to 309), which can interact with RNAPII, stimulated elongation to levels similar to those for full-length TFIIS when Ccr4-Not was absent from the reactions (Fig. 7E). However, Ccr4-Not was less effective at stimulating elongation by mutant 2 TFIIS than wild-type TFIIS (Fig. 7E). Though Ccr4-Not failed to increase the recruitment of mutant 2 into ECs in a binding assay (Fig. 7D), there was a low level of stimulation by Ccr4-Not. A likely explanation is that Ccr4-Not may bind weakly to mutant 2 in solution under transcription conditions but this interaction cannot withstand the wash conditions used in the binding assays. Nonetheless, the data are consistent with the conclusion that Ccr4-Not recruits TFIIS into ECs to enhance the rescue of arrested RNAPII and that domain I of TFIIS is important for this function.
DISCUSSION
RNA polymerase II (RNAPII) encounters many types of barriers as it transcribes across the genome, resulting in pausing, arrest, or backtracking (5, 6). Each type of stoppage of elongation is accompanied by different structural changes in RNAPII and the integrity of the RNA-DNA duplex in the active site (51). For example, backtracking leads to a displacement of the 3′ end of the transcript from the active site and its insertion into the funnel or secondary channel of RNAPII, while pausing does not. Resumption of elongation of backtracked polymerase requires the restoration of the 3′ end of the transcript within the active site, which can occur by cleavage of the displaced transcript or by forward tracking of RNA polymerase along the template (1, 40, 51). Reactivation of RNAPII under these conditions is enhanced by multiple elongation factors, which can restore elongation through complementary and synergistic mechanisms (7, 8).
In the current study, we show that Ccr4-Not and TFIIS cooperate to rescue arrested RNAPII and stimulate elongation. This mechanism works, at least in part, by Ccr4-Not-dependent recruitment of TFIIS into elongation complexes. Interestingly, the synergy between these two elongation factors is dependent on the interaction between Ccr4-Not and the N terminus, or domain I, of TFIIS. Domain I has recently been shown to be important for TFIIS to synergistically stimulate elongation in combination with TFIIF (12). While it was not tested in the study, the authors' interpretation of their results was that TFIIF stabilized the association of TFIIS with RNAPII, which promoted transcript cleavage. Together with independent experiments showing that TFIIF cross-links to TFIIS in preinitiation complexes, it seems likely that TFIIS is recruited to RNAPII by an interaction between its N terminus and a subunit of TFIIF (47). In light of these and our own results, it appears that the N-terminal domain of TFIIS plays an important role in the ability of TFIIS to synergize with other elongation factors. This would be consistent with biochemical and structural studies suggesting that the N terminus is free to interact with other factors when TFIIS is bound to RNAPII in ECs (39, 40, 48, 49). We speculate that the N terminus of TFIIS is a conserved recruitment platform required for it to function with multiple elongation factors, including Ccr4-Not.
We cannot rule out the possibility that Ccr4-Not makes other contributions in promoting the TFIIS-dependent cleavage of the transcript and the rescue of RNAPII. Ccr4-Not interacts with the N terminus of TFIIS, and this interaction is required to recruit it into the EC and stimulate elongation (this work). However, Ccr4-Not could, additionally, facilitate the insertion of domain III into the active site, thus promoting cleavage in this fashion. Cocrystal structures of RNAPII and TFIIS suggest the potential for a steric clash between TFIIS and the displaced RNA in the funnel of RNAPII (3, 39, 40, 51). Furthermore, in the presence of TFIIS, longer RNAs in backtracked RNAPII are cleaved more slowly than shorter RNAs (51). Extensively backtracked RNAPII containing a long displaced transcript in the funnel might inhibit the insertion of the acidic loop of TFIIS into the active site. Ccr4-Not could stimulate transcript cleavage and TFIIS binding to the EC by promoting the forward translocation of RNAPII down the template, reducing the length of RNA in the funnel. This would accelerate the cleavage of the transcript and/or reduce the potential for a steric clash in the funnel.
