Abstract
The lactococcal abortive phage infection mechanism AbiQ recently was classified as a type III toxin-antitoxin system in which the toxic protein (ABIQ) is regulated following cleavage of its repeated noncoding RNA antitoxin (antiQ). In this study, we investigated the role of the antitoxin in antiphage activity. The cleavage of antiQ by ABIQ was characterized using 5′ rapid amplification of cDNA ends PCR and was located in an adenine-rich region of antiQ. We next generated a series of derivatives with point mutations within antiQ or with various numbers of antiQ repetitions. These modifications were analyzed for their effect on the antiphage activity (efficiency of plaquing) and on the endoribonuclease activity (Northern hybridization). We observed that increasing or reducing the number of antiQ repeats significantly decreased the antiphage activity of the system. Several point mutations had a similar effect on the antiphage activity and were associated with changes in the digestion profile of antiQ. Interestingly, a point mutation in the putative pseudoknot structure of antiQ mutants led to an increased AbiQ antiphage activity, thereby offering a novel way to increase the activity of an abortive infection mechanism.
INTRODUCTION
Lactococcus lactis is a Gram-positive bacterium used by the dairy industry to transform milk into fermented products such as cheese and yogurt. Many virulent phages specific to L. lactis strains have emerged over years of production, and despite numerous control strategies, they still represent one of the major risks of productivity loss in cheese factories (1). The constant threat of phage infection led to the selection of strains with robust natural antiphage systems. Antiphage mechanisms can prevent phage adsorption, block the entry of phage DNA, cleave foreign nucleic acids using restriction-modification systems or CRISPR-Cas systems, or abort infection through altruistic suicide (2). The latter group of antiphage mechanisms are known as abortive infection systems (Abi). Globally, they act at various steps of the phage replication cycle, from DNA replication to bacterial lysis (3, 4), but their common characteristic is inducing cell death in phage-infected bacteria, seemingly to favor the survival of the bacterial population (3).
To date, an impressive number of distinct Abi systems have been identified in L. lactis (3–6). These 23 systems are effective, at various degrees, against some or all prevalent groups of lactococcal phages (936, c2, and P335) found in dairy plants (3). Nevertheless, only a few Abi systems have been characterized at the molecular level. In the lactococcal AbiD1 system, the phage protein ORF1 (bIL66) activates abiD1 translation, and the effective bacterial protein AbiD1 reduces transcription of a phage gene coding for an RuvC-like resolvase that is essential for replication and maturation of viral DNA (7–10). In the AbiK system, the AbiK protein has a template-independent reverse transcriptase activity that generates random cDNA fragments, which likely prevents viral protein translation (11, 12). In AbiP+ cells, phage DNA replication is stopped by the accumulation of early transcripts that prevent transcription of the middle/late phage genes (13). It has been suggested that this phenotype is caused by direct binding of the AbiP membrane protein to RNA and single-stranded DNA (14). Like AbiP, AbiV causes a significant reduction in transcription of the middle- and late-expressed genes (15). During the infection, the phage protein SaV directly interacts with AbiV and inhibits the translational machinery of the cell (15, 16). Finally, AbiQ was recently identified as a type III toxin-antitoxin system (17).
A toxin-antitoxin (TA) system is typically a bicistronic operon that codes for a toxic protein and its cognate antitoxin, which is more prone to degradation under stress conditions (18). Originally described for their role in postsegregational killing (plasmid stabilization) with the characterization of the CcdA/CcdB system (19), many other functions now have been associated with TA systems: protection against phages, persistence, biofilm formation, global cell regulation, and stabilization of mobile genetic elements (20–25). TA systems currently are divided into five groups (types I to V) based on the nature of the antitoxin and the mode of regulation of its cognate toxin (18, 20).
Type I TA systems involve an antisense RNA that regulates translation of the toxic protein (26). In type II systems, an antitoxic protein interacts directly with a toxic protein to inhibit its activity (27). The type III systems involve a noncoding RNA that regulates the toxin through protein-RNA complex formation (25, 28). Like type II systems, type IV systems involve two proteins, but the antitoxin interacts with the target of the toxin, rather than the toxin itself, to prevent toxin activity (29, 30). Finally, the antitoxin of the type V system has specific endoribonuclease activity that regulates toxic gene transcription (31).
