Abstract
In mouse models of atherosclerosis, normalization of hyperlipidemia promotes macrophage emigration and regression of atherosclerotic plaques in part by liver X receptor (LXR)-mediated induction of the chemokine receptor CCR7. Here we report that LXRα serine 198 (S198) phosphorylation modulates CCR7 expression. Low levels of S198 phosphorylation are observed in plaque macrophages in the regression environment where high levels of CCR7 expression are observed. Consistent with these findings, CCR7 gene expression in human and mouse macrophages cell lines is induced when LXRα at S198 is nonphosphorylated. In bone marrow-derived macrophages (BMDMs), we also observed induction of CCR7 by ligands that promote nonphosphorylated LXRα S198, and this was lost in LXR-deficient BMDMs. LXRα occupancy at the CCR7 promoter is enhanced and histone modifications associated with gene repression are reduced in RAW264.7 cells expressing nonphosphorylated LXRα (RAW-LXRα S198A) compared to RAW264.7 cells expressing wild-type (WT) phosphorylated LXRα (RAW-LXRα WT). Expression profiling of ligand-treated RAW-LXRα S198A cells compared to RAW-LXRα WT cells revealed induction of cell migratory and anti-inflammatory genes and repression of proinflammatory genes. Modeling of LXRα S198 in the nonphosphorylated and phosphorylated states identified phosphorylation-dependent conformational changes in the hinge region commensurate with the presence of sites for protein interaction. Therefore, gene transcription is regulated by LXRα S198 phosphorylation, including that of antiatherogenic genes such as CCR7.
INTRODUCTION
Liver X receptors (LXRs) are oxysterol-responsive transcription factors that manage cholesterol absorption, transport, and elimination. In macrophages, LXR signaling initiates the homeostatic response to cellular lipid loading. Macrophage uptake of normal and oxidized low-density lipoprotein (LDL) leads to increased cellular concentrations of cholesterol and oxysterols. Activation of LXRs by oxysterols induces the expression of genes involved in cellular cholesterol trafficking and efflux (1). However, in the face of persistent high cholesterol levels (hyperlipidemia), the LXR-regulated cholesterol homeostatic mechanisms in macrophages are overwhelmed (2). This results in the accumulation of cholesterol in the cytoplasm of macrophages, promoting their differentiation into foam cells that become retained in the subendothelial space and contributing to the formation and growth of an atherosclerotic plaque. In some mouse models, normalization of cholesterol levels promotes macrophage emigration from plaques and the regression of atherosclerosis (3–6). This is mediated in part by the induction of the chemokine receptor CCR7 via LXRs (7).
LXRα (NR1H3) and LXRβ (NR1H2) belong to the nuclear receptor (NR) superfamily of transcription factors (1). LXRβ is ubiquitously expressed, while LXRα is more tissue selective and is preferentially expressed in macrophages and tissues involved in lipid metabolism, such as the liver. LXRs form obligate heterodimers with retinoid X receptor (RXR) and are activated by ligands that are oxysterol cholesterol derivatives or cholesterol precursors (8–10). In the absence of ligand, the LXR/RXR heterodimer is nuclear and is bound to LXR response elements (LXREs) in the promoter of many (but not all) target genes in a complex with corepressors, such as silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) and nuclear receptor corepressor (N-CoR) (11). Upon ligand binding, the receptors undergo a conformational change that dismisses corepressors and recruits coactivators to induce gene activation. In macrophages, upon ligand binding, LXR upregulates the expression of genes involved in cholesterol transport and efflux (12, 13), including the ATP-binding cassette (ABC) transporters (ABCA1 and ABCG1) and extracellular cholesterol acceptors, such as apolipoprotein E (APOE) (14). These processes contribute to the stimulation of the reverse cholesterol transport (RCT) by LXRs. Accordingly, systemic administration of LXR agonists not only reduces atherosclerosis progression in LDL receptor−/− and ApoE−/− mice (15) but also promotes the atherosclerosis regression of plaques (7, 16). Both LXRα and LXRβ are necessary for full regression of plaques, though the requirement for LXRα is greater (15, 17).
LXRα is modified by phosphorylation at serine 198 (S198), which affects transcriptional regulatory activities (18–20). Phosphorylation of LXRα at S198 is conserved across species but is not conserved in LXRβ, suggesting that common signaling pathways modulate LXRα in rodents and humans, while different signals impact LXRβ (20). Foam cells in progressing plaques, as well as cholesterol-loaded cultured macrophages, showed increased phosphorylation of LXRα at S198 (20). Treatment with synthetic LXR agonist T0901317 (here referred to as T) also increased LXRα S198 phosphorylation (20). Casein kinase 2α (CK2) phosphorylates LXRα at S198 in macrophages, which selectively affects LXRα target gene expression (20). Changes in expression of certain LXR-dependent genes, including CCL24 (20), are markedly enhanced in cultured macrophages expressing a LXRα phosphorylation-deficient mutant (S198A) or by treatment with RXR agonists, such as 9-cis-retinoic acid (9cRA), that reduce the level of T-dependent S198 phosphorylation (20). This suggests that LXRα phosphorylation at S198 negatively regulates gene expression.
Our previous results suggested that LXRs contribute to expression of CCR7 via a promoter-proximal regulatory element (7). In the present studies, we examined the regulation of CCR7 gene expression by LXRα S198 phosphorylation in human and mouse macrophage cells and compared the results to LXRα S198-P levels in mouse progressing and regressing plaques. In addition, expression profiles from wild-type (WT) RAW-LXRα (RAW-LXRα WT) and RAW-LXRα S198A cells were used to identify genes selectively affected by LXRα phosphorylation upon ligand treatment. We also performed molecular modeling of LXRα/RXR on DNA to identify the location of S198 on the structure of LXRα in the absence and presence of phosphorylation. Our results suggest that LXRα phosphorylation may be an important determinant of atherosclerosis regression, for example, by regulating CCR7 expression through local conformational alterations, and that novel LXRα pharmacology could be achieved by targeting the phosphorylation of this receptor.
MATERIALS AND METHODS
Cell culture and transfections.
THP1 cells were obtained from the ATCC. RAW264.7 cells expressing FLAG-tagged WT human LXRα (hLXRα) (RAW-LXRα WT) or mutant human LXRα S198A (RAW-LXRα S198A) have been described previously (20). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). For ligand treatments, RAW cells were cultured in DMEM supplemented with 1% FBS and the indicated amounts of LXR agonist T0901317 (T; Sigma-Aldrich), 9-cis-retinoic acid (9cRA; Sigma-Aldrich), or 24(S),25 epoxycholesterol (EC; Sigma-Aldrich). THP1 cells were maintained in RPMI medium supplemented with 10% FBS. After differentiation in the presence of phorbol myristate acetate (PMA) (5 ng/ml) for 3 days, cells were cultured in RPMI medium with 1% FBS and subjected to ligand treatment as done with the RAW cells.
Mouse primary macrophage isolation and culture.
LXRα−/−/β−/− mice were generated as previously described (21). Bone marrow-derived macrophages (BMDMs) were prepared from wild-type and knockout mice as described previously (22). Tibiae and femurs were isolated from 10-to-20-week-old C57BL/6 (WT) or SV129 × C57BL/6 (LXRα−/−/β−/−) mice, and the bone marrow was flushed out with DMEM. Cells were seeded into petri dishes in RPMI medium–20% FBS supplemented with 10 ng/ml macrophage colony-stimulating factor (M-CSF) (Peprotech). After 7 days in culture, BMDMs were scraped off, split into 6-well plates, and treated as described in the figure legends.
qPCR.
Total RNAs from RAW and THP1 cells were extracted with TRIzol (Invitrogen) as described by the manufacturer. RNA from BMDMs was prepared using a Qiagen RNeasy minikit. The total RNA was converted into cDNA using enhanced avian reverse transcriptase (USB) and random primer hexamers by following the manufacturer's instructions. cDNAs were amplified using a SYBR green quantitative PCR (qPCR) kit (USB) on an iCycler instrument (Bio-Rad). The following primers were used: for mouse CCR7, 5′-TGTACGAGTCGGTGTGCTTC-3′ and 5′-GGTAGGTATCCGTCATGGTCTTG-3′; for human CCR7, TGAGGTCACGGACGATTACAT-3′ and 5′-GTAGGCCCACGAAACAAATGAT-3′ and 5′-GACGGAGCTTGTGTCCAAGAT-3′; for human GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-CATGAGAAGTATGACAACAGCCT-3′ and 5′-AGTCCTTCCACGATACCAAAGT-3′; and for mouse cyclophilin, 5′-GGCCGATGACGAGCCC-3′ and 5′-TGTCTTTGGAACTTTGTCTGCAA-3′.
