Abstract
Bone metastasis is the hallmark of progressive and castration-resistant prostate cancers. MicroRNA 1 (miR-1) levels are decreased in clinical samples of primary prostate cancer and further reduced in metastases. SRC has been implicated as a critical factor in bone metastasis, and here we show that SRC is a direct target of miR-1. In prostate cancer patient samples, miR-1 levels are inversely correlated with SRC expression and a SRC-dependent gene signature. Ectopic miR-1 expression inhibited extracellular signal-regulated kinase (ERK) signaling and bone metastasis in a xenograft model. In contrast, SRC overexpression was sufficient to reconstitute bone metastasis and ERK signaling in cells expressing high levels of miR-1. Androgen receptor (AR) activity, defined by an AR output signature, is low in a portion of castration-resistant prostate cancer. We show that AR binds to the miR-1-2 regulatory region and regulates miR-1 transcription. Patients with low miR-1 levels displayed correlated low canonical AR gene signatures. Our data support the existence of an AR–miR-1–SRC regulatory network. We propose that loss of miR-1 is one mechanistic link between low canonical AR output and SRC-promoted metastatic phenotypes.
INTRODUCTION
Bone metastasis is one of the most clinically important features of prostate cancer (PC), and understanding the signaling networks that promote bone metastasis is central to improving outcomes for patients (1, 2). Metastatic PC is treated with androgen deprivation therapy, which initially reduces symptoms and tumor cell growth, although castration-resistant prostate cancer (CRPC) almost always recurs. Reconstitution of androgen receptor (AR)-mediated signaling is a central mechanism leading to CRPC (3, 4). While AR signaling is almost always observed in prostate cancer, substantial intertumoral heterogeneity exists in the level of AR output, measured by expression of AR target gene signatures, especially for CRPC (5–8). MicroRNAs (miRs) are pleiotropic regulators of gene expression. The altered expression of miRs in cancer leading to loss of tumor suppressor or gain of oncogenic activities has been well documented. Given the central role of AR in prostate cancer biology and treatment, the targets of androgen-regulated miRs are of interest (9).
After the development of castration resistance, a reduction in the level of an AR gene signature commonly expressed in prostate cancer suggests either a change in AR-dependent transcriptional specificity or decreased AR transcriptional activity (6, 8). It is expected that tumors with low rather than high canonical AR activity are dependent upon a distinct signaling milieu for survival and growth. Interestingly, an analysis of PC clinical samples for AR output in combination with various signaling pathways determined that decreasing predicted AR activity correlated with increasing predicted SRC activity (5). It has not been established whether an AR-dependent transcriptional mechanism influences SRC activity. A functional link between AR and SRC activities is potentially clinically significant for predictive purposes in selecting and monitoring responses to therapies (10).
SRC is a nonreceptor tyrosine kinase that is thought to integrate numerous signaling pathways, including AR (11, 12). SRC is activated downstream of various receptor families, notably receptor tyrosine kinases (13). Several lines of evidence support a role for SRC in prostate cancer, particularly CRPC. SRC is not a mutational target in prostate cancer, but rather, levels of expression and kinase activation are thought to determine functionality. Importantly, histopathological analyses of clinical samples have shown that markers of SRC family kinase activity are increased relative to primary PC in a proportion of CRPCs, with associated poor prognosis and reduced overall survival (14). In experimental models, inhibition of SRC signaling decreased proliferation, invasion, and migration of prostate cancer cells (15).
The mechanisms whereby SRC may contribute to CRPC survival and growth are of obvious interest. Experimental models have shown that SRC interacts with and phosphorylates AR, resulting in enhanced AR activity (11, 16). In addition, SRC is downstream of receptors, such as epidermal growth factor receptor (EGFR) and fibroblast growth factor receptor (FGFR), and upstream of signaling molecules, such as AKT and extracellular signal-regulated kinase (ERK), that have been implicated in PC survival in the absence of sufficient AR signaling (13, 17–19). Thus, SRC appears to be able to amplify low levels of AR activity and possibly replace AR-dependent survival signals.
We previously described the role of miR-1 in the progression of the aggressive Pten- and TP53-null prostate cancer mouse model, where we determined that loss of miR-1 contributes to mesenchymal commitment, invasiveness, and tumorigenesis (20). Investigations into the composition of miRs in clinical samples of prostate cancers have shown that miR-1 is significantly decreased in primary tumors compared to normal tissue and that low miR-1 expression is prognostic for progression (21–23).
To investigate the mechanistic role of miR-1 in prostate cancer progression, we have used a xenograft model of human prostate cancer bone metastasis (24, 25). We previously showed that RAS-dependent RAL effector pathway activation in DU145 cells leads to the acquisition of bone metastasis (24). Although RAS mutations are rare in prostate cancer (26, 27), RAS pathway activation correlates with prostate cancer progression, as demonstrated by analyses of patient samples for Ras transcriptomic signatures (28, 29). Consistent with this, downstream pathway biomarkers of the RAS pathway activation, phospho-ERK and phosphoinositol 3-kinase (PI3K)/phospho-AKT signaling, increase during progression (18). Also, in genetically engineered mouse models of prostate cancer, the addition of activated RAS signaling in the context of local disease gives rise to metastasis (28, 30).
We show here that miR-1 expression suppresses experimental bone metastasis and that miR-1 transcription is directly and positively modulated by AR. We determined that SRC is a direct target of miR-1 and, importantly, that ectopic SRC expression is sufficient to overcome miR-1 tumor suppression, implying a dominant biological function for SRC in the miR-1 target network. These data define one regulatory mechanism whereby low AR output tumors increase SRC expression as a result of decreased miR-1 levels, and provide a rationale for SRC activity as a biomarker in patients undergoing androgen deprivation therapy.
MATERIALS AND METHODS
Cell lines and plasmids and reagents.
The cell line DU145RasV12G37 (RasG37) (24) and a bone metastasis-derived subline, DU145/RasB1 (RasB1) (25), were derived and maintained as described previously. Cells expressing miR-1 or control miR were generated as described previously (20). The anti-miR inhibitor was from GeneCopoeia (MD), and its sequence is AUACAUACUUCUUUACAUUCCA. Red fluorescent protein (RFP) reporter vectors were constructed using the Clone-it enzyme-free lentivector kit (System Biosciences, CA). Human ETS1, KIF2A, SRC, EGFR, and SNAI2 full-length 3′ untranslated region (3′-UTR) reporters were constructed using the psiHECK-2 vector (Promega, WI). The positions of AR binding sites located upstream of human primary hsa-miR-1-2 are as follows: ARE1, 18:19417275, and ARE2, 18:19417513. SRC and AR were separately subcloned into the pFUGW lentiviral vector with a TRE3G promoter and with an IRES-mCherry reporter and a puromycin-selectable marker, respectively. To achieve controlled expression of target genes, DU145 or RasB1 cells stably expressing a Tet-On 3G transactivator (Tet-on TA) were established, and the Tet-on TA cells were further infected with TRE3G-driven constructs. Stable SRC-expressing cell lines were established by fluorescence-activated cell sorting (FACS) of mCherry-positive cells following doxycycline induction. All primers used for these constructs are listed in Table 1. All constructs were verified by DNA sequence analysis. Transient transfections were carried out using Lipofectamine RNAiMAX (Invitrogen, CA).
TABLE 1.
