Abstract
Daptomycin produced by Streptomyces roseosporus is an important lipopeptide antibiotic used to treat human infections caused by Gram-positive pathogenic bacteria, including drug-resistant strains. The genetic basis for regulatory mechanisms of daptomycin production is poorly known. Here, we characterized the dptR3 gene, which encodes a MarR family transcriptional regulator located adjacent to the known daptomycin biosynthetic (dpt) genes. Deletion of dptR3 reduced daptomycin production significantly and delayed aerial mycelium formation and sporulation on solid media. Dissection of the mechanism underlying the function of DptR3 in daptomycin production revealed that it stimulates daptomycin production indirectly by altering the transcription of dpt structural genes. DptR3 directly activated the transcription of its own gene, dptR3, but repressed the transcription of the adjacent, divergent gene orf16 (which encodes a putative ABC transporter ATP-binding protein). A 66-nucleotide DptR3-binding site in the intergenic region of dptR3-orf16 was determined by DNase I footprinting, and the palindromic sequence TCATTGTTACCTATGCTCACAATGA (underlining indicates inverted repeats) in the protected region was found to be essential for DptR3 binding. orf16, the major target gene of DptR3, exerted a positive effect on daptomycin biosynthesis. Our findings indicate that DptR3 functions as a global regulator that positively controls daptomycin production and morphological development in S. roseosporus.
INTRODUCTION
Streptomycetes are soil-dwelling filamentous bacteria characterized by complex morphological differentiation and the ability to produce a variety of antibiotics. The production of these antibiotics is a complex process that is usually accompanied by morphological differentiation and is controlled by multiple regulatory proteins that respond to nutritional status, population density, and a variety of environmental conditions (1–3). Generally, the lowest level of the regulatory network involves pathway-specific regulatory genes that are found within the respective antibiotic biosynthesis gene cluster and affect only a single antibiotic biosynthetic pathway.
Daptomycin is a cyclic lipopeptide antibiotic used clinically to treat complex skin infections caused by Gram-positive pathogens, including 15 genera and 35 species, notably, penicillin-resistant Streptococcus pneumoniae, vancomycin-resistant Enterococcus spp., glycopeptide-insensitive Staphylococcus aureus, and methicillin-resistant S. aureus (4). Daptomycin is a minor component of the A21978C complex produced by Streptomyces roseosporus via a nonribosomal peptide synthetase (NRPS) mechanism. Components of the complex have in common a 10-membered cyclic peptide nucleus and a three-amino-acid tail with various fatty acid moieties attached to the N-terminal tryptophan (Trp) (5). The fatty acid portion of daptomycin is straight-chain decanoic acid, and daptomycin production can be increased by adding the precursor decanoic acid or sodium decanoate to the fermentation broth (6, 7).
In view of the wide medical application of daptomycin, there have been many studies of its biosynthesis. The dpt gene cluster has been cloned and consists of at least 12 known genes (5, 8, 9). Among these, dptA, dptBC, and dptD encode the three subunits of an NRPS; dptE and dptF are responsible for the activation of fatty acids that are then catalyzed by the N-terminal C domain of DptA to form N-acylation of Trp; and dptG, dptH, dptI, dptJ, dptM, dptN, and dptP are involved in precursor supply, resistance, or transport. Many open reading frames (ORFs) flanking the dpt genes encode multicomponent transporters or hypothetical proteins. Three regulatory genes (dptR1, dptR2, dptR3) located adjacent to the known dpt genes encode LuxR, DeoR, and MarR family regulators, respectively. Because of their location, these three genes were presumed to encode the pathway-specific regulators of daptomycin production (5). However, dptR2 was shown recently to be required for daptomycin production but not for expression of the dpt gene cluster, suggesting that the DptR2 protein does not function as a pathway-specific regulator (10). To our knowledge, dptR2 is the only regulatory gene reported for the regulation of daptomycin production in S. roseosporus. The functions of dptR1 and dptR3 remain unknown. Identification and characterization of regulatory genes involved in daptomycin production are important for elucidation of the regulatory networks of daptomycin biosynthesis and for practical construction of strains with high daptomycin production levels.
The MarR (multiple antibiotic resistance regulator) family of transcriptional regulators is named for the almost completely characterized Escherichia coli MarR protein, a repressor of genes that regulate multiple antibiotic resistance operons (11, 12). MarR homologs are widely distributed in bacteria and archaea and control a wide range of cellular activities, including antibiotic resistance, stress responses, virulence, and catabolism of aromatic compounds (13–15). They are winged helix-turn-helix (HTH) DNA-binding proteins that exist as dimers and bind palindromic sequences within cognate promoters. They typically act as transcriptional repressors, although a few act as activators or dual repressors-activators. A common regulatory mechanism involves MarR homolog binding to the intergenic region between the marR gene and a divergently oriented gene (or operon), regulating the transcription of both genes positively or negatively. Such transcriptional regulation is blocked by conformational changes upon the binding of small-molecule ligands to MarR proteins. Over 12,000 MarR-like proteins have been annotated to date in bacterial and archaeal genomes (14), but only a few have been studied in Streptomyces. Streptomyces coelicolor OhrR acts as either a repressor or an activator in response to organic hydroperoxides (16). The orthologous proteins PenR and PntR serve as pathway-specific activators of the biosynthesis of the antibiotic phenalinolactone in Streptomyces exfoliatus and Streptomyces arenae, respectively (17). S. coelicolor TamR controls the expression of the gene encoding trans-aconitate methyltransferase (tam) and is important for metabolic flux through the citric acid cycle during oxidative stress (18). S. coelicolor PecS was proposed to function under oxidative stress conditions by responding to the ligand urate (19). S. coelicolor PcaV controls the transcription of genes encoding β-ketoadipate pathway enzymes (which are essential for aromatic catabolism) through its interaction with the pathway substrate protocatechuate (PCA) (20). The crystal structures of the PcaV-PCA complex (20) and S. coelicolor SCO5413 (21) have been determined.
The present study addressed the positive regulatory role of the MarR family regulator DptR3 in daptomycin production and morphological differentiation of S. roseosporus. We found that DptR3 regulates daptomycin biosynthesis indirectly at the transcriptional level.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. S. roseosporus wild-type (WT) strain NRRL11379 is a producer of daptomycin. S. roseosporus strains were grown at 28°C on solid DA1 medium (0.4% glucose, 1% malt extract, 0.4% yeast extract, 0.2% CaCO3, 1.5% agar) for sporulation or in liquid YEME medium (22) containing 25% sucrose for growth of mycelia. RM14 medium (23) was used for regeneration of protoplasts and for selection of transformants. R2YE and MM agar (22) were used for S. roseosporus phenotype observation. Seed medium and fermentation medium [1.1% yeast extract, 0.086% Fe(NH4)2(SO4)2·6H2O, 1.07% glucose, 7.2% potato dextrin, 0.72% cane molasses, pH 7.0] were used for daptomycin production. The primary and secondary seed media contained 2.5% dextrin and 3% Trypticase soy broth. Fermentation medium was also used to cultivate mycelia for RNA isolation.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| S. roseosporus | ||
| NRRL11379 | WT strain | 7 |
| DR3 | dptR3 deletion mutant | This study |
| CR3 | dptR3-complemented strain | This study |
| WT/pKC1139-ermpR3 | dptR3 overexpression strain | This study |
| WT/pKC1139 | WT strain carrying empty vector pKC1139 | This study |
| D16 | dpt16 deletion mutant | This study |
| E. coli | ||
| JM109 | General cloning host for plasmid manipulation | Laboratory stock |
| ET12567 | Methylation-deficient strain | 23 |
| BL21(DE3) | Host for protein overexpression | Novagen |
| Plasmids | ||
| pMD18-T | TA cloning vector | TaKaRa |
| pKC1139 | Multiple-copy, temperature-sensitive E. coli-Streptomyces shuttle vector | 36 |
| pSET152 | Integrative E. coli-Streptomyces shuttle vector | 36 |
| pET-28a(+) | Vector for protein overexpression in E. coli | Novagen |
| pJL117 | pIJ2925 derivative carrying Streptomyces strong constitutive promoter ermE*p | 37 |
| pDR3 | dptR3 deletion vector based on pKC1139 | This study |
| pD16 | orf16 deletion vector based on pKC1139 | This study |
| pKC1139-ermpR3 | dptR3 overexpression vector based on pKC1139 | This study |
| pSET152-ermpR3 | dptR3 complemented vector based on pSET152 | This study |
| pET28-R3 | dptR3 overexpression vector based on pET-28a(+) | This study |
E. coli strains JM109 and BL21(DE3) (Novagen, Germany) were used as cloning and expression hosts, respectively. E. coli ET12567 (dam dcm hsdS) (23) was used to propagate nonmethylated DNA for transformation into S. roseosporus. The antibiotics used were described previously (24).
