Abstract
The requirement for alternative splicing during adipogenesis is poorly understood. The Sam68 RNA binding protein is a known regulator of alternative splicing, and mice deficient for Sam68 exhibit adipogenesis defects due to defective mTOR signaling. Sam68 null preadipocytes were monitored for alternative splicing imbalances in components of the mTOR signaling pathway. Herein, we report that Sam68 regulates isoform expression of the ribosomal S6 kinase gene (Rps6kb1). Sam68-deficient adipocytes express Rps6kb1-002 and its encoded p31S6K1 protein, in contrast to wild-type adipocytes that do not express this isoform. Sam68 binds an RNA sequence encoded by Rps6kb1 intron 6 and prevents serine/arginine-rich splicing factor 1 (SRSF1)-mediated alternative splicing of Rps6kb1-002, as assessed by cross-linking and immunoprecipitation (CLIP) and minigene assays. Depletion of p31S6K1 with small interfering RNAs (siRNAs) partially restored adipogenesis of Sam68-deficient preadipocytes. The ectopic expression of p31S6K1 in wild-type 3T3-L1 cells resulted in adipogenesis differentiation defects, showing that p31S6K1 is an inhibitor of adipogenesis. Our findings indicate that Sam68 is required to prevent the expression of p31S6K1 in adipocytes for adipogenesis to occur.
INTRODUCTION
Src-associated substrate during mitosis of 68 kDa (Sam68) is an RNA binding protein that belongs to the conserved STAR (signal transduction activator of RNA) family (1, 2). Sam68 is a sequence-specific RNA binding protein that binds repeats of U(U/A)AA sequences in single-stranded RNA (3, 4). The binding of Sam68 near alternative splice junctions in pre-mRNAs has been shown to regulate splice site selection and regulate the usage of alternative exons (1). Sam68 promotes the inclusion of CD44 variable exon 5 (v5), and interaction of Sam68 with SND1 (staphylococcal nuclease domain 1) enhances v5 inclusion (5, 6). The alternative splicing of Bcl-x is regulated by Sam68 and its interaction with hnRNPA1 and FBI-1, affecting prosurvival and apoptotic pathways (7, 8). Sam68 regulates the epithelial-to-mesenchymal transition by decreasing the presence of an alternative serine/arginine-rich splicing factor 1 gene (Srsf1) transcript degraded by nonsense-mediated mRNA decay (9). Sam68 has been shown to regulate alternative splicing of mRNAs during neurogenesis (10) and in cerebellar neurons (11). Stimulation of cerebellar neurons using the glutamate receptor agonist kainic acid was dramatically attenuated without Sam68, indicating that Sam68 is required for activity-dependent alternative splicing of Nrxn1 in vivo (11).
The role of Sam68 in alternative splicing has implications for spinal muscular atrophy (SMA) and fragile X-associated tremor/ataxia syndrome (FXTAS). Sam68 promotes the skipping of exon 7, leading to a nonfunctional SMN2 protein, and it was shown that the inhibition of Sam68 enhanced exon 7 inclusion in endogenous SMN2 and increased survival motor neuron (SMN) levels in SMA patient cells (12). Expanded CGG repeats in the 5′ untranslated region (UTR) of the FMR1 gene causes FXTAS, and Sam68 association with these repeats in RNA aggregates blocks it from fulfilling its splicing functions (13). The inhibition of Sam68 phosphorylation prevents Sam68 from aggregating with RNA, suggesting that it may be a therapeutic option for FXTAS patients (13).
Sam68 null mice have revealed numerous unexpected physiological roles for Sam68. Male Sam68−/− mice are infertile, with defects in spermatogenesis, a process where Sam68 has been shown to regulate alternative splicing (14) and the polysomal recruitment of specific mRNAs in germ line cells (15). Ablation of Sam68 leads to increased energy expenditure, decreased numbers of early adipocyte progenitors, and defective adipogenic differentiation, resulting in mice having a lean phenotype protected against dietary-induced obesity (16). The lack of Sam68 results in mTOR (mammalian target of rapamycin) intron 5 retention and the production of a short transcript (named mTORi5), leading to reduced mTOR protein levels, which results in defects in insulin-stimulated S6 and Akt phosphorylation (16).
mTOR signaling plays a major role in the regulation of mRNA translation, cell growth, metabolism, and autophagy (17–19). The tuberous sclerosis complex (TSC; tuberous sclerosis 1 and 2 heterodimer) acts as a GTPase-activating protein (GAP) on the Ras-like protein Rheb, which activates the mTOR complex 1 (mTORC1) (20–22), and PRAS40 (proline-rich Akt substrate of 40 kDa) is an inhibitory mediator of mTORC1 signaling. The phosphorylation and inhibition of the TSC and PRAS40 by the upstream kinase Akt (serine/threonine protein kinase B) activate mTORC1 signaling (23–25). Activated mTOR signaling results in phosphorylation of 4EBP1 (initiation factor 4E-binding protein 1) and S6K1 (S6 kinase 1) (18, 19, 26). Active S6K1 phosphorylates the 40S ribosomal protein S6, thereby facilitating mRNA translation, while phosphorylated 4EBP1 promotes the release of eIF4E (eukaryotic translation initiation factor 4E) and initiates translation (26).
In the present manuscript, we identify Sam68 as an RNA binding protein that prevents the production of the alternative short isoform of Rps6kb1, encoding p31S6K1, in mouse preadipocytes and white adipose tissue (WAT). The binding of Sam68 to an Rps6kb1 intronic RNA sequence counteracted the alternative splicing effects of the SR protein, SRSF1. Expression of p31S6K1 in preadipocytes inhibited differentiation, while the depletion of p31S6K1 in Sam68-deficient preadipocytes partially restored the adipogenic differentiation defects in a p70S6K1-independent manner. Our findings show that Sam68 is a regulator of Rps6kb1 alternative splicing during adipogenesis.
MATERIALS AND METHODS
Alternative splicing assessment and real-time PCR.
Total RNA was isolated using TRIzol reagent according to the manufacturer's instructions (Invitrogen). Four micrograms of RNA was incubated at 65°C for 5 min and then at 42°C for 1 h with 100 pmol of oligo(dT) primer and 100 U of Moloney murine leukemia virus (M-MLV) reverse transcriptase (catalog no. M1701; Promega) according to the manufacturer's protocol. cDNAs were then amplified by PCR. Endogenous Rps6kb1 and Rps6kb1-002 were amplified with the common forward primer 5′-GCA ATG ATA GTG AGG AAT GCT AAG-3′ located in exon 5. The reverse primer for Rps6kb1 was 5′-GCT GTG TCT TCC ATG AAT ATT CC-3′ located in exon 6, and for Rps6kb1-002 the reverse primer was 5′-GAA TAG GAG GGC AGA TCC CAT CC-3′ located in exon 6b. For TSC1 and TSC1-003 amplification, the common forward primer 5′-GTG GAA GAC ATT AGA AAC TCA TG-3′ was used. The reverse primer sequence for TSC1 was 5′-AGG TGG ACT GAA CAA CAT CAG C-3′, and for TSC1-003 it was 5′-TCA ACT ACA AGT AGT ATG TTA TG-3′. The common forward primer for TSC1 and TSC1-006 amplification was 5′-GTG GAA GAC ATT AGA AAC TCA TG-3′. The reverse primer sequence for TSC1 was 5′-AGG TGG ACT GAA CAA CAT CAG C-3′, and for TSC1-006 it was 5′-ACC CAG CGG TCC ACA CTG ATT TG-3′. For Rheb and Rheb-002 amplification the common forward primer 5′-GAA AGT CCT CAT TGA CAA TTC AG-3′ was used. The reverse primer sequence for Rheb was 5′-CTG CCC CGC TGT GTC TAC AAG C-3′, and for Rheb-002 it was 5′-GTG AGT GTC AGC CCT CAC TCT AC-3′. Endogenous Rheb and Rheb-003 were amplified with the common forward primer 5′-GAT CAG CTA TGA AGA AGG AAA GGC-3′. The reverse primer sequence for Rheb was 5′-TTG GAC AGA GTC AGA CGT TAA C-3′, and for Rheb-003 it was 5′-CAT CAC CGA GCA CGA AGA CTT TC-3′. For Akt1 and Akt1-003 amplification, the forward primer sequence for Akt1 was 5′-GGA GGG CTG GCT GCA CAA ACG AG-3′, and that for Akt1-003 was 5′-GCC GCT GCG TGA CCT TGG GTG G-3′. The common reverse sequence for both Akt1 and Akt1-003 was 5′-CCG CTC TGT CTT CAT CAG CTG GC-3′. For Rps6kb1 and Rps6kb1-005 amplification, the common forward sequence used was 5′-TCT CAG AAA CTA GTG TGA ACA G-3′. The reverse primer sequence for Rps6kb1 was 5′-CAC TGA GAT ACT CGA GGA TGA GG-3′, and for Rps6kb1-005 it was 5′-ATT AAG ATA TAG CAT AGA GTG AG-3′. For Deptor and Deptor-002 amplification, the common forward primer 5′-ATT GTT GGT GAC GCA GTT GGC TG-3′ was used. The reverse primer sequence for Deptor was 5′-AGA TAT GTA ACC TGG TTC TTC CAC-3′, and for Deptor-002 it was 5′-ACC CAC CTT CCC TCC CAT TAG GTC-3′.