Primer sequences of 3′-UTR reporter and has-mir-1-2 promoter reporter constructs
| Construct and primer | Sequence |
|---|---|
| 3′-UTR reporter | |
| hETS1/psi-2 3′UTR(Xhol)F | CTCGAGGGAAACCCTGCTGAGACCTT |
| hETS1/psi-2 3′UTR(Pmel)R | GTTTAAACTCTCCAGCAAAATGATGTGC |
| hKif2a/psi-2 3′UTR(Xhol)F | CTCGAGACCGGCATTTGCTGCTAAAG |
| hKif2a/psi-2 3′UTR(Xhol)R | GTTTAAACCAGAATTTCATTCGTTTTTATTAT |
| hSRC/psi-2 3′UTR(Xhol)F | CTCGAGCTTCTCGGCTTGGATCCTG |
| hSRC/psi-2 3′UTR(Pmel)R | GTTTAAACATGTCGTGGCCAGAGTTGAC |
| hEGFR/psi-2 3′UTR(Xhol)F | CTCGAGCGGAGGATAGTATGAGCCCTAA |
| hEGFR/psi-2 3′UTR(Pmel)R | GTTTAAACACAATGCTGTAGGGGCTCTG |
| hSNAI2/psi-2 3′UTR(Xhol)F | CTCGAGGTGACGCATCAATGTTTACTCG |
| hSNAI2/psi-2 3′UTR(Pmel)R | GTTTAAACTTTTCTTGTTAACAAAGAATTC |
| hsa-miR-1-2 promoter reporter | |
| hsa-miR-1-2 P1 | TGATGTCTCTCCACCACCAA |
| hsa-miR-1-2 P2 | AGCAATGGTAAATCATTTGCAG |
| hsa-miR-1-2 P3 | GAGGCAGCAGAGACCGTTATGTCTCTCCACCACCAA |
| hsa-miR-1-2 P4 | CGAACAGAGAGAGACCGAGCAATGGTAAATCATTTGCAG |
Invasion assay and in vitro growth curves.
Invasion assays were performed as described previously (20). Cells were added to the upper chamber of Matrigel-coated Transwells (cell culture insert 8-micron pore; Corning). After 6 and 24 h incubation at 37°C, membranes were fixed and stained in Diff-Quick (Thermo Fisher Scientific, MA), and the invaded cells on the underside of the membrane were quantified. Five medium-power fields were counted for each replicate. The in vitro growth rate was measured as described previously using a starting density of 2 × 103 cells per well (25).
Promoter analysis and microRNA assay.
Promoter function was analyzed using FACS, and relative median fluorescent intensity (MFI) values were measured as described previously (31). MFI was determined from the first peak of fluorescence. Cells were treated with or without dihydrotestosterone (DHT) (10 nM; Sigma) and MDV3100 (10 μM; Selleck) for 48 h. The MFI value for RFP was measured by FACS using FACSDiva software and normalized to the value of the vehicle. For miR target reporter assays, cells were transfected with 1 μg of the appropriate 3′-UTR reporter and 1 μg precursor miR or 50 nM anti-miR. Cell extracts were prepared 48 h after transfection, and luciferase activities were measured using the dual-luciferase reporter assay system (Promega, WI). Three independent experiments were done with triplicate samples. MicroRNA binding sites were identified using the Computational Biology Center, Memorial Sloan Kettering Cancer Center, website (microRNA.org).
Western blot analysis and real-time RT-PCR.
For Western blot analysis, cells were lysed with radioimmunoprecipitation assay (RIPA) buffer containing complete protease inhibitors (Thermoscientific) and phosphatase inhibitors (Boston Bioproducts). Primary antibodies were incubated overnight at 4°C using the dilutions listed in Table 2. For real-time RT-PCR analysis, total RNA was isolated using a mirVana Paris RNA isolation system (Ambion). Reverse transcription of cDNA and PCR was performed as described previously (32). All reactions were normalized to human GAPDH and run in triplicate using primers listed in Table 3. MicroRNA reverse transcription and PCRs were performed using a TaqMan microRNA assay kit (Applied Biosystems). All values were normalized to the value for a human SNORD48 endogenous control, and all reactions were run in triplicate.
TABLE 2.
Antibodies used for Western blot analysisa
| Primary-antibody target | Source | Dilution |
|---|---|---|
| p-ERK1/2 | Cell Signaling (4376) | 1/2,000 |
| ERK2 | Cell Signaling (9102) | 1/1,000 |
| SRC | Cell Signaling (2109) | 1/2,000 |
| ZEB1 | Novus (NBP1-05987) | 1/500 |
| ETS1 | Epitomics (T3123-1) | 1/2,000 |
| GAPDH | Novus (NB300-221) | 1/4,000 |
| p-Erk2 | Cell Signaling (4370) | 1/1,000 |
| Erk2 | Santa Cruz (1647) | 1/1,000 |
| α-Tubulin | Sigma (T8203) | 1:2000 |
Horseradish peroxidase-linked anti-mouse IgG (NXA931; 1:2,000) or anti-rabbit (NA934; 1:2,000) IgG (GE Health Care) was used for protein detection.
TABLE 3.
qRT-PCR primer sequences
| Primer | Sequence |
|---|---|
| hGapdh F | GGACTCATGACCACAGTCCA |
| hGapdh R | CCAGTAGAGGCAGGGATGAT |
| hSRC F | CCAGGCTGAGGAGTGGTATT |
| hSRC R | TTCGTGGTCTCAGTTTCTCG |
| hAR F | TCTTGTCGTCTTCGGAAATG |
| hAR R | TCTGGGTTGTCTCCTCAGTG |
| hFkbp5 F | TAGAAATCCACCTGGAAGGC |
| hFkbp5 R | CCAGAGCTTTGTCAATTCCA |
Chromatin immunoprecipitation (ChIP).
Cells were treated with DHT for 2 h, and ChIP assays were performed using the EZ magna ChIP A kit (Millipore) with a modified protocol (32). ChIP antibodies and PCR primers are listed in Table 4. Predictions for transcription factor binding sites within promoter regions were adopted from the AliBaba 2.1 program (http://www.gene-regulation.com/pub/programs.html).
TABLE 4.
ChIP assay antibodies and primer sequences
| Antibody or primer | Source or sequence |
|---|---|
| Primary antibodies | |
| AR | Epitomics (3184-1) |
| FOXA1 | Abcam (ab23738) |
| OCT1 | Novus (NB100-91899) |
| GAPDH | Novus (NB300-221) |
| Rabbit IgG | Santa Cruz (sc-2027) |
| pRNApol2 | Abcam (ab5408) |
| Primers | |
| 1-2ARE1 F | GCTTACAGGTAACAGAGCACCA |
| 1-2ARE1 R | TTGAATTGTGAAATTAGGCTATGG |
| 1-2ARE2 F | TGTTTGCTCGTTTGTTTTTCA |
| 1-2ARE2 R | GCAGCCATCTCATCTTGATTG |
| non-ARE F | GCATCAAAGTGCAAACTTCAGG |
| non-ARE R | AAACCAGGATGAGTGTGGGT |
| FKBP5 ARE6/7 F | CCCCCCTATTTTAATCGGAGTAC |
| FKBP5 ARE6/7 R | TTTTGAAGAGCACAGAACACCCT |
All antibodies were used at a 1/50 dilution.
Animal studies.
All animal studies were performed in accordance with a protocol approved by the NIH Animal Care and Use Committee. To analyze tumorigenesis, 5-week-old male nude mice (NCI, Frederick, MD) were injected with 1 × 106 tumor cells in 50% Matrigel (Falcon, NJ) subcutaneously. Tumor size was measured weekly with calipers, and tumor volume was calculated using the following formula: tumor volume = (4/3π) × (L/2) × (W/2)2, where L is length and W is width. Results are means ± standard errors (SE) for each experimental group. To investigate metastasis, 1 × 105 cells were inoculated via an intracardiac route, and metastases were monitored by bioluminescent imaging (BLI) (25). Bone metastases were evaluated on magnified (3×) radiographs taken with a Faxitron MX-20 radiography system. Each bone metastasis was scored based on the following criteria: 0, no metastasis; 1, bone lesion covers less than 1/4 of the bone width; 2, bone lesion involves 1/4 to 1/2 of bone width; 3, bone lesion across 1/2 to 3/4 of bone width; 4, bone lesion is more than 3/4 of bone width. The bone metastasis score for each mouse represents the sum of scores of all bone lesions from four limbs. Doxycycline-containing chow (Bio-Serv) was used to induce SRC expression. Bones and subcutaneous tumors were processed for histological analyses as described previously (24). SRC immunohistochemistry was performed on tissue following antigen retrieval at pH 6.0 with polyclonal rabbit antibody (Cell Signaling) used at a 1:200 dilution and overnight incubation conditions. Mice were euthanized when one of the following situations applied: 10% loss of body weight, paralysis, or head tilting.
Clinical outcome and correlation analyses using human data sets.
