Abstract
Burkholderia cenocepacia is an emerging opportunistic pathogen causing life-threatening infections in immunocompromised individuals and in patients with cystic fibrosis, which are often difficult, if not impossible, to treat. Understanding the genetic basis of virulence in this emerging pathogen is important for the development of novel treatment regimes. Generation of deletion mutations in genes predicted to encode virulence determinants is fundamental to investigating the mechanisms of pathogenesis. However, there is a lack of appropriate selectable and counterselectable markers for use in B. cenocepacia, making its genetic manipulation problematic. Here we describe a Gateway-compatible allelic exchange system based on the counterselectable pheS gene and the I-SceI homing endonuclease. This system provides efficiency in cloning homology regions of target genes and allows the generation of precise and unmarked gene deletions in B. cenocepacia. As a proof of concept, we demonstrate its utility by deleting the Bcam1349 gene, encoding a cyclic di-GMP (c-di-GMP)-responsive regulator protein important for biofilm formation.
INTRODUCTION
Burkholderia cenocepacia is a member of a group of closely related Gram-negative bacteria referred to as the Burkholderia cepacia complex (Bcc). The Bcc contains at least 18 different species that thrive in diverse ecological niches, including clinical, industrial, and natural environments. These bacteria possess very large genomes separated into multiple replicons and hence are considered one of the most versatile groups of Gram-negative bacteria (1, 2). Some Bcc species have biotechnological potential for use in processes such as the enhancement of plant growth or breakdown of pollutants, while others are opportunistic pathogens causing life-threatening infections in immunocompromised individuals and in patients with cystic fibrosis (CF) (3). Although all members of the Bcc have been isolated from CF patients, B. cenocepacia accounts for the majority of these isolates, comprising the most virulent and transmissible strains associated with a poor clinical course and a high mortality rate (4). Therefore, research on the virulence mechanisms of Bcc bacteria has focused largely on B. cenocepacia.
The genomes of several B. cenocepacia strains have recently been sequenced (5–7), enabling bioinformatics-based predictions of virulence determinants in this pathogen. Although a number of genes associated with virulence in B. cenocepacia have been identified (4, 8, 9) and tested in various infection models (10, 11), it seems likely that the list of genes implicated in virulence is far from complete and will expand with genetic tools becoming available to manipulate B. cenocepacia strains. The deletion of genes potentially associated with virulence is a powerful way to investigate their function in bacterial physiology and pathogenesis. Most of the virulence traits of B. cenocepacia, such as antibiotic resistance, motility, biofilm formation, cell invasion, and intracellular survival, are multifactorial, involving more than one gene; thus, multiple gene deletions may need to be generated in one strain to fully assess the genetic basis of a particular virulence trait. This requires an efficient method to generate gene deletions, which are preferably not marked with antibiotic resistance cassettes, as this would prevent the ability to mutate more than a single gene in one particular strain and moreover may cause polar effects on adjacent genes. During the past few years, a number of elegant systems have been developed for the generation of unmarked gene deletions in B. cenocepacia (12, 13) as well as in other Burkholderia species (14–16). In these systems, regions of homology containing a mutant allele of a target gene are cloned into a suicide vector. These vectors are then transferred into the bacterial host by conjugation. The integration of the plasmid into the chromosome by homologous recombination is selected by antibiotic resistance encoded by a gene on the plasmid, leading to the formation of merodiploids, which contain both the mutant and wild-type alleles of the target gene. The resolution of merodiploids by the excision of the integrated plasmid in a second homologous recombination event results in a population of cells of which a significant fraction contains the desired gene deletion. The latter step usually requires counterselection for the integrated plasmid since the second homologous recombination can be an exceptionally rare event.
Sucrose counterselection based on the sacB gene (15, 17) and an engineered counterselectable marker based on the Burkholderia pseudomallei pheS gene encoding the α-subunit of phenylalanyl tRNA synthase (14) have been used in some Burkholderia species. However, they appear to be inappropriate and leaky counterselectable markers for the generation of B. cenocepacia gene deletions in our laboratory. Another way to stimulate the second homologous recombination event and, consequently, the resolution of merodiploids is based on the yeast homing endonuclease I-SceI, which recognizes a specific 18-bp sequence (12, 15). After an allelic exchange vector carrying the I-SceI recognition site has integrated into the chromosome, a replicative second plasmid constitutively expressing the I-SceI enzyme is introduced into the merodiploid bacteria. The I-SceI enzyme creates a double-stranded DNA break at the I-SceI site within the integrated plasmid, which stimulates a second homologous recombination event by the host's DNA repair system. The excision of the integrated plasmid results in a population of cells carrying either the wild-type or the mutant allele, which can be identified by PCR and partial sequencing.
Another major limitation of allelic exchange vectors for Burkholderia species is their dependence on restriction and ligation enzymes for cloning. Restriction-free cloning based on Gateway recombineering technology (18) is an alternative method that can expedite the construction of gene replacement vectors containing mutant alleles.