Chromatin immunoprecipitation (ChIP) assays.
Assays were performed as described previously with the following modifications. After treatments (2 h), the cells were cross-linked with 1% formaldehyde at room temperature for 10 min, followed by neutralization with 0.125 M glycine for 5 min. The cells then were washed twice in ice-cold phosphate-buffered saline (PBS) and resuspended in Farnham lysis buffer {5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 8.0), 85 mM KCl, 0.5% NP-40, proteinase inhibitor cocktail (Sigma-Aldrich)} for 10 min. The fractions of nuclei were separated by spinning and resuspended in sonication buffer (PBS supplemented with 1% NP-40, 0.5% sodium deoxycholate, 0.5% sodium lauryl sulfate, and proteinase inhibitor cocktail). The sonication was done in a Bioruptor sonication system (Diagenode). The lysates were cleared by centrifugation and diluted to 0.1% sodium lauryl sulfate. Specific antibodies (anti-LXRα [PPZ0412; Perseus Proteomics], anti-H3K9me3 [ab8898; Abcam], and anti-H3K27me3 [07-449; Millipore]) or matching IgGs were coupled to Dynabeads (protein G or protein A; Invitrogen) and then added to cell lysates. After immunoprecipitation, beads were washed 5 times with LiCl buffer (Tris [100 mM; pH 7.5], 500 mM LiCl, 1% NP-40, 1% sodium deoxycholate) and once with TE buffer (Tris [10 mM; pH 7.5], 0.1 mM EDTA) and then resuspended in elution buffer (1% sodium lauryl sulfate, 0.1 M NaHCO3) and incubated at 65°C for 1 h. The supernatants were transferred and kept at 65°C overnight, followed by purification with a PrepEase DNA cleanup kit (USB). qPCR was done to measure binding on DNA with the following primer pair for mCCR7 −120 to −15: 5′-TCCTATGACAGCCGAATGTG-3′ and 5′-GCCCCTTTTAAGTTGTTCCA-3′.
Immunohistochemistry.
The grafted arches from progressing and regressing plaques were removed, embedded in OCT, and frozen. Serial sections (6 μm thick) were obtained using a cryostat. Sections were fixed in ice-cold acetone for 10 min and rinsed in PBS three times for 3 min each time. Sections were incubated in 3% H2O2 for 10 min to quench the peroxidase activity and blocked with 5% normal horse serum–PBS for 1 h. Fixed sections were stained overnight at 4°C with the primary antibody to LXRα (ab3585; Abcam) at 10 μg/ml or LXRα-S198-P (affinity-purified rabbit 1135 [20]) at 15 μg/ml in 2% BSA–PBS. Sections were washed five times in PBS for 3 min each time and incubated with a secondary biotinylated anti-rabbit IgG (H+L) antibody at a 1:1,000 dilution (BA-1000; Vector Laboratories) for 1 h at room temperature. Sections were washed five times in PBS for 3 min each time, incubated with horseradish peroxidase streptavidin (SA-5704; Vector Laboratories) for 30 min, and washed again five times in PBS for 3 min each time. Staining was visualized using a DAB (3,3′-diaminobenzidine) peroxidase substrate kit (SK-4100; Vector Laboratories) following the manufacturer's protocol, and the reaction mixture was counterstained with hematoxylin for 2 min (H3401; Vector Laboratories) and rinsed with tap water until the water ran clear. Sections were imaged on a Zeiss microscope.
Microarray.
Total RNA was isolated from a murine macrophage RAW 264.7 cell line expressing wild-type (WT) human LXRα or human LXRα with a serine-to-alanine mutation (S198A) (20) (1.1 × 106 cells) cultured in DMEM–10% FBS, washed twice with PBS, and treated with T (1 μM) in phenol-red free DMEM–10% lipoprotein-deficient serum for 24 h. A murine 430A 2.0 Affymetrix GeneChip was used for analysis of gene expression.
Normalization and analysis.
CEL files representative of four different conditions and duplicates (a total of 8) were normalized to account for technical variations between the arrays using Robust Multichip Average (RMA) Express for Windows. Primary data were analyzed by using MultiExperiment Viewer (MeV) for significance and hierarchical clustering. The analysis of variance (ANOVA) test was performed under each set of conditions to find those probe sets that were considered significant (P < 0.05). Genes were considered significant if (i) they were identified in the ANOVA test as significant and (ii) they displayed a fold change in expression of >1.5 when the logarithmic values determined in the presence and absence of hormone treatment were subtracted.
The L2L Microarray Database (L2L MDB), a collection of published microarray data, in the form of a set of lists, was used to determine gene ontology (GO) classes and associations.
Molecular modeling.
A three-dimensional (3D) structural model of LXRα was built by homology based on the LXRα sequence (Uniprot sequence accession no. Q13133). The published crystallographic structure of LXRβ in a liganded hLXRβ-hRXRα heterodimer on DNA (PDB accession no. 4NQA) was used as the template (23). The DNA binding domain (DBD) and ligand binding domain (LBD) as well as the hinge region connecting the two are present in both hRXRα and hLXRβ in the “b” chain of the LXRβ template structure, so this specific chain from the crystal structure was used as a template. The DBD, LBD, and hinge region were thus all modeled in LXRα in the context of hRXRα and DNA. A previously published homology modeling method was used (24, 25). The sequence similarities between the LXRα and LXRβ DBDs and LBDs were sufficiently high to unambiguously assign all of the amino acids in those domains and build an initial model via sequence alignment. The LXRα hinge loop region was first modeled in the absence of the hRXRα and DNA. The lowest-energy loop conformation was then used as the starting conformation for the subsequent loop sampling in the context of the hRXRα and DNA, with and without a serine phosphorylation at LXRα amino acid position 198 (S198), via a previously published loop modeling method (26). The loop was subjected to a biased-probability Monte-Carlo conformational search with local minimization for 50 million steps/calls (27). ICM-Pro software (Molsoft, LLC, La Jolla, CA) was used to perform all modeling manipulations.
RESULTS
LXRα S198 phosphorylation is enhanced in atherosclerosis progression versus regression.
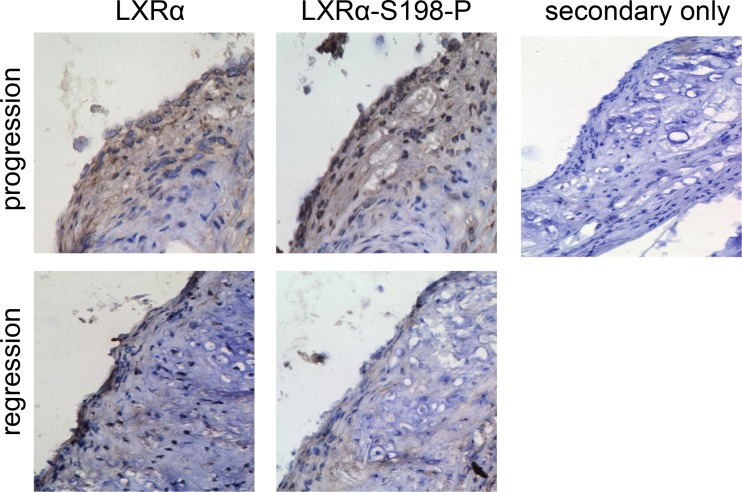
To address the relevance of LXRα phosphorylation in atherosclerosis, we examined the protein levels of LXRα-S198-P and LXRα from aortic arches of ApoE-deficient mice undergoing atherosclerosis progression and regression by immunohistochemistry. We found that LXRα-S198-P was higher in progressing plaques than in regressing plaques (Fig. 1). This correlates with previous studies showing increased CCR7 mRNA and protein levels in regressing plaques (4, 7) and suggests that the nonphosphorylated form of LXRα S198 in the regression environment is associated with CCR7 expression.