We used mRNA expression data from a public human prostate cancer data set (29). The study was conducted under Memorial Sloan-Kettering Cancer Center (MSKCC) Institutional Review Board approval on 28 normal, 151 primary, and 19 metastatic samples. The expression data (and resulting Z scores) were log2 normalized. Additionally, microRNA expression was determined for 113 tumors and 28 matched normal samples with Agilent microRNA V2 arrays. Androgen-responsive (33) and SRC-dependent (34) gene signatures were used to determine correlations with miR-1 levels. Gene sets were scored by summing the expression Z score per tumor within the cohort. Tumors were mean stratified by miR-1 expression, and the mean Z scores were determined in each group.
Statistical analysis.
All data are presented as means and standard errors of the means (SEM). Statistical calculations were performed with GraphPad Prism analysis tools. Differences between individual groups were analyzed by one way or two-way analysis of variance (ANOVA). Bonferroni's posttest was used for comparisons among 3 or more groups. P values less than 0.05 were considered statistically significant.
RESULTS
To investigate the contribution of miR-1 and its potential targets to human prostate cancer metastasis, we analyzed miR-1 expression levels and correlative mRNAs in a publicly available data set of 28 normal prostate, 98 primary tumor, and 13 distant metastasis samples collected and analyzed at MSKCC. As shown in Fig. 1A and elsewhere (22), miR-1 expression levels relative to those in normal prostate were reduced in primary tumors and further dramatically decreased in metastases. In a smaller study of primary prostate cancer only, decreased miR-1 levels were correlated with non-organ-confined disease (35).
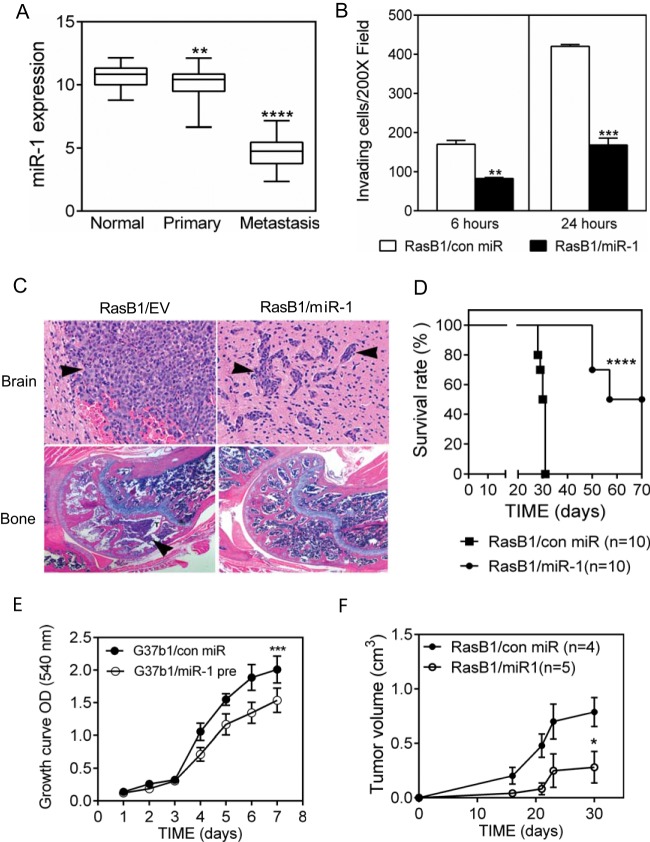
FIG 1.
miR-1 expression decreases in clinical metastatic PC samples and inhibits metastasis in an experimental model. (A) Relative expression levels of miR-1 in normal epithelium (n = 28), primary PC (n = 98), and metastatic PC (n = 13), analyzed in the MSKCC PC data set. **, P < 0.01, and ****, P < 0.0001, versus normal epithelium. (B) Invasion of RasB1 cells expressing either a control pre-miR or pre-miR-1 through Matrigel-coated Transwells at two time points. Data are means and SEM (n = 5). **, P < 0.01, and ***, P < 0.001, versus control miR. (C) Representative histology of brain metastases (top) and bone metastases (bottom) in tumor-bearing mice inoculated with the indicated cell lines. Tumor cells are marked by arrow heads. (D) Survival rate for mice bearing DU145/RasB1 tumors expressing either a control pre-miR (n = 10) or pre-miR-1 (n = 10). ****, P < 0.0001 for comparison of the two groups. (E) In vitro growth rate of DU145/RasB1 and DU145/RasB1/miR-1 cells. n = 6 per group. OD, optical density. (F) Tumorigenesis of DU145/RasB1 and DU145/RasB1/miR-1 cells after subcutaneous injection. *, P < 0.05, and ***, P < 0.001, versus controls.
To determine the potential role of miR-1 in metastases, we analyzed the phenotypic consequences of ectopic miR-1 precursor expression in a xenograft model of prostate cancer metastasis. DU145 cells expressing the activated RASV12G37 effector mutant gain bone metastatic capability (24), and DU145/RasB1 (RasB1) cells are a line expanded from a bone metastasis (25). As shown in Fig. 1B, increased miR-1 levels in RasB1 cells inhibited their invasion in vitro, consistent with our results in an independent model and with the results of others (20, 22). Importantly, miR-1 expression inhibited the experimental brain and bone metastases that are observed in this model following intracardiac inoculation (Table 5). Metastases that did develop were delayed in the time to detection, from 3 weeks to 10 weeks, diminished in size, and mainly found in brain (Fig. 1C). This is reflected in a doubling of the time to morbidity in mice receiving RasB1 expressing miR-1 compared to RasB1 expressing a control miR (Fig. 1D). miR-1 expression partially inhibited the in vitro and subcutaneous growth rates of prostate cancer cells (Fig. 1E and F) (20, 22), suggesting that the inhibition of metastasis involves targets that regulate both invasion and proliferation.
TABLE 5.
Metastasis formation is inhibited by ectopic miR-1 expressiona
| Cell line | No. of mice with metastasis/total no. |
|||
|---|---|---|---|---|
| Week 3 |
Week 10 |
|||
| Brain | Bone | Brain | Bone | |
| DU145/RasB1 | 9/10 | 10/10 | NA | NA |
| DU145/RasB1/miR-1 | 0/10 | 0/10 | 8/10 | 2/10 |
The presence of metastasis was assessed every 2 weeks using BLI in mice inoculated with the indicated cell lines by intracardiac injection. Mice demonstrating persistent BLI signals were counted. Metastases were verified by X-ray (bone) and pathological analysis (bone and brain). NA, not applicable.
To gain a better understanding of the phenotypic consequences of miR-1 expression, we assayed signaling pathways anticipated to be of functional relevance in DU145/RasB1 cells. We observed an increase in mesenchymal markers such as ZEB1 and ETS1 in DU145 expressing RASV12G37, both in the polyclonal population (RasG37 in Fig. 2A) and in the derivative in vivo passaged RasB1 line. Ectopic miR-1 inhibited the expression of these EMT markers, consistent with our previous results showing that miR-1 inhibits SNAI2 expression and the EMT phenotype (20) and consistent with decreased invasion (Fig. 1B). In addition, we observed slightly elevated pERK levels associated with RasV12G37 expression and noticeably decreased pERK levels correlated with elevated miR-1 expression (Fig. 2A).