Fermentation and high-performance liquid chromatography (HPLC) analysis of daptomycin.
Spores from various S. roseosporus strains cultured on DA1 plates for 10 days were added to 250-ml flasks containing 50 ml of primary seed medium and incubated at 28°C for 48 h on a rotary shaker (250 rpm). Cultures in primary seed medium were inoculated at 6% (vol/vol) into 50 ml of secondary seed medium and incubated at 28°C with shaking for 30 h. Then a 6% (vol/vol) inoculation volume of cultures in secondary seed medium was transferred into 50 ml of fermentation medium and cultured further for 10 days. After 48 h of fermentation, sodium decanoate (final concentration, 0.02%, wt/vol) was added every 12 h until the end of the fermentation.
HPLC analysis of daptomycin production in fermentation culture was performed as described previously, with modification (7). In brief, the fermentation broth (1.0 ml) was centrifuged at 12,000 × g for 10 min, the supernatant was filtered (membrane pore size, 2.2 μm), and the filtrate was applied directly to an HPLC system (model 600; Waters, Milford, CT) with a C18 column (4.6 [inside diameter] by 250 mm). A mobile phase of 0.1% (vol/vol) trifluoroacetic acid in water and acetonitrile (55:45, vol/vol) was used at a flow rate of 1.0 ml · min−1. Daptomycin was detected by measuring UV absorption at 218 nm and using authentic daptomycin samples (Shanghai Qiao Chemical Science, China) to draw calibrated standard curves.
Gene deletion, complementation, and overexpression.
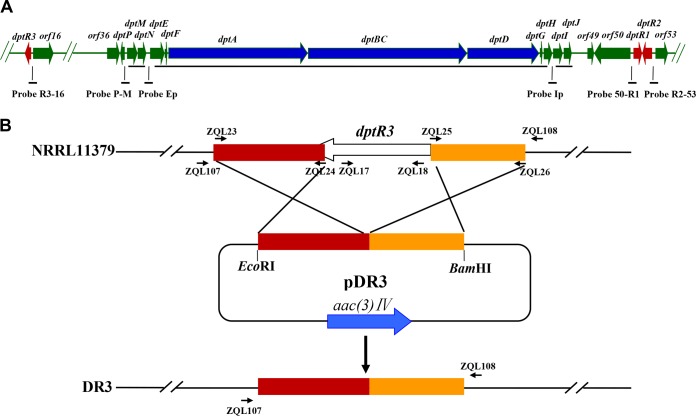
To construct a dptR3 deletion mutant, two fragments flanking dptR3 were prepared by PCR from genomic DNA of NRRL11379. A 481-bp 5′ flanking region (positions −413 to +68 relative to the dptR3 start codon) was amplified with primers ZQL25 and ZQL26, and a 544-bp 3′ flanking region (positions −104 to +440 relative to the dptR3 stop codon) was amplified with primers ZQL23 and ZQL24 (complementary to ZQL25). The two fragments were fused together by PCR with primers ZQL23 and ZQL26 and then ligated into EcoRI/BamHI-digested pKC1139 to generate dptR3 deletion vector pDR3. Transformation of pDR3 into NRRL11379 and selection of double-crossover recombinant strains were performed as described previously (24). S. roseosporus does not sporulate on RM14 medium; transformants regenerated on RM14 plates were transferred to DA1 medium for sporulation. Spores of the transformants were collected and spread on DA1 plates containing apramycin. The plates were incubated initially at 28°C for 48 h and then at 39°C for 7 days. Because pKC1139 cannot replicate itself in Streptomyces when the temperature is higher than 34°C, only mutants in which pDR3 was integrated into the S. roseosporus chromosome by a single crossover can grow at 39°C on DA1 plates. Single-crossover mutants were transferred to DA1 plates under nonselection conditions for three rounds of sporulation, and double crossover took place only in colonies sensitive to apramycin. The putative dptR3 deletion mutants were confirmed by PCR analysis with primers ZQL107, ZQL108, ZQL17, and ZQL18, followed by DNA sequencing. A 1.23-kb band was detected when primers ZQL107 and ZQL108, which flank the exchange regions, were used, whereas a 1.6-kb band was detected when genomic DNA of NRRL11379 was used as the template. When primers ZQL17 and ZQL18, located within the deletion region of dptR3, were used, only NRRL11379 produced a 405-bp PCR fragment, as predicted (data not shown). We thus obtained dptR3 gene deletion mutant DR3, in which dptR3 was mostly deleted by double-crossover recombination (Fig. 1B).
FIG 1.
Genetic organization of 12 known dpt genes and the genes adjacent to them, from dptR3 to orf53, in S. roseosporus WT strain NRRL11379 (A) and a schematic strategy for deletion of dptR3 (B). (A) Long black lines, transcriptional units. Short lines at the bottom, probes of the promoter regions used for EMSAs. (B) Large arrows, genes and their directions. Short arrows, positions of primers used for cloning exchange regions and confirming gene deletion as described in Materials and Methods. Rectangles, homologous exchange regions used for deletion of dptR3.
For complementation of DR3, a 590-bp DNA fragment carrying the dptR3 ORF was amplified by PCR with primers ZQL21 and ZQL22. The PCR product was digested with HindIII/XbaI and ligated simultaneously with the EcoRI/HindIII ermE*p fragment from pJL117 and EcoRI/XbaI-digested pSET152 to generate the dptR3-complemented vector pSET152-ermpR3, in which the dptR3 gene was controlled by the ermE*p promoter. pSET152-ermpR3 was introduced into DR3 to obtain complemented strain CR3. The same 590-bp dptR3 ORF and ermE*p fragments were cloned into pKC1139 to produce dptR3 overexpression vector pKC1139-ermpR3, which was then introduced into NRRL11379 to obtain dptR3 overexpression strain WT/pKC1139-ermpR3.
To construct an orf16 deletion mutant, a 664-bp 5′ flanking region (positions −472 to +192 relative to the orf16 start codon) and a 605-bp 3′ flanking region (positions −181 to +424 relative to the orf16 stop codon) were amplified with primer pairs CQ37/CQ38 and CQ39/CQ40, respectively. The two fragments were fused together by PCR with primers CQ37 and CQ40 and then ligated into EcoRI/XbaI-digested pKC1139 to generate orf16 deletion vector pD16, which was transformed into NRRL11379 protoplasts. The resulting orf16 deletion mutant D16 was confirmed by PCR analysis with primers CQ41, CQ42, ZQL113, and ZQL114 (see Fig. 6A). A 1.54-kb band was detected when primers CQ41 and CQ42, which flank the exchange regions, were used, whereas a 3.0-kb band was detected from genomic DNA of NRRL11379. When primers ZQL113 and ZQL114, located within the deletion region of orf16, were used, only NRRL11379 produced a 151-bp PCR fragment, as predicted (data not shown). All of the primers used in this study are listed in Table 2.
FIG 6.