Here we present a Gateway-compatible allelic exchange system for Burkholderia species that utilizes the I-SceI homing endonuclease and pheS-based counterselection. We further describe the application of this system for generating in-frame and unmarked gene deletions in B. cenocepacia H111. As a proof of concept, we describe the deletion and complementation of the Bcam1349 gene, which is a regulator of biofilm formation in B. cenocepacia H111. In addition, we also provide evidence that this system can be used to make gene deletions in Burkholderia thailandensis, indicating that it may be used in other Burkholderia species as well.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All B. cenocepacia and Escherichia coli strains were grown at 37°C. Luria broth (LB) medium was used for overnight batch cultivation of all bacteria unless otherwise stated. Solid media were prepared with 2% (wt/vol) agar. Eighty micrograms tetracycline (Tet) ml−1 (liquid medium), 120 μg μg Tet ml−1 (solid medium), 25 μg gentamicin sulfate (Gm) ml−1, 100 μg kanamycin sulfate (Km) ml−1, and 100 μg trimethoprim (Tp) ml−1 were used for B. cenocepacia strains, and 20 μg Tet ml−1, 10 μg Gm ml−1, 50 μg Km ml−1, 50 μg Tp ml−1, 100 μg ampicillin (Ap) ml−1, and 25 μg chloramphenicol (Cm) ml−1 were used for E. coli strains where appropriate. After conjugal transfer of plasmids into B. cenocepacia, AB agar medium (19) supplemented with 10 mmol liter−1 Na-citrate and appropriate antibiotics was used to select for B. cenocepacia transconjugants. For self-curing of plasmid pDAI-SceI-pheS, 0.1% (wt/vol) p-chlorophenylalanine (cPhe) (dl-4-chlorophenylalanine; Sigma-Aldrich) was autoclaved together with B-salts solution and A-salts solution (19), and the carbon source of choice was added thereafter.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Laboratory identification no. | Relevant characteristic(s) | Reference or source |
|---|---|---|---|
| Strains | |||
| B. cenocepacia H111 | MF108 | Clinical isolate from a cystic fibrosis patient | 7 |
| B. thailandensis CDC2721121 | Clinical isolate from a patient with pleural infection | 27 | |
| E. coli DH5α | TTN322 | Used for standard DNA manipulations | Invitrogen |
| E. coli DB3.1 | TTN312 | Host for the Gateway-compatible gene replacement vectors | Invitrogen |
| Plasmids | |||
| pBBR1MCS5 | MF528 | Broad-host-range cloning vector; Gmr | 22 |
| pBBR1MCS2 | MF124 | Broad-host-range cloning vector; Kmr | 22 |
| pMF564 | MF564 | Bcam1349 gene cloned into pBBR1MCS5 | This study |
| pYedQ | MF202 | E. coli yedQ (yhcK) gene cloned into pRK404A | 21 |
| pYedQ2 | MF217 | yedQ gene cloned into the HindIII/BamHI site in pBBR1MCS5 | This study |
| pRK600 | TTN365 | Helper plasmid in triparental conjugations; Cmr ori-ColE1 RK-mob+ RK-tra+ | 28 |
| pDONR221 | TTN313 | Source of Gateway donor site, Gateway donor vector; Kmr | Invitrogen |
| pBBR1MCS-Km-pheS | MF138 | Engineered pheS cloned into pBBR1MCS2; Kmr | 14 |
| pEX18Tp-pheS | MF322 | Gene replacement vector based on pheS and Tpr | 14 |
| pEX18Gm-pheS | MF320 | Gene replacement vector based on pheS and Gmr | 14 |
| pEX18Km-pheS | MF321 | Gene replacement vector based on pheS and Kmr | 14 |
| pUC57-pheS | MF130 | Cloning vector containing engineered pheS; Apr | 14 |
| pDAI-SceI | MF339 | Cloning vector containing the I-SceI endonuclease; Tetr | 12 |
| pDONRPEX18Tp-SceI-pheS | MF415 | ∼2.6-kb Gateway donor site cloned into the XbaI/HindIII site of pEX18Tp-pheS; Tpr | This study |
| pDONRPEX18Gm-SceI-pheS | MF356 | ∼2.6-kb Gateway donor site cloned into the XbaI/HindIII site of pEX18Gm-pheS; Gmr | This study |
| pDONRPEX18Km-SceI-pheS | MF414 | ∼2.6-kb Gateway donor site cloned into the XbaI/HindIII site of pEX18Km-pheS; Kmr | This study |
| pENTRPEX18Tp-SceI-pheS-Bcam1349 | MF455 | Gene replacement vector containing the Bcam1349 deletion allele; Tpr | This study |
| pENTRPEX18Tp-SceI-pheS-phzF | MF450 | Gene replacement vector containing the phzF deletion allele; Tpr | This study |
| pDAI-SceI-pheS | MF355 | ∼1.2-kb XbaI/SphI pheS fragment from pUC57-pheS cloned into the XbaI/SphI site of pDAI-SceI | This study |
Construction of Gateway-compatible allelic exchange vectors.