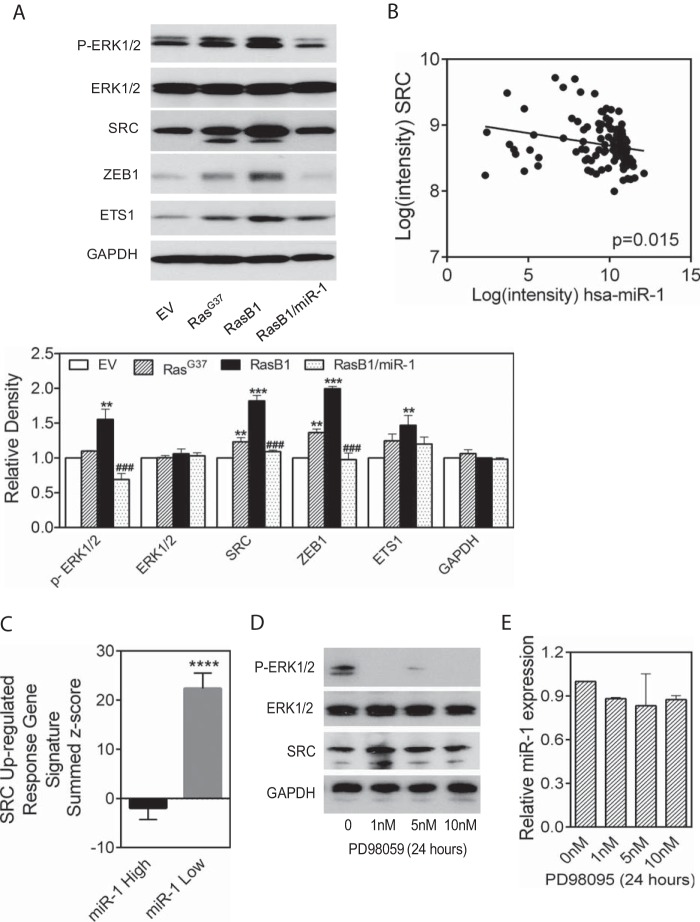
FIG 2.
miR-1 and SRC levels are anticorrelated. (A) Western blot (top) and analysis (bottom) of various proteins in whole-cell lysates, including DU145 cells infected with a control retrovirus (EV) and with RasV12G37 retrovirus (RasG37). DU145/RasB1 (RasB1) cells were derived from a RasG37 bone metastasis and later modified to express pre-miR-1 (RasB1/miR-1) (n = 3). **, P < 0.01, and ***, P < 0.001, versus EV; ###, P < 0.001, versus RasB1. (B) Correlation analysis (Pearson) of miR-1 and SRC levels in individual PC clinical samples (n = 111) taken from the MSKCC PC data set. r = −0.2305; P < 0.05 (two-tailed). (C) SRC upregulated signature genes expressed as a summed Z score for samples separated on the basis of miR-1 expression above the median (miR-1 High) or below the median (miR-1 Low) in the MSKCC data set. ****, P < 0.0001. (D) Western blot of the displayed proteins in whole-cell lysates from RasB1 cells treated with PD98059 at various concentrations for 24 h. (E) Relative miR-1 expression in RasB1 cells treated with various concentrations of PD98095 for 24 h.
We analyzed the relationship of potential miR-1 target mRNAs and miR-1 levels in the MSKCC prostate cancer data set (29). We identified SRC as a promising candidate based upon the presence of a miR-1 homology site, anticorrelated miR-1 and SRC mRNA levels (Fig. 2B), and a proposed role for SRC in prostate cancer bone metastasis (14, 36). Consistent with a correlation between low miR-1 expression and elevated SRC, patient tumors with miR-1 levels below the median demonstrated enrichment for a SRC-dependent gene signature, while tumors with miR-1 levels above the median did not (Fig. 2C). Interestingly, SRC protein levels increased in the metastatic derivatives of DU145 (RasG37 and RasB1) and, as would be expected for a miR-1 target, decreased in pre-miR-1 expressing RasB1 (Fig. 2A). Finally, because SRC and pERK expression correlated, we analyzed the effect of treating RasB1 cells with the MEK inhibitor PD98059 and observed no apparent effect on SRC levels (Fig. 2D), suggesting that pERK is not upstream but is either downstream of or in a pathway parallel to SRC. Consistent with this, MEK inhibition did not affect miR-1 levels (Fig. 2E).
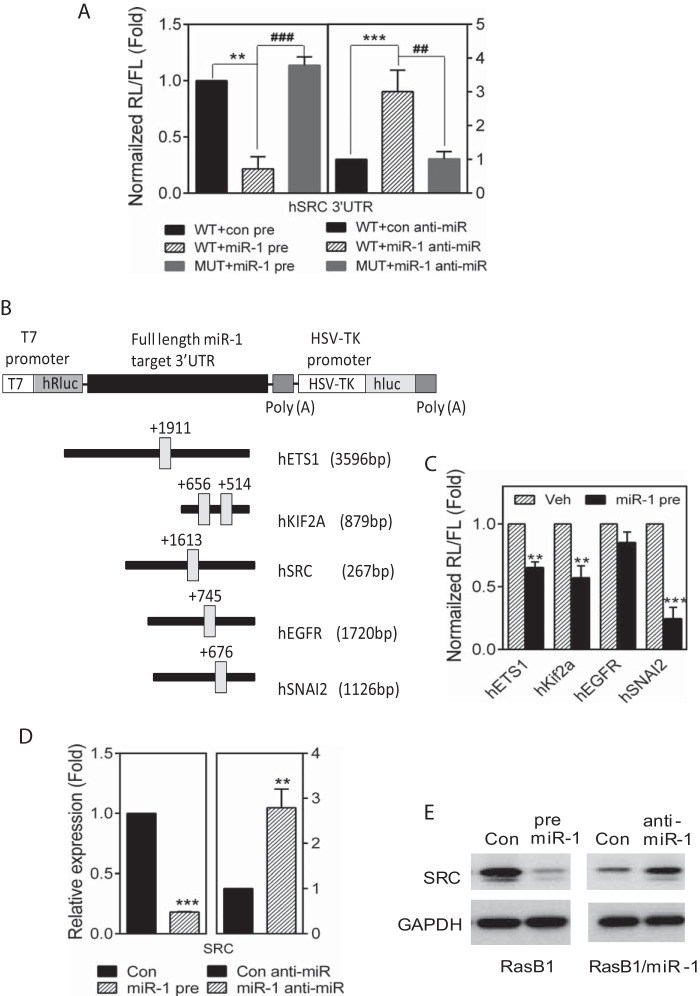
To address whether SRC levels can be directly regulated by miR-1, a full-length SRC 3′-UTR reporter containing either the wild-type or a miR-1 mutated homology site were assayed in DU145/RasB1 cells (Fig. 3A). As anticipated for direct targeting of SRC by miR-1, coexpression of a miR-1 precursor led to reduced reporter activity, while coexpression of a miR-1-antagonistic miR (anti-miR) led to increased reporter activity. miR-1-dependent reporter activity was not observed if the miR-1 homology site was mutated, demonstrating a direct and specific effect. The direct regulation of various other known and predicted miR-1 target reporters was analyzed (Fig. 3B) and showed the expected specificity (Fig. 3C), demonstrating the fidelity of the model. For endogenous SRC, the anticipated pattern of miR-1 dependent regulation was observed for RNA and protein levels in pre-miR-1 and anti-miR-1 expressing DU145/RasB1 cells (Fig. 3D and E).
FIG 3.
miR-1 directly regulates SRC levels in DU145/RasB1 cells. (A) Normalized reporter activity of the wt SRC 3′-UTR (2,671 bp) or a mutant containing a modified miR-1 binding site after transient transfection of control or miR-1 precursor miRs (left) and control or miR-1 anti-miRs (right). Data are means and SEM from separate transfections (n = 3). **, P < 0.01; ###, P < 0.001. (B) Schematic of predicated miR-1 binding sites in full-length human ETS1, KIF2A, SRC, EGFR and SNAI2 3′-UTR reporter constructs. (C) Normalized reporter activity of miR-1 target reporters containing the full-length human ETS1, KIF2A, EGFR, or SNAI2 3′-UTR in DU145/RasB1 and DU145/RasB1/miR-1 precursor-overexpressing cells. **, P < 0.01, and ***, P < 0.001, versus vehicle. (D) qPCR analysis of endogenous SRC RNA levels relative to GAPDH for the conditions described for panel A. (E) Representative Western blots of whole-cell extracts from RasB1 cells transiently transfected with control or miR-1 precursor miRs (left) and RasB1/miR-1 cells transiently transfected with control and miR-1 anti-miRs (right).