For real-time reverse transcription-PCR (RT-PCR), mouse Rps6kb1 was amplified with 5′-CGT GGA GTC TGC GGC G-3′ (located in exon 1) and 5′-CAT ATG GTC CAA CTC CCC CA-3′ (located in exon 2), mouse Rps6kb1-002 was amplified with 5′-TAT GCC TTT CAG ACC GGA GG-3′ (located in exon 5) and 5′-ACC TCC CTA AGA CTG CAC CT-3′ (located in exon 6b), 18S rRNA was amplified with 5′-GTA ACC CGT TGA ACC CCA TT-3′ and 5′-CCA TCC AAT CGG TAG TAG CG-3′, C/EBPα was amplified with 5′-CGC AAG AGC CGA GAT AAA GC-3′ and 5′-GCG GTC ATT GTC ACT GGT CA-3′, GLUT4 was amplified with 5′-TCG TGG CCA TAT TTG GCT TTG TGG-3′ and 5′-TAA GGA CCC ATA GCA TCC GCA ACA-3′, peroxisome proliferator-activated receptor γ (PPARγ) was amplified with 5′-GAA CGT GAA GCC CAT CGA GGA C-3′ and 5′-CTG GAG CAC CTT GGC GAA CA-3′, as previously described (27), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified with 5′-AGC CAC ATC GCT CAG ACA C-3′ and 5′-GCC CAA TAC GAC CAA ATC C-3′. Sam68 was amplified with 5′-GTG GAG ACC CCA AAT ATG CCC A-3′ and 5′-AAA CTG CTC CTG ACA GAT ATC A-3′. Moreover, primers for mouse GAPDH, Sam68, C/EBPα, and PPARγ were purchased from Qiagen (Valencia, CA). Real-time quantitative RT-PCR (RT-qPCR) was performed on a 7500 Fast real-time PCR system (Applied Biosystems, Foster City, CA) using SYBR green PCR Mastermix (Qiagen, Valencia, CA). The primer efficiency test using dilutions confirmed that the efficiencies were close to 1.0 for PPARγ, C/EBPα, GLUT4, GAPDH, and 18S rRNA.
Plasmid constructions.
The GFP-Sam68 expression vector encoding an N-terminal green fluorescent protein (GFP) was described previously (28). The GFP-SRSF1 expression vector was obtained from Addgene (catalog no. 17990; Cambridge, MA). Gene Rps6kb1 exon 6, intron 6, and exon 7 were amplified from mouse genomic DNA by PCR using the forward primer 5′-GGG GGA TCC GGA GGA GAA CTA TTT ATG CAG TTA-3′, containing a BamHI site, and the reverse primer 5′-GGG CTC GAG CTT GGT GAT TAA GCA TGA TGT TCT-3′, containing an XhoI site. The DNA fragment was then subcloned in the corresponding site of pcDNA3.1 containing a FLAG epitope tag. The mutation of the Sam68 binding site (SBS) in intron 6 of the minigene was performed in a two-step PCR using primers 5′-ATG ATT CAT GTA ATT CCA AGC AAA ACC ACC TT-3′ (forward primer) and 5′-AAG GTG GTT TTG CTT GGA ATT ACA TGA ATC AT-3′ (reverse primer). The plasmids encoding full-length p31S6K1 were purchased from IDT and subcloned in pcDNA3.1. An expression vector encoding p31S6K1 was kindly provided by Rotem Karni (Hebrew University-Hadassah Medical School). The common forward primer for RT-PCR and RT-qPCR detection of the Rps6kb1 minigene transcripts was F1 (5′-GAT TAC AAG GAT GAC GAC GAT AAG-3′). The reverse primers for RT-PCR detection were as follows: R1, 5′-AGG ATG GAG GGT GTG TCC TAG AGG-3′; R2, 5′-CTT GGT GAT TAA GCA TGA TGT TCT-3′. The reverse primers for RT-qPCR detection were the following: R3, 5′-CAA TTC AAG GAA ATT CTG CAG TG-3′; R4, 5′-GCC ATG GAG ATT TCA GCC AAG-3′.
Synthetic RNA oligonucleotides.
The RNA oligonucleotides with 3′ biotin tags were synthesized and purchased from IDT. The sequences of these oligonucleotides are as follows: Rps6kb1-SBS, 5′-CAU GAU UCA UGU AAU UAA AAG CAA AAC CAC CUU C-3′-biotin; Rps6kb1-SBSmut, 5′-CAU GAU UCA UGU AAU UCC AAG CAA AAC CAC CUU C-3′-biotin.
Preadipocyte differentiation and WAT.
Sam68-deficient 3T3-L1 cells were generated using pRetrosuper harboring a short hairpin RNA (shRNA) targeting Sam68 (Sam68sh), and pRetrosuper 3T3-L1 cells were used as a control, as described previously (16). Preadipocyte 3T3-L1 adipogenic differentiation was performed as described previously (29). The cells were fixed with 3% formaldehyde and 0.025% glutaraldehyde and incubated with Oil Red O solution (Sigma-Aldrich, St. Louis, MO). Cell extracts were prepared and analyzed as described previously (16). Antibodies for Sam68 (Millipore), p70 S6K (BD Transduction Laboratories, Cell Signaling), GFP (Roche), SRSF1 (Santa Cruz), FLAG M2, β-actin, and β-tubulin (Sigma) were purchased.
Stable 3T3-L1 clones overexpressing p31S6K1 were generated as follows. Cells were transfected with either pcDNA3.1 or pcDNA3.1 FLAG-p31 plasmid constructs. At 48 h posttransfection, G418 was added to the medium, and individual clones were selected several weeks later. The expression level of p31S6K1/FLAG was assessed by immunoblotting.
RNA interference and transfection.
The following siGENOME SMARTpool small interfering RNAs (siRNAs) were ordered from Dharmacon/Thermo Scientific: human KHDRBS1 (Sam68) (catalog no. M-020019-00-0010), mouse KHDRBS1 (Sam68) (catalog no. M-065115-01-0010), human SRSF1 (catalog no. M-018672-00-0005), and mouse SRSF1 (catalog no. M-040886-01-0005). Mouse RPS6KB1 (p70/p31) siGENOME set of four siRNAs (catalog no. MQ-040893-02-0002) was ordered from Dharmacon/Thermo Scientific. The following siRNAs were also ordered from Dharmacon/Thermo Scientific: mouse p31 siRNA-A sense sequence (5′-GCU CUU CAC UGC AGA AUU UUU-3′) and antisense sequence (5′-AAA UUC UGC AGU GAA GAG CUU-3′), mouse p31 siRNA-B sense sequence (5′-ACA CAG AAG CUG CAU UUA AUU-3′) and antisense sequence (5′-UUA AAU GCA GCU UCU GUG UUU-3′), and siRNA targeting GFP (siGFP) sense sequence (5′-AAC ACU UGU CAC UAC UUU CUC UU-3′) and antisense sequence (5′-GAG AAA GUA GUG ACA AGU GUU UU-3′).
For siRNA transfections, typically cells were plated in six-well plates and transfected with 100 nM siRNA using Lipofectamine RNAiMAX (siRNA), as recommended by the manufacturer (Invitrogen). HEK293 cells plated in six-well plates were transfected using Lipofectamine 2000. Each well received a total of 5 μg with GFP-SRSF1 (0, 0.25, 1, and 2 μg), GFP-Sam68 (1 μg), and 2 μg of the indicated minigene; an empty vector was used to compensate the amount of transfected DNA in Fig. 7C and D.