The attB1- and attB2-flanked Gateway donor site was amplified by PCR from pDONR221 by using primers GWE-SceI-F (flanked by HindIII and I-SceI restriction sites) and GWE-R (flanked by the XbaI site). The resulting 2.6-kb PCR product was digested with HindIII and XbaI and cloned into HindIII/XbaI-digested plasmids pEX18Tp-pheS, pEX18Gm-pheS, and pEX18Km-pheS (14), resulting in the allelic exchange vectors pDONRPEX18Tp-SceI-pheS, pDONRPEX18Gm-SceI-pheS, and pDONRPEX18Km-SceI-pheS, respectively (Fig. 1). The insertion of the Gateway donor site was confirmed by restriction analysis and partial sequencing of the newly generated vectors. These vectors are maintained in E. coli strain DB3.1, which contains a gyrA462 mutation (Invitrogen).
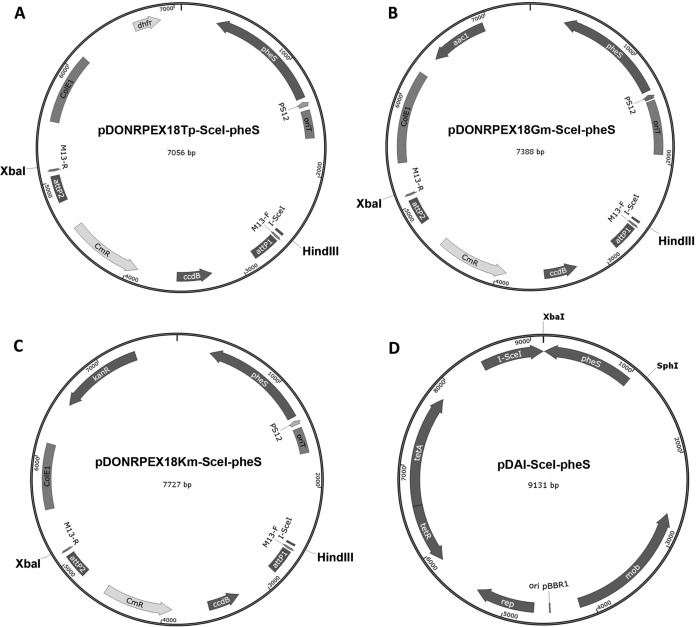
FIG 1.
Maps of the allelic exchange vectors and the I-SceI expression vector constructed in this study. (A to C) Gene replacement vectors, each containing a different antibiotic resistance marker, were constructed by cloning the Gateway donor site into the XbaI/HindIII site of a set of pEX family vectors based on the mutant pheS gene (14). attP1 and attP2, lambda recombination sites; CmR, chloramphenicol acetyltransferase; ccdB, gene encoding a gyrase-modifying enzyme; dhfr, dihydrofolate reductase-encoding gene; aac1, Gm acetyltransferase-encoding gene; kanR, gene conferring resistance to kanamycin; pheS, mutant gene for the α-subunit of phenylalanyl tRNA synthase; PS12, B. pseudomallei rpsL gene promoter; I-SceI, I-SceI endonuclease recognition site; ColE1, origin of replication; oriT, conjugal origin of transfer; M13-F and M13-R, primer binding sites for partial sequencing of the DNA sequence cloned into attP1-attP2 sites. (D) pDAI-SceI-pheS was constructed by cloning the pheS gene into the XbaI/SphI site of plasmid pDAI-SceI (12). tetA and tetR, genes encoding the tetracycline-specific efflux protein and repressor protein, respectively; mob, region facilitating conjugal transfer; I-SceI, I-SceI endonuclease; ori pBBR1, origin of replication; rep, gene encoding the pBBR1 replication protein.
Construction of the I-SceI expression vector pDAI-SceI-pheS.
To construct pDAI-SceI-pheS (Fig. 1), an ∼1.2-kb fragment containing the pheS gene was excised from pUC57-pheS (14) by restriction with XbaI and SphI and was ligated into XbaI/SphI-digested plasmid pDAI-SceI (12). The presence of the insertion was verified by restriction analysis.
Construction of the gene replacement vector pENTRPEX18Tp-SceI-pheS-Bcam1349.