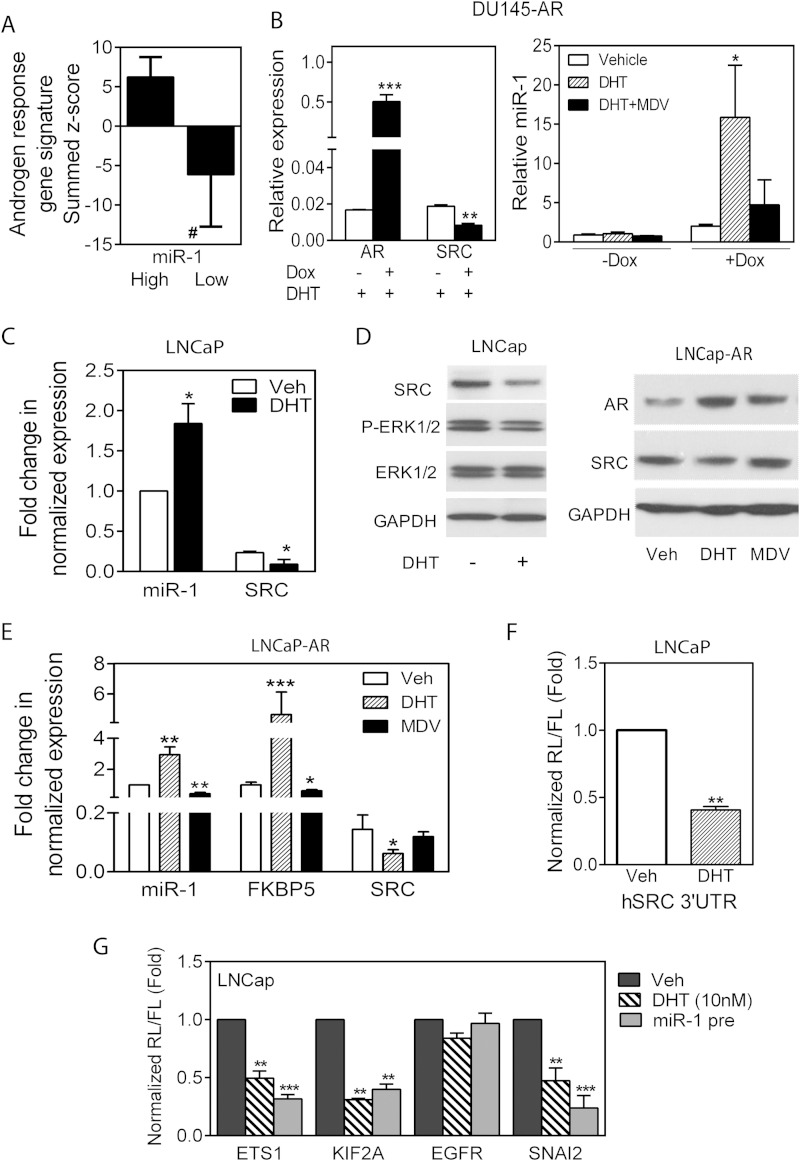
An important phenotypic property of individual prostate cancers is the AR-dependent output, which is measurable by a transcription-based signature (5, 33). Heterogeneity of AR output exists in different patient samples, especially in conjunction with the development of castration-resistant prostate cancer (5–7). Following the development of castration resistance, a decrease in the level of an AR gene signature commonly expressed in prostate cancer indicates either reduced AR transcriptional activity or a shift in AR-dependent transcriptional specificity (6, 8). We asked whether miR-1 expression levels were related to AR output in the data set from the MSKCC clinical samples. As shown in Fig. 4A, tumors expressing levels of miR-1 greater than the median demonstrate significantly greater AR activity (shown as a summed Z score) than those expressing less than median miR-1 levels. Of particular interest, previous studies have reported that decreasing predicted canonical AR activity correlates with increasing predicted SRC levels (5), although a mechanism relating the two phenotypes has not been described. The positive correlation of AR activity and miR-1 levels may represent coexisting nonrelated phenotypes or may suggest a causal relationship. Because miR-1 is widely expressed in most tissues, there clearly exists AR independent transcriptional regulation of miR-1. However, we sought to determine whether AR signaling modulates miR-1 and SRC levels in prostate epithelial cells.
FIG 4.
AR positively regulates miR-1 and negatively regulates SRC levels. (A) AR regulated signature genes expressed as a summed Z score for samples separated on the basis of miR-1 expression above the median (High) or below the median (Low) in the MSKCC data set. #, P < 0.0001. (B) qPCR determination of AR and SRC levels in DU145-AR cells without (−) or with (+) doxycycline (Dox) in the presence of 10 nM DHT (left) and relative miR-1 levels in control or doxycycline-induced cells treated with vehicle, DHT (10 nM), or DHT and MDV3100 (10 μM) (right). (C) Fold change in normalized miR-1 and SRC RNA levels in LNCaP cells treated with vehicle or DHT (10 nM). (D) Western blot analysis of various proteins in extracts from LNCap cells treated with DHT (left) and LNCap/AR cells treated with DHT or MDV3100 (right). (E) Fold change in normalized miR-1, FKBP5, and SRC RNA levels in LNCaP-AR cells treated with vehicle, DHT, or MDV3100. (F) Normalized reporter activity for the SRC 3′-UTR following transfection into LNCaP cells and treatment with vehicle or DHT. (G) Normalized activity in LNCaP cells of miR-1 target reporters containing a full-length human ETS1, KIF2A, EGFR, or SNAI2 3′-UTR. Cells were treated with or without DHT (10 nM) for 48 h or transfected with pre-miR-1. For panels B to G, all treatments were for 48 h. Data are means and SEM for separate treatments (n = 3). *, P < 0.05, **, P < 0.01, and ***, P < 0.001, versus vehicle.
We used three human cell models to address AR-initiated regulation of miR-1: inducible AR expression in the AR-negative DU145 cells, endogenously AR-positive LNCaP, and highly AR-expressing LNCaP-AR (37). As shown in Fig. 4B, doxycycline induction of AR in DU145 led to significantly increased miR-1 levels associated with ligand-activated AR, as demonstrated by DHT-mediated increases, which were reversed with the AR antagonist MDV3100. SRC RNA levels decreased as would be predicted. Similarly, DHT treatment and activation of endogenous AR in LNCaP cells or overexpressed AR in LNCaP-AR cells resulted in 2- to 3-fold-increased miR-1 and >50%-decreased SRC levels (Fig. 4C and D). In addition, inhibition of AR with MDV3100 in LNCaP-AR cells decreased expression of the AR target FKBP5 and also reduced miR-1 expression (Fig. 4E). Changes in SRC RNA levels initiated by modulating AR activity correlated with changes in SRC protein amounts (Fig. 4E). miR-1-dependent transcription modulated by AR was investigated by assaying the activity of a SRC 3′-UTR reporter, which decreased following DHT treatment of LNCaP (Fig. 4F), demonstrating posttranscriptional regulation of SRC consistent with miR-1 regulation. Additional 3′-UTR reporters for other predicted miR-1 targets tested in LNCaP cells under various conditions of AR activity were regulated parallel to SRC, further validating a miR-1-dependent effect of AR activities (Fig. 4G).