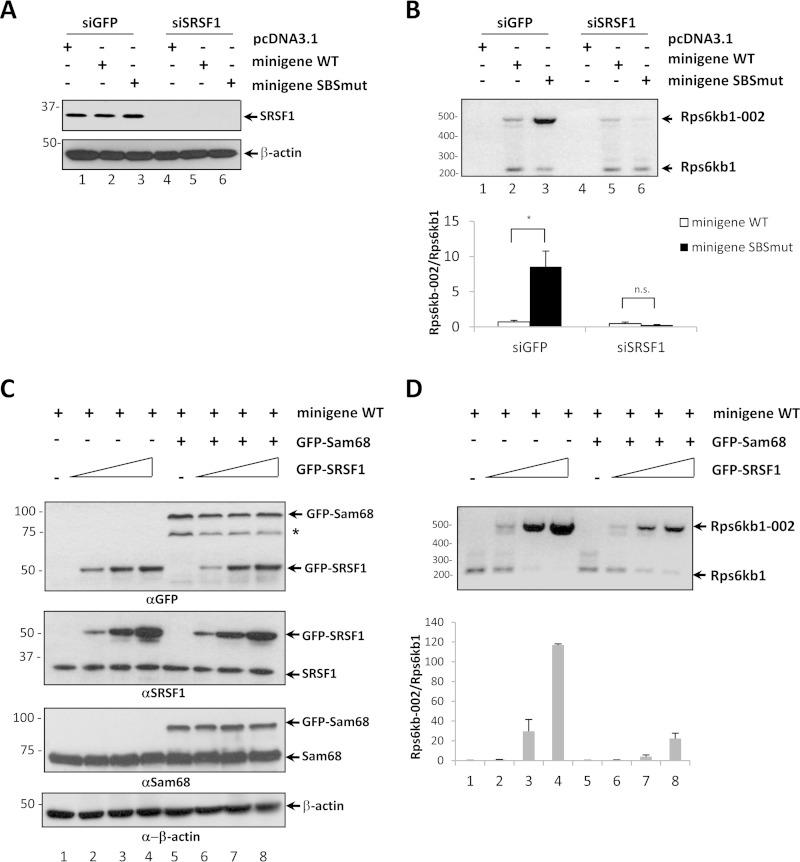
FIG 7.
Sam68 competes with SRSF1 for the positive regulation of Rps6kb1 splicing. (A) HEK293 cells were transfected with siRNAs targeting either GFP or human SRSF1. After 24 h, cells were transfected with either pcDNA3.1, the Rps6kb1 wild-type minigene plasmid, or the Rps6kb1 SBSmut minigene plasmid. The cells were harvested after 48 h. Protein extracts were immunoblotted with the indicated antibodies. β-Actin is shown as the loading control, and the asterisk denotes an unknown protein. Molecular mass markers are shown on the left in kilodaltons. (B) Total RNA was isolated and treated with RQ1 DNase, and the mRNA levels of Rps6kb1 and Rps6kb1-002 were assessed by RT-PCR using the primer pairs indicated in Fig. 5A. Densitometry analysis was performed from two independent experiments, and fold induction of Rps6kb1-002 was normalized to the level of Rps6kb1. Error bars represent ± standard deviations of the means (*, P < 0.05; n.s., not significant). (C) HEK293 cells were cotransfected with either the Rps6kb1 minigene plasmid alone or with GFP-Sam68 or with increasing amounts of GFP-SRSF1. The cells were harvested after 48 h. The protein extracts were immunoblotted with the indicated antibodies. β-Actin is shown as the loading control. (D) Total RNA was isolated and treated with RQ1 DNase, and the mRNA levels of Rps6kb1 and Rps6kb1-002 were assessed by RT-PCR. Densitometry analysis was performed from two independent experiments, and fold induction of Rps6kb1-002 was normalized to the level of Rps6kb1. Error bars represent ± standard deviations of the means.
RNA binding assays.
3T3-L1 cells were lysed in 1 ml of cell lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 40 units/ml RNaseOUT, supplemented with Roche Complete Mini, EDTA-free protease inhibitor) and incubated for 15 min at 4°C. Lysates were cleared by centrifugation, and 5 μl of 100 μM biotinylated RNA was added to the lysates and incubated at 4°C for 60 min with constant end-over-end mixing with streptavidin-Sepharose beads. The beads were washed three times with lysis buffer and once with 1× phosphate-buffered saline (PBS). Protein samples were analyzed on SDS-polyacrylamide gels and transferred to nitrocellulose membranes for immunoblotting.
Immunoprecipitation.
Transfected HEK293 cells were lysed in buffer containing 1% Triton X-100, 150 mM NaCl, 20 mM Tris (pH 7.5), and proteinase inhibitor (Roche) for 15 min on ice. Total cell lysates were clarified by centrifugation for 10 min at 10,000 × g at 4°C. The lysates were incubated with the indicated antibodies (see Fig. 4D) for 1 h or overnight at 4°C, and then 20 μl of 50% protein A/G slurry was added. The mixture was incubated for 30 min at 4°C. The protein A/G-Sepharose beads were washed three times with lysis buffer and once with 1× PBS. The samples were boiled and subjected to standard Western blot analysis.
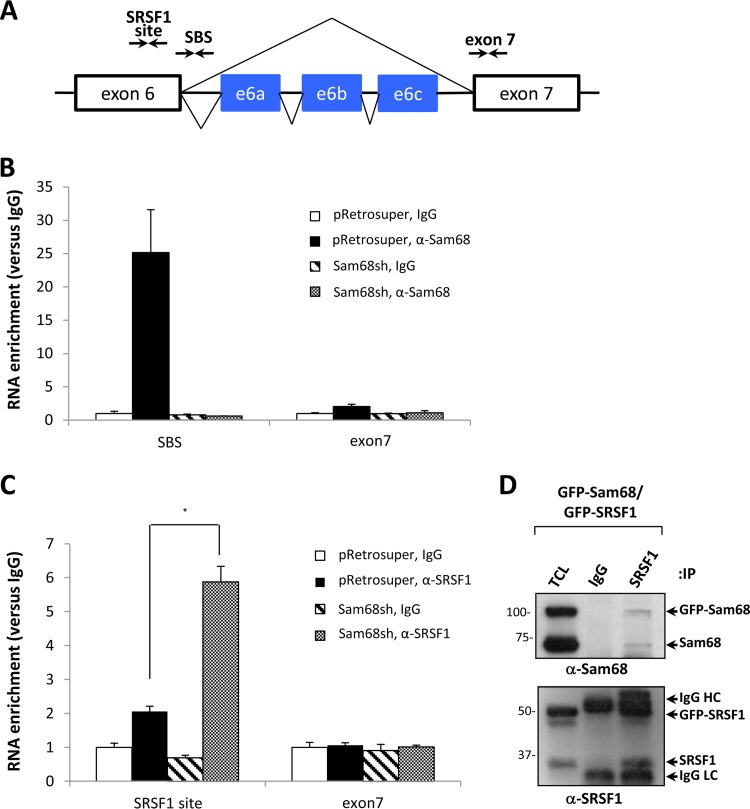
FIG 4.
Sam68 binds an intronic SBS and prevents the binding of SRSF1 to its consensus site in Rps6kb1 exon 6. (A) Schematic of the genomic architecture of Rps6kb1 spanning exons 6 and 7 with three alternative exons located in intron 6. Arrows depict the primer pairs used in RT-qPCR to detect the RNA-bound Sam68 binding site (SBS), the exon 6 SRSF1 binding site, and exon 7 as a negative control. (B and C) CLIP assays were performed using anti-Sam68 antibodies and anti-SRSF1 antibodies or control IgGs. Bound RNA was analyzed in triplicate by RT-qPCR with the primers shown in panel A. The levels of bound RNA of the SBS, SRSF1 binding site, and exon 7 in immunoprecipitates were normalized to the levels of the total RNA in the input. Mean values are expressed as fold enrichment. Error bars represent ± standard deviations of the means (*, P < 0.05). (D) HEK293 cells were transfected with expression vectors encoding GFP-Sam68 and GFP-SRSF1. After 48 h, the cells were lysed and subjected to immunoprecipitation (IP) and immunoblotting. The migration of GFP-Sam68, Sam68, GFP-SRSF1, SRSF1, and the heavy chain (HC) and light chains (LC) of IgG is indicated. TCL, total cell lysate. Molecular mass markers are shown on the left in kilodaltons.
UV CLIP.