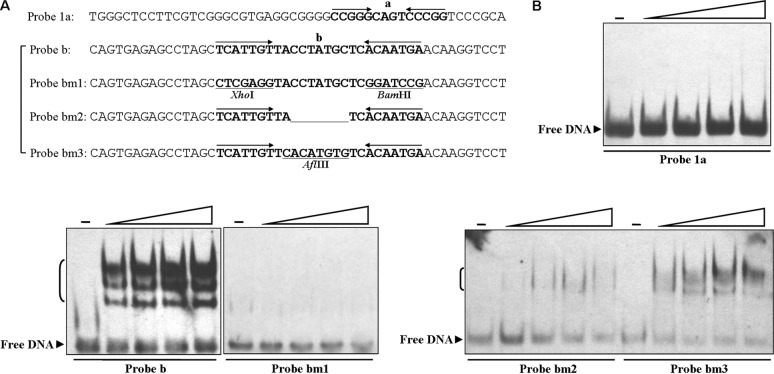
Mutational analysis of the DptR3-binding site. (A) DNA probes containing an intact palindromic sequence in the DptR3-binding site or mutated sequences. Probes 1a and b, 50-bp WT DNA containing intact palindromic sequences a and b, respectively. Mutations were introduced into probe b to produce mutated probes bm1, bm2, and bm3, respectively. Altered nucleotides are underlined. (B) EMSAs with various DNA probes. Each probe was incubated with 50, 100, 150, and 200 ng of His6-DptR3 (concentrations increase from left to right in each series of four lanes, as indicated by the triangles above the blots).
TABLE 2.
Primers used in this study
| Application(s) and primer | DNA sequence (5′–3′)a | Use(s) |
|---|---|---|
| Gene disruption, complementation, and overexpression | ||
| ZQL23 | CCGGAATTCTCGTCCAGCACCTCCACG (EcoRI) | dptR3 gene deletion |
| ZQL24 | CTCTCGCGATTCCCGGGACCGCCGCAGAGGAGATCC | |
| ZQL25 | GGATCTCCTCTGCGGCGGTCCCGGGAATCGCGAGAG | |
| ZQL26 | CGGGATCCGATGAGGGTCTGGAGCACG (BamHI) | |
| ZQL21 | CCCAAGCTTCTCCAGCAGCGGCATGTC (HindIII) | Complementation of DR3, overexpression of dptR3 in S. roseosporus |
| ZQL22 | GCTCTAGACGCACAGTGAGAGCCTAGC (XbaI) | |
| ZQL17 | TCCGGCATTCCGTCGAC | Confirmation of dptR3 deletion mutant DR3 |
| ZQL18 | GCTCGCCGAACAGCTGC | |
| ZQL107 | CGGCAGCACGAAGGTGAG | |
| ZQL108 | CAGGTCGTGCATGACGCG | |
| CQ37 | CCGGAATTCCCCTCGTCGGTGATCTCG (EcoRI) | Deletion of orf16 gene |
| CQ38 | GCGATGGTGAGCGTGGTCGAGGATCTCGCGCAGCAG | |
| CQ39 | CTGCTGCGCGAGATCCTCGACCACGCTCACCATC GC | |
| CQ40 | TGCTCTAGACGCCTGGATCTTCACCACG (XbaI) | |
| CQ41 | CAGCTCGCCGAACACCTC | Confirmation of orf16 deletion mutant D16 |
| CQ42 | GGAACTCCGCGTCCCACT | |
| ZQL113 | CTTCGGCGTGCTCCAGAC | |
| ZQL114 | GTTGGCGATGCGGGACTG | |
| ZQL35 | GGAATTCCATATGGACGCCCCGGATTCCCCC (NdeI) | Overexpression of His6-tagged DptR3 protein in E. coli |
| ZQL36 | CCGCTCGAGTCAGCAGTGGCGCTCCGG (XhoI) | |
| EMSA | ||
| ZQL37 | TCCCGGGAATCGCGAGAG | Probe R3-16 |
| ZQL38 | GAAGAGGCGGAGGATGCG | |
| ZQL39 | GGCGTGGAACATACTGGCG | Probe Ep |
| ZQL40 | GCACAGCGGCTCTCACTC | |
| ZQL47 | GCGCGGTCAACAAGATTCTT | Probe Ip |
| ZQL48 | CCTTGACGGTCACGTGGTAG | |
| ZQL49 | TGACCTGGTCCGGCCGAT | Probe 50-R1 |
| ZQL50 | CCCCACCGGCGGAATTGT | |
| ZQL51 | CCGTTCCGATGCGAGTGC | Probe R2-53 |
| ZQL52 | CGTGCAGGAAGGTGTTCGC | |
| ZQL57 | CGGTCCGAACCGGCTCTTG | Probe P-M |
| ZQL58 | AGGCGCTGCGGATCGATG | |
| GJ91 | CCAAGGGCTACAAGTTCTCC | Probe hrdB |
| GJ92 | TTGATGACCTCGACCATGTG | |
| 5′ RACE | ||
| ZQL59 | GGATCTCCTCTGCGGCGG | Identification of TSP of dptR3 |
| ZQL60 | GGAACTGGGCCGGAGTGATGT | |
| ZQL62 | CAGCTGGCGGCGCTGGAT | |
| ZQL64 | GTTGGCGATGCGGGACTG | Identification of TSP of orf16 |
| ZQL65 | GAGATGAGGGTCTGGAGCACG | |
| ZQL67 | GAAGAGGCGGAGGATGCGG | |
| Real-time RT-PCR | ||
| ZQL75 | TCCCGGGAATCGCGAGAG | dptR3 ORF |
| ZQL76 | CCCGGATTCCCCCGACTC | |
| ZQL79 | GAACAGACCACCCTCCTCG | dptBC ORF |
| ZQL80 | CTGTGGCCGATGGGGTAG | |
| ZQL81 | CGGCGTACATCATCCAGACC | dptA ORF |
| ZQL82 | GTCATGCTCAGTTCGGAGACG | |
| ZQL83 | GAGTGAGAGCCGCTGTGC | dptE ORF |
| ZQL84 | GGTGTCCCGTACGAGAACC | |
| ZQL85 | CCAGATCCTCTCGACGGTG | dptM ORF |
| ZQL86 | CCGATACAGGCGCGTACC | |
| ZQL87 | TTCCGGTACGAGCGGCTG | dptD ORF |
| ZQL88 | CGTCAGATCGAAGCGGCG | |
| ZQL89 | TCCGCACCATCACGTTCAC | dptH ORF |
| ZQL90 | GACTTCCTGGGCCACCTG | |
| ZQL91 | GAGATCGCCCTCCTGGAC | dptR1 ORF |
| ZQL92 | GTGGAAGCTCAGCCCCTC | |
| ZQL93 | GGCCAGGATCGTGACGTC | dptR2 ORF |
| ZQL94 | CGGAACGGCAGGAGTTCATC | |
| ZQL113 | CTTCGGCGTGCTCCAGAC | orf16 ORF |
| ZQL114 | GTTGGCGATGCGGGACTG | |
| ZQL115 | GACAGATCGCCGAGTTCGT | dptF ORF |
| ZQL116 | GACGCTCCACAGCAGCTC | |
| ZQL117 | CTACCACGTGACCGTCAAGG | dptI ORF |
| ZQL118 | GTCGTCGAACTCGTTTCCCTC | |
| ZQL119 | CAACGACGGCAGCTACCTC | dptG ORF |
| ZQL120 | GGCGCATATCGGTCCAGTTC | |
| ZQL121 | CAATCCGCCGTACCAGGC | dptP ORF |
| ZQL122 | GGACACGCCCGCTGTTGG | |
| DNase I footprinting assay | ||
| FAM-ZQL103 | GAAGAGGCGGAGGATGCG | dptR3-orf16 intergenic region |
| ZQL104 | GGGCCGGAGTGATGTCGAT |
The restriction site in parentheses is underlined.
Overexpression and purification of recombinant His6-tagged DptR3 protein.
A DNA fragment encoding the predicted 166 amino acids of the DptR3 protein was generated by PCR with primers ZQL35 and ZQL36. The PCR fragment was digested with NdeI/XhoI and inserted between the corresponding sites in expression vector pET-28a(+) to generate pET28-R3, which was confirmed by DNA sequencing and then introduced into E. coli BL21(DE3) for protein overexpression. Following induction with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG), bacteria harboring pET28-R3 were incubated for 8 h at 16°C. Cells were collected, resuspended in lysis buffer (20 mM Tris-Cl [pH 8.0], 200 mM NaCl, 1 mM EDTA [pH 8.0], 0.5% NP-40), and disrupted by sonication. After centrifugation, the supernatant was recovered and the resulting recombinant His6-tagged DptR3 protein was purified on a Ni2+-nitrilotriacetic acid spin column (Qiagen, Germany) according to the manufacturer's instructions. The purified protein was used for electrophoretic mobility shift assays (EMSAs) and DNase I footprinting assays as described below.