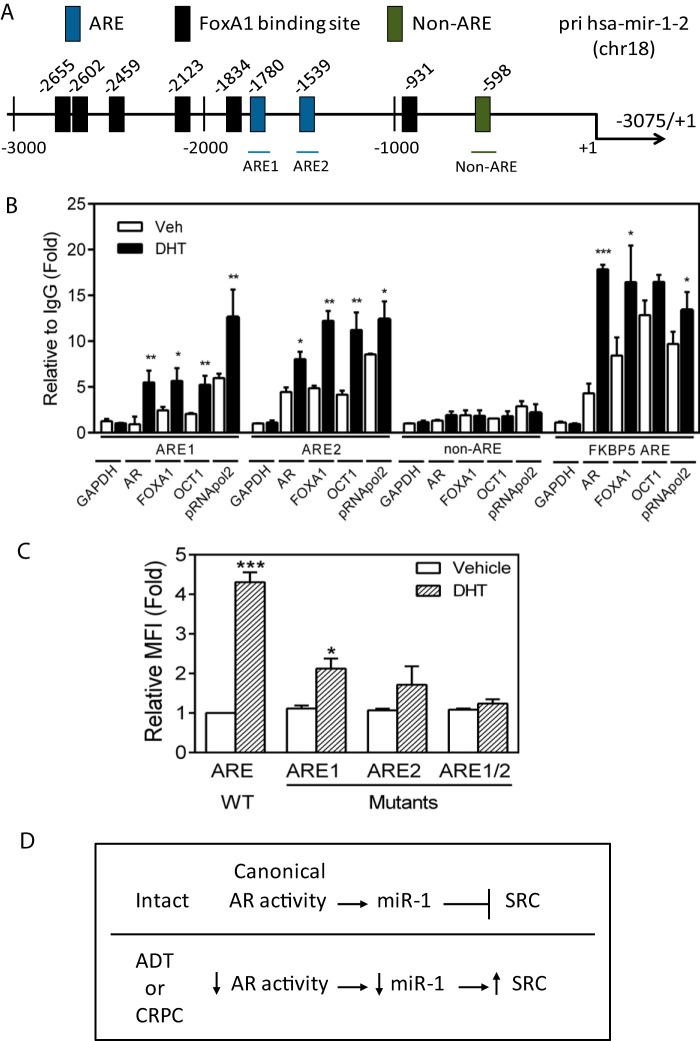
To address whether AR directly binds to and regulates the primary miR (pri-miR) encoding human miR-1-2 on chromosome 18, putative AR binding sites (AREs) were mapped and are shown in context with FOXA1 binding sites (Fig. 5A). Chromatin immunoprecipitation assays were performed in LNCaP cells with antibodies directed against AR. As shown in Fig. 5B, AR activated by DHT increased binding within 2 h to ARE1 and ARE2, in complex with the known AR cofactors FOXA1 and OCT1. The increased binding of POL2 further suggested a direct transcriptional effect of ligand-bound AR. To determine whether the ARE elements in the pri-miR-1-2 upstream region are functional, the region between −3075 and the transcription start site was cloned into an RFP reporter construct and assayed with and without AR activation. As shown in Fig. 5C, DHT induced a 4-fold increase in reporter activity, which was partially inhibited by mutation of either ARE1 or ARE2 and fully inhibited by mutation of both AR-binding elements. The data suggest that pri-miR-1-2 is positively transcriptionally regulated by AR binding at both identified AREs in prostate cancer cells and support a causal relationship for AR-mediated miR-1 regulation and anticorrelated SRC levels, as schematically depicted in Fig. 5D.
FIG 5.
Primary miR-1 RNA levels are directly and positively regulated by AR binding to the pri-miR-1-2 promoter. (A) Schematic of predicted AR (AREs) and FOXA1 binding sites as well as a control non-ARE region in the hsa-miR-1-2 primary miR (pri-miR) promoter. (B) ChIP analysis of AR, FOXA1, OCT1, and POL2 binding to predicted AR response elements (AREs) in the pri-miR-1-2 promoter region measured in LNCaP cells treated with DHT (10 nM) for 2 h. The binding activity of each protein to each site is given as a percentage of total input that was then normalized to each IgG. Data are means and SEM for separate treatments (n = 3). **, P < 0.01 for DHT versus vehicle. (C) MFI for the pri-miR-1-2 promoter-RFP reporter and ARE1, ARE2, and ARE1 and -2 site-specific mutants assayed in LNCaP cells treated with vehicle or DHT for 48 h. Data are means and SEM (n = 3). *, P < 0.05, and ***, P < 0.001, for DHT versus vehicle. (D) Schematic model summarizing the experimental data shown in Fig. 3 and 4 and this figure combined with the finding of others that canonical AR activity is decreased in a subset of CRPC clinical samples.
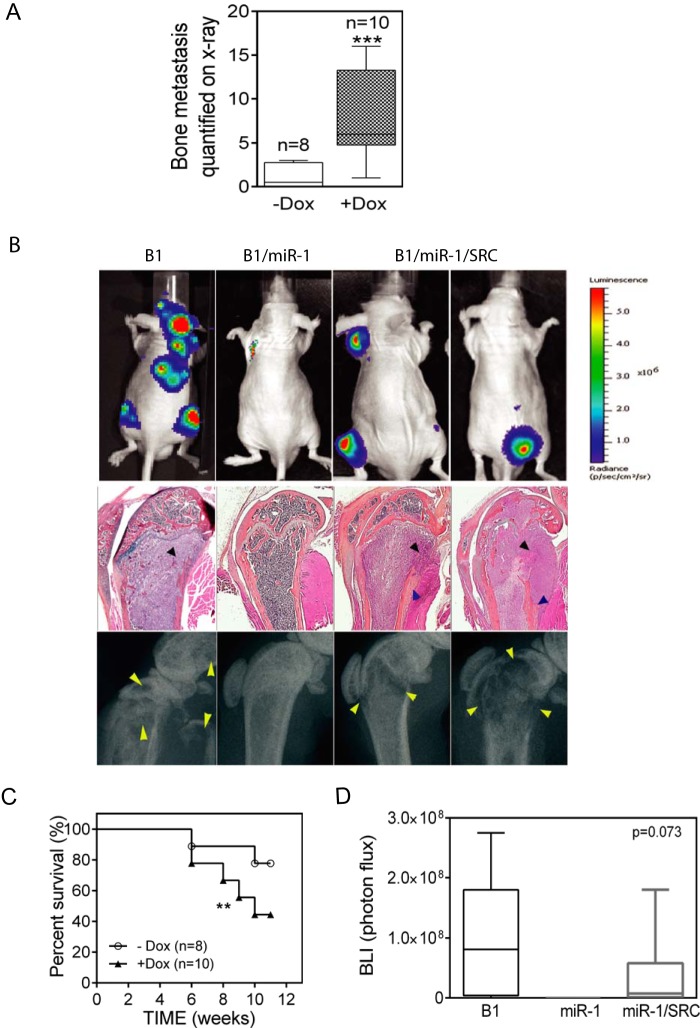
SRC, a miR-1 target, has been implicated in the development of prostate cancer bone metastasis. On the other hand, miR-1 has been shown to target several different gene transcripts (Fig. 3B and C) (22, 38, 39). Having demonstrated that ectopic pre-miR-1 expression decreased SRC expression and the development of bone metastasis, we asked whether reconstitution of SRC levels in miR-1 cells would be sufficient to restore bone metastatic activity. We infected DU145/RasB1/miR-1 with lentivirus encoding doxycycline-inducible SRC (Fig. 6A). As expected, in the absence of doxycycline, SRC protein expression was low due to ectopic miR-1 expression. Interestingly, induced SRC expression increased pERK signaling even in the presence of high miR-1 expression, implying that that SRC is downstream of miR-1 and upstream of pERK, consistent with Fig. 2A and D. We confirmed that miR-1 expression remained elevated in SRC-induced DU145/RasB1/miR-1/SRC cells (Fig. 6B). Immunohistochemical staining results for SRC in doxycycline-induced DU145/RasB1/miR-1/SRC and DU145/RasB1 tumors were approximately similar (Fig. 6C). As shown in Fig. 7A and B, SRC induction significantly reconstituted bone metastases in pre-miR-1-expressing cells. Histological analyses of reconstituted metastases revealed aggressive tumor growth in the bone accompanied predominantly by osteolysis but with a clear presence of osteoblastic elements as well (Fig. 7B). Finally, doxycycline-inducible SRC expression in inoculated mice was associated with decreased survival, as anticipated for reconstituted metastasis induction (Fig. 7C). Taken together, these data suggest that SRC is a physiologically important miR-1 target that functions in the development of bone metastases. Compared with DU145/RasB1, SRC overexpression only partially rescued bone metastases (Fig. 7C and D), suggesting the existence of multiple miR-1 targets. However, the fact that robust SRC expression alone was sufficient to reverse the inhibitory effects of high levels of miR-1 on bone metastasis implies that SRC is a dominant miR-1 target for determining the bone-metastatic phenotype.
FIG 6.

Characterization of SRC reconstitution in miR-1-expressing DU145/RasB1 cells. (A) Western blot analysis of extracts from DU145/RasB1/miR-1 cells expressing TRE3G-regulated SRC with and without doxycycline induction. (B) miR-1 levels in the parental and SRC-rescued DU145/RasB1/miR-1 cells used for panel A. ****, P < 0.0001 versus control miR. (C) Representative IHC images of SRC expression in brain metastases from DU145/RasB1-bearing mice and doxycycline-fed mice bearing DU145/RasB1/miR-1/Src.