The upstream fragment of the Bcam1349 gene was amplified by using primers Bcam1349-UpF-GWR and Bcam1349-UpR-tail, and the downstream fragment of the Bcam1349 gene was amplified by using primers Bcam1349-DnF and Bcam1349-DnR-GWL (Table 2). Both fragments were amplified by using Phusion high-fidelity DNA polymerase (Thermo Scientific) according to the manufacturer's instructions and under the following thermal cycling conditions: 98°C for 2 min; 25 cycles of 98°C for 15 s, 64°C for 30 s, and 72°C for 1 min; and a final extension step of 72°C for 7 min. The PCR fragments were purified by using the Wizard SV gel and PCR Clean-Up system (Promega), and their concentrations were determined spectrophotometrically. The up- and downstream fragments were fused together and amplified by using primers GW-attB1 and GW-attB2 (Table 2) in splicing by overlap extension PCR (SOE PCR) (20) to generate the Bcam1349 mutant allele as follows. Equal amounts (50 ng) of each up- and downstream fragment and the other components of the PCR mixture except primers GW-attB1 and GW-attB2 were mixed. PCR was carried out under the following thermal cycling conditions: 98°C for 2 min; 3 cycles of 98°C for 15 s, 64°C for 30 s, and 72°C for 1 min; and a final extension step of 72°C for 1 min. The final extension step was paused at 30 s, primers GW-attB1 and GW-attB2 were added, and thermal cycling was continued, with 27 cycles of 98°C for 15 s, 64°C for 30 s, and 72°C for 2 min and a final extension step of 72°C for 7 min. The PCR product was then purified and verified by restriction analysis.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| Gene specific | |
| Bcam1349-UpF-GWLa | TACAAAAAAGCAGGCTAACGGGGATTTCGCACGAT |
| Bcam1349-UpR-tailb | GGACATCGACTGCATCGTCAAGCTCGAGTGAAGATGAAGCA |
| Bcam1349-DnF | TGACGATGCAGTCGATGTCC |
| Bcam1349-DnR-GWRa | TACAAGAAAGCTGGGTGAGATTGATCGCCGGCAT |
| Commonc | |
| GW-attB1 | GGGGACAAGTTTGTACAAAAAAGCAGGCT |
| GW-attB2 | GGGGACCACTTTGTACAAGAAAGCTGGGT |
| Amplification of the Gateway donor site | |
| GWE-SceI-Fd | TACTACAAGCTTTAGGGATAACAGGGTAATAGCATGGATGTTTTCCCAGT |
| GWE-Rd | TACTACTCTAGATCAGAGATTTTGAGACACGGG |
| Othere | |
| Bcam1349-F | TACTACCCCGGGTAAATCGCTTATTCGGGCTG |
| Bcam1349-R | TACTACTCTAGACATTCGTTCCACCGGACAT |
| Bcam1349-RBS-F | TACTACTCTAGAATTGTCCGGAAATGGATTGGT |
| Bcam1349-RBS-R | TACTACCCCGGGATTCGTTCCACCGGACAT |
Double-underlined sequences are common for all genes amplified and overlap the GW-attB primer sequences (29).
The sequence in boldface type overlaps the gene-specific DnF primer.
Sequences were obtained from reference 29.
Restriction enzyme sites are single underlined, and the I-SceI endonuclease recognition site is in boldface type and double underlined.
Restriction enzyme sites are underlined.
A BP clonase reaction for the recombinational transfer of the mutant allele into the allelic exchange vector pDONRPEX18Tp-SceI-pheS was performed at 25°C overnight according to the Gateway cloning manual (Invitrogen), using only one-half the recommended amount of BP clonase II enzyme mix (Invitrogen). The BP clonase reaction product was transferred into chemically competent E. coli DH5α cells. The transformants growing on LB agar plates containing 50 μg Tp ml−1 were screened by colony PCR using primers GWE-SceI-F and GWE-R for insertion of the deletion allele. A number of positive clones were streaked onto LB agar plates containing 50 μg Tp ml−1 for purification, plasmid isolation, and partial sequencing.
Construction of pYedQ2 and complementation plasmid pMF564.
Plasmid pYedQ2, which was used to elevate intracellular cyclic di-GMP (c-di-GMP) levels, was constructed as follows. The yedQ expression cassette was excised from plasmid pYedQ (21) by restriction with BamHI and HindIII and was inserted into the BamHI/HindIII-digested broad-host-range cloning vector pBBR1MCS-5 (22). The presence of the insertion was confirmed by restriction analysis.
Complementation plasmid pMF564 was constructed as follows. The vector pBBR1MCS-5 was digested with SphI and blunt ended by T4 DNA polymerase. The linearized vector was further digested with XbaI and dephosphorylated with shrimp alkaline phosphatase. The SphI/XbaI digestion removed the Plac promoter and the related regulatory sequences from the plasmid. An ∼1.5-kb fragment containing the Bcam1349 gene and its ∼0.7-kb upstream DNA sequence was PCR amplified by using primers Bcam1349-RBS-F and Bcam1349-RBS-R, which were flanked by SmaI and XbaI restriction sites, respectively. The PCR fragment was digested with SmaI and XbaI and cloned into the previously linearized vector, yielding complementation plasmid pMF564. The presence of the insertion was confirmed by restriction analysis.
Mutagenesis of B. cenocepacia H111.
The gene replacement vector pENTRPEXTp-SceI-pheS-Bcam1349 was introduced by conjugation into B. cenocepacia via triparental mating, as described previously (23). The cointegrants were selected for Tp resistance on AB-citrate agar plates containing 100 μg Tp ml−1. Four Tp-resistant colonies were streaked onto the same selective plates, and the growing colonies were screened for the integration of the plasmid by colony PCR using primers Bcam1349-F and Bcam1349-R (Table 2). A single positive merodiploid clone was transformed with pDAI-SceI-pheS by triparental mating to stimulate the second homologous recombination event and resolve the merodiploid state. The transconjugants were screened for Tet resistance on AB-citrate agar plates containing 120 μg Tet ml−1. Batches of 10 Tet-resistant colonies were screened for the loss of the wild-type allele and the presence of the desired gene deletion by colony PCR using primers Bcam1349-F and Bcam1349-R. Two positive clones were purified by streaking and growing the clones on an AB-citrate agar plate. Thereafter, a single colony for each clone was picked and grown in 1 ml AB-glucose medium containing 0.1% (wt/vol) cPhe at 37°C overnight in order to stimulate the loss of pDAI-SceI-pheS via the counterselectable marker pheS on the plasmid. Tenfold serial dilutions of the cultures grown overnight were plated onto LB agar plates without any antibiotic, and 20 of the growing colonies of each clone were patched onto LB agar plates with or without tetracycline by using a sterile toothpick to screen for Tet sensitivity, which indicated the loss of plasmid pDAI-SceI-pheS. A single positive colony for each clone was selected and stored at −80°C.