FIG 1.
LXRα and LXRα-S198-P levels in atherosclerotic plaques. Aortic arches from donor ApoE−/− mice fed a high-fat diet for 16 weeks were transplanted into WT (regression) or ApoE−/− (progression) recipients. At 6 days posttransplant, the grafts were harvested. Sections were immunostained for LXRα and LXRα-S198-P. Dark brown areas indicate areas of staining. A representative section is shown. No staining above the background level is evident in the presence of the secondary antibody alone (secondary only). Magnification, ×200.
Expression of CCR7 in macrophages is associated with nonphosphorylated LXRα-S198.
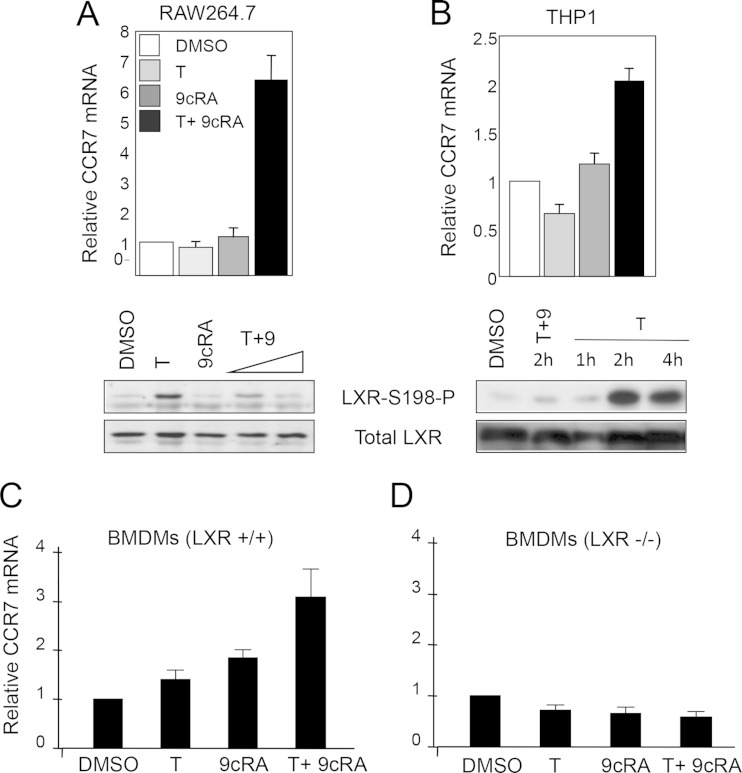
We next examined the impact of LXRα S198 phosphorylation on Ccr7 expression in mouse macrophage RAW264.7 cells ectopically expressing human LXRα (RAW-LXRα WT) (20) and in human differentiated THP1 cells expressing endogenous LXRα. When either RAW-LXRα cells (Fig. 2A) or THP1 cells (Fig. 2B) were stimulated with both T0901317 (T) and 9-cis-retinoic acid (9cRA), the combination of which reduces the level of LXRα S198 phosphorylation compared to that seen with T treatment alone, we observed an increase in CCR7 mRNA expression. In contrast, treatment with either ligand alone elicited little Ccr7 expression. In parental RAW cells that contained abundant LXRβ but lacked LXRα, we did not see induction of Ccr7 mRNA upon T or 9cRA or T-plus-9cRA treatment (not shown). Thus, the S198 nonphosphorylated form of LXRα is associated with Ccr7 expression in macrophage cell lines.
FIG 2.
RXR ligand 9-cis-retinoic acid inhibits LXRα phosphorylation and induces Ccr7 expression. (A) (Top panel) Ccr7 expression was analyzed by qPCR in RAW264.7 cells expressing WT LXRα in the absence of ligand (DMSO) and in the presence of T0901317 (T; 5 μM) or 9-cis-retinoic acid (9cRA; 1 μM) or upon cotreatment with both T (5 μM) and 9cRA (1 μM) for 24 h. The values indicate expression levels normalized to cyclophilin A mRNA levels and are presented relative to the expression in vehicle-treated cells, which was set as 1. Error bars represent standard deviations (SD). (Bottom panel) Nuclear extracts were prepared from RAW-LXRα WT cells cultured for 4 h in the absence (DMSO) or presence of T (5 μM) or 9cRA (1 μM) or a combination of T (5 μM) and increasing (0.1 and 1 μM) concentrations of 9cRA. LXRα S198 phosphorylation and the total level of LXRα were detected by Western blotting. (B) (Top panel) Ccr7 expression was analyzed by qPCR in differentiated THP1 cells treated as described for panel A. Values were normalized to GAPDH gene levels. (Bottom panel) Nuclear extracts from THP1 cells were analyzed for T-dependent LXRα S198 phosphorylation over a 4-h time course and in the presence of a 2-h 9cRA (1 μM) cotreatment. (C and D) LXR functions as an activator of Ccr7 mRNA expression. Bone marrow-derived macrophages were isolated from Lxr+/+ and Lxrα/β−/− mice. Cells were treated in culture with the vehicle (DMSO), T (5 μM), 9cRA (1 μM), or both ligands for 18 h, and Ccr7 mRNA expression normalized to cyclophilin A levels was analyzed by qPCR; data are presented relative to the expression in vehicle-treated cells, which was set as 1. Error bars represent SD.
We also tested the LXR dependency of Ccr7 expression in primary bone-derived macrophages (BMDMs). We isolated BMDMs from wild-type and Lxrα/Lxrβ double-knockout mice (here termed Lxr−/−) (21) and treated them in culture with vehicle (dimethyl sulfoxide [DMSO]), T alone, 9cRA alone, or T in combination with 9cRA and examined Ccr7 mRNA expression. Treatment with T alone or 9cRA alone resulted in a small induction of Ccr7 mRNA expression, whereas the combination of T plus 9cRA resulted in a 3-fold increase in the induction of Ccr7 mRNA (Fig. 2C). Treatment with GW3965, a synthetic LXR agonist that induces S198 phosphorylation, did not induce the expression of Ccr7 mRNA (not shown). Importantly, the induction of Ccr7 expression was LXR dependent as there was no induction by ligand of Ccr7 mRNA from Lxr−/− macrophages (Fig. 2D). Thus, the ligand-dependent induction of Ccr7 requires LXR.
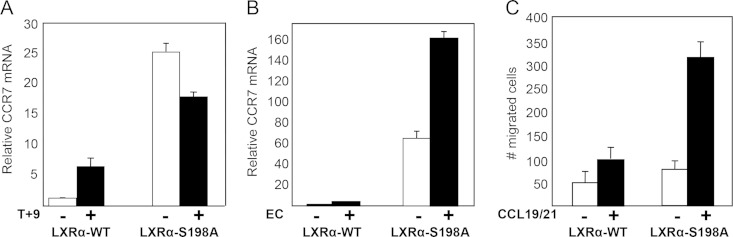
We next determined the impact of inhibiting LXR S198 phosphorylation through mutation of the site from a serine to an alanine in RAW cells (RAW-LXRα S198A). We found that, in those cells, LXRα S198A markedly enhanced basal levels of Ccr7 mRNA compared to the results seen with phosphorylated WT LXRα (Fig. 3A, DMSO). Whereas Ccr7 expression was slightly decreased by LXR S198A upon treatment with T plus 9cRA (or treatment with T alone; not shown), the natural ligand 24(S),25-epoxycholesterol further increased the expression of Ccr7 (Fig. 3B), suggesting that additional changes imparted by ligand affect LXRα-mediated transcription of CCR7. In addition, the chemotactic potential of cells with respect to CCR7-specific chemokines (CCL21 and CCL19) was higher in LXRα-S198A cells than in LXRα WT cells (Fig. 3C). These results demonstrate that, in RAW cells, Ccr7 expression and chemotaxis are modulated by LXRα S198 phosphorylation.
FIG 3.