FIG 7.
SRC expression reconstitutes metastasis in miR-1 expressing DU145/RasB1 cells. (A) Bone metastasis scores per mouse in tumor-bearing mice fed normal or doxycycline diets. Mice were inoculated with DU145/RasB1/miR-1 cells expressing Tet-regulated SRC. ***, P < 0.001. (B) Representative bioluminescence images (top), histology sections (middle), and radiographs of bone metastases (bottom) in the mice used for panel A compared with DU145/RasB1. Bone metastases are indicated by arrowshead. Black arrowheads, adjacent osteolysis; blue arrowheads, adjacent new bone formation. (C) Survival rates of the tumor-bearing mice used for panel A. **, P < 0.01. (D) BLI analysis of tumor burden in bones of the tumor-bearing mice used for panel A compared with DU145/RasB1.
DISCUSSION
miR-1 is a prostate cancer tumor suppressor, and increased SRC activity has been implicated in PC progression (14, 36). In the current study, we provide evidence for a molecular mechanism linking the two phenotypes. We show that SRC is a direct miR-1 target and that SRC RNA and protein levels are modulated by miR-1 in various prostate cancer model systems. SRC expression reverses ectopic-miR-1-mediated inhibition of bone metastasis, implying that SRC levels are dominant in determining miR-1 tumor suppressor activity. Importantly, in clinical samples, increased SRC levels as well as a SRC-dependent transcription signature correlate with decreased miR-1 levels. Prior transcriptomic analyses have shown an inverse relationship between canonical AR output and SRC-dependent transcription as well as predicted responsiveness to the SRC inhibitor dasatanib (5, 16). We show here that in experimental models, AR directly and positively contributes to the regulation of miR-1 transcription. The current study suggests one molecular mechanism whereby low canonical AR output contributes to reduced miR-1 and increased SRC levels.
Various independent analyses of clinical samples have determined that miR-1 is present at decreased abundance in prostate cancer compared to benign prostate tissue. Of note, the data set reported by Taylor et al. (29) and the study by Martens-Uzunova et al. (23) showed that miR-1 levels decrease further relative to primary tumors with metastatic progression. miR-1 contributes to a predictive signature for progressive prostate cancer (23), and consistent with this, primary tumors in the data set of Taylor et al. with lower-than-median miR-1 levels recurred earlier than those with miR-1 levels above the median (22). Of additional interest, using a rank-based method comparing matched normal and tumor tissue, miR-1 was found to be one of the 10 most attenuated miRs in a variety of cancers, suggesting that the miR-1 regulatory network may influence fundamental processes in transformation (40).
The underlying cancer and tissue-specific mechanisms regulating miR-1 expression are not fully known. Although there is evidence for CpG island methylation within the miR-1-1 promoter in hepatocellular, colon, and prostate cancer cell lines (22, 38), the methylation pattern in primary tumors is more limited and heterogeneous. Thus, methylation may be a contributing factor in miR-1 expression levels in some tumors. We present here an additional mechanism, AR-dependent regulation of miR-1-2, of specific significance for prostate cancer progression, which is characterized by heterogeneity of canonical AR-driven gene signatures (5, 7).
The data presented here emphasize the dominance of SRC as a miR-1 target and determinant of bone metastatic activity, even though a variety of other miR-1 targets have been described (22, 38, 39). Additionally, miR-23b was recently described as targeting SRC and AKT in prostate cancer, and it is reasonable to anticipate that miR-1 and miR-23b both contribute to regulating SRC levels (41). SRC is of particular interest with respect to bone metastasis, the predominant site for metastatic prostate cancer. Recent investigations in triple-negative breast cancer suggest a rationale for the correlation between SRC activity and the bone-metastatic phenotype (42). It was shown that SRC enhances PI3K-AKT pathway activity initiated by insulin-like growth factor 1 (IGF1) and CXCL12, survival and growth factors that are expressed in limiting amounts in primary tumor stroma and in the bone microenvironment.
Importantly, SRC family inhibition has been advanced as a strategy for the treatment of metastatic CRPC (43, 44). In addition to an implied role for SRC in CRPC, SRC signaling is involved in bone turnover, which is thought to be a determinant of metastatic bone colonization (36, 45). Thus, dasatinib, a potent oral inhibitor of SRC family and other kinases (45), has been used in the treatment of metastatic prostate cancer. Unfortunately, the READY trial, a randomized, double-blind phase 3 trial, did not demonstrate a survival benefit upon the addition of dasatinib to docetaxel in chemotherapy-naive CRPC patients (46). One concern is that the heterogeneity of CRPC may require the preselection of specific categories of patients likely to benefit from SRC family kinase inhibition. A more complete mechanistic understanding of the role of SRC family kinases in the context of other growth and survival signaling pathways may be needed. The data presented here provide one molecular rationale linking reduced AR activity in CRPC with increased SRC signaling.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research; the National Health Research Institutes (NHRI-EX103-10308BC); and the Ministry of Science and Technology (MOST 103-2314-B-038-051) of Taiwan.
REFERENCES
- 1.Roudier MP, True LD, Higano CS, Vesselle H, Ellis W, Lange P, Vessella RL. 2003. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol 34:646–653. doi: 10.1016/S0046-8177(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 2.Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar GR, Varambally S, Harwood J, Bismar TA, Kim R, Rubin MA, Pienta KJ. 2004. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res 64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 3.Scher HI, Sawyers CL. 2005. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol 23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 4.Zong Y, Goldstein AS. 2013. Adaptation or selection—mechanisms of castration-resistant prostate cancer. Nat Rev Urol 10:90–98. doi: 10.1038/nrurol.2012.237. [DOI] [PubMed] [Google Scholar]
- 5.Mendiratta P, Mostaghel E, Guinney J, Tewari AK, Porrello A, Barry WT, Nelson PS, Febbo PG. 2009. Genomic strategy for targeting therapy in castration-resistant prostate cancer. J Clin Oncol 27:2022–2029. doi: 10.1200/JCO.2008.17.2882. [DOI] [PubMed] [Google Scholar]
- 6.Sharma NL, Massie CE, Ramos-Montoya A, Zecchini V, Scott HE, Lamb AD, MacArthur S, Stark R, Warren AY, Mills IG, Neal DE. 2013. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell 23:35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, Shah RB, Chinnaiyan AM. 2007. Integrative molecular concept modeling of prostate cancer progression. Nat Genet 39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, Wu T, Regan MM, Meyer CA, Carroll JS, Manrai AK, Janne OA, Balk SP, Mehra R, Han B, Chinnaiyan AM, Rubin MA, True L, Fiorentino M, Fiore C, Loda M, Kantoff PW, Liu XS, Brown M. 2009. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maugeri-Sacca M, Coppola V, De Maria R, Bonci D. 2013. Functional role of microRNAs in prostate cancer and therapeutic opportunities. Crit Rev Oncog 18:303–315. doi: 10.1615/CritRevOncog.2013007206. [DOI] [PubMed] [Google Scholar]
- 10.Wang XD, Reeves K, Luo FR, Xu LA, Lee F, Clark E, Huang F. 2007. Identification of candidate predictive and surrogate molecular markers for dasatinib in prostate cancer: rationale for patient selection and efficacy monitoring. Genome Biol 8:R255. doi: 10.1186/gb-2007-8-11-r255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Z, Dai B, Jiang T, Xu K, Xie Y, Kim O, Nesheiwat I, Kong X, Melamed J, Handratta VD, Njar VC, Brodie AM, Yu LR, Veenstra TD, Chen H, Qiu Y. 2006. Regulation of androgen receptor activity by tyrosine phosphorylation. Cancer Cell 10:309–319. doi: 10.1016/j.ccr.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Yeatman TJ. 2004. A renaissance for SRC. Nat Rev Cancer 4:470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 13.Bromann PA, Korkaya H, Courtneidge SA. 2004. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene 23:7957–7968. doi: 10.1038/sj.onc.1208079. [DOI] [PubMed] [Google Scholar]