3T3-L1 cells (pRetrosuper and Sam68sh cells) were treated with 4-thiouridine to a final concentration of 100 μM added directly to the cell culture medium 8 h prior to cross-linking. The cells were washed with ice-cold PBS and irradiated with 0.15 J/cm2 of 365-nm UV light at 4°C. The cells were collected by centrifugation at 514 × g for 1 min at 4°C. The cell pellets were resuspended in cross-linking and immunoprecipitation (CLIP) lysis buffer supplemented with protease inhibitors (Roche) and 0.5 U/μl RNasin (Promega) and sonicated twice with 10-s bursts (30). The lysates were mixed with 10 μl of a 1:250 dilution of RNase I (Life Technologies) and 2 μl of Turbo DNase (Life Technologies) with shaking at 37°C for 3 min. The lysates were then cleared and immunoprecipitated with 2 μg of anti-Sam68 or anti-SRSF1 antibody and control mouse/rabbit IgGs (Santa Cruz). Proteinase K buffer (containing 1.2 mg/ml proteinase K) was added to the immunoprecipitates and incubated for 20 min at 37°C. RNA was isolated through TRIzol reagent and subjected to RT-qPCR. The reverse primers listed below were used for the reverse transcription reaction. qPCR was performed with the following primers for Rps6kb1: intron 6 SBS, 5′-GAT TCA GGT CAT GAT TCA TG-3′ (forward) and 5′-CAG TGG GAA GGT GGT TTT GC-3′ (reverse); exon 6 (SRSF1 site), 5′-GAG GAG AAC TAT TTA TGC AG-3′ (forward) and 5′-GAA TAT TCC CTC TCT TTC TAA-3′ (reverse); exon 7, 5′-TTT ACT TGG CTG AAA TCT CC-3′ (forward) and 5′-CTT GGT GAT TAA GCA TGA TG-3′ (reverse).
RESULTS
Sam68 regulates the alternative splicing of Rps6kb1 in preadipocytes and mouse white adipose tissue (WAT).
Sam68-deficient preadipocytes are unable to differentiate into mature adipocytes (16). We reported that Sam68-deficient preadipocytes have decreased mTOR expression as they increase the production of a short mTORi5 isoform rather than synthesizing the full-length mTOR mRNA (16). The Sam68-deficient preadipocyte defect is partially rescued by the ectopic expression of the full-length mTOR expression, suggesting that there may be other splicing events regulated by Sam68 in the mTOR signaling pathway. To identify these alternative splicing events that contribute to the differentiation defects of Sam68-deficient preadipocytes, we monitored the presence of spliced isoforms in the mTOR signaling pathway. Using the Ensembl database, we identified the existence of spliced isoforms for the murine Rps6kb1, TSC1, Rheb, Akt1, and Deptor genes but not for IRS1, TSC2, 4EBP1, or eIF4E. Among the candidate isoforms tested, we observed that the mRNA levels of isoform Rps6kb1-002 were dramatically increased in Sam68-deficient cells (Sam68sh) (Fig. 1A) compared to levels in control pRetrosuper 3T3-L1 cells (Fig. 1B). We also noted a slight to moderate upregulation of isoforms TSC1-003, Rheb-003, Akt1-003, and Rps6kb1-005 in Sam68sh 3T3-L1 cells, and we did not observe significant fluctuations with the following isoforms in Sam68sh 3T3-L1 cells: TSC1-006, Rheb-002, and Deptor-002 (see Fig. S1 in the supplemental material).
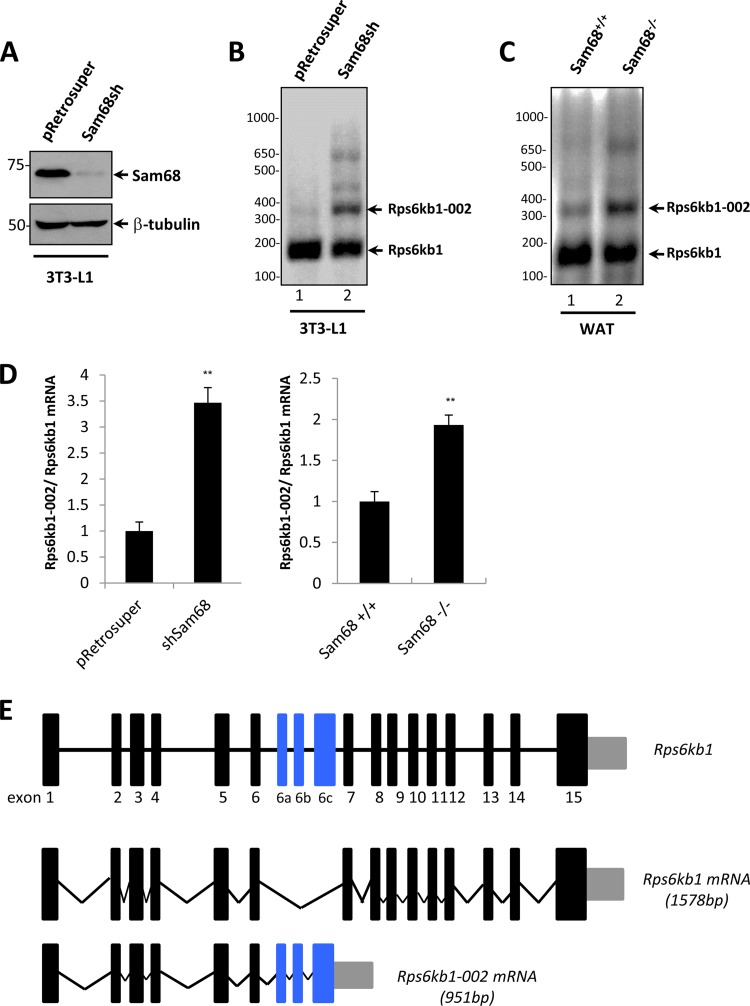
FIG 1.
Sam68 regulates the alternative splicing of Rps6kb1 in mouse preadipocytes and WAT. (A) Mouse 3T3-L1 preadipocytes stably transfected with pRetrosuper or Sam68sh pRetrosuper were lysed and immunoblotted with anti-Sam68 and anti-β-tubulin antibodies. The molecular mass markers are shown on the left in kilodaltons. (B and C) Total RNA from undifferentiated pRetrosuper and Sam68sh 3T3-L1 cells and mouse WAT was isolated and analyzed using a three-primer RT-PCR strategy with a common forward primer in exon 5 and reverse primers in exons 6 and 6b. The DNA markers are shown on the left in base pairs. (D) Total RNA from pRetrosuper and Sam68sh 3T3-L1 cells and mouse WAT was isolated and subjected to RT-qPCR. The presence of Rps6kb1-002 is expressed as a ratio of total Rps6kb1 transcripts. Error bars represent ± standard deviations of the means (**, P < 0.01). (E) Schematic representation of Rps6kb1 gene, the wild-type isoform Rps6kb1, and alternative spliced isoform Rps6kb1-002. Constitutive exons are shown as black boxes, and alternative exons are shown in blue. Introns are shown as horizontal lines, and splicing events are indicated by angled lines. The 3′ UTRs are shown as gray boxes.
We next examined the levels of isoform Rps6kb1-002 in white adipose tissue (WAT) of wild-type and Sam68 null mice. The level of Rps6kb1-002 was more abundant in white adipose tissue of Sam68 deficient mice than in the tissue of the littermate control mice (Fig. 1C). The increase in the mRNA ratio of Rps6kb1-002 to Rps6kb1 was also confirmed by RT-qPCR in Sam68-deficient preadipocytes and WAT isolated from Sam68 null mice (Fig. 1D). Thus, the loss of Sam68 promotes the production of splicing variant Rps6kb1-002.
Sam68 deficiency increases the expression of p31S6K1.
Rps6kb1 encodes p85/p70 S6K1, and the inclusion of alternative exons 6a, 6b, and 6c leads to the generation of the Rps6kb1-002 isoform (Fig. 1E). The Rps6kb1 transcript generates two proteins due to alternative mRNA translation start sites resulting in p70S6K1 and p85S6K1, whereas the shorter Rps6kb1-002 isoform harbors only the first six exons with alternative exons 6a, 6b, and 6c; and its alternative splicing was shown to be positively regulated by SRSF1 (31). The presence of a stop codon in exon 6c generates a truncated protein of 31 kDa, termed p31 or p31S6K1, that expresses the S6K1 N-terminal domain followed by a truncated kinase domain. The increase of Rps6kb1-002 mRNA was reflected at the protein level since we observed the presence of p31S6K1, as well as p70S6K1, in Sam68-deficient preadipocytes by immunoblotting with an N-terminal S6K1 antibody (BD Transduction Lab, Inc.) (Fig. 2A). We observed similar results in mouse WAT isolated from Sam68−/− mice using a different anti-S6K1 antibody (Cell Signaling, Inc.) that detects p31S6K1 in addition to p70S6K1 and p85S6K1 (Fig. 2B). As SRSF1 is a known regulator of p31S6K1 (31) and as Sam68 has been shown to regulate the alternative splicing of Srsf1 associated with nonsense-mediated decay (9), we performed immunoblotting to examine SRSF1 levels in the absence of Sam68. The depletion of Sam68 in preadipocytes or Sam68-deficient WAT did not affect the SRSF1 protein levels (Fig. 2A and B). These findings show that Sam68-deficient preadipocytes have increased p31S6K1 expression using two different N-terminal S6K1 antibodies with little to no effect on the global expression of p70S6K1, p85S6K1, and SRSF1.