LXRα phosphorylation regulates Ccr7 gene expression and chemotaxis. (A) RAW-LXRα WT and LXRα-S198A-expressing cells were incubated with DMSO vehicle (−) or T (1 μM) and 9cRA (1 μM) (T+9) for 24 h, and transcripts were analyzed by qPCR. Values indicate expression of CCR7 normalized to expression of cyclophilin A, and levels are presented as fold induction relative to the expression in vehicle-treated LXRα WT cells, which was arbitrarily set as 1. (B) RAW-LXRα WT cells or the RAW-LXRα S198A cells were incubated for 24 h with DMSO vehicle (−) or 24(S),25-epoxycholesterol (5 μM) (EC). Transcripts were analyzed as described above. Error bars represent SD. The experiment was repeated at least 3 times with similar results. (C) RAW-LXRα WT or RAW-LXRα S198A cells (4 × 104) were added to the upper chambers of a Transwell (Neuroprobe) (5-μm-diameter pore) plate. Chemotaxis buffer (DMEM–10% FBS) without (−) or with (+) CCR7 ligands (100 ng/ml CCL19 and 100 ng/ml CCL21) was added to the lower chambers, and plates were placed at 37°C for 3 h. The cells that migrated toward the lower chamber were counted. Assays were performed in triplicate, and the results were averaged. Error bars represent SD.
Nonphosphorylated LXRα S198 relieves histone H3-repressive chromatin at CCR7.
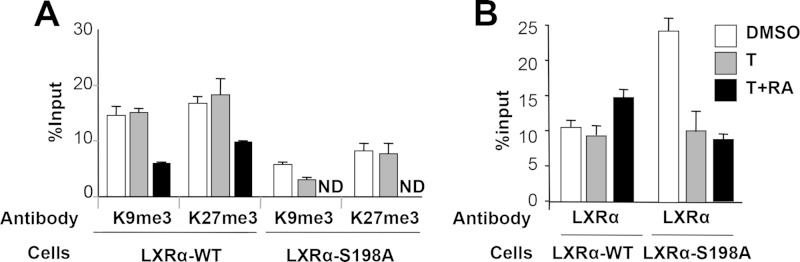
We next examined chromatin modifications at the endogenous CCR7 promoter as a function of LXRα S198 phosphorylation. As CCR7 expression is repressed in most cell types, we analyzed the histone-repressive modifications, H3K9me3 and H3K27me3 (28, 29), of the mouse CCR7 promoter by ChIP analysis as previously described (7). In RAW-LXRα WT cells, compared to basal and T treatment conditions, CCR7 regulatory regions showed a reduction in the levels of H3K9me3 and H3K27me3 upon cotreatment with T-plus-9cRA (Fig. 4A, LXRα-WT). The levels of H3K9me3 and H3K27me3 at the CCR7 promoter were substantially lower in RAW-LXRα S198A cells, which express CCR7 at high levels, than in RAW-LXRα WT cells (Fig. 4A, LXRα-S198A).
FIG 4.
Unphosphorylated LXRα decreases the repressive chromatin state and increases LXRα occupancy at the CCR7 promoter. H3K9me3 and H3K27me3 binding was assessed by ChIP. RAW-LXRα WT and RAW-LXRα S198A cells were incubated with vehicle and 5 μM T (T) alone or in combination with 1 μM 9cRA for 2 h, and chromatin was prepared and processed as described in Materials and Methods. ND, not determined. (A) Modified histones were immunoprecipitated using antibodies against H3K9me3 and H3K27me3. Precipitated DNA was amplified by qPCR using primers located between residues −120 and −15 of the CCR7 promoter and normalized to input chromatin levels; data are reported as percent input. (B) LXRα occupancy at CCR7 as assessed by ChIP. RAW-LXRα WT and RAW-LXRα S198A cells were treated as indicated; chromatin was prepared and precipitated using an LXRα-specific monoclonal antibody. DNA was amplified by qPCR using primers located between residues −120 and −15. Samples were measured in triplicate, and the results were averaged. Error bars represent SD.
We also examined the recruitment of LXRα to the CCR7 promoter and found a higher level of LXRα occupancy upon stimulation with T plus 9cRA in RAW-LXRα WT cells than in control and T-treated cells (Fig. 4B, LXRα-WT). LXRα displayed greater occupancy at the CCR7 promoter in the S198A cells than in the cells expressing the wild-type LXRα under basal (DMSO) conditions, while LXRα binding was reduced upon treatment with T plus 9cRA (Fig. 4B, LXRα-S198A). This mirrors the expression pattern of Ccr7 in the S198A line, where Ccr7 expression was high under basal conditions and was reduced upon T-plus-9cRA treatment. Thus, it appears that both a decrease in repressive histone modifications and increased occupancy of the unphosphorylated LXRα contribute to Ccr7 expression.
LXRα S198 phosphorylation affects changes in global gene expression in RAW cells.
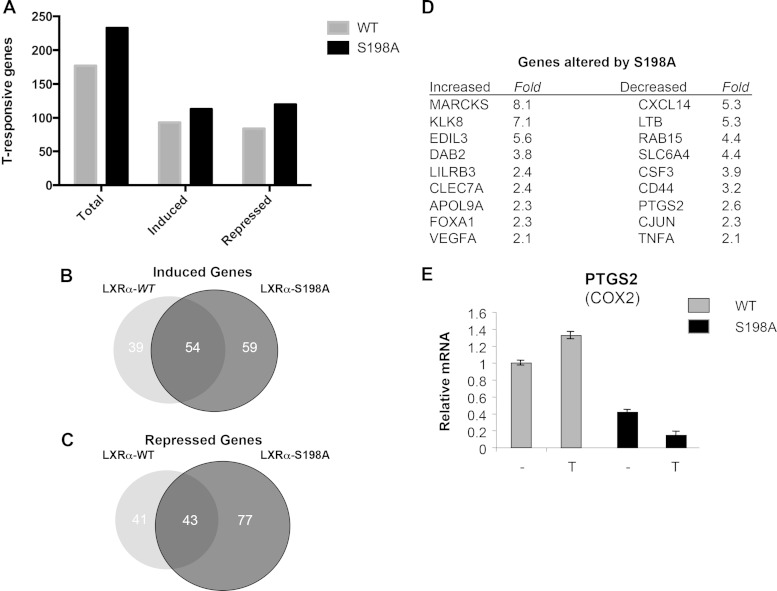
To assess global gene expression changes related to LXRα phosphorylation, microarray analysis was performed on RNA extracted from RAW-LXRα WT and RAW-LXRα S198A cells upon T treatment. At a differential gene expression threshold of 1.5-fold, a total of 177 genes were induced (93 genes) or repressed (84 genes) by T treatment in the RAW-LXRα WT cells, whereas a total of 233 genes were induced (113 genes) or repressed (120 genes) by T treatment in the cells expressing LXRα S198A (Fig. 5A). Of the 113 genes that were induced in the S198A cells, 59 were activated by T treatment only when the S198 site was mutated to alanine (Fig. 5B). Similarly, of the 120 repressed genes, 77 were suppressed by T treatment only in the S198A-expressing cells (Fig. 5C). There were also genes induced and repressed in the RAW-LXRα WT cells that were neither enhanced nor suppressed in the S198A cells (Fig. 5B and C). This suggests that phosphorylation affected the LXRα ligand-dependent gene expression.
FIG 5.
The effects of LXRα S198 phosphorylation on gene expression. (A) Numbers of genes induced and repressed upon T treatment from RAW264.7 cells expressing LXRα WT versus LXRα S198A (<1.5-fold change). (B and C) Venn diagrams of the induced (B) or repressed (C) genes from RAW-LXRα WT and RAW-LXRα S198A cells. (D) Fold change in the expression of selected genes upon T treatment in the S198A-expressing cells compared to the WT-expressing cells from the array data. (E) LXRα-mediated repression of PTGS2 (Cox2) by nonphosphorylated LXR S198. RAW-LXRα S198A cells were incubated with DMSO (−) or T (1 μM) for 24 h, and levels of Ptgs2 (Cox2) mRNA were analyzed by qPCR. Values indicate expression normalized to cyclophilin A levels, and the RNA levels are presented relative to the expression in vehicle-treated (−) RAW-LXRα WT cells, which was set to 1.