EMSAs.
EMSAs were performed with a DIG Gel Shift kit, 2nd generation (Roche) according to the manufacturer's instructions. The probes were amplified by PCR and labeled at the 3′ ends with nonradioactive digoxigenin (DIG). The reaction mixture (20 μl) contained the probes, proteins, and 1 μg of poly(dI-dC) (vial 9) in the binding buffer (vial 5). The mixture was incubated at 25°C for 30 min, and then 5 μl of loading buffer with bromophenol blue (vial 13) was added. Protein-bound and free DNAs were separated by electrophoresis on 5% native polyacrylamide gels with 0.5× Tris-borate-EDTA as the running buffer and then transferred to positively charged nylon membranes by electroblotting. The membranes were baked at 80°C for 10 min, and the DNA fragments were cross-linked by exposure to UV radiation for 10 min. Chemiluminescence detection was performed according to the manufacturer's instructions, and the membranes were exposed to X-ray film for 15 to 30 min.
DNase I footprinting assay.
A nonradiochemical capillary electrophoresis method was used for DNase I footprinting (25). To characterize the DptR3 protein binding sites in the dptR3-orf16 intergenic region, a 6-carboxyfluorescein (FAM) fluorescence-labeled DNA fragment was synthesized by PCR with primers FAM-ZQL103/ZQL104. The resulting 378-bp DNA fragments covered the entire intergenic region. Following purification with a Midi Purification kit (Tiangen, China), labeled DNA fragments (400 ng) and appropriate concentrations of His6-tagged DptR3 protein were added to a final reaction volume of 50 μl and the mixture was incubated for 30 min at 25°C in binding buffer [20 mM HEPES (pH 7.6), 10 mM (NH4)2SO4, 1 mM dithiothreitol, 0.2% Tween 20, 30 mM KCl]. Digestion with DNase I (0.016 U) was performed for 40 s at 37°C and stopped by adding EDTA to a final concentration of 60 mM. The reaction mixture was heated to 80°C for 10 min to totally inactivate DNase I. The samples were subjected to phenol-chloroform extraction, ethanol precipitation, and capillary electrophoresis by loading into a DNA genetic analyzer (model 3730) together with the internal-lane size standard ROX-500 (Applied Biosystems). Electropherograms were analyzed with the GeneMarker program, v1.8 (Applied Biosystems).
Identification of TSPs by 5′ RACE.
To determine the transcriptional start points (TSPs) of dptR3 and orf16, total RNA was extracted from a 96-h culture of NRRL11379 grown in fermentation medium. Total RNA (2 μg) was used for reverse transcription (RT) with 20 pmol of gene-specific primer ZQL59 or ZQL64 with a 5′/3′ rapid amplification of cDNA ends (RACE) kit (2nd generation; Roche). The sample was purified with a PCR Product Purification kit (Beijing HT-Biotech Co. Ltd., China). An oligo(dA) tail was added to the 3′ end of the cDNA by using terminal deoxynucleotidyl transferase, followed by direct amplification of the tailed cDNA with an oligo(dT) anchor primer (GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTTV) and the second inner gene-specific primer ZQL60 or ZQL65. An additional round of PCR was performed with a 1,000-fold dilution of the original PCR product as the template, an anchor primer (GACCACGCGTATCGATGTCGAC), and nested primer ZQL62 or ZQL67 to obtain a single specific band. The final PCR products were cloned into the vector pMD18-T (TaKaRa, Japan) for sequencing. The first nucleotide following the oligo(dA) sequence was considered the TSP.
RNA preparation and real-time RT-PCR analysis.
Mycelia of S. roseosporus grown in fermentation medium were collected, frozen in liquid nitrogen, and ground to a fine powder. RNA extractions were performed with TRIzol reagent (Tiangen) according to the manufacturer's instructions. To remove chromosomal DNA contamination, each RNA sample was treated with DNase I and tested by PCR to confirm the absence of chromosomal DNA. The treated RNA sample (2 μg) was reverse transcribed with Moloney murine leukemia virus (RNase H−; TaKaRa), random hexamers (25 μM), and a deoxynucleoside triphosphate mixture (10 mM each). The cDNAs obtained were used as templates for real-time RT-PCR analysis with the primers listed in Table 2. The experiments were performed with FastStart Universal SYBR green master mix (ROX; Roche) and analysis with an ABI 7900HT sequence detection system with optical-grade 96-well plates. Template cDNA, 10 μl of FastStart Universal SYBR green master mix, and forward and reverse primers (each at 300 nM) were mixed in each reaction system (total volume, 20 μl). The PCR protocol was 95°C for 10 min and 40 cycles of 95°C for 10 s and 60°C for 30 s. The DNase I-treated RNA samples without RT were used as negative controls to rule out chromosomal DNA contamination. The hrdB gene (SAV2444), which encodes the major sigma factor in Streptomyces, was used as a positive internal control to normalize samples. Relative expression levels were calculated by the comparative cycle threshold method. Each experiment was performed in triplicate.
RESULTS
DptR3 is a positive regulator of morphological differentiation and daptomycin production.
Miao et al. (5) reported that S. roseosporus WT strain NRRL11379 contains the dpt gene cluster involved in daptomycin biosynthesis. The dptR3 gene contains 501 nucleotides (nt) and encodes a 166-amino-acid putative MarR family transcriptional regulator, including a conserved HTH DNA-binding motif homologous to MarR. There are 21 ORFs between dptR3 and dptP at the left end of the dpt gene cluster (Fig. 1A). The divergently transcribed gene orf16 is located upstream of dptR3 and encodes a putative ABC transporter ATP-binding protein. The nucleotide sequences and deduced amino acid sequences of orf16-dptR3 are highly homologous to those of SCO1144-1145 in S. coelicolor and SAV1564-1565 in S. avermitilis (DptR3, 60 and 65% identical to SCO1145 and SAV1565; ORF16, 81 and 83% identical to SCO1144 and SAV1564, respectively).
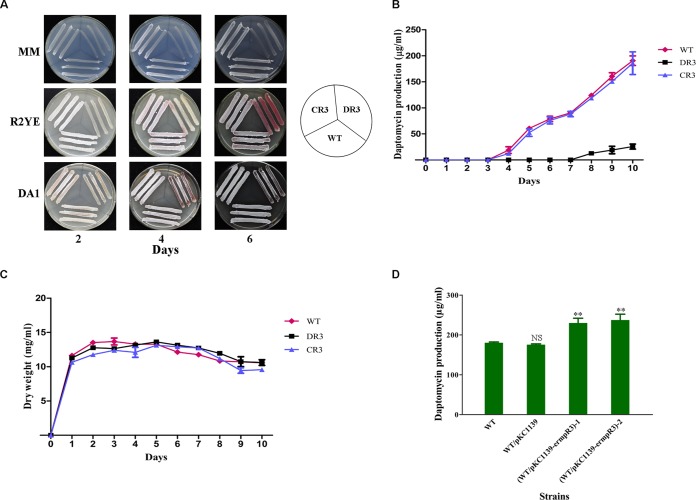
To elucidate the role of dptR3 in S. roseosporus, we deleted the dptR3 gene from the WT strain by homologous recombination (Fig. 1B). The deletion mutant DR3 showed strikingly delayed formation of aerial mycelia and spores on the solid media MM agar, R2YE, and DA1 (Fig. 2A). Deletion of dptR3 also led to markedly delayed and reduced daptomycin production in comparison with that of the WT strain. Daptomycin production in DR3 was not detectable by HPLC until fermentation day 7, and the final yield after day 10 was only ∼13% of that of the WT (Fig. 2B).
FIG 2.