FIG 2.
Sam68 regulates the expression of p31S6K1 but not that of SRSF1 in preadipocytes and WAT. Protein extracts from pRetrosuper and Sam68sh 3T3-L1 cells or from mouse WAT were immunoblotted with anti-S6K1 antibodies from BD Transduction Labs (A) or Cell Signaling, Inc. (B). Anti-Sam68 and anti-β-tubulin antibodies were used to monitor the levels of Sam68 and β-tubulin, respectively. Molecular mass markers are shown on the left in kilodaltons.
Sam68 binds an RNA element within intron 6 that diminishes SRSF1 binding to Rps6kb1 exon 6.
Sam68 binds RNA with U(U/A)AA motifs with high affinity (3, 4). Sam68 binding sites (SBSs) often reside near splice sites within pre-mRNAs, acting either as splice enhancers or silencers to regulate neighboring splice site usage (1). Since the absence of Sam68 increases the splicing of Rps6kb1-002, we searched intron 6 for repeats of the U(U/A)AA motif. The Rps6kb1 gene sequence is shown in Fig. 3A with the Rps6kb1-002 alternative exons in blue. We identified a putative Sam68 binding site within Rps6kb1 intron 6 with an encoded sequence of 5′-UAAUUAAA-3′, termed the SBS, 68 nucleotides downstream of a putative SRSF1 binding site in exon 6 (Fig. 3A). To determine whether Sam68 binds the SBS sequence, we synthesized a biotinylated RNA of the SBS, as well as a biotinylated control RNA (SBSmut) that has the 5′-UAAUUAAA-3′ sequence replaced with 5′-UAAUUCCA-3′ (substitutions are underlined) (Fig. 3B). The RNA oligonucleotides harboring either a wild-type or mutated SBS were used to perform affinity pulldown assays. Cell lysates from undifferentiated wild-type 3T3-L1 cells were incubated with the biotinylated RNAs, and complexes were purified with streptavidin-Sepharose beads. The bound proteins were separated by SDS-PAGE and immunoblotted for Sam68. Wild-type SBS bound Sam68 with a much higher affinity than the mutated SBS, as assessed by increasing the concentration of salt in the wash buffer (Fig. 3B). These findings show that Sam68 associates in vitro with RNA sequences within intron 6 of the Rps6kb1 pre-mRNA.
FIG 3.
Sam68 associates in vitro with RNA elements in Rps6kb1 intron 6. (A) Sequence spanning mouse Rps6kb1 exon 6, intron 6, which contains the three alternative exons (6a, 6b, and 6c) and exon 7, taken from the Ensembl browser. Underlined sequences represent the Sam68 binding site (SBS) within intron 6 and the SRSF1 binding site in exon 6. (B) The sequences of intron 6 and SBS and of the mutated version (SBSmut) of the synthetic RNAs generated. Affinity pulldown assays were performed with biotinylated RNA and streptavidin beads using 3T3-L1 cells and immunoblotted with anti-Sam68 antibodies. TCL, total cell lysate. A molecular mass marker is shown on the left.
We next examined whether endogenous Sam68 bound the intron 6 SBS in vivo using UV cross-linking and immunoprecipitation (CLIP) with a dilution of RNase I (1:250) that digests RNAs into fragments of 50 to 300 nucleotides in length (30). Preadipocytes (pRetrosuper and Sam68sh cells) were prepared for CLIP, as described in Materials and Methods, and immunoprecipitated with either control immunoglobulin G (IgG), anti-Sam68 antibodies, or anti-SRSF1 antibodies. The putative binding sites were mapped by using the primers indicated in Fig. 4A. Anti-Sam68 immunoprecipitations compared to those with IgG were enriched ∼25-fold for the Rps6kb1 intron 6 region spanning the SBS but not for an RNA region spanning Rps6kb1 exon 7 in pRetrosuper 3T3-L1 cells (Fig. 4B). In contrast, there was no RNA enrichment detected in anti-Sam68 immunoprecipitations in Sam68sh 3T3-L1 cells, as expected (Fig. 4B). These findings suggest that Sam68 associates in vivo with the SBS site of the Rps6kb1 intron 6. CLIP with anti-SRSF1 antibodies revealed a modest 2-fold enrichment of the Rps6kb1 exon 6 fragment encompassing the putative SRSF1 binding site over levels in control in pRetrosuper 3T3-L1 cells (Fig. 4C). Interestingly, cells depleted of Sam68 contained an ∼6-fold increase in SRSF1 at this site (Fig. 4C), suggesting that Sam68 occupancy at the SBS prevents SRSF1 binding to exon 6. Taken together, these data suggest that Sam68 directly associates with the SBS and that the presence of Sam68 influences SRSF1 binding to exon 6.
Sam68 and SRSF1 are known to interact as endogenous Sam68 immunoprecipitations contain SRSF1, as detected by mass spectrometry analysis (32). To confirm the interaction, HEK293 cells were cotransfected with GFP-Sam68 and GFP-SRSF1. The cells were lysed and immunoprecipitated with control IgG or anti-SRSF1 antibodies. The bound proteins were separated by SDS-PAGE and immunoblotted with anti-Sam68 antibodies. Both GFP-Sam68 and endogenous Sam68 coimmunoprecipitated with SRSF1 but not with control IgG (Fig. 4D, upper panel). Immunoblotting with anti-SRSF1 antibodies confirmed that GFP-SRSF1 and endogenous SRSF1 were immunoprecipitated (Fig. 4D, lower panel). These data confirm that Sam68 interacts with SRSF1.
Minigene assays indicate that Sam68 suppresses the alternative splicing of Rps6kb1-002.
A splicing minigene was constructed with a cytomegalovirus (CMV) promoter driving the expression of a 1.6-kb genomic fragment encompassing Rps6kb1 exon 6, intron 6, and exon 7 (Fig. 5A). The minigene transcription start site was located in the plasmid upstream of Rps6kb1 exon 6, followed by the sequence of a FLAG epitope tag. A forward primer complementary to the FLAG cDNA sequence (F1) and reverse primers in exons 6c (R1), exon 6a (R3), and 7 (R2 and R4) recognize fragments corresponding to Rps6kb1-002 and Rps6kb1, respectively. pRetrosuper and Sam68sh 3T3-L1 cells were transfected with either pcDNA3.1, the wild-type Rps6kb1, or the SBS mutated (SBSmut) minigene. Total RNA was isolated 48 h after transfection, treated with RQ1 DNase, and monitored for Rps6kb1-002 and Rps6kb1 transcripts using three-primer RT-PCR (Fig. 5B) and RT-qPCR (Fig. 5C). There was little expression of the Rps6kb1-002 fragment from the wild-type minigene in pRetrosuper cells (Fig. 5B, lane 2); however, the presence of the Rps6kb1-002 transcript increased (∼6- to 8-fold) when the SBS was mutated or when Sam68 was ablated in the cells (Fig. 5B, lanes 3 and 5, and C). Interestingly, the deletion of the SBS in addition to the ablation of Sam68 in 3T3-L1 cells led to an ∼40-fold increase in the Rps6kb1-002 transcript (Fig. 5B, lane 6, and C). These findings show that Sam68 and the SBS negatively regulate Rps6kb1-002.
FIG 5.
Rps6kb1 minigene assay defines intron 6 as the minimal requirement. (A) Mouse genomic fragment encompassing Rps6kb1 exons 6 and 7 was cloned in pcDNA3.1 with an N-terminal FLAG epitope tag. The SBS is shown in the proximity of the 5′ splice site within intron 6. The same mouse genomic fragment containing CC replacing AA in the Sam68 binding site was also constructed in pcDNA3.1 with an N-terminal FLAG epitope tag. The splicing of the wild-type Rps6kb1 and SBSmut Rps6kb1 minigenes leads to two different transcripts. Arrows indicate the primers used in the minigene assay. (B and C) pRetrosuper and Sam68sh 3T3-L1 cells were transfected with either pcDNA3.1 alone, Rps6kb1 (WT), or the Rps6kb1 (SBSmut) minigene. Total RNA was isolated and digested with RQ1 DNase, and the levels of Rps6kb1-002 and Rps6kb1 transcripts were assessed by semiquantitative RT-PCR using primer F1 with R1 and R2 (B) or by RT-qPCR using F1 with R3 and R4 (C). In panel B the mRNA levels of Sam68 and GAPDH were also assessed. Error bars represent ± standard deviations of the means (**, P < 0.01). (D) HEK293 cells were cotransfected with either pcDNA3.1 or GFP-Sam68 alone with the Rps6kb1 minigene or Rps6kb1 minigene SBSmut. The cells were harvested after 48 h, and the cellular extracts were immunoblotted with anti-GFP, anti-Sam68, and anti-β-actin antibodies. The asterisk denotes a nonspecific protein recognized with anti-GFP antibodies. (E) The total RNA from the cells indicated in panel D was isolated and treated with RQ1 DNase, and the mRNA levels of Rps6kb1 and Rps6kb1-002 were assessed by RT-qPCR. The mRNA expression level of Rps6kb1-002 was normalized to Rps6kb1 levels. Error bars represent ± standard deviations of the means (*, P < 0.05; n.s., not significant).