Is the only function of Ccr4-Not in elongation to recruit TFIIS and promote transcript cleavage? The results described here and published elsewhere suggest not. Ccr4-Not stimulates elongation in vitro without TFIIS (24). Furthermore, double mutants of Ccr4-Not subunits and DST1 display enhanced sensitivity to elongation inhibitors and slower growth and are more impaired than the single dst1Δ mutant in completing the transcription of long open reading frames (Fig. 1). If the only function of Ccr4-Not in elongation were to recruit TFIIS, we would expect them to show an epistatic genetic interaction and the single mutants would display phenotypes similar to those of the double mutants. Biochemical experiments indicate that both Ccr4-Not and TFIIS rescue backtracked RNAPII, yet their mechanisms are different (24, 34). TFIIS promotes the cleavage of the displaced transcript by RNAPII, reregistering the 3′ end in the active site (9, 13). Ccr4-Not, on the other hand, does so without stimulating transcript cleavage (24) (Fig. 4), possibly by promoting the diffusion of RNAPII downstream on the template. Since these two mechanisms are distinct, it is tempting to speculate that they are used to resolve different forms of arrested RNAPII throughout the genome. Ccr4-Not, like TFIIF, may prevent pausing or resolve RNAPII that has backtracked a few nucleotides (11, 12). However, when extensive backtracking takes place, such as when RNAPII encounters a very formidable barrier, this condition can be resolved rapidly only by cleaving the transcript. Here, Ccr4-Not could recruit TFIIS into the EC to perform this important step in the rescue of RNAPII. While we do not address the possibility here, Ccr4-Not can then promote the resumption of elongation by RNAPII after TFIIS-induced cleavage. It will be interesting to test such a model in future studies, possibly using single-molecule techniques.
Genome-wide studies have shown that TFIIS is localized across the open reading frame of genes, a distribution similar to that of RNAPII (52). It is tempting to speculate that TFIIS cooperates with different elongation factors during the stages of transcription. For example, TFIIF acts primarily at promoter clearance at the beginning of genes and then leaves RNAPII as it makes its way down the open reading frame (53–56). The synergy observed between TFIIF and TFIIS may be important early in the transcription cycle. Ccr4-Not cross-links across the open reading frame of genes (24, 57) and may synergize with TFIIS to resolve arrested RNAPII later in the transcription cycle or under a specialized condition. Ccr4-Not has been implicated as a regulator of stress pathways and could promote elongation during stress-induced transcriptional arrest.
ACKNOWLEDGMENTS
Jesper Svejstrup is acknowledged for providing the expression vectors for TFIIS. The members of the Center for Eukaryotic Gene Regulation, J. Brooks Crickard, and Craig Kaplan are recognized for their comments and feedback during the completion of this work.
This research was supported by funds from the National Institutes of Health to J.C.R. (GM58672) and to J.F. (GM100997).
REFERENCES
- 1.Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, Jawhari A, Jennebach S, Kamenski T, Kettenberger H, Kuhn CD, Lehmann E, Leike K, Sydow JF, Vannini A. 2008. Structure of eukaryotic RNA polymerases. Annu Rev Biophys 37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 2.Nudler E. 2009. RNA polymerase active center: the molecular engine of transcription. Annu Rev Biochem 78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Rucobo FW, Cramer P. 2013. Structural basis of transcription elongation. Biochim Biophys Acta 1829:9–19. doi: 10.1016/j.bbagrm.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan CD. 2013. Basic mechanisms of RNA polymerase II activity and alteration of gene expression in Saccharomyces cerevisiae. Biochim Biophys Acta 1829:39–54. doi: 10.1016/j.bbagrm.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomez-Herreros F, de Miguel-Jimenez L, Millan-Zambrano G, Penate X, Delgado-Ramos L, Munoz-Centeno MC, Chavez S. 2012. One step back before moving forward: regulation of transcription elongation by arrest and backtracking. FEBS Lett 586:2820–2825. doi: 10.1016/j.febslet.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 6.Svejstrup JQ. 2007. Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem Sci 32:165–171. doi: 10.1016/j.tibs.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Arndt KM, Kane CM. 2003. Running with RNA polymerase: eukaryotic transcript elongation. Trends Genet 19:543–550. doi: 10.1016/j.tig.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Sims RJ III, Belotserkovskaya R, Reinberg D. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev 18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 9.Wind M, Reines D. 2000. Transcription elongation factor SII. Bioessays 22:327–336. doi:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fish RN, Kane CM. 2002. Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta 1577:287–307. doi: 10.1016/S0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 11.Ishibashi T, Dangkulwanich M, Coello Y, Lionberger TA, Lubkowska L, Ponticelli AS, Kashlev M, Bustamante C. 2014. Transcription factors IIS and IIF enhance transcription efficiency by differentially modifying RNA polymerase pausing dynamics. Proc Natl Acad Sci U S A 111:3419–3424. doi: 10.1073/pnas.1401611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schweikhard V, Meng C, Murakami K, Kaplan CD, Kornberg RD, Block SM. 2014. Transcription factors TFIIF and TFIIS promote transcript elongation by RNA polymerase II by synergistic and independent mechanisms. Proc Natl Acad Sci U S A 111:6642–6647. doi: 10.1073/pnas.1405181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cipres-Palacin G, Kane CM. 1994. Cleavage of the nascent transcript induced by TFIIS is insufficient to promote read-through of intrinsic blocks to elongation by RNA polymerase II. Proc Natl Acad Sci U S A 91:8087–8091. doi: 10.1073/pnas.91.17.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Guermah M, Roeder RG. 2010. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palangat M, Renner DB, Price DH, Landick R. 2005. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc Natl Acad Sci U S A 102:15036–15041. doi: 10.1073/pnas.0409405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C, Yan H, Burton ZF. 2003. Combinatorial control of human RNA polymerase II (RNAP II) pausing and transcript cleavage by transcription factor IIF, hepatitis delta antigen, and stimulatory factor II. J Biol Chem 278:50101–50111. doi: 10.1074/jbc.M307590200. [DOI] [PubMed] [Google Scholar]
- 17.Elmendorf BJ, Shilatifard A, Yan Q, Conaway JW, Conaway RC. 2001. Transcription factors TFIIF, ELL, and Elongin negatively regulate SII-induced nascent transcript cleavage by non-arrested RNA polymerase II elongation intermediates. J Biol Chem 276:23109–23114. doi: 10.1074/jbc.M101445200. [DOI] [PubMed] [Google Scholar]
- 18.Izban MG, Luse DS. 1992. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem 267:13647–13655. [PubMed] [Google Scholar]
- 19.Miller JE, Reese JC. 2012. Ccr4-Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol 47:315–333. doi: 10.3109/10409238.2012.667214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denis CL, Chen J. 2003. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol 73:221–250. doi: 10.1016/S0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 21.Collart MA, Panasenko OO. 2012. The Ccr4-Not complex. Gene 492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 22.Denis CL, Chiang YC, Cui Y, Chen J. 2001. Genetic evidence supports a role for the yeast CCR4-NOT complex in transcriptional elongation. Genetics 158:627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaillard H, Tous C, Botet J, Gonzalez-Aguilera C, Quintero MJ, Viladevall L, Garcia-Rubio ML, Rodriguez-Gil A, Marin A, Arino J, Revuelta JL, Chavez S, Aguilera A. 2009. Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-Not in transcription-coupled repair. PLoS Genet 5:e1000364. doi: 10.1371/journal.pgen.1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC. 2011. The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev 25:581–593. doi: 10.1101/gad.2020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babbarwal V, Fu J, Reese JC. 2014. The Rpb4/7 module of RNA polymerase II is required for carbon catabolite repressor protein 4-negative on TATA (Ccr4-Not) complex to promote elongation. J Biol Chem 289:33125–33130. doi: 10.1074/jbc.C114.601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132. doi:. [DOI] [PubMed] [Google Scholar]
- 27.Morillo-Huesca M, Vanti M, Chavez S. 2006. A simple in vivo assay for measuring the efficiency of gene length-dependent processes in yeast mRNA biogenesis. FEBS J 273:756–769. doi: 10.1111/j.1742-4658.2005.05108.x. [DOI] [PubMed] [Google Scholar]
- 28.Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell 25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 29.Ginsburg DS, Govind CK, Hinnebusch AG. 2009. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol Cell Biol 29:6473–6487. doi: 10.1128/MCB.01033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reese JC, Green MR. 2003. Functional analysis of TFIID components using conditional mutants. Methods Enzymol 370:415–430. doi: 10.1016/S0076-6879(03)70036-6. [DOI] [PubMed] [Google Scholar]
- 31.Suh MH, Ye P, Zhang M, Hausmann S, Shuman S, Gnatt AL, Fu J. 2005. Fcp1 directly recognizes the C-terminal domain (CTD) and interacts with a site on RNA polymerase II distinct from the CTD. Proc Natl Acad Sci U S A 102:17314–17319. doi: 10.1073/pnas.0507987102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. 2010. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell 38:202–210. doi: 10.1016/j.molcel.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Wu CH, Gilmour DS. 2004. Analysis of polymerase II elongation complexes by native gel electrophoresis. Evidence for a novel carboxyl-terminal domain-mediated termination mechanism. J Biol Chem 279:23223–23228. doi: 10.1074/jbc.M402956200. [DOI] [PubMed] [Google Scholar]