Daptomycin production and growth of S. roseosporus WT and dptR3 mutant strains. (A) Effect of dptR3 deletion on morphological development. The WT strain, the dptR3 deletion mutant (DR3), and the complemented strain (CR3) were grown on MM agar, R2YE, and DA1 plates at 28°C, and the plates were photographed every 48 h. (B) Daptomycin yields of the WT (⧫), DR3 (■), and CR3 (▲). (C) Growth curves of the WT, DR3, and CR3. Cell growth was measured in cell dry weight. (D) Comparison of daptomycin production in the WT, vector control strain WT/pKC1139, and dptR3 overexpression strains WT/pKC1139-ermpR3-1 and -2 grown in fermentation medium for 10 days. S. roseosporus was cultivated in 250-ml shaking flasks containing 50 ml of seed medium or fermentation medium. Apramycin was added to primary and secondary seed medium of WT/pKC1139 and WT/pKC1139-ermpR3 for plasmid selection. Error bars show the standard deviation of three replicate flasks. Statistical significance was determined by comparing the mutant values to those of the WT strain. **, P < 0.01; NS, not significant.
To demonstrate that these changes were due solely to the deletion of dptR3, we constructed dptR3-complemented strain CR3. The morphological phenotype and daptomycin production were restored in CR3 (Fig. 2A and B). To investigate whether the reduced daptomycin production of DR3 was due to altered cell growth, we analyzed the growth of the WT, DR3, and CR3 strains in fermentation medium. The results showed that deletion of dptR3 had no notable effect on cell growth (Fig. 2C). These findings indicate that DptR3 plays a positive regulatory role in morphological differentiation and daptomycin biosynthesis.
In general, overexpression of a positive regulator can increase the production of the corresponding antibiotic. To examine this possibility, we introduced dptR3 overexpression vector pKC1139-ermpR3 into the WT strain and evaluated its effects on daptomycin production. HPLC analysis of the fermentation products following 10 days of culture in fermentation medium showed that a transformant containing the empty vector pKC1139 produced nearly the same amount of daptomycin as the host strain, whereas the daptomycin yield was ∼28 and 31% higher in dptR3 overexpression strains WT/pKC1139-ermpR3-1 and -2, respectively (Fig. 2D). These findings further confirmed the role of dptR3 as a daptomycin biosynthesis activator gene.
DptR3 activates dpt genes but represses the adjacent gene orf16.
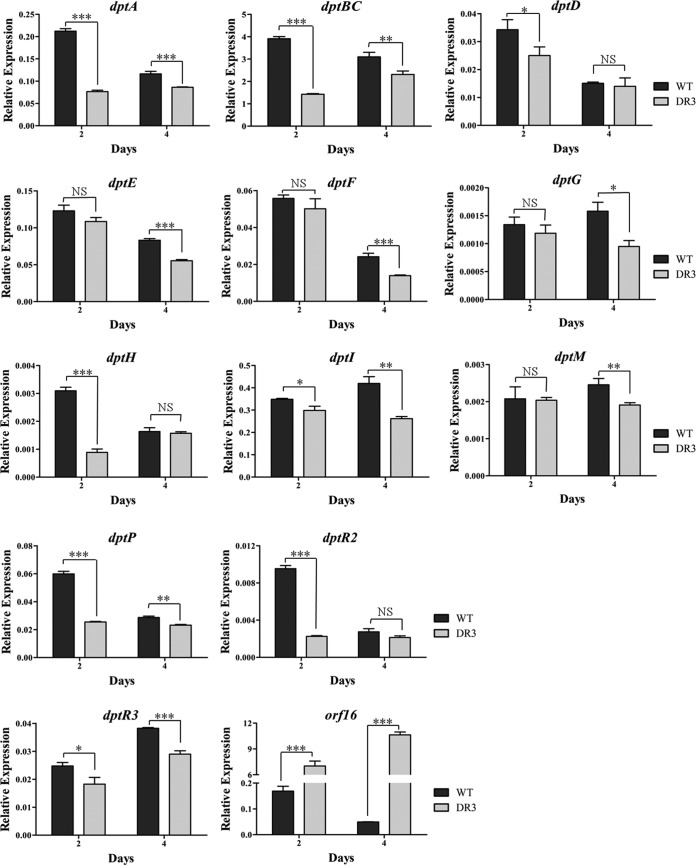
To further elucidate the role of DptR3 in daptomycin biosynthesis, we performed a real-time RT-PCR analysis with RNAs isolated from the WT and DR3 strains grown in fermentation medium for 2 (late exponential phase) or 4 (stationary phase) days. The transcription levels of structural genes dptA, dptBC, dptD, dptE, dptF, dptG, dptH, dptI, dptM, and dptP in the dpt gene cluster were reduced in DR3 at either both time points or one time point (Fig. 3), indicating that DptR3 affects daptomycin production by stimulating the transcription of dpt structural genes.
FIG 3.
Real-time RT-PCR analysis of the dpt and orf16 gene transcription levels in the WT and DR3 strains. RNA samples were isolated from 2- and 4-day fermentation cultures. The relative transcription levels of each gene were obtained after normalization against the internal reference hrdB at corresponding time points. dptR3, 60-bp transcript amplified from the remainder ORF of dptR3 in DR3 with primers ZQL75 and ZQL76. Error bars show the standard deviation of three independent experiments. Statistical significance was determined by Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant.
Given the common regulatory mechanism of MarR proteins (14, 15), we predicted that DptR3 regulates the expression of itself and of the adjacent, divergently transcribed gene orf16. The transcription levels of dptR3 and orf16 were thus tested with the same RNA preparations. The dptR3 transcription level in DR3, measured with a 60-bp transcript amplified from the remainder ORF of dptR3, was lower than that in the WT, whereas orf16 expression was strikingly upregulated in DR3 (Fig. 3). These results indicate that DptR3 functions as an autoactivator, as well as a repressor, of orf16.
The regulatory genes dptR1 and dptR2 adjacent to the dpt cluster were also subjected to transcription analysis. dptR1 transcription was undetectable in the WT and DR3 strains by real-time RT-PCR with various primer pairs, suggesting either that the level was too low to be detected or that the dptR1 gene was not expressed under our culture conditions. dptR2 expression was downregulated in DR3 (Fig. 3), suggesting that DptR3 positively regulates dptR2 transcription, either directly or indirectly.
DptR3 binds specifically to the bidirectional dptR3-orf16 promoter region.
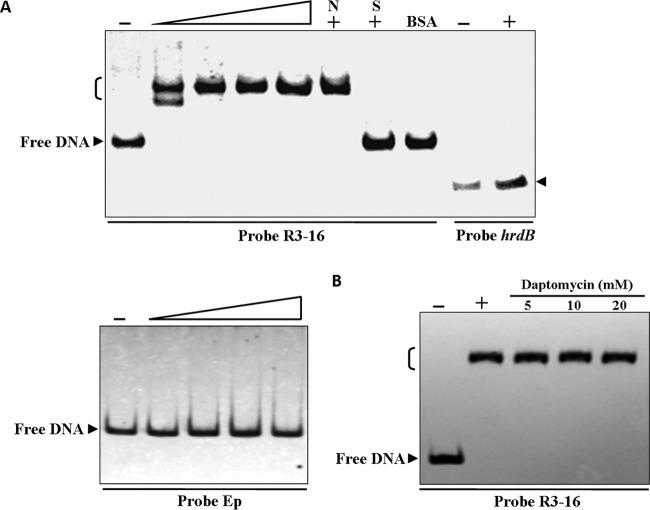
To determine whether DptR3 directly regulates the genes mentioned in the preceding section, we performed EMSAs with a full-length recombinant, His6-tagged DptR3 protein expressed in E. coli. In the dpt cluster, the core genes are oriented in the same direction (Fig. 1A). The contiguous genes from dptE to dptH were reported to be transcribed on a large polycistronic transcript (26). Given that dptM overlaps dptN and that dptI overlaps dptJ, it is possible that dptM and dptN are cotranscribed and that dptI-dptJ forms another transcriptional unit. The dptP-dptM, orf50-dptR1, dptR2-orf53, and dptR3-orf16 intergenic regions lie between two divergently transcribed genes and presumably contain bidirectional promoters. The promoter regions of dptE and dptI were therefore designated probes Ep and Ip, respectively. Probes P-M, 50-R1, R2-53, and R3-16 were designed to cover the corresponding intergenic regions containing two divergent promoters (Fig. 1A). Results showed that purified His6-DptR3 did not bind to probe Ep, Ip, P-M, 50-R1, or R2-53, even when the protein amount was large (200 ng) (only probe Ep is shown in Fig. 4A), indicating that the positive regulatory effect of DptR3 on daptomycin production and on dptR2 expression is indirect and that DptR3 does not function as a pathway-specific regulator of the dpt gene cluster.