We next overexpressed Sam68 to examine its influence on the wild-type and SBSmut Rps6kb1 minigenes. The expression of GFP-Sam68 in HEK293 cells was confirmed by immunoblotting (Fig. 5D). Mutation of the SBS led to an increase in Rps6kb1-002 in control pcDNA3.1-transfected cells (Fig. 5E). However, the expression of GFP-Sam68 completely quenched the expression of Rps6kb1-002 production from both minigenes (Fig. 5E). These findings suggest that Sam68 is a potent repressor of Rps6kb1-002 and also has SBS-independent functions.
Sam68 counteracts the positive effects of SRSF1 for Rps6kb1-002 expression.
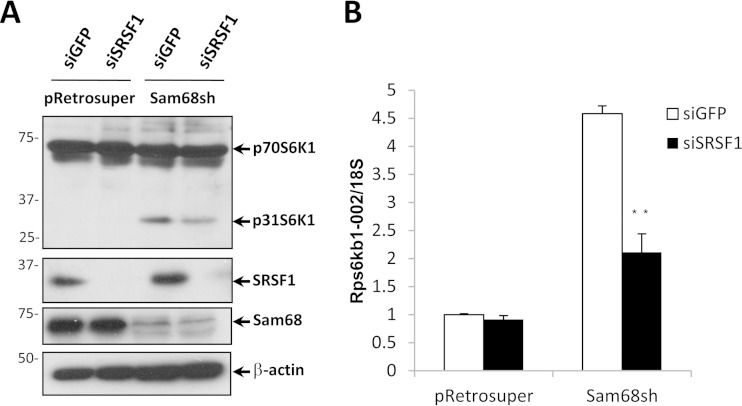
We next confirmed that SRSF1 was responsible for regulating the p31S6K1 levels in Sam68sh 3T3-L1 cells. Depletion of SRSF1 using siRNA reduced the levels of p31S6K1 protein (Fig. 6A) and decreased Rps6kb1-002 mRNA in Sam68sh but not pRetrosuper 3T3-L1 cells, as assessed by RT-qPCR (Fig. 6B). These findings confirm that SRSF1 is required for the production of Rps6kb1-002 in adipocytes.
FIG 6.
The presence of the Rps6kb1-002 isoform in Sam68-depleted preadipocytes requires SRSF1. (A) pRetrosuper or Sam68sh 3T3-L1 cells were transfected with siGFP or siSRSF1. The protein extracts were prepared 48 h after and immunoblotted with the indicated antibodies. β-Actin is shown as the loading control. Molecular mass markers are shown on the left in kilodaltons. (B) The total RNA was isolated from pRetrosuper or Sam68sh 3T3-L1 cells transfected with siGFP or siSRSF1. The mRNA levels of Rps6kb1-002 were assessed by RT-qPCR and normalized to the level of 18S rRNA. Error bars represent ± standard deviations of the means (**, P < 0.01).
To examine the influence of SRSF1 and Sam68 on the alternative splicing of Rps6kb1, we transfected the Rps6kb1 minigenes in HEK293 cells. SRSF1 was efficiently depleted using siRNAs in these cells (Fig. 7A). The presence of the Rps6kb1-002 isoform was increased with the mutation of the SBS in siGFP-transfected but not siSRSF1-transfected HEK293 cells (Fig. 7B). These findings suggest that SRSF1 is a positive regulator of Rps6kb1-002. We next cotransfected GFP-SRSF1 and GFP-Sam68 in HEK293 cells and assayed for the presence of the Rps6kb1-002 transcript generated from the wild-type Rps6kb1 minigene (Fig. 7C and D). GFP-SRSF1 increased the appearance of Rps6kb1-002 in a dose-dependent manner (Fig. 7D), and this increase was attenuated with the overexpression of GFP-Sam68 (Fig. 7D, lanes 5 to 8). These data show that Sam68 counteracts the positive effects of SRSF1 in regulating the production of Rps6kb1-002.
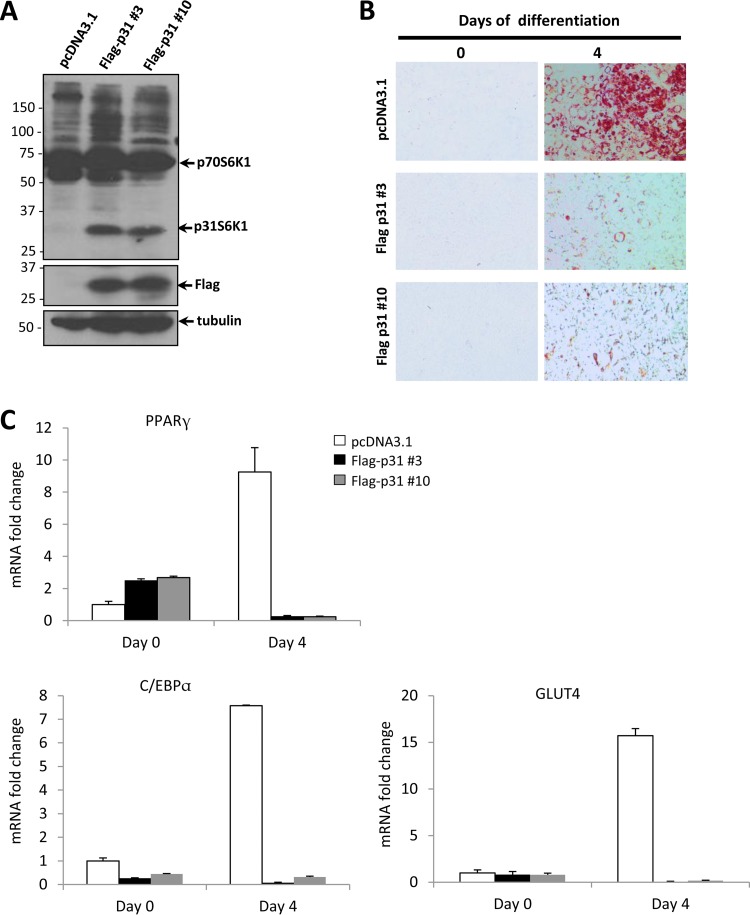
The ectopic expression of p31S6K1 suppresses adipogenesis.
The direct role of Rps6kb1-002 in adipogenesis was investigated. Preadipocytes were stably transfected with pcDNA3.1 (control) or an expression vector encoding FLAG-p31S6K1. Two stable clones (FLAG-p31 clones 3 and 10) ectopically expressing p31S6K1 were selected (Fig. 8A), and their expression was comparable to levels observed in Sam68-deficient cells (data not shown). p31S6K1-expressing cells were monitored for lipid accumulation following differentiation for 4 days. We observed a notable decrease of lipid accumulation in both p31S6K1-overexpressing 3T3-L1 cell lines compared to levels in control cells, as visualized by Oil Red O staining (Fig. 8B). We subsequently examined the expression pattern of adipose-specific markers at differentiation day 0 and day 4 (Fig. 8C). The mRNA levels of PPARγ, C/EBPα, and GLUT4 were increased dramatically in control cells (pcDNA3.1) upon differentiation, as expected, while in clones 3 and 10, the expression of these markers was largely absent after differentiation day 4 (Fig. 8C). These results show that p31S6K1 is a repressor of adipogenesis.
FIG 8.
Ectopic expression of p31S6K1 suppresses adipogenesis. (A) 3T3-L1 cells were stably transfected with pcDNA3.1 or FLAG-p31S6K1. A polyclonal population of pcDNA3.1 and two individual clones (FLAG-p31 3 and FLAG-p31 10) was selected for analysis by immunoblotting using the indicated antibodies. β-Tubulin is shown as the loading control. Molecular mass markers are shown on the left in kilodaltons. (B) The cells indicated in panel A were induced to differentiate for 4 days. Adipocyte differentiation was assessed by Oil Red O staining. (C) The mRNA levels of PPARγ, C/EBPα, and GLUT4 normalized to 18S rRNA were assessed by RT-qPCR. The data are expressed as relative values from differentiation days 0 and 4. Error bars represent ± standard deviations of the means.
Depletion of p31S6K1 in Sam68-deficient preadipocytes partially rescues the adipogenesis defect.