Only a few type III TA systems have been characterized to date (17, 25, 28, 32). ToxIN, an Abi system from Pectobacterium atrosepticum (ToxINPa), was the first to be studied and is the model for type III systems (25). The antitoxin (ToxI) is a 5.5 repeat of a 36-nucleotide (nt) noncoding RNA that is cleaved specifically by the toxin (ToxN) (25, 28). The mature small RNA fragment (one repeat of 36 nt) interacts directly with ToxN, forming a triangular heterohexameric (3ToxI:3ToxN) complex that inhibits toxicity (28). It also has been shown that the secondary structure (pseudoknot) of ToxI RNA is essential for the antitoxic activity (28). Under stress conditions, ToxN is free and can target essential mRNAs, leading to cell growth arrest and preventing phage replication (25, 28, 32).
The protein ToxN has 31% identity with the ABIQ protein from the L. lactis AbiQ system, suggesting a similar mode of action (25, 33). The antitoxin, antiQ, is a 2.8 repeat of 35 nt located downstream of a rho-independent terminator and the abiQ gene, respectively (17, 33). Similar to ToxINPa, the toxic ABIQ protein is an endoribonuclease that specifically cleaves its cognate antitoxin (17). The superimposed three-dimensional (3D) structures of these two systems suggest that they share a similar mechanism of regulation (17). Interestingly, the endoribonuclease activity is not necessarily associated with the antiphage activity, as key amino acid residues of ABIQ protein were different for both activities (17). AbiQ was described first as a defense mechanism against lactococcal phages of the c2 and 936 phage groups, and its activity resulted in the accumulation of concatemeric viral DNA (33). It was recently shown that phage P008 (936 group) can become resistant to AbiQ through mutations within its orf38 gene or ribosome binding site, suggesting that the phage protein ORF38 is playing an essential role in the activity of AbiQ (34).
In this study, we investigated the role of the antiQ antitoxin region and the effect of mutations in this region on the antiphage activity of the lactococcal abortive infection mechanism AbiQ.
MATERIALS AND METHODS
Bacterial strains and phage propagation.
Phages, bacteria, and plasmids used in this study are listed in Table 1. All L. lactis strains were grown at 30°C in M17 medium (Oxoid) supplemented with 0.5% glucose (GM17). Escherichia coli was grown in LB medium at 37°C with agitation. When needed, ampicillin (100 μg/ml) or chloramphenicol (5 μg/ml) was added to the medium. For phage propagation, L. lactis IL1403 was grown at 30°C to an optical density at 600 nm (OD600; Spectronic 20) of 0.1 before the addition of 105 to 107 PFU/ml and CaCl2 to a final concentration of 10 mM. After complete lysis, the clear lysate was passed through a 0.45-μm filter and kept at 4°C until use. Phage titer was determined by the double agar overlay plaque assay (35), in triplicate, on a lawn of L. lactis IL1403 on GM17 plates, and the contents were incubated overnight. The efficiency of plaquing (EOP) was calculated by dividing phage titer on a resistant strain (AbiQ+) by phage titer on a sensitive strain (AbiQ−). To increase titer, phages were purified on a discontinuous CsCl gradient (36).
TABLE 1.