FIG 4.
EMSAs of DptR3 binding to the dptR3-orf16 intergenic region. (A) EMSAs of the interaction of promoter regions with purified His6-DptR3 protein. Each lane contained 0.15 nM labeled probe. EMSAs with 50-fold unlabeled specific probe (S) or nonspecific (N) competitor DNA were performed to confirm the specificity of band shifts. Labeled probe hrdB was used to eliminate nonspecific binding of DptR3 protein. BSA was used as a negative control for DptR3 protein. The labeled probes were incubated in the absence (−) or presence of various amounts (50, 100, 150, 200 ng) of His6-DptR3. Two hundred nanograms of His6-DptR3 was used for control probe hrdB and competition experiments. (B) EMSAs of His6-DptR3 (150 ng) interaction with daptomycin. Arrowhead, free probe; bracket, DptR3-DNA complex.
His6-DptR3 was observed to bind to the dptR3-orf16 intergenic region (probe R3-16) and generated significantly shifted bands. The binding specificity was evaluated by the addition of excess unlabeled specific probe R3-16 (lane S), which competed strongly with labeled probe R3-16 for binding to DptR3, and by the addition of excess unlabeled nonspecific competitor DNA (lane N), which could not attenuate or abolish the retarded signal. The labeled nonspecific probe hrdB and bovine serum albumin (BSA) were used as negative controls (Fig. 4A). Our findings indicate that DptR3 directly regulates the transcription of its own gene, dptR3, and the adjacent gene orf16 through interaction with their promoter regions.
The DNA-binding activity of MarR proteins is typically attenuated by the binding of small-molecule ligands (14, 15). Because DptR3 regulates daptomycin production, we examined the possibility that daptomycin functions as the ligand of DptR3. This possibility was ruled out by the finding that daptomycin did not induce dissociation of DptR3 from probe R3-16 even at a high concentration (20 mM) (Fig. 4B).
Identification of the DptR3-binding site in the dptR3-orf16 intergenic region.
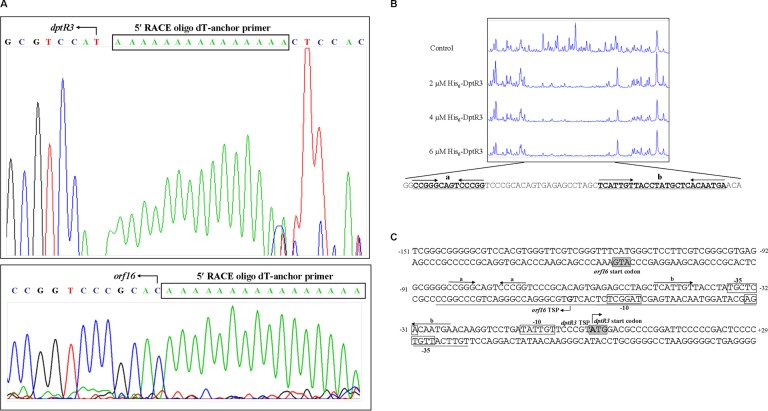
To elucidate the mechanism whereby DptR3 regulates dptR3 and orf16, we determined the promoter structures of the two genes and the specific binding site of DptR3 in their intergenic region. The TSPs of the genes were determined by 5′ RACE analysis. The dptR3 TSP coincides with the first A of the putative ATG translational start codon of dptR3 (Fig. 5A and C), which is used frequently in Streptomyces. This finding indicates that dptR3 is transcribed as a leaderless mRNA, as are many other Streptomyces genes (27–29). The orf16 TSP was mapped to G at a position 50 nt upstream of the translational start codon of orf16. Determination of these TSPs led to the proposal of the putative −10 and −35 promoter sequences indicated by boxes in Fig. 5C.
FIG 5.
dptR3 and orf16 promoter structures and DptR3-binding site in the dptR3-orf16 intergenic region. (A) Determination of TSPs of dptR3 and orf16 by 5′ RACE PCR. Boxed areas, complementary sequences of 5′ RACE oligo(dT) anchor primers. Bent arrows, complementary bases of TSPs. (B) DNase I footprinting assay of DptR3 in the dptR3-orf16 intergenic region. Fluorograms correspond to control DNA (10 μM BSA) and to protection reactions with increasing concentrations of His6-DptR3 protein. (C) Nucleotide sequences of the dptR3-orf16 promoter region and the DptR3-binding site. The numbers are distances (in nucleotides) from the TSP of dptR3. Solid line, DptR3-binding site. Straight arrows, inverted repeats. Bent arrows, TSP and transcription orientation. Boxed areas, putative −10 and −35 regions. Shaded areas, translational start codon.
The precise binding sequence of DptR3 was determined by DNase I footprinting experiments with a 378-bp FAM-labeled DNA probe comprising the dptR3-orf16 intergenic region, in the presence or absence of His6-DptR3 protein. A 66-nt protected region was found on the coding strand of dptR3 (Fig. 5B), extending from −87 to −22 nt relative to the dptR3 TSP and from −42 to +24 nt relative to the orf16 TSP (Fig. 5C). The 66-nt region overlaps the potential −10 and −35 regions of the orf16 promoter, suggesting that DptR3 negatively regulates orf16 transcription by blocking the access of RNA polymerase to its promoter region. The protection region is located upstream but overlaps the potential −35 region of the dptR3 promoter. DptR3 may activate dptR3 by stabilizing the RNA polymerase or by promoting the recruitment of RNA polymerase to the dptR3 promoter.
MarR proteins typically form symmetric dimers and bind to palindromic sequences (14, 15). Analysis of the DptR3-binding region with the DNAMAN program revealed two perfect palindromic sequences termed a and b (Fig. 5B and C). To estimate the relative contributions of these two sequences to DptR3 binding, we performed EMSAs with a probe that contained either the intact palindromic sequence or a mutated sequence, as illustrated in Fig. 6A. DptR3 did not retard probe 1a containing palindromic sequence a (Fig. 6B), indicating that sequence a is not essential for DptR3 binding. The affinity of DptR3 for mutated probe bm1, which lacked inverted repeats, was abolished completely, whereas strong shifted signals were observed between DptR3 and corresponding WT probe b containing palindromic sequence b (Fig. 6B). Seven base pairs were deleted from the spacer region between the inverted repeats in probe bm2. For probe bm3, base changes were introduced into the spacer sequence. The affinity of DptR3 for mutated probes bm2 and bm3 was still present but very weak. These findings indicate that the inverted repeats in sequence b are essential for DptR3 binding and that the spacer sequence and length are also important for effective interaction with DptR3.
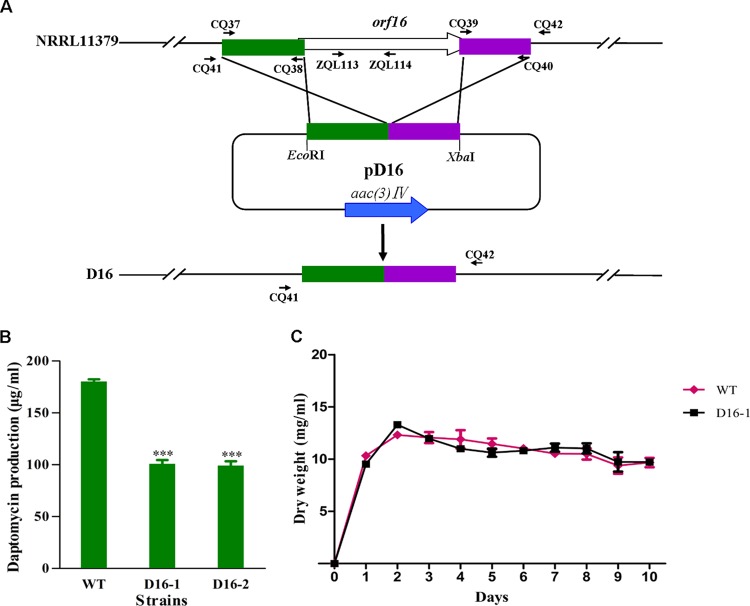
Effect of orf16 deletion on daptomycin production.