We performed gene ontology (GO) analysis of the genes preferentially regulated by T treatment in the S198A cells. We focused our analysis on the 59 genes upregulated and the 77 genes downregulated selectively in S198A versus WT cells upon T treatment, as these represented the broadest LXRα S198 phosphorylation states (completely nonphosphorylated LXRα in the RAW LXRα-S198A cells compared to the fully S198-phosphorylated LXRα in the T-treated RAW LXRα WT cells) in order to uncover key processes associated with LXRα S198 phosphorylation. This analysis revealed that the GO classes of T-dependent S198A-upregulated genes were associated with membrane organization, endocytosis, cell adhesion, phagocytosis, and vesicle transport, whereas the GO classes of the downregulated genes include genes linked to the inflammatory response, cell proliferation, and cytokine production (Table 1).
TABLE 1.
GO analysis of genes upregulated or downregulated upon T treatment in RAW-LXRα S198A-versus RAW-LXRα WT-expressing cellsa
| Biological process | No. of genes | Fold enrichment | P value |
|---|---|---|---|
| Upregulated in S198A cells | |||
| Membrane organization | 7 | 9 | 8.40 × 10−05 |
| Endocytosis | 6 | 11 | 1.46 × 10−04 |
| Immune response | 7 | 5 | 1.57 × 10−03 |
| Cell adhesion | 7 | 4 | 3.76 × 10−03 |
| Phagocytosis | 3 | 22 | 7.83 × 10−03 |
| Vesicle-mediated transport | 6 | 5 | 8.18 × 10−03 |
| Downregulated in S198A cells | |||
| Regulation of cell proliferation | 7 | 5 | 2.28 × 10−03 |
| Inflammatory response | 5 | 8 | 2.56 × 10−03 |
| Cell activation | 5 | 8 | 3.53 × 10−03 |
| Cytokine production | 3 | 26 | 5.45 × 10−03 |
Fold enrichment of actual over expected probe sets is shown along with P values.
Because the basal level of Ccr7 is high in S198A-expressing cells and because T treatment does not enhance its expression, Ccr7 was not included in the genes induced by T treatment in the S198A cells. Additionally, genes encoding products such as AIM and LPL are not included in the set of genes selectively induced upon T treatment in RAW-LXRα S198A cells, since these genes are also induced in RAW-LXRα WT cells (20).
Examples of specific gene products induced by T treatment in RAW-LXRα S198A but not RAW-LXRα WT cells (Fig. 5D) include MARCKS, a factor that promotes macrophage motility (30); KLK8, a serine protease involved in collagen IV degradation (31); EDIL3, an integrin ligand and leukocyte adhesion inhibitor (32); DAB2, a protein involved in endocytosis (33); and LILRB3, a suppressor of M1 macrophage activation (34).
Genes repressed in RAW-LXRα S198A compared to RAW-LXRα WT cells (Fig. 5E) include those encoding products such as the CXCL14 chemokine, a chemoattractant for monocytes (35); lymphotoxin beta (LTB; tumor necrosis factor [TNF] superfamily, member 3), an inducer of the inflammatory response in atherosclerosis (36); RAB15, a GTPase involved in endocytosis (37); SLC6A4, a serotonin transporter expressed in neurons and macrophages (38); and the proinflammatory factors PTGS2 (COX2) and TNF-α. The repression of PTGS2 mRNA in the RAW-LXRα S198A cells compared to the RAW-LXRα WT cells was verified by qPCR and shows that, consistent with the gene array studies, repression of the level of Ptgs2 mRNA by LXR signaling is a function of the nonphosphorylated receptor (Fig. 5E). Therefore, ligand-treated S198A cells enhance the expression of genes involved in cell motility and anti-inflammatory responses while also reducing the expression of proinflammatory genes, suggesting a role for LXRα phosphorylation in macrophage inflammatory responses.
Molecular modeling of LXRα S198.
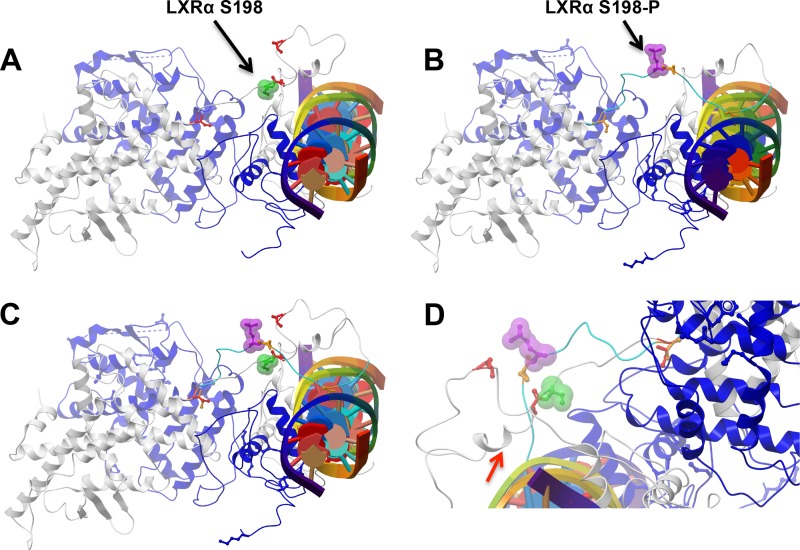
We next determined the location of S198 on the structure of LXRα. Recently, the crystal structure of LXRβ in complex with RXRα on the canonical DR4 DNA element has been solved (23). From this, we built a homology model for LXRα, which included conformational sampling of the hinge region between the DNA and the ligand binding domains in the context of bound RXRα and DNA (Fig. 6). S198 does not contact DNA, the RXRα DNA binding domain, or either the LXRα or RXRα ligand binding domains in our model (Fig. 6A). Moreover, phosphorylation of S198 did not introduce any contacts with the other domains (Fig. 6B). Interestingly, phosphorylation of S198 alters the orientation of the residue such that unphosphorylated S198 is buried whereas phosphorylated S198 is exposed on the surface of the complex, which is consistent with a site for protein interaction (Fig. 6C and D). In addition, a segment containing unphosphorylated S198 adopts a short alpha helical stretch which is not observed in the phosphorylated S198 simulations, suggesting an alternative surface for protein interaction in the S198 nonphosphorylated state (Fig. 6D, arrow). Hence, phosphorylated S198 and nonphosphorylated S198 are predicted to impart distinct structural characteristics to the LXRα hinge domain which we hypothesize could influence cofactor recruitment, ultimately modulating LXRα activity in a gene-specific manner.
FIG 6.
Molecular modeling of the DNA-bound LXRα-RXRα heterodimer reveals alterations in the orientation of the S198 residue upon phosphorylation. Simulated conformations of the hLXRα-hRXRα heterodimer bound to DNA (multicolored cylinder with central plates) in the context of nonphosphorylated S198 (green, arrow) (A) and phosphorylated S198 (purple, arrow) (B) are shown. RXRα is shown in blue, and LXRα is depicted in gray. Three serines (red) located at residues 191, 197, and 207 in the hinge do not contact any elements of the hRXRα-hLXRα heterodimer. (C) Superimposition of the images in panels A and B, showing the difference in the orientations of S198 in its unphosphorylated (green) and phosphorylated (purple) conformations. (D) Closeup view showing the conformations of unphosphorylated (green) and phosphorylated (purple) S198. Note the helical tendency of residues 191 to 195 in the LXRα S198 nonphosphorylated state (arrow).