- 14.Tatarov O, Mitchell TJ, Seywright M, Leung HY, Brunton VG, Edwards J. 2009. SRC family kinase activity is up-regulated in hormone-refractory prostate cancer. Clin Cancer Res 15:3540–3549. doi: 10.1158/1078-0432.CCR-08-1857. [DOI] [PubMed] [Google Scholar]
- 15.Tatarov O, Edwards J. 2007. The role of SRC family kinases in prostate cancer. Transl Oncogenomics 2:67–77. [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Karaca M, Zhang Z, Gioeli D, Earp HS, Whang YE. 2010. Dasatinib inhibits site-specific tyrosine phosphorylation of androgen receptor by Ack1 and Src kinases. Oncogene 29:3208–3216. doi: 10.1038/onc.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, Scardino PT, Rosen N, Sawyers CL. 2011. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinkade CW, Castillo-Martin M, Puzio-Kuter A, Yan J, Foster TH, Gao H, Sun Y, Ouyang X, Gerald WL, Cordon-Cardo C, Abate-Shen C. 2008. Targeting AKT/mTOR and ERK MAPK signaling inhibits hormone-refractory prostate cancer in a preclinical mouse model. J Clin Invest 118:3051–3064. doi: 10.1172/JCI34764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, Plaisier S, Garraway IP, Huang J, Graeber TG, Wu H. 2011. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu YN, Yin JJ, Abou-Kheir W, Hynes PG, Casey OM, Fang L, Yi M, Stephens RM, Seng V, Sheppard-Tillman H, Martin P, Kelly K. 2013. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 32:296–306. doi: 10.1038/onc.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, Yfantis HG, Stephens RM, Croce CM. 2008. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res 68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hudson RS, Yi M, Esposito D, Watkins SK, Hurwitz AA, Yfantis HG, Lee DH, Borin JF, Naslund MJ, Alexander RB, Dorsey TH, Stephens RM, Croce CM, Ambs S. 2012. MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res 40:3689–3703. doi: 10.1093/nar/gkr1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Moller S, Trapman J, Bangma CH, Litman T, Visakorpi T, Jenster G. 2012. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene 31:978–991. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 24.Yin J, Pollock C, Tracy K, Chock M, Martin P, Oberst M, Kelly K. 2007. Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol 27:7538–7550. doi: 10.1128/MCB.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin JJ, Zhang L, Munasinghe J, Linnoila RI, Kelly K. 2010. Cediranib/AZD2171 inhibits bone and brain metastasis in a preclinical model of advanced prostate cancer. Cancer Res 70:8662–8673. doi: 10.1158/0008-5472.CAN-10-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gumerlock PH, Poonamallee UR, Meyers FJ, DeVere White RW. 1991. Activated ras alleles in human carcinoma of the prostate are rare. Cancer Res 51:1632–1637. [PubMed] [Google Scholar]
- 27.Wang XS, Shankar S, Dhanasekaran SM, Ateeq B, Sasaki AT, Jing X, Robinson D, Cao Q, Prensner JR, Yocum AK, Wang R, Fries DF, Han B, Asangani IA, Cao X, Li Y, Omenn GS, Pflueger D, Gopalan A, Reuter VE, Kahoud ER, Cantley LC, Rubin MA, Palanisamy N, Varambally S, Chinnaiyan AM. 2011. Characterization of KRAS rearrangements in metastatic prostate cancer. Cancer Discov 1:35–43. doi: 10.1158/2159-8274.CD-10-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aytes A, Mitrofanova A, Kinkade CW, Lefebvre C, Lei M, Phelan V, LeKaye HC, Koutcher JA, Cardiff RD, Califano A, Shen MM, Abate-Shen C. 2013. ETV4 promotes metastasis in response to activation of PI3-kinase and Ras signaling in a mouse model of advanced prostate cancer. Proc Natl Acad Sci U S A 110:E3506–E3515. doi: 10.1073/pnas.1303558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. 2010. Integrative genomic profiling of human prostate cancer. Cancer Cell 18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, Gleave M, Wu H. 2012. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res 72:1878–1889. doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Day CP, Carter J, Bonomi C, Esposito D, Crise B, Ortiz-Conde B, Hollingshead M, Merlino G. 2009. Lentivirus-mediated bifunctional cell labeling for in vivo melanoma study. Pigment Cell Melanoma Res 22:283–295. doi: 10.1111/j.1755-148X.2009.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu YN, Abou-Kheir W, Yin JJ, Fang L, Hynes P, Casey O, Hu D, Wan Y, Seng V, Sheppard-Tillman H, Martin P, Kelly K. 2012. Critical and reciprocal regulation of KLF4 and SLUG in transforming growth factor beta-initiated prostate cancer epithelial-mesenchymal transition. Mol Cell Biol 32:941–953. doi: 10.1128/MCB.06306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, Maloney KN, Clardy J, Hahn WC, Chiosis G, Golub TR. 2006. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, Olson JA Jr, Marks JR, Dressman HK, West M, Nevins JR. 2006. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 35.Katz B, Reis ST, Viana NI, Morais DR, Moura CM, Dip N, Silva IA, Iscaife A, Srougi M, Leite KR. 2014. Comprehensive study of gene and microRNA expression related to epithelial-mesenchymal transition in prostate cancer. PLoS One 9:e113700. doi: 10.1371/journal.pone.0113700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Araujo J, Logothetis C. 2009. Targeting Src signaling in metastatic bone disease. Int J Cancer 124:1–6. doi: 10.1002/ijc.23998. [DOI] [PubMed] [Google Scholar]
- 37.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. 2004. Molecular determinants of resistance to antiandrogen therapy. Nat Med 10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 38.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, Ghoshal K, Jacob ST. 2008. Methylation mediated silencing of microRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res 68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Leone V, D'Angelo D, Rubio I, de Freitas PM, Federico A, Colamaio M, Pallante P, Medeiros-Neto G, Fusco A. 2011. MiR-1 is a tumor suppressor in thyroid carcinogenesis targeting CCND2, CXCR4, and SDF-1alpha. J Clin Endocrinol Metab 96:E1388–E1398. doi: 10.1210/jc.2011-0345. [DOI] [PubMed] [Google Scholar]
- 40.Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. 2009. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One 4:e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majid S, Dar AA, Saini S, Arora S, Shahryari V, Zaman MS, Chang I, Yamamura S, Tanaka Y, Deng G, Dahiya R. 2012. miR-23b represses proto-oncogene Src kinase and functions as methylation-silenced tumor suppressor with diagnostic and prognostic significance in prostate cancer. Cancer Res 72:6435–6446. doi: 10.1158/0008-5472.CAN-12-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang XH, Jin X, Malladi S, Zou Y, Wen YH, Brogi E, Smid M, Foekens JA, Massague J. 2013. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell 154:1060–1073. doi: 10.1016/j.cell.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherji D, Eichholz A, De Bono JS. 2012. Management of metastatic castration-resistant prostate cancer: recent advances. Drugs 72:1011–1028. doi: 10.2165/11633360-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 44.Saylor PJ, Armstrong AJ, Fizazi K, Freedland S, Saad F, Smith MR, Tombal B, Pienta K. 2013. New and emerging therapies for bone metastases in genitourinary cancers. Eur Urol 63:309–320. doi: 10.1016/j.eururo.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montero JC, Seoane S, Ocana A, Pandiella A. 2011. Inhibition of SRC family kinases and receptor tyrosine kinases by dasatinib: possible combinations in solid tumors. Clin Cancer Res 17:5546–5552. doi: 10.1158/1078-0432.CCR-10-2616. [DOI] [PubMed] [Google Scholar]
- 46.Araujo JC, Trudel GC, Saad F, Armstrong AJ, Yu EY, Bellmunt J, Wilding G, McCaffrey J, Serrano SV, Matveev VB, Efstathiou E, Oudard S, Morris MJ, Sizer B, Goebell PJ, Heidenreich A, de Bono JS, Begbie S, Hong JH, Richardet E, Gallardo E, Paliwal P, Durham S, Cheng S, Logothetis CJ. 2013. Docetaxel and dasatinib or placebo in men with metastatic castration-resistant prostate cancer (READY): a randomised, double-blind phase 3 trial. Lancet Oncol 14:1307–0316. doi: 10.1016/S1470-2045(13)70479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]