Because orf16 was identified as the target gene of DptR3, we constructed orf16 deletion mutant D16 (Fig. 7A) and further investigated the role of this gene in daptomycin production and morphological development. D16 displayed normal growth on the solid media DA1, R2YE, and MM agar (data not shown). In comparison with that of the WT strain, daptomycin production was reduced to ∼56 and 55% in deletion mutants D16-1 and D16-2, respectively (Fig. 7B). Deletion of orf16 did not affect biomass accumulation in fermentation medium (Fig. 7C) but attenuated expression of daptomycin biosynthetic genes (see Fig. S1 in the supplemental material). The transcription levels of dpt structural genes in the WT and D16 were consistent with the daptomycin yield data for these strains. These results indicate that orf16 has a positive effect on daptomycin biosynthesis. The expression level of orf16 in the WT strain was low. Deletion of dptR3 led to a striking increase in the orf16 transcription level (Fig. 3) and reduced daptomycin production (Fig. 2B). These findings suggest that the alteration of daptomycin production in the DR3 mutant did not result from varying expression of orf16 and that other DptR3 targets may affect daptomycin biosynthesis.
FIG 7.
Effect of orf16 deletion on daptomycin production and cell growth. (A) Strategy used for deletion of orf16. (B) Comparison of daptomycin production in WT and orf16 deletion mutants D16-1 and -2 grown in fermentation medium for 10 days. ***, P < 0.001. (C) WT and D16-1 growth curves.
DISCUSSION
Little is known regarding the regulation of daptomycin production at the transcriptional level, although the biosynthetic pathway (5, 9), combinational biosynthesis (30), metabolic engineering (31, 32), and fermentation optimization (6, 7) of this lipopeptide antibiotic have been extensively studied. We have shown here that DptR3 functions as a global activator of daptomycin production and morphological differentiation in S. roseosporus. Our findings extend the limited knowledge of the complex regulation of daptomycin biosynthesis.
Transcription and binding experiments have shown that DptR3 stimulates daptomycin production at the transcriptional level by altering the expression of structural genes in the dpt gene cluster; however, this stimulatory effect is indirect, indicating that DptR3 does not function as a pathway-specific regulator of daptomycin biosynthesis. Another group demonstrated that, of the three regulatory genes (dptR1, dptR2, dptR3) adjacent to the known dpt structural genes, dptR2 is not a pathway-specific regulatory gene (10). We found that dptR1 transcription under fermentation conditions is not detectable by real-time RT-PCR and that deletion of dptR1 has no notable effect on daptomycin production (unpublished data), suggesting that dptR1 is also not a pathway-specific regulatory gene. It therefore appears that the dpt cluster, similar to the erythromycin biosynthetic (ery) cluster in Saccharopolyspora erythraea (33), lacks a pathway-specific regulatory gene. BldD, a key developmental regulator whose gene resides apart from the ery cluster on the S. erythraea chromosome, was shown to directly control the expression of the ery cluster (34). The altered and coordinated expression of nearly all of the dpt genes suggests the existence of a common direct regulator of the dpt cluster. BldD homologs are present throughout the sporulating actinomycetes, including S. roseosporus. The consensus target site of BldD in S. coelicolor is 5′-NTNCNC(A/T)GNGTNAN-3′ (3). Similar binding motifs are present in some promoter regions of the dpt cluster, i.e., promoter regions of dptE and dptI, and the bidirectional promoter region of dptP-dptM, suggesting that the dpt genes may be direct targets of a BldD homolog in S. roseosporus. Further studies are necessary to identify the direct regulator(s) of the dpt cluster in order to clarify the daptomycin biosynthesis regulatory network.
Previous studies showed that most MarR homolog genes are oriented divergently from their neighboring genes and that the transcription of both genes is tightly controlled by binding of MarR proteins to the intergenic region (13, 14). The case of dptR3 and orf16 is similar. We found that DptR3 directly regulates the transcription of its own gene and orf16 by binding to the 25-bp palindromic sequence TCATTGTTACCTATGCTCACAATGA (the underlining indicates inverted repeats) in the dptR3-orf16 intergenic region. We used this palindromic sequence to scan the S. roseosporus genome in a search for new putative target genes of DptR3. No such sequence was found in other promoter regions. However, other DptR3 target genes may be present in S. roseosporus. The opposite effects of dptR3 deletion on the orf16 transcription level and of orf16 deletion on the daptomycin production level and the positive regulation of morphological development by DptR3 suggest the existence of other DptR3 targets besides dptR3 and orf16. Future studies using chromatin immunoprecipitation-DNA sequencing and bacterial one-hybrid systems will lead to the identification of other DptR3 target genes, and the roles of the new target genes in daptomycin production and morphological development can then be investigated.
We found that dptR3 is positively autoregulated. Overexpression of dptR3 in the WT strain promoted daptomycin production, indicating that endogenous DptR3 is not present in saturating amounts. Thus, enhancement of the dptR3 expression level is a practical strategy for increasing daptomycin production. Although DptR3 regulates daptomycin biosynthesis, daptomycin is not the ligand of DptR3, and this may be due to the indirect control of daptomycin production by DptR3. The physiological roles of ∼100 MarR proteins have been characterized in detail to date. Because the natural ligands of ligand-responsive MarR family proteins are unknown in most cases (13, 14, 21), our knowledge of the molecular regulatory mechanisms of these proteins remains limited. orf16 encodes a putative ABC transporter ATP-binding protein, which is one of the components of the ABC transport system and is repressed directly by DptR3. In the present study, deletion of orf16 resulted in reduced daptomycin production, suggesting that this gene plays an important role in the uptake of compounds necessary for daptomycin biosynthesis. SCO1144, the orf16 homolog gene in S. coelicolor, encodes a protein belonging to a lipid exporter family in the ABC-type export system. The substrates of this exporter family include lipoproteins, lipopolysaccharides, lipid A, multiple drugs, and peptides (35). Another possibility is that the orf16 product is involved in the expulsion of harmful or toxic by-products generated from the biosynthesis of daptomycin or that it has an effect on daptomycin resistance through the expulsion of daptomycin. orf16 is not a regulatory gene and may not influence gene expression directly. Therefore, the effect of orf16 deletion, i.e., reducing the expression of daptomycin biosynthetic genes, is indirect. The as-yet-unknown substrate of the Orf16 protein may be the ligand of a regulatory gene(s) involved in daptomycin biosynthesis. More experiments are needed to test these possibilities.
The mechanism whereby DptR3 regulates morphological differentiation is unclear. AdpA and BldD are global regulators of secondary metabolism and morphological differentiation in S. coelicolor (3). We found that DptR3 could bind to promoter regions of adpA and bldD homologous genes in S. roseosporus (see Fig. S2 in the supplemental material), raising the possibility that DptR3 regulates development by controlling the expression of adpA and bldD, and this requires further detailed investigation.
This is the first report that a transcriptional regulator in S. roseosporus controls both daptomycin production and morphological development. Improved knowledge of DptR3 target genes and their functions and the specific ligand of DptR3 will lead to more effective strategies for increasing daptomycin production.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Chinese Universities Scientific Fund (grant 2014QT018).
We are grateful to Yinghua Lu (Xiamen University, Xiamen, China) for providing S. roseosporus NRRL11379 and to S. Anderson for editing the English of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00057-15.