We examined whether the elevated level of p31S6K1 is a contributing factor for the adipogenesis defect of Sam68-deficient 3T3-L1 cells. To test this possibility, we decreased the expression of p31S6K1 using siRNA specific to this isoform. pRetrosuper and Sam68sh 3T3-L1 cells were transfected with either siGFP or two different p31S6K1 siRNAs, designated sip31-A and sip31-B. The elevated expression of p31S6K1 in Sam68sh 3T3-L1 cells was depleted in sip31-A- and sip31-B-transfected cells (Fig. 9A). Sam68sh 3T3-L1 cells exhibited reduced adipogenesis compared to levels in pRetrosuper cells, as previously reported (16), and the presence of transfected siGFP had no influence on adipogenesis as visualized by Oil Red O staining (Fig. 9B). The transfection of p31S6K1 siRNA increased lipid accumulation in Sam68-deficient cells, suggesting that the loss of p31S6K1 expression partially rescues the adipogenesis defect observed in Sam68sh cells (Fig. 9B, compare staining of siGFP with that of sip31-A and sip31-B). We also examined the expression pattern of adipose-specific markers at differentiation days 0 and 4 of 3T3-L1 cells. The mRNA levels of PPARγ, C/EBPα, and GLUT4 were increased in pRetrosuper cells upon differentiation, as expected (Fig. 9C). Strikingly, the attenuated expression of differentiation markers (PPARγ, C/EBPα, and GLUT4) in Sam68sh cells was partially derepressed with the depletion of p31S6K1 (Fig. 9C). These results indicate that expression of p31S6K1 represses adipogenesis and is a contributing factor for the observed defects in Sam68-deficient preadipocytes.
FIG 9.
The expression of p31S6K1 contributes to the adipogenesis defects of Sam68-deficient mouse preadipocytes. (A) pRetrosuper or Sam68sh 3T3-L1 cells were transfected with siGFP (control), sip31-A, and sip31-B. The protein extracts were immunoblotted with the indicated antibodies. β-Tubulin is shown as the loading control. Molecular mass markers are shown on the left in kilodaltons. (B) The cells indicated in panel A were induced to differentiate for 4 days. Adipocyte differentiation was assessed by Oil Red O staining. (C) The mRNA levels of PPARγ, C/EBPα, and GLUT4 normalized to GAPDH were assessed by RT-qPCR. The data are expressed as relative values from differentiation days 0 and 4. Error bars represent ± standard deviations of the means (*, P < 0.05).
To answer the question of whether there was a specific change in the levels of the Rps6kb1-002 isoform or just a general change in the expression levels of the whole gene (including both isoforms) that contributes to adipogenesis, we abrogated total S6K1 expression using siRNAs. pRetrosuper 3T3-L1 cells were transfected with either siGFP or siRNA targeting both p70/p31 S6K1 isoforms (Fig. 10A). Deletion of total S6K1 in pRetrosuper 3T3-L1 cells had no influence on adipogenesis as cells differentiated normally (Fig. 10B) and expressed high levels of PPARγ, C/EBPα, and GLUT4 at day 4 of differentiation (Fig. 10C). Since p31S6K1 is absent in pRetrosuper cells, these findings demonstrate that deletion of p70S6K1 alone does not affect 3T3-L1 differentiation, as reported previously (27). Next, we examined whether Sam68sh 3T3-L1 cells were partially rescued with siRNAs targeting p70/p31S6K1, and indeed this was the case (Fig. 10B and C). The partial derepression we observed was similar to decreasing the levels of p31S6K1 alone (compare Fig. 9 and 10). These observations indicate that p31S6K1 inhibits adipogenesis independent of the p70S6K1 isoform.
FIG 10.
p31S6K1 contributes to the adipogenesis defects independently of the p70S6K1 isoform. (A) pRetrosuper or Sam68sh 3T3-L1 cells were transfected with siGFP (control) and sip70/p31. The protein extracts were immunoblotted with the indicated antibodies. β-Actin is shown as the loading control. Molecular mass markers are shown on the left in kilodaltons. (B) The cells indicated in panel A were induced to differentiate for 4 days. Adipocyte differentiation was assessed by Oil Red O staining. (C) The mRNA levels of PPARγ, C/EBPα, and GLUT4 normalized to 18S rRNA were assessed by RT-qPCR. The data are expressed as relative values from differentiation days 0 and 4. Error bars represent ± standard deviations of the means (*, P < 0.05).
DISCUSSION
Alternative splicing leads to the generation of key isoforms required for cellular differentiation and proliferation (33). In the present manuscript, we report that the Sam68 RNA binding protein exerts a suppressive effect on the alternative splicing of the ribosomal S6 kinase gene (Rps6kb1) in adipocytes. Consequently, Sam68-depleted 3T3-L1 preadipocytes harbor elevated levels of the short isoform 2 of S6K1 (Rps6kb1-002) and its encoded protein p31S6K1. Mechanistically, Sam68 binds an RNA element (Sam68 binding site, SBS) in intron 6 near the 5′ splice site and prevents the usage of alternative exons 6a, 6b, and 6c that generate Rps6kb1-002 by counteracting the positive effects of the serine/arginine-rich splicing factor 1 (SRSF1) that binds within exon 6. The ectopic expression of p31S6K1 in wild-type preadipocytes inhibited adipogenesis, and the depletion of p31S6K1 using two separate siRNAs partially restored the adipogenesis defects in Sam68-deficient preadipocytes. These findings demonstrate that the expression of Sam68 in adipocytes is required to prevent the expression of the short isoform 2 of S6K1, a potent suppressor of adipogenesis.
We identified an A/U-rich intronic sequence bound by Sam68, 46 nucleotides downstream of the 5′ splice site of Rps6kb1 exon 6. Deletion of this element promoted the skipping of Rps6kb1 exons 6a, 6b, and 6c, thus preventing the expression of Rps6kb1-002. Sam68 is an established regulator of alternative splicing, and it is known to function by directly associating with A/U-rich elements near 5′ splice sites (5, 10, 12, 16). The Rps6kb1 intron 6 UAAUUAAA bipartite sequence is recognized by Sam68 with relatively high affinity. Mutation of the SBS to UAAUUCCA (mutated residues underlined) diminished Sam68 association with this RNA sequence. By performing CLIP assays, we confirmed that Sam68 localizes directly at the SBS in vivo as anti-Sam68 immunoprecipitations enriched the SBS RNA sequence 25-fold over levels of control immunoprecipitations (Fig. 4B). The presence of Sam68 suppressed the positive effects of SRSF1 on the production of Rps6kb1-002, as measured using a minigene. SRSF1 is a known positive regulator of Rps6kb1-002 (31), but its Rps6kb1 binding site(s) and how it regulates the production of Rps6kb1-002 remained unknown. We show that SRSF1 displayed reduced binding to its Rps6kb1 exon 6 GAAAGAGAGGGAA site in the presence of Sam68 by CLIP assays. Using the Rps6kb1 minigene, mutation of the SBS increased Rps6kb1-002 production, and the increase was observed in the presence or absence of Sam68 (knockdown cells), suggesting that the SBS also has Sam68-independent functions. It is possible that the residual Sam68 levels in the knockdown cells play this role or that the SBS is scavenged by other A/U-rich RNA binding repressors in the absence of Sam68. Alternatively, the SBS may regulate local RNA secondary structure that influences Rps6kb1-002 alternative splicing by SRSF1.
Several possible mechanisms have been proposed to explain how RNA binding proteins suppress neighboring SR proteins (33–35). Sam68 could compete directly with SRSF1 for the splicing machinery for intron 6 definition. Sam68 may also alter the neighboring RNA secondary structure and/or the rate of transcription, as proposed by Batsche and coworkers (36). The rate of transcription may influence the binding of SRSF1, thus influencing exon selection (33–35).
We show that Rps6kb1-002 and its encoded protein, p31S6K1, are present in Sam68-depleted preadipocytes and mouse white adipose tissue of Sam68 null mice. Sam68 protein expression increases during adipogenesis (16), and its role may be, in part, to ensure the suppression of Rps6kb1-002. Using an anti-S6K1 antibody that recognizes all the isoforms sharing the common N terminus, we observed that Sam68 deficiency leads to increased p31S6K1 expression, without the apparent reduction in p70S6K1 and p85S6K1 expression. The latter is probably due to the fact that p70/p85 S6K1 are considerably more abundant than p31S6K1 (31). The cellular role of p31S6K1 is unknown; however, it does have oncogenic properties. The expression of p31S6K1 is sufficient to induce transformation in NIH 3T3 cells (31). Unlike mice, which express only one short isoform, humans generate two short isoforms (h6A and h6C) of S6K1, and their expression is elevated in breast cancer cell lines (37). Depletion of these isoforms in breast cancer cell lines decreases their proliferation (37). These short isoforms lack kinase activity because their kinase domains are truncated; however, they retain the ability to bind mTORC1 as they contain the Raptor binding motif (38). p31S6K1 has been shown to associate with mTOR and increase its activity (37); however, this activity in Sam68-deficient preadipocytes is difficult to assess since these cells have reduced mTOR levels (16). Indeed, Sam68-deficient preadipocytes exhibit decreased phosphorylation of rpS6 and AKT during adipogenesis (16). p31S6K1 has also been shown to be nuclear, unlike p85S6K1 and p70S6K1 (39); therefore, it may also fulfill other functions.