DISCUSSION
We have examined the regulation of CCR7 in macrophages and found that phosphorylation of LXRα at S198 is an important determinant. The regulation of expression of CCR7 appears multifaceted and distinct from that of canonical LXR targets such as ABCA1. For example, reduction of LXRα S198 phosphorylation is associated with enhanced Ccr7 expression (this study) but has little effect on Abca1 expression (20). In addition, analysis of the ENCODE database (39) reveals that, in cells where CCR7 is expressed (such as the GM12878 lymphoblastoid cell line), an open chromatin configuration is observed, and a host of transcription factors (including PU.1, an Ets family member transcription factor that determines LXR occupancy and gene expression in macrophages [40, 41]) and RNA polymerase II occupy the promoter, whereas in cells that do not express CCR7 (such as hepatoma HEPG2 cells), a closed chromatin configuration is detected, and a lack of transcription factors and RNA polymerase II at the promoter is observed. This is in contrast to ABCA1, which has an open chromatin configuration independent of its expression (not shown). Such a preaccessible chromatin pattern at ABCA1 is also consistent with the observation that, in NIH 3T3 fibroblasts and 3T3-F422A preadipocytes with ectopic LXRα expression (13), Abca1 expression is induced upon LXR ligand treatment. However, expression of APOE, another LXR target gene, is induced by LXR ligands only in preadipocytes and not fibroblasts, suggesting cell type-dependent regulation of certain LXR target genes. Thus, our findings suggest that for LXR to induce the expression of Ccr7 it would have to overcome a repressive chromatin environment. We speculate that such a change in chromatin in the CCR7 locus occurs in vivo during atherosclerosis regression, where the phenotypic state of macrophages changes from M1 to M2 upon normalization of hyperlipidemia (5, 29, 42). Consistent with this idea, treatment of RAW-LXRα WT cells with the M2 polarizing cytokine interleukin-4 (IL-4) resulted in the T- and PU.1-dependent induction of Ccr7 mRNA expression (B. Pourcet and I. Pineda-Torra, unpublished data). Additional studies, including the generation of an LXRα S198A knock-in mouse, will be required to determine the impact of LXRα phosphorylation on regulation of CCR7 in macrophages upon atherosclerosis progression and regression in vivo.
Global gene expression analysis in regressing versus progressing plaque macrophages identified arginase 1 (ARG1) as one of the most highly upregulated genes in macrophages from regressing plaques (42). Interestingly, ARG1 is also upregulated by LXR signaling in RAW-LXRα WT cells. The induction of ARG1 upon LXR signaling is indirect and involves the activation of interferon regulatory factor 8 (IRF8) (41). However, this regulatory mechanism does not underlie the LXR-dependent induction of CCR7 since depletion of IRF8 had no effect on Ccr7 expression (Pourcet and Pineda-Torra, unpublished). ARG1 is a marker of M2 (anti-inflammatory or tissue repair) macrophages, and other accepted M2 markers, such as mannose receptor, are also upregulated in both RAW-LXRα S198A (not shown) and regressing plaque macrophages (5, 29). This might represent an association of nonphosphorylated LXRα S198 with a more reparative M2 macrophage phenotype. Although CCR7 is considered an M1 marker, our findings suggest a complexity in the regulation of gene expression by LXR S198 phosphorylation that is not specific to either an M1 or M2 lineage. Such a phenotypic spectrum is also reflected in regressing plaque macrophages that express M2 (ARG1) and M1 (CCR7) determinants (3, 4, 42, 43).
Insight into how phosphorylation of LXRα at S198 might impart such gene selectivity comes from the recently determined structure of the LXRβ-RXRα heterodimer on a DR4 DNA element (23). Judging of the basis of our LXRα-RXRα model, functional effects of S198 in the hinge region are unlikely to occur through contacting either the DNA binding or ligand binding domains. This is in contrast to the peroxisome proliferator-activated receptor γ (PPARγ)/RXRα structure on a DR1 element, which shows the hinge region contacting the minor groove of the DNA (44). Whereas the phosphorylated S198 is surface exposed, the nonphosphorylated S198 assumes the opposite orientation. Importantly, the nonphosphorylated form promotes a short α-helix in the hinge region that is not observed in the phosphorylated S198 simulation, suggesting a unique surface for protein interaction in the S198 nonphosphorylated state (Fig. 6D). Interestingly, mutations in LXRα have been identified at S198 and P199 in ovarian and intestinal cancers, respectively, suggesting roles for these residues in LXRα function (45, 46).
CCR7 does not contain a classical DR4. Indeed, recent genome-wide studies identifying LXR binding sites in mouse liver and human macrophage THP1 cells revealed that less than 10% of LXR binding site sequences contain a DR4 element, implying indirect or non-DR4 element binding of LXRα to its target genes (47, 48). Interrogation of chromatin immunoprecipitation sequencing (ChIP-seq) data from RAW-LXRα WT cells did not reveal an association of LXRα within 50 kb of the CCR7 locus (M. A. Hussein, D. Savic, M. J. Garabedian, and R. M. Myers, unpublished data). This might reflect weak or transient binding of LXRα to the CCR7 promoter and the inability of ChIP-seq to reveal low-occupancy sites because of limitations in the number of sequence reads (49). Consistent with this interpretation, we find that LXRα occupancy can be detected at the CCR7 promoter by directed ChIP-qPCR, an approach not constrained by read depth (7) (Fig. 4B). Given the complexity of CCR7 regulation, it would seem that additional functional analyses of the CCR7 regulatory regions are warranted to reveal the LXR-responsive region(s) in the context of the gene's endogenous chromatin environment using genome-editing approaches (50).
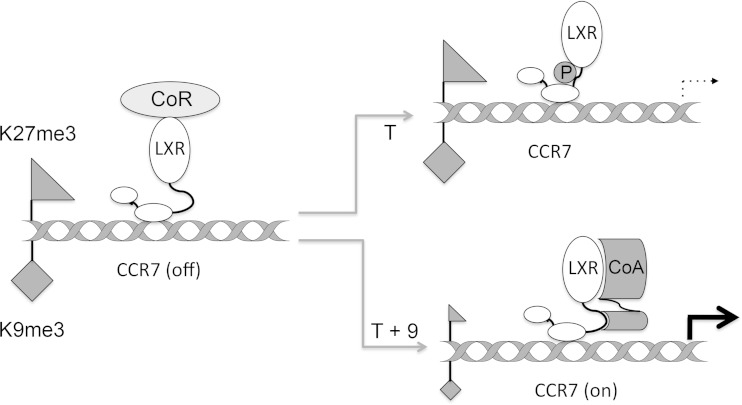
We propose a model for LXRα S198 phosphorylation-dependent gene regulation using CCR7 as an example (Fig. 7). We speculate that, in the absence of ligand, LXRα is associated with the promoter in a repressed state. We further envision that T (or GW9365)-dependent LXRα S198 phosphorylation results in a conformation that is not compatible with coactivator binding to relieve repression. Thus, T treatment alone would not elicit a robust transcriptional response. In contrast, upon T-plus-9cRA treatment, by promoting the S198 nonphosphorylated state and an alternative conformation, LXRα is competent for coactivator assembly, thereby relieving repression and inducing transcription. The requirement for 9cRA-liganded RXR would be to recruit a phosphatase (or dismiss a kinase) to reduce LXRα S198 phosphorylation and promote a conformation compatible with coactivator binding.
FIG 7.
Model for LXRα-S198 phosphorylation-dependent regulation. A schematic representation of LXR regulation of CCR7 is shown. In the absence of ligand, nonphosphorylated LXR is bound to CCR7 in a repressive chromatin state (high H3K9me3 [diamond] and high H3K27me3 [triangle]). Upon T treatment, S198 becomes hyperphosphorylated; this conformation is not compatible with the coactivator recruitment that would reverse the repressive chromatin condition. Upon T-plus-9cRA treatment, S198 is nonphosphorylated, and this promotes the assembly of an as-yet-unidentified coregulator complex (CoR) that relieves the repressive chromatin environment and promotes gene expression. The model excludes RXR for simplicity, although we propose that the requirement of 9cRA and RXR would be to recruit a phosphatase to reduce LXRα S198 phosphorylation.
Recent studies also indicate that tumors secrete a sulfotransferase 2B1b (SULT2B1)-sensitive product that inhibits Ccr7 expression in maturing dendritic cells to dampen the antitumor response (51). Given that SULT2B1 inactivates endogenous LXR oxysterol ligands (52) and that deletion of LXRα in bone marrow-derived cells restores CCR7 to control tumor growth (51), this suggests that Ccr7 expression is suppressed by LXRα signaling in tumor-infiltrating dendritic cells. Whether alterations in LXRα S198 phosphorylation contribute to Ccr7 expression in tumor-associated dendritic cells remains unknown. However, this observation is consistent with our finding that basal expression of Ccr7 is high when LXRα is in the S198 nonphosphorylated state and that, under these conditions, certain LXR ligands suppress Ccr7 expression.