Phenotypic characterization of the B. cenocepacia Bcam1349 deletion mutant.
Colony morphology, pellicle formation, and flow cell biofilm formation assays were performed as described previously (23).
RESULTS AND DISCUSSION
Features of the Gateway-compatible allelic exchange vectors.
The allelic exchange vectors pEX18Tp-pheS, pEX18Gm-pheS, and pEX18Km-pheS, which contain different antibiotic resistance markers, were first described by Barrett and colleagues (14). These vectors are derivatives of a set of pEX family vectors (25), in which the counterselectable marker gene sacB was replaced with a mutant allele of the B. pseudomallei pheS gene. Here we modified these vectors for use as Gateway-compatible donor vectors to clone regions of homology containing the deleted allele of a target gene. This was carried out by cloning the Gateway donor site from pDONR221 into the multicloning site of the above-mentioned vectors. The 18-bp I-SceI recognition site was incorporated into the vectors as a tail to the forward primer during PCR amplification of the donor cassette. The resulting vectors (Fig. 1) contain attP1 and attP2 sequences required for recombination-based cloning and the ccdB gene as a counterselectable marker, which kills gyrA+ host cells, such as E. coli DH5α cells, by inducing gyrase-mediated double-stranded DNA breaks, providing positive selection for E. coli clones bearing plasmids with cloned inserts. Additionally, the vectors contain the counterselectable pheS gene (14) driven by the PS12 promoter of the B. pseudomallei rpsL gene (26) and the I-SceI recognition site for downstream resolution of merodiploids. Although the mutant pheS gene was shown to be efficient in killing Burkholderia thailandensis cells in the presence of cPhe when expressed as a single copy from the gene replacement vector integrated on the chromosome (14), it was inefficient in killing B. cenocepacia H111 cells, and the resolution of merodiploids was almost impossible when the cells were grown in the presence of cPhe. Therefore, we incorporated the I-SceI site into the gene replacement vectors for downstream resolution of merodiploids. We preferred to keep the pheS gene on the gene replacement vectors, as it can efficiently be utilized as a counterselectable marker in strains such as B. thailandensis (14).
Features of the I-SceI expression vector pDAI-SceI-pheS.
The vector pDAI-SceI-pheS (Fig. 1), which constitutively expresses the I-SceI endonuclease, is a derivative of the vector pDAI-SceI, the features of which were previously described by Flannagan and colleagues (12). Although the mutant pheS gene was not efficient in killing B. cenocepacia cells in the presence of cPhe when expressed as a single copy on the chromosome, it effectively killed almost all B. cenocepacia cells when expressed from the multicopy plasmid pBBR1MCS-Km-pheS (14) (see Fig. S1 in the supplemental material), indicating that the mutant pheS gene has to be present in multiple copies in the cells to provide effective counterselection in B. cenocepacia. Based on this finding, we modified pDAI-SceI by cloning the mutant B. pseudomallei pheS gene from pUC57-pheS (14) into the multicloning site of pDAI-SceI to expedite self-curing of the plasmid. In the presence of 0.1% cPhe, the mutant pheS gene enables efficient killing of B. cenocepacia cells containing pDAI-SceI-pheS and curing of the B. cenocepacia deletion mutants from the plasmid once they are obtained after the resolution of merodiploids. In this way, the deletion mutants become ready for subsequent rounds of mutagenesis.
Construction of the B. cenocepacia Bcam1349 deletion mutant.
Using the allelic exchange system described here, we have successfully generated gene deletions in both B. cenocepacia H111 and B. thailandensis (see the supplemental material). As a proof of concept, we present a procedure that was used to delete the Bcam1349 gene. This gene encodes a c-di-GMP-responsive CRP/FNR superfamily transcription factor and regulates biofilm formation in B. cenocepacia H111 (23, 24). We previously showed that elevated intracellular levels of c-di-GMP promoted wrinkled-colony formation on solid medium, robust pellicle formation at the air-liquid interface of static liquid cultures, and increased biofilm formation in flow cells. However, despite having high intracellular c-di-GMP levels, a transposon insertion mutant of Bcam1349 did not form wrinkled colonies, pellicles, or thick flow cell biofilms (23).