- 34.Reese JC. 2013. The control of elongation by the yeast Ccr4-Not complex. Biochim Biophys Acta 1829:127–133. doi: 10.1016/j.bbagrm.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riles L, Shaw RJ, Johnston M, Reines D. 2004. Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast 21:241–248. doi: 10.1002/yea.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mason PB, Struhl K. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 37.Dedrick RL, Chamberlin MJ. 1985. Studies on transcription of 3′-extended templates by mammalian RNA polymerase II. Parameters that affect the initiation and elongation reactions. Biochemistry 24:2245–2253. [DOI] [PubMed] [Google Scholar]
- 38.Sluder AE, Price DH, Greenleaf AL. 1988. Elongation by Drosophila RNA polymerase II. Transcription of 3′-extended DNA templates. J Biol Chem 263:9917–9925. [PubMed] [Google Scholar]
- 39.Kettenberger H, Armache KJ, Cramer P. 2003. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114:347–357. doi: 10.1016/S0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 40.Cheung AC, Cramer P. 2011. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 41.Reines D. 1992. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem 267:3795–3800. [PMC free article] [PubMed] [Google Scholar]
- 42.Gu W, Reines D. 1995. Variation in the size of nascent RNA cleavage products as a function of transcript length and elongation competence. J Biol Chem 270:30441–30447. doi: 10.1074/jbc.270.51.30441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu W, Powell W, Mote J Jr, Reines D. 1993. Nascent RNA cleavage by arrested RNA polymerase II does not require upstream translocation of the elongation complex on DNA. J Biol Chem 268:25604–25616. [PMC free article] [PubMed] [Google Scholar]
- 44.Izban MG, Luse DS. 1993. SII-facilitated transcript cleavage in RNA polymerase II complexes stalled early after initiation occurs in primarily dinucleotide increments. J Biol Chem 268:12864–12873. [PubMed] [Google Scholar]
- 45.Jeon C, Yoon H, Agarwal K. 1994. The transcription factor TFIIS zinc ribbon dipeptide Asp-Glu is critical for stimulation of elongation and RNA cleavage by RNA polymerase II. Proc Natl Acad Sci U S A 91:9106–9110. doi: 10.1073/pnas.91.19.9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Awrey DE, Shimasaki N, Koth C, Weilbaecher R, Olmsted V, Kazanis S, Shan X, Arellano J, Arrowsmith CH, Kane CM, Edwards AM. 1998. Yeast transcript elongation factor (TFIIS), structure and function. II: RNA polymerase binding, transcript cleavage, and read-through. J Biol Chem 273:22595–22605. [DOI] [PubMed] [Google Scholar]
- 47.Murakami K, Elmlund H, Kalisman N, Bushnell DA, Adams CM, Azubel M, Elmlund D, Levi-Kalisman Y, Liu X, Gibbons BJ, Levitt M, Kornberg RD. 2013. Architecture of an RNA polymerase II transcription pre-initiation complex. Science 342:1238724. doi: 10.1126/science.1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diebold ML, Koch M, Loeliger E, Cura V, Winston F, Cavarelli J, Romier C. 2010. The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A and Med26. EMBO J 29:3979–3991. doi: 10.1038/emboj.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fish RN, Ammerman ML, Davie JK, Lu BF, Pham C, Howe L, Ponticelli AS, Kane CM. 2006. Genetic interactions between TFIIF and TFIIS. Genetics 173:1871–1884. doi: 10.1534/genetics.106.058834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim B, Nesvizhskii AI, Rani PG, Hahn S, Aebersold R, Ranish JA. 2007. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc Natl Acad Sci U S A 104:16068–16073. doi: 10.1073/pnas.0704573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. 2009. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science 324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghavi-Helm Y, Michaut M, Acker J, Aude JC, Thuriaux P, Werner M, Soutourina J. 2008. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev 22:1934–1947. doi: 10.1101/gad.471908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan Q, Moreland RJ, Conaway JW, Conaway RC. 1999. Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J Biol Chem 274:35668–35675. doi: 10.1074/jbc.274.50.35668. [DOI] [PubMed] [Google Scholar]
- 54.Ujvari A, Pal M, Luse DS. 2011. The functions of TFIIF during initiation and transcript elongation are differentially affected by phosphorylation by casein kinase 2. J Biol Chem 286:23160–23167. doi: 10.1074/jbc.M110.205658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng B, Price DH. 2007. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem 282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 56.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 57.Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, Pugh BF. 2011. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell 41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]