REFERENCES
- 1.Bibb MJ. 2005. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 2.van Wezel GP, McDowall KJ. 2011. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep 28:1311–1333. doi: 10.1039/c1np00003a. [DOI] [PubMed] [Google Scholar]
- 3.Liu G, Chater KF, Chandra G, Niu G, Tan H. 2013. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baltz RH. 2009. Daptomycin: mechanisms of action and resistance, and biosynthetic engineering. Curr Opin Chem Biol 13:144–151. doi: 10.1016/j.cbpa.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 5.Miao V, Coëffet-Legal MF, Brian P, Brost R, Penn J, Whiting A, Martin S, Ford R, Parr I, Bouchard M, Silva CJ, Wrigley SK, Baltz RH. 2005. Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 151:1507–1523. doi: 10.1099/mic.0.27757-0. [DOI] [PubMed] [Google Scholar]
- 6.Huber FM, Pieper RL, Tietz AJ. 1988. The formation of daptomycin by supplying decanoic acid to Streptomyces roseosporus cultures producing the antibiotic complex A21978C. J Biotechnol 7:283–292. doi: 10.1016/0168-1656(88)90040-5. [DOI] [Google Scholar]
- 7.Ng IS, Ye C, Zhang Z, Lu Y, Jing K. 2014. Daptomycin antibiotic production processes in fed-batch fermentation by Streptomyces roseosporus NRRL11379 with precursor and medium optimization. Bioprocess Biosyst Eng 37:415–423. doi: 10.1007/s00449-013-1007-2. [DOI] [PubMed] [Google Scholar]
- 8.Liao G, Shi T, Xie J. 2012. Regulation mechanisms underlying the biosynthesis of daptomycin and related lipopeptides. J Cell Biochem 113:735–741. doi: 10.1002/jcb.23414. [DOI] [PubMed] [Google Scholar]
- 9.Wittmann M, Linne U, Pohlmann V, Marahiel MA. 2008. Role of DptE and DptF in the lipidation reaction of daptomycin. FEBS J 275:5343–5354. doi: 10.1111/j.1742-4658.2008.06664.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang F, Ren NN, Luo S, Chen XX, Mao XM, Li YQ. 2014. DptR2, a DeoR-type auto-regulator, is required for daptomycin production in Streptomyces roseosporus. Gene 544:208–215. doi: 10.1016/j.gene.2014.04.044. [DOI] [PubMed] [Google Scholar]
- 11.Martin RG, Rosner JL. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci U S A 92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin RG, Nyantakyi PS, Rosner JL. 1995. Regulation of the multiple antibiotic resistance (mar) regulon by marORA sequences in Escherichia coli. J Bacteriol 177:4176–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkinson SP, Grove A. 2006. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol 8:51–62. [PubMed] [Google Scholar]
- 14.Perera IC, Grove A. 2010. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J Mol Cell Biol 2:243–254. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- 15.Grove A. 2013. MarR family transcription factors. Curr Biol 23:R142–R143. doi: 10.1016/j.cub.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Oh SY, Shin JH, Roe JH. 2007. Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J Bacteriol 189:6284–6292. doi: 10.1128/JB.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu DQ, Wang YP, Zhang MM, Ikeda H, Deng ZX, Gene DE. 2013. Product-mediated regulation of pentalenolactone biosynthesis in Streptomyces species by the MarR/SlyA family activators PenR and PntR. J Bacteriol 195:1255–1266. doi: 10.1128/JB.02079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H, Grove A. 2013. The transcriptional regulator TamR from Streptomyces coelicolor controls a key step in central metabolism during oxidative stress. Mol Microbiol 87:1151–1166. doi: 10.1111/mmi.12156. [DOI] [PubMed] [Google Scholar]
- 19.Huang H, Mackel BJ, Grove A. 2013. Streptomyces coelicolor encodes a urate-responsive transcriptional regulator with homology to PecS from plant pathogens. J Bacteriol 195:4954–4965. doi: 10.1128/JB.00854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis JR, Brown BL, Page R, Sello JK. 2013. Study of PcaV from Streptomyces coelicolor yields new insights into ligand-responsive MarR family transcription factors. Nucleic Acids Res 41:3888–3900. doi: 10.1093/nar/gkt009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holley TA, Stevenson CE, Bibb MJ, Lawson DM. 2013. High resolution crystal structure of Sco5413, a widespread actinomycete MarR family transcriptional regulator of unknown function. Proteins 81:176–182. doi: 10.1002/prot.24197. [DOI] [PubMed] [Google Scholar]
- 22.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics: a laboratory manual. John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 23.MacNeil DJ, Klapko LM. 1987. Transformation of Streptomyces avermitilis by plasmid DNA. J Ind Microbiol 2:209–218. doi: 10.1007/BF01569542. [DOI] [Google Scholar]
- 24.Zhao J, Wen Y, Chen Z, Song Y, Li JL. 2007. An adpA homologue in Streptomyces avermitilis is involved in regulation of morphogenesis and melanogenesis. Chin Sci Bull 52:623–630. doi: 10.1007/s11434-007-0105-4. [DOI] [Google Scholar]
- 25.Zianni M, Tessanne K, Merighi M, Laguna R, Tabita F. 2006. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J Biomol Tech 17:103–113. [PMC free article] [PubMed] [Google Scholar]
- 26.Coëffet-Legal MF, Thurston L, Rich P, Miao V, Baltz RH. 2006. Complementation of daptomycin dptA and dptD deletion mutations in trans and production of hybrid lipopeptide antibiotics. Microbiology 152:2993–3001. doi: 10.1099/mic.0.29022-0. [DOI] [PubMed] [Google Scholar]
- 27.Hoskisson PA, Rigali S, Fowler K, Findlay KC, Buttner MJ. 2006. DevA, a GntR-like transcriptional regulator required for development in Streptomyces coelicolor. J Bacteriol 188:5014–5023. doi: 10.1128/JB.00307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu D, Seghezzi N, Esnault C, Virolle MJ. 2010. Repression of antibiotic production and sporulation in Streptomyces coelicolor by overexpression of a TetR family transcriptional regulator. Appl Environ Microbiol 76:7741–7753. doi: 10.1128/AEM.00819-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romero DA, Hasan AH, Lin YF, Kime L, Ruiz-Larrabeiti O, Urem M, Bucca G, Mamanova L, Laing EE, Wezel GP, Smith CP, Kaberdin VR, McDowall KJ. 30 September 2014. A comparison of key aspects of gene regulation in Streptomyces coelicolor and Escherichia coli using nucleotide-resolution transcription maps produced in parallel by global and differential RNA sequencing. Mol Microbiol doi: 10.1111/mmi.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen KT, Ritz D, Gu JQ, Alexander D, Chu M, Miao V, Brian P, Baltz RH. 2006. Combinatorial biosynthesis of novel antibiotics related to daptomycin. Proc Natl Acad Sci U S A 103:17462–17467. doi: 10.1073/pnas.0608589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao G, Wang L, Liu Q, Guan F, Huang Y, Hu C. 2013. Manipulation of kynurenine pathway for enhanced daptomycin production in Streptomyces roseosporus. Biotechnol Prog 29:847–852. doi: 10.1002/btpr.1740. [DOI] [PubMed] [Google Scholar]
- 32.Huang D, Wen J, Wang G, Yu G, Jia X, Chen Y. 2012. In silico aided metabolic engineering of Streptomyces roseosporus for daptomycin yield improvement. Appl Microbiol Biotechnol 94:637–649. doi: 10.1007/s00253-011-3773-6. [DOI] [PubMed] [Google Scholar]
- 33.Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. 2007. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol 25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- 34.Chng C, Lum AM, Vroom JA, Kao CM. 2008. A key developmental regulator controls the synthesis of the antibiotic erythromycin in Saccharopolyspora erythraea. Proc Natl Acad Sci U S A 105:11346–11351. doi: 10.1073/pnas.0803622105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Getsin I, Nalbandian GH, Yee DC, Vastermark A, Paparoditis PC, Reddy VS, Saier MH Jr. 2013. Comparative genomics of transport proteins in developmental bacteria: Myxococcus xanthus and Streptomyces coelicolor. BMC Microbiol 13:279. doi: 10.1186/1471-2180-13-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bierman M, Logan R, Obrien K, Seno ET, Rao RN, Schoner BE. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 37.Li LL, Guo J, Wen Y, Chen Z, Song Y, Li JL. 2010. Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains. J Ind Microbiol Biotechnol 37:673–679. doi: 10.1007/s10295-010-0710-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.