S6K1−/− mice have decreased adipose tissue mass, increased energy expenditure, and are resistant to dietary-induced obesity (40). S6K1 participates in the upregulation of transcription factors during the commitment phase of adipogenesis (27). Adipocytes normally express p70/p85S6K1 but not p31S6K1 (Fig. 2). The expression of p31S6K1 in 3T3-L1 cells prevented adipogenesis. Depletion of both p70S6K1 and p31S6K1 rescued the adipogenesis defects of Sam68-deficient cells to a similar extent as depletion of p31S6K1 alone, indicating that the negative effects of p31S6K1 in adipogenesis are independent of p70S6K1. Therefore, p31S6K1 contributes to the Sam68 deficiency-induced adipogenesis defects observed.
Sam68 likely regulates many alternative spliced events that contribute to the observed lean phenotype of Sam68-deficient mice (16). Sam68 regulates the splicing of Bcl-x (8), as well as mTOR, tripeptidyl peptidase II (Tpp2), and Tubby (Tub) (16). Sam68 has also been shown to regulate the alternative splicing of the Srsf1 transcript in colon cancer cells to influence the epithelial-to-mesenchymal transition (9). Indeed, we also detected an increase in the Srsf1 NMD transcript in 3T3-L1 cells (data not shown), but this did not affect SRSF1 protein levels.
In conclusion, we show that the alternative splicing of the mouse Rps6kb1 gene is negatively regulated by Sam68 as it antagonizes the positive effects of SRSF1. We also show that the short isoform of Rps6kb1, namely, p31S6K1, is a potent repressor of adipogenesis, and its presence in Sam68-deficient preadipocytes contributes to the adipogenesis defects observed in these cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhenbao Yu and Gillian Vogel for helpful discussions and for critically reading the manuscript.
The work was supported by a grant from the Canadian Institute of Health Canada (MOP-123531) to S.R.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01488-14.
REFERENCES
- 1.Bielli P, Busà R, Paronetto MP, Sette C. 2011. The RNA-binding protein Sam68 is a multifunctional player in human cancer. Endocr Relat Cancer 18:R91-102. doi: 10.1530/ERC-11-0041. [DOI] [PubMed] [Google Scholar]
- 2.Richard S. 2010. Reaching for the stars: linking RNA binding proteins to diseases. Adv Exp Med Biol 693:142–157. doi: 10.1007/978-1-4419-7005-3_10. [DOI] [PubMed] [Google Scholar]
- 3.Galarneau A, Richard S. 2009. The STAR RNA binding proteins GLD-1, QKI, SAM68 and SLM-2 bind bipartite RNA motifs. BMC Mol Biol 10:47. doi: 10.1186/1471-2199-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Q, Taylor SJ, Shalloway D. 1997. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J Biol Chem 272:27274–27280. [DOI] [PubMed] [Google Scholar]
- 5.Matter N, Herrlich P, Konig H. 2002. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 6.Cappellari M, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Saarikettu J, Silvennoinen O, Sette C. 2014. The transcriptional co-activator SND1 is a novel regulator of alternative splicing in prostate cancer cells. Oncogene 33:3794–3802. doi: 10.1038/onc.2013.360. [DOI] [PubMed] [Google Scholar]
- 7.Bielli P, Busà R, Di Stasi SM, Munoz MJ, Botti F, Kornblihtt AR, Sette C. 2014. The transcription factor FBI-1 inhibits SAM68-mediated BCL-X alternative splicing and apoptosis. EMBO Rep 15:419–427. doi: 10.1002/embr.201338241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. 2007. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol 176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valacca C, Bonomi S, Buratti E, Pedrotti S, Baralle FE, Sette C, Ghigna C, Biamonti G. 2010. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J Cell Biol 191:87–99. doi: 10.1083/jcb.201001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chawla G, Lin CH, Han A, Shiue L, Ares MJ, Black DL. 2009. Sam68 regulates a set of alternatively spliced exons during neurogenesis. Mol Cell Biol 29:201–213. doi: 10.1128/MCB.01349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iijima T, Wu K, Witte H, Hanno-Iijima Y, Glatter T, Richard S, Scheiffele P. 2011. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell 147:1601–1614. doi: 10.1016/j.cell.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedrotti S, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Stamm S, Manley JL, Sette C. 2010. The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy. EMBO J 29:1235–1247. doi: 10.1038/emboj.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, Hagerman PJ, Charlet-Berguerand N. 2010. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J 29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paronetto MP, Messina V, Barchi M, Geremia R, Richard S, Sette C. 2011. Sam68 marks the transcriptionally active stages of spermatogenesis and modulates alternative splicing in male germ cells. Nucleic Acids Res 39:4961–4974. doi: 10.1093/nar/gkr085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paronetto MP, Messina V, Bianchi E, Barchi M, Vogel G, Moretti C, Palombi F, Stefanini M, Geremia R, Richard S, Sette C. 2009. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J Cell Biol 185:235–249. doi: 10.1083/jcb.200811138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huot ME, Vogel G, Zabarauskas A, Ngo CT, Coulombe-Huntington J, Majewski J, Richard S. 2012. The Sam68 STAR RNA-binding protein regulates mTOR alternative splicing during adipogenesis. Mol Cell 46:187–199. doi: 10.1016/j.molcel.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dann SG, Selvaraj A, Thomas G. 2007. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med 13:252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Shimobayashi M, Hall M. 2014. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 15:155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- 20.Inoki K, Zhu T, Guan KL. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115:577–590. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 21.Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol 5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 22.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. 2003. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 13:1259–1268. doi: 10.1016/S0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Harris TE, Roth RA, Lawrence JCJ. 2007. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem 282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 24.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. 2007. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol 9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 25.Thedieck K, Polak P, Kim ML, Molle KD, Cohen A, Jenö P, Arrieumerlou C, Hall M. 2007. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS One 2:e1217. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carnevalli LS, Masuda K, Frigerio F, Le Bacquer O, Um SH, Gandin V, Topisirovic I, Sonenberg N, Thomas G, Kozma SC. 2010. S6K1 plays a critical role in early adipocyte differentiation. Dev Cell 18:763–774. doi: 10.1016/j.devcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen T, Boisvert FM, Bazett-Jones DP, Richard S. 1999. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Biol Cell 10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y, Ma YC, Huang J, Chen KY, McGarrigle DK, Huang XY. 2005. Requirement of SRC-family tyrosine kinases in fat accumulation. Biochemistry 44:14455–14462. doi: 10.1021/bi0509090. [DOI] [PubMed] [Google Scholar]
- 30.Huppertz I, Attig J, D'Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, König J, Ule J. 2014. iCLIP: protein-RNA interactions at nucleotide resolution. Methods 65:274–287. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. 2007. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol 14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huot ME, Vogel G, Richard S. 2009. Identification of a Sam68 ribonucleoprotein complex regulated by epidermal growth factor. J Biol Chem 284:31903–31913. doi: 10.1074/jbc.M109.018465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu XD, Ares MJ. 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Witten JT, Ule J. 2011. Understanding splicing regulation through RNA splicing maps. Trends Genet 27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das S, Krainer AR. 2014. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res 12:1195–1204. doi: 10.1158/1541-7786.MCR-14-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Batsche E, Yaniv M, Muchardt C. 2006. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol 13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 37.Ben-Hur V, Denichenko P, Siegfried Z, Maimon A, Krainer A, Davidson B, Karni R. 2013. S6K1 alternative splicing modulates its oncogenic activity and regulates mTORC1. Cell Rep 3:103–115. doi: 10.1016/j.celrep.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schalm SS, Blenis J. 2002. Identification of a conserved motif required for mTOR signaling. Curr Biol 12:632–639. doi: 10.1016/S0960-9822(02)00762-5. [DOI] [PubMed] [Google Scholar]
- 39.Rosner M, Hengstschläger M. 2011. Nucleocytoplasmic localization of p70 S6K1, but not of its isoforms p85 and p31, is regulated by TSC2/mTOR. Oncogene 30:4509–4522. doi: 10.1038/onc.2011.165. [DOI] [PubMed] [Google Scholar]
- 40.Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, Thomas G. 2004. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.