We created the Bcam1349 mutant allele in two consecutive PCR rounds using three primer pairs (Table 2). Two of these primer pairs were gene specific, and one of them was common and can be used routinely. Gene-specific primers were designed to amplify fragments ranging from 0.8 to 1 kb in size. The fragments were chosen so that the gene-specific UpF-GWR primer is placed within 10 to 100 bp after the gene start and the gene-specific primer DnR-GWL is placed within 10 to 100 bp before the stop codon. The gene-specific primers were compared to the B. cenocepacia H111 genome to make sure that they would not fully anneal to unspecific regions in the genome. In the first PCR round, the gene-specific primers were used to amplify up- and downstream homology regions of the target gene. We usually obtained single major PCR products of the correct size, which were subsequently purified with a PCR cleanup kit and used in the second PCR round. However, if there are multiple bands, the entire PCR mixtures should be loaded onto an agarose gel, and fragments with the correct size should be gel extracted. In the second PCR round, equal amounts of up- and downstream PCR fragments were fused together and amplified with the common primers GW-attB1 and GW-attB2 (Table 2), incorporating the attB1 and attB2 recombination sites at either end of the deletion allele. We usually obtained a single major PCR product of the correct size (∼2 kb) at this step.
We recombined the Bcam1349 mutant allele into pDONRPEX18Tp-SceI-pheS using BP clonase and transferred the entire BP reaction product into E. coli DH5α cells. Tp-resistant transformants were selected, and the presence of the correct plasmid was checked by colony PCR using primers GWE-SceI-F and GWE-R. Alternatively, M13-F and M13-R primers can be used. Plasmids were isolated from a number of positive clones, and the presence of the deletion allele was verified by restriction analysis and partial sequencing.
The resulting gene replacement vector, pENTRPEX18Tp-SceI-pheS-Bcam1349, was transferred into B. cenocepacia by triparental mating, giving rise to Tp-resistant merodiploids (Fig. 2A). The integration of the nonreplicative vector into the chromosome can normally be verified by colony PCR using gene-specific primers UpF-GWR and DnR-GWL, often resulting in two PCR products corresponding to the wild-type and deletion alleles (Fig. 2B). However, we had to use another pair of primers, Bcam1349-F and Bcam1349-R, to verify the integration of the vector, as the former primer pair did not result in any PCR products. During the generation of deletion mutants of other genes, we also noticed that it is not always possible to see a PCR product corresponding to the wild-type allele, as its amplification may not be favored due to its relatively large size compared to that of the deletion allele. A single merodiploid clone was selected and transformed with pDAI-SceI-pheS by conjugation to stimulate the second homologous recombination event via the generation of a double-stranded DNA break by I-SceI endonuclease expressed from the plasmid. Depending on the location of the second recombination event, the resolution of the merodiploid state either restored the wild-type allele or generated the desired gene deletion (Fig. 2A). Eight Tet-resistant colonies were selected and verified for Bcam1349 deletion by colony PCR. In our experience, at least one colony always contained the desired gene deletion (Fig. 2B). Finally, the deletion mutant was cured from plasmid pDAI-SceI-pheS by growing the mutant in liquid medium containing 0.1% cPhe, as described in Materials and Methods. The counterselection medium with cPhe should not contain any competing phenylalanine for efficient counterselection. We therefore prefer to use AB minimal medium supplemented with glucose as a carbon source. However, in the case of deleting genes essential for growth in minimal medium, the mutants can alternatively be cured from plasmid pDAI-SceI-pheS by growing them in serial passages in rich medium without cPhe and Tet, which is required for the maintenance of the plasmid.
FIG 2.
Schematic diagram depicting the gene replacement procedure in B. cenocepacia H111 (A) and gel image (B). In step 1, the gene replacement vector pENTRPEX18Tp-SceI-pheS-Bcam1349 (derivative of pDONRPEX18Tp-SceI-pheS) contains regions of homology flanking the Bcam1349 gene. The vector was transferred into B. cenocepacia by conjugation and integrated into the chromosome by the first homologous recombination event, resulting in trimethoprim-resistant merodiploids, which were verified by colony PCR (B, lane 1). In step 2, the merodiploid was transformed with pDAI-SceI-pheS. The I-SceI endonuclease expressed from the plasmid introduces a double-stranded DNA break at the I-SceI recognition site on the chromosome. In step 3, The DNA break stimulates the second homologous recombination event through the host DNA repair system. Depending on the location of the second recombination event, the resolution of the merodiploid state either generates the desired gene deletion (step 3A [A] and lane 3A [B]) or restores the wild-type allele (step 3B [A] and lane 3B [B]), which is identified by colony PCR.
Phenotypic characterization of the Bcam1349 deletion mutant.