Alterations in gene expression between the phosphorylated LXRα WT and LXRα-S198A cells are evident in the RAW cell model and suggest that the changes elicited by the nonphosphorylated LXRα promote a more motile and less inflammatory macrophage state, features that are consistent with an antiatherogenic phenotype. Thus, it would be interesting to identify LXR ligands that selectively promote the nonphosphorylated form of LXRα and test their effects as potential anti-inflammatory and antiatherogenic compounds (53). In fact, PPARγ ligands that block phosphorylation of the hinge region have been shown to selectively affect gene expression, resulting in antidiabetes activity without the side effects that promote fat accumulation (54, 55). This suggests that novel pharmacology can be achieved by targeting nuclear receptor phosphorylation.
ACKNOWLEDGMENTS
This work was supported by NIH grants HL084312 and P01 HL098055 (E.A.F.), HL117226 (M.J.G. and E.A.F.), and 4DP2OD004631 (T.C.), NIH training grants T32GM007238 (E.S.) and T32AI07180 (M.H.), and Medical Research Council New Investigator grant G0801278 (I.P.-T.).
We thank the members of the Garabedian, S. K. Logan, and Fisher laboratories for critically evaluating the manuscript.
REFERENCES
- 1.Calkin AC, Tontonoz P. 2012. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol 13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Im SS, Osborne TF. 2011. Liver x receptors in atherosclerosis and inflammation. Circ Res 108:996–1001. doi: 10.1161/CIRCRESAHA.110.226878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore KJ, Sheedy FJ, Fisher EA. 2013. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol 13:709–721. doi: 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trogan E, Feig JE, Dogan S, Rothblat GH, Angeli V, Tacke F, Randolph GJ, Fisher EA. 2006. Gene expression changes in foam cells and the role of chemokine receptor CCR7 during atherosclerosis regression in ApoE-deficient mice. Proc Natl Acad Sci U S A 103:3781–3786. doi: 10.1073/pnas.0511043103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feig JE, Rong JX, Shamir R, Sanson M, Vengrenyuk Y, Liu J, Rayner K, Moore K, Garabedian M, Fisher EA. 2011. HDL promotes rapid atherosclerosis regression in mice and alters inflammatory properties of plaque monocyte-derived cells. Proc Natl Acad Sci U S A 108:7166–7171. doi: 10.1073/pnas.1016086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randolph GJ. 2014. Mechanisms that regulate macrophage burden in atherosclerosis. Circ Res 114:1757–1771. doi: 10.1161/CIRCRESAHA.114.301174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feig JE, Pineda-Torra I, Sanson M, Bradley MN, Vengrenyuk Y, Bogunovic D, Gautier EL, Rubinstein D, Hong C, Liu J, Wu C, van Rooijen N, Bhardwaj N, Garabedian M, Tontonoz P, Fisher EA. 2010. LXR promotes the maximal egress of monocyte-derived cells from mouse aortic plaques during atherosclerosis regression. J Clin Invest 120:4415–4424. doi: 10.1172/JCI38911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, Mangelsdorf DJ. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell 93:693–704. doi: 10.1016/S0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 9.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. 1996. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature 383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 10.Yang C, McDonald JG, Patel A, Zhang Y, Umetani M, Xu F, Westover EJ, Covey DF, Mangelsdorf DJ, Cohen JC, Hobbs HH. 2006. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J Biol Chem 281:27816–27826. doi: 10.1074/jbc.M603781200. [DOI] [PubMed] [Google Scholar]
- 11.Wagner BL, Valledor AF, Shao G, Daige CL, Bischoff ED, Petrowski M, Jepsen K, Baek SH, Heyman RA, Rosenfeld MG, Schulman IG, Glass CK. 2003. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol Cell Biol 23:5780–5789. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calkin AC, Tontonoz P. 2010. Liver x receptor signaling pathways and atherosclerosis. Arterioscler Thromb Vasc Biol 30:1513–1518. doi: 10.1161/ATVBAHA.109.191197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkateswaran A, Laffitte BA, Joseph SB, Mak PA, Wilpitz DC, Edwards PA, Tontonoz P. 2000. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc Natl Acad Sci U S A 97:12097–12102. doi: 10.1073/pnas.200367697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laffitte BA, Repa JJ, Joseph SB, Wilpitz DC, Kast HR, Mangelsdorf DJ, Tontonoz P. 2001. LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc Natl Acad Sci U S A 98:507–512. doi: 10.1073/pnas.98.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin N, Bischoff ED, Daige CL, Thomas D, Vu CT, Heyman RA, Tangirala RK, Schulman IG. 2005. Macrophage liver X receptor is required for antiatherogenic activity of LXR agonists. Arterioscler Thromb Vasc Biol 25:135–142. doi: 10.1161/01.ATV.0000150044.84012.68. [DOI] [PubMed] [Google Scholar]
- 16.Verschuren L, de Vries-van der Weij J, Zadelaar S, Kleemann R, Kooistra T. 2009. LXR agonist suppresses atherosclerotic lesion growth and promotes lesion regression in apoE*3Leiden mice: time course and mechanisms. J Lipid Res 50:301–311. doi: 10.1194/jlr.M800374-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Bischoff ED, Daige CL, Petrowski M, Dedman H, Pattison J, Juliano J, Li AC, Schulman IG. 2010. Non-redundant roles for LXRalpha and LXRbeta in atherosclerosis susceptibility in low density receptor knockout mice. J Lipid Res 51:900–906. doi: 10.1194/jlr.M900096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen M, Bradley MN, Beaven SW, Tontonoz P. 2006. Phosphorylation of the liver X receptors. FEBS Lett 580:4835–4841. doi: 10.1016/j.febslet.2006.07.074. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto T, Shimano H, Inoue N, Nakagawa Y, Matsuzaka T, Takahashi A, Yahagi N, Sone H, Suzuki H, Toyoshima H, Yamada N. 2007. Protein kinase A suppresses sterol regulatory element-binding protein-1C expression via phosphorylation of liver X receptor in the liver. J Biol Chem 282:11687–11695. doi: 10.1074/jbc.M611911200. [DOI] [PubMed] [Google Scholar]
- 20.Torra IP, Ismaili N, Feig JE, Xu CF, Cavasotto C, Pancratov R, Rogatsky I, Neubert TA, Fisher EA, Garabedian MJ. 2008. Phosphorylation of liver X receptor alpha selectively regulates target gene expression in macrophages. Mol Cell Biol 28:2626–2636. doi: 10.1128/MCB.01575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alberti S, Schuster G, Parini P, Feltkamp D, Diczfalusy U, Rudling M, Angelin B, Bjorkhem I, Pettersson S, Gustafsson JA. 2001. Hepatic cholesterol metabolism and resistance to dietary cholesterol in LXRbeta-deficient mice. J Clin Invest 107:565–573. doi: 10.1172/JCI9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reily MM, Pantoja C, Hu X, Chinenov Y, Rogatsky I. 2006. The GRIP1:IRF3 interaction as a target for glucocorticoid receptor-mediated immunosuppression. EMBO J 25:108–117. doi: 10.1038/sj.emboj.7600919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lou X, Toresson G, Benod C, Suh JH, Philips KJ, Webb P, Gustafsson JA. 2014. Structure of the retinoid X receptor alpha-liver X receptor beta (RXRalpha-LXRbeta) heterodimer on DNA. Nat Struct Mol Biol 21:277–281. doi: 10.1038/nsmb.2778. [DOI] [PubMed] [Google Scholar]
- 24.Abagyan R, Batalov S, Cardozo T, Totrov M, Webber J, Zhou Y. 1997. Homology modeling with internal coordinate mechanics: deformation zone mapping and improvements of models via conformational search. Proteins (Suppl 1) 1997:29–37. [DOI] [PubMed] [Google Scholar]
- 25.Cardozo T, Totrov M, Abagyan R. 1995. Homology modeling by the ICM method. Proteins 23:403–414. doi: 10.1002/prot.340230314. [DOI] [PubMed] [Google Scholar]
- 26.Arnautova YA, Abagyan RA, Totrov M. 2011. Development of a new physics-based internal coordinate mechanics force field and its application to protein loop modeling. Proteins 79:477–498. doi: 10.1002/prot.22896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abagyan R, Totrov M. 1994. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol 235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 28.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feig JE, Parathath S, Rong JX, Mick SL, Vengrenyuk Y, Grauer L, Young SG, Fisher EA. 2011. Reversal of hyperlipidemia with a genetic switch favorably affects the content and inflammatory state of macrophages in atherosclerotic plaques. Circulation 123:989–998. doi: 10.1161/CIRCULATIONAHA.110.984146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green TD, Park J, Yin Q, Fang S, Crews AL, Jones SL, Adler KB. 2012. Directed migration of mouse macrophages in vitro involves myristoylated alanine-rich C-kinase substrate (MARCKS) protein. J Leukoc Biol 92:633–639. doi: 10.1189/jlb.1211604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajapakse S, Ogiwara K, Takano N, Moriyama A, Takahashi T. 2005. Biochemical characterization of human kallikrein 8 and its possible involvement in the degradation of extracellular matrix proteins. FEBS Lett 579:6879–6884. doi: 10.1016/j.febslet.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 32.Choi EY, Chavakis E, Czabanka MA, Langer HF, Fraemohs L, Economopoulou M, Kundu RK, Orlandi A, Zheng YY, Prieto DA, Ballantyne CM, Constant SL, Aird WC, Papayannopoulou T, Gahmberg CG, Udey MC, Vajkoczy P, Quertermous T, Dimmeler S, Weber C, Chavakis T. 2008. Del-1, an endogenous leukocyte-endothelial adhesion inhibitor, limits inflammatory cell recruitment. Science 322:1101–1104. doi: 10.1126/science.1165218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris SM, Arden SD, Roberts RC, Kendrick-Jones J, Cooper JA, Luzio JP, Buss F. 2002. Myosin VI binds to and localises with Dab2, potentially linking receptor-mediated endocytosis and the actin cytoskeleton. Traffic 3:331–341. doi: 10.1034/j.1600-0854.2002.30503.x. [DOI] [PubMed] [Google Scholar]
- 34.Ma G, Pan PY, Eisenstein S, Divino CM, Lowell CA, Takai T, Chen SH. 2011. Paired immunoglobin-like receptor-B regulates the suppressive function and fate of myeloid-derived suppressor cells. Immunity 34:385–395. doi: 10.1016/j.immuni.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hara T, Tanegashima K. 2012. Pleiotropic functions of the CXC-type chemokine CXCL14 in mammals. J Biochem 151:469–476. doi: 10.1093/jb/mvs030. [DOI] [PubMed] [Google Scholar]
- 36.Grandoch M, Feldmann K, Gothert JR, Dick LS, Homann S, Bayer JK, Klatt C, Waldheim JN, Rabausch B, Nagy N, Oberhuber A, Deenen R, Kohrer K, Lehr S, Homey B, Pfeffer K, Fischer JW. 4 March 2015, posting date Deficiency in lymphotoxin beta receptor protects from atherosclerosis in apoE-deficient mice. Circ Res doi: 10.1161/CIRCRESAHA.116.305723. [DOI] [PubMed] [Google Scholar]
- 37.Zuk PA, Elferink LA. 2000. Rab15 differentially regulates early endocytic trafficking. J Biol Chem 275:26754–26764. [DOI] [PubMed] [Google Scholar]
- 38.Rudd ML, Nicolas AN, Brown BL, Fischer-Stenger K, Stewart JK. 2005. Peritoneal macrophages express the serotonin transporter. J Neuroimmunol 159:113–118. doi: 10.1016/j.jneuroim.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 39.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O'Geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pourcet B, Feig JE, Vengrenyuk Y, Hobbs AJ, Kepka-Lenhart D, Garabedian MJ, Morris SM Jr, Fisher EA, Pineda-Torra I. 2011. LXRalpha regulates macrophage arginase 1 through PU.1 and interferon regulatory factor 8. Circ Res 109:492–501. doi: 10.1161/CIRCRESAHA.111.241810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feig JE, Vengrenyuk Y, Reiser V, Wu C, Statnikov A, Aliferis CF, Garabedian MJ, Fisher EA, Puig O. 2012. Regression of atherosclerosis is characterized by broad changes in the plaque macrophage transcriptome. PLoS One 7:e39790. doi: 10.1371/journal.pone.0039790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams HJ, Fisher EA, Greaves DR. 2012. Macrophage differentiation and function in atherosclerosis: opportunities for therapeutic intervention? J Innate Immun 4:498–508. doi: 10.1159/000336618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. 2008. Structure of the intact PPAR-gamma-RXR-alpha nuclear receptor complex on DNA. Nature 456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pehkonen P, Welter-Stahl L, Diwo J, Ryynanen J, Wienecke-Baldacchino A, Heikkinen S, Treuter E, Steffensen KR, Carlberg C. 2012. Genome-wide landscape of liver X receptor chromatin binding and gene regulation in human macrophages. BMC Genomics 13:50. doi: 10.1186/1471-2164-13-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boergesen M, Pedersen TA, Gross B, van Heeringen SJ, Hagenbeek D, Bindesboll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, Gustafsson JA, Stunnenberg HG, Staels B, Mandrup S. 2012. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol 32:852–867. doi: 10.1128/MCB.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, Chen Y, DeSalvo G, Epstein C, Fisher-Aylor KI, Euskirchen G, Gerstein M, Gertz J, Hartemink AJ, Hoffman MM, Iyer VR, Jung YL, Karmakar S, Kellis M, Kharchenko PV, Li Q, Liu T, Liu XS, Ma L, Milosavljevic A, Myers RM, Park PJ, Pazin MJ, Perry MD, Raha D, Reddy TE, Rozowsky J, Shoresh N, Sidow A, Slattery M, Stamatoyannopoulos JA, Tolstorukov MY, White KP, Xi S, Farnham PJ, Lieb JD, Wold BJ, Snyder M. 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mali P, Esvelt KM, Church GM. 2013. Cas9 as a versatile tool for engineering biology. Nat Methods 10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villablanca EJ, Raccosta L, Zhou D, Fontana R, Maggioni D, Negro A, Sanvito F, Ponzoni M, Valentinis B, Bregni M, Prinetti A, Steffensen KR, Sonnino S, Gustafsson JA, Doglioni C, Bordignon C, Traversari C, Russo V. 2010. Tumor-mediated liver X receptor-alpha activation inhibits CC chemokine receptor-7 expression on dendritic cells and dampens antitumor responses. Nat Med 16:98–105. doi: 10.1038/nm.2074. [DOI] [PubMed] [Google Scholar]
- 52.Chen W, Chen G, Head DL, Mangelsdorf DJ, Russell DW. 2007. Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice. Cell Metab 5:73–79. doi: 10.1016/j.cmet.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong C, Tontonoz P. 2014. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat Rev Drug Discov 13:433–444. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- 54.Choi JH, Banks AS, Kamenecka TM, Busby SA, Chalmers MJ, Kumar N, Kuruvilla DS, Shin Y, He Y, Bruning JB, Marciano DP, Cameron MD, Laznik D, Jurczak MJ, Schurer SC, Vidovic D, Shulman GI, Spiegelman BM, Griffin PR. 2011. Antidiabetic actions of a non-agonist PPARgamma ligand blocking Cdk5-mediated phosphorylation. Nature 477:477–481. doi: 10.1038/nature10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amato AA, Rajagopalan S, Lin JZ, Carvalho BM, Figueira AC, Lu J, Ayers SD, Mottin M, Silveira RL, Souza PC, Mourao RH, Saad MJ, Togashi M, Simeoni LA, Abdalla DS, Skaf MS, Polikparpov I, Lima MC, Galdino SL, Brennan RG, Baxter JD, Pitta IR, Webb P, Phillips KJ, Neves FA. 2012. GQ-16, a novel peroxisome proliferator-activated receptor gamma (PPARgamma) ligand, promotes insulin sensitization without weight gain. J Biol Chem 287:28169–28179. doi: 10.1074/jbc.M111.332106. [DOI] [PMC free article] [PubMed] [Google Scholar]