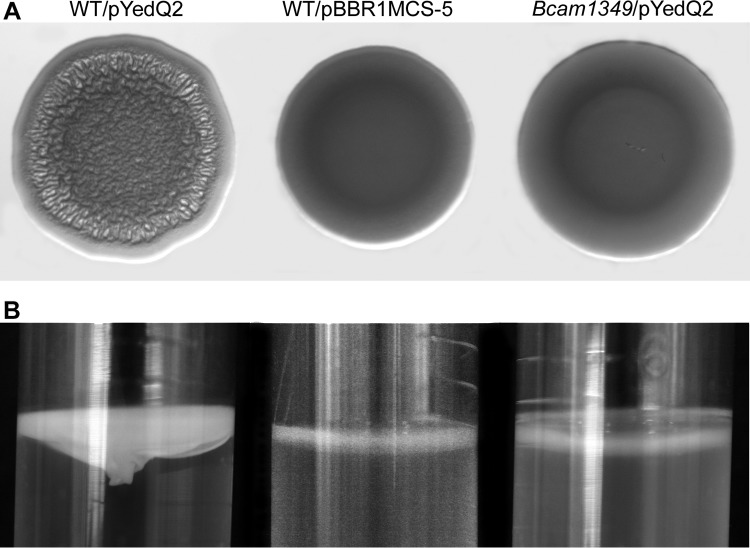
We previously demonstrated that a transposon insertion mutant of Bcam1349 did not form wrinkled colonies, robust pellicles, or thick flow cell biofilms despite having high intracellular c-di-GMP levels (23). To characterize the Bcam1349 deletion mutant obtained here, we first transformed it with plasmid pYedQ2, which contains the E. coli diguanylate cyclase protein YedQ and leads to elevated intracellular levels of c-di-GMP in B. cenocepacia (23). Unlike the pYedQ2-containing wild type, the pYedQ2-containing Bcam1349 mutant formed smooth colonies on AB agar medium (Fig. 3A) and did not form robust pellicles in static liquid culture (Fig. 3B). Furthermore, we tested the biofilm formation ability of the Bcam1349 mutant in a flow cell biofilm system. In accordance with the above-described results, the Bcam1349 mutant was markedly impaired in biofilm formation compared to the wild-type strain (Fig. 4). To rule out the possibility that the observed biofilm defect was due to a secondary mutation obtained during the mutagenesis procedure, we genetically complemented the mutant strain with an intact copy of the Bcam1349 gene and its 0.7-kb upstream DNA sequence on a replicative plasmid (pMF564). After complementation of the mutant strain, the biofilm formation ability was restored to wild-type levels (Fig. 4), indicating that the biofilm defect was indeed a result of the Bcam1349 deletion.
FIG 3.
Phenotypic characterization of the Bcam1349 deletion mutant. Shown are colony morphology on AB-glucose agar medium (A) and pellicle formation in static LB liquid culture (B) of the wild-type (WT) and Bcam1349 mutant strains carrying pYedQ2 and the wild-type strain carrying pBBR1MCS-5 (vector control).
FIG 4.

Flow cell biofilm formation by the wild type (WT), the Bcam1349 mutant and its complemented counterpart, and vector control strains. Confocal laser scanning microscopy images were acquired after 24 h of incubation at 37°C.
Conclusion.
The Gateway-compatible allelic exchange system described here takes advantage of bacteriophage lambda-based site-specific recombination instead of the traditional cloning procedures based on restriction enzymes and ligase and provides flexibility and efficiency. With proper primer design, the system allows precise in-frame deletion of open reading frames without generating truncated genes, reducing the risk of undesired polar effects. Moreover, the unmarked nature of the deletion procedure enables repetitive rounds of gene deletions in a single strain. We believe that the allelic exchange system described here will be useful in understanding the genetic basis of virulence in B. cenocepacia and in systematic analyses of the functions of genes in the physiology of this emerging pathogen and other Burkholderia species with medical relevance or potential biotechnological use. Furthermore, this allelic exchange system may enable the engineering of Burkholderia strains that retain their biotechnologically useful functions but are attenuated for virulence.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Lundbeck Foundation (to M.F.) and the Danish Council for Independent Research (to T.T.-N).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03909-14.
REFERENCES
- 1.Coenye T, Vandamme P. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol 5:719–729. doi: 10.1046/j.1462-2920.2003.00471.x. [DOI] [PubMed] [Google Scholar]
- 2.Mahenthiralingam E, Baldwin A, Dowson CG. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J Appl Microbiol 104:1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x. [DOI] [PubMed] [Google Scholar]
- 3.Chiarini L, Bevivino A, Dalmastri C, Tabacchioni S, Visca P. 2006. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol 14:277–286. doi: 10.1016/j.tim.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Drevinek P, Mahenthiralingam E. 2010. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect 16:821–830. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 5.Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeño-Tárraga AM, Thomson NR, Bason N, Quail MA, Sharp S, Cherevach I, Churcher C, Goodhead I, Hauser H, Holroyd N, Mungall K, Scott P, Walker D, White B, Rose H, Iversen P, Mil-Homens D, Rocha EP, Fialho AM, Baldwin A, Dowson C, Barrell BG, Govan JR, Vandamme P, Hart CA, Mahenthiralingam E, Parkhill J. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol 191:261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varga JJ, Losada L, Zelazny AM, Kim M, McCorrison J, Brinkac L, Sampaio EP, Greenberg DE, Singh I, Heiner C, Ashby M, Nierman WC, Holland SM, Goldberg JB. 2013. Draft genome sequences of Burkholderia cenocepacia ET12 lineage strains K56-2 and BC7. Genome Announc 1(5):e00841-13. doi: 10.1128/genomeA.00841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlier A, Agnoli K, Pessi G, Suppiger A, Jenul C, Schmid N, Tümmler B, Pinto-Carbo M, Eberl L. 2014. Genome sequence of Burkholderia cenocepacia H111, a cystic fibrosis airway isolate. Genome Announc 2(2):e00298-14. doi: 10.1128/genomeA.00298-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suppiger A, Schmid N, Aguilar C, Pessi G, Eberl L. 2013. Two quorum sensing systems control biofilm formation and virulence in members of Burkholderia cepacia complex. Virulence 4:400–409. doi: 10.4161/viru.25338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fazli M, Almblad H, Rybtke ML, Givskov M, Eberl L, Tolker-Nielsen T. 2014. Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ Microbiol 16:1961–1981. doi: 10.1111/1462-2920.12448. [DOI] [PubMed] [Google Scholar]
- 10.Uehlinger S, Schwager S, Bernier SP, Riedel K, Nguyen DT, Sokol PA, Eberl L. 2009. Identification of specific and universal virulence factors in Burkholderia cenocepacia strains by using multiple infection hosts. Infect Immun 77:4102–4110. doi: 10.1128/IAI.00398-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwager S, Agnoli K, Köthe M, Feldmann F, Givskov M, Carlier A, Eberl L. 2013. Identification of Burkholderia cenocepacia strain H111 virulence factors using nonmammalian infection hosts. Infect Immun 81:143–153. doi: 10.1128/IAI.00768-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flannagan RS, Linn T, Valvano MA. 2008. A system for construction of targeted unmarked gene deletions in the genus Burkholderia. Environ Microbiol 10:1652–1660. doi: 10.1111/j.1462-2920.2008.01576.x. [DOI] [PubMed] [Google Scholar]
- 13.Aubert DF, Hamad MA, Valvano MA. 2014. A markerless deletion method for genetic manipulation of Burkholderia cenocepacia and other multidrug-resistant gram-negative bacteria. Methods Mol Biol 1197:311–327. doi: 10.1007/978-1-4939-1261-2_18. [DOI] [PubMed] [Google Scholar]
- 14.Barrett AR, Kang Y, Inamasu KS, Son MS, Vukovich JM, Hoang TT. 2008. Genetic tools for allelic replacement in Burkholderia species. Appl Environ Microbiol 74:4498–4508. doi: 10.1128/AEM.00531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López CM, Rholl DA, Trunck LA, Schweizer HP. 2009. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl Environ Microbiol 75:6496–6503. doi: 10.1128/AEM.01669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamad MA, Zajdowicz SL, Holmes RK, Voskuil MI. 2009. An allelic exchange system for compliant genetic manipulation of the select agents Burkholderia pseudomallei and Burkholderia mallei. Gene 430:123–131. doi: 10.1016/j.gene.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logue CA, Peak IR, Beacham IR. 2009. Facile construction of unmarked gene deletion mutants in Burkholderia pseudomallei using sacB counter-selection in sucrose-resistant and sucrose-sensitive isolates. J Microbiol Methods 76:320–323. doi: 10.1016/j.mimet.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Katzen F. 2007. Gateway recombinational cloning: a biological operating system. Expert Opin Drug Dicov 2:571–589. doi: 10.1517/17460441.2.4.571. [DOI] [PubMed] [Google Scholar]
- 19.Clark DJ, Maaløe O. 1967. DNA replication and division cycle in Escherichia coli. J Mol Biol 23:99–112. doi: 10.1016/S0022-2836(67)80070-6. [DOI] [Google Scholar]
- 20.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 21.Ausmees N, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Lindberg M. 2001. Genetic data indicate that proteins containing GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol Lett 204:163–167. doi: 10.1111/j.1574-6968.2001.tb10880.x. [DOI] [PubMed] [Google Scholar]
- 22.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 23.Fazli M, O'Connell A, Nilsson M, Niehaus K, Dow JM, Givskov M, Ryan RP, Tolker-Nielsen T. 2011. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol Microbiol 82:327–341. doi: 10.1111/j.1365-2958.2011.07814.x. [DOI] [PubMed] [Google Scholar]
- 24.Fazli M, McCarthy Y, Givskov M, Ryan RP, Tolker-Nielsen T. 2013. The exopolysaccharide gene cluster Bcam1330–Bcam1341 is involved in Burkholderia cenocepacia biofilm formation, and its expression is regulated by c-di-GMP and Bcam1349. Microbiologyopen 2:105–122. doi: 10.1002/mbo3.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 26.Yu M, Tsang JS. 2006. Use of ribosomal promoters from Burkholderia cenocepacia and Burkholderia cepacia for improved expression of transporter protein in Escherichia coli. Protein Expr Purif 49:219–227. doi: 10.1016/j.pep.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Peano C, Chiaramonte F, Motta S, Pietrelli A, Jaillon S, Rossi E, Consolandi C, Champion OL, Michell SL, Freddin L, Falciola L, Basilico F, Garlanda C, Mauri P, De Bellis G, Landini P. 2014. Gene and protein expression in response to different growth temperatures and oxygen availability in Burkholderia thailandensis. PLoS One 9:e93009. doi: 10.1371/journal.pone.0093009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kessler B, de Lorenzo V, Timmis KN. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of Pm promoter in the TOL plasmid studied with all controlling elements in monocopy. Mol Gen Genet 233:293–301. doi: 10.1007/BF00587591. [DOI] [PubMed] [Google Scholar]
- 29.Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol 5:30. doi: 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.