Strains, plasmids, and phages used in this study
| Bacterial strain, plasmid, or phage | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| Lactococcus lactis | ||
| IL1403 | Laboratory strain, plasmid free; host of phage P008 | 48 |
| MG1363 | Laboratory strain, plasmid free; cloning strain | 49 |
| Escherichia coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIq ZΔM15 Tn10 (Tetr)] | Stratagene |
| MG1655 | F− λ− ilvG-rfb-50 rph-1 | 50 |
| Plasmids | ||
| pBS-KS | Cloning vector suitable for blue-white screening, 3.0 kb, Ampr | Stratagene |
| pNZ123 | Shuttle vector (L. lactis and E. coli), 2.5 kb, Cmr | 51 |
| pNZ123-AbiQ | AbiQ-wt operon cloned in pNZ123 at EcoRI site, Cmr | 17 |
| pNZ-AbiQ (1.8r) | AbiQ operon in pNZ123, antiQ of 1.8 repeat (Mut 1.8r), Cmr | This study |
| pNZ-AbiQ (3.8r) | AbiQ operon in pNZ123, antiQ of 3.8 repeat (Mut 3.8r), Cmr | This study |
| pNZ-AbiQ (A13C) | AbiQ operon in pNZ123, mutation A13C in the first antiQ repeat, Cmr | This study |
| pNZ-AbiQ (A24C) | AbiQ operon in pNZ123, mutation A24C in the first antiQ repeat, Cmr | This study |
| pNZ-AbiQ (T25C) | AbiQ operon in pNZ123, mutation T25C in the first antiQ repeat, Cmr | This study |
| pNZ-AbiQ (A26C) | AbiQ operon in pNZ123, mutation A26C in the first antiQ repeat, Cmr | This study |
| pNZ-AbiQ (A28C) | AbiQ operon in pNZ123, mutation A28C in the first two antiQ repeats, 3.8 repeat, Cmr | This study |
| pNZ-AbiQ (G32A-3.8) | AbiQ operon in pNZ123, mutation G32A in the first two antiQ repeats, 3.8 repeat, Cmr | This study |
| pNZ-AbiQ (G32A) | AbiQ operon in pNZ123, mutation G32A in the first antiQ repeat, Cmr | This study |
| Phages | ||
| P008 | Siphoviridae, 936 group, sensitive to AbiQ | 52 |
| P008-Q12 | P008 mutated in ORF38 (Pro38Leu), resistant to AbiQ | 34 |
Cloning and DNA manipulation.
Primers used in this study can be found in Table S1 in the supplemental material. Plasmids (Table 1) were purified using a QIAprep spin Miniprep kit according to the manufacturer's instructions. For L. lactis strains, a lysozyme treatment (30 mg/ml in P1 buffer at 37°C for 20 min) was added to weaken the cell wall. To construct plasmids of interest, an endonuclease-based cloning strategy was carried out as described elsewhere (36). Briefly, DNA fragments were amplified by standard PCR, digested using restriction enzymes, ligated to a dephosphorylated restricted plasmid (overnight [O/N] at 16°C), and then introduced into competent cells. The following enzymes and commercial kits were used as recommended by the manufacturers: restriction enzymes (Roche), Taq DNA polymerase (Feldan), Antarctic phosphatase (New England Biolabs), T4 DNA ligase (Invitrogen), and QIAquick PCR purification (Qiagen). DNA was sequenced at the Plateforme de Séquençage et de Génotypage des Génomes of the CHUL Center. Sequences were analyzed using BioEdit (37), Staden (38), or Geneious software (39).
5′ Rapid amplification of cDNA ends (5′ RACE PCR).
Approximately 109 cells (O/N culture) of L. lactis IL1403 pNZ123-AbiQ were centrifuged, and the pellets were flash frozen (ice-cold 80% isopropanol) at −80°C. After pretreating the cells with lysozyme (60 mg/ml in 25% sucrose solution) for 10 min at 37°C, total RNA was extracted using TRIzol reagent, as recommended by the manufacturer (Invitrogen). The extracts then were treated with DNase I (Roche) and protected with RNase inhibitors (Roche) for 30 min at 37°C before being purified again using RNA clean-up protocols (RNeasy kit) as recommended (Qiagen). Total RNA was retrotranscribed (primer JS2) in accordance with the manufacturer's instructions using Superscript III (Invitrogen) and then treated with RNase H (Roche). The resulting double-stranded cDNA was purified using a PCR cleanup kit (Qiagen). A 5′ poly(G) tail was added using terminal transferase as recommended (Invitrogen). The cDNA was amplified by PCR using primers PolyC/AbiQRev or AbiQFwd/AbiQRev (control) before being directionally cloned into pBluescript II KS+ (XhoI and EcoRI restriction sites) and transferred into E. coli XL1-Blue for blue/white screening (40). Plasmids from clones were extracted and the insert regions sequenced (primers M13Fwd/M13Rev).
Site-directed mutagenesis.
Mutagenesis was carried out as described elsewhere (17). Briefly, the plasmids were isolated from L. lactis IL1403 and then transferred into E. coli MG1655 by chemical transformation (36). Plasmid DNA was extracted before being amplified by PCR (16 cycles) using mutated reverse-complementary primers in combination with the high-fidelity enzyme Pwo (Roche). The residual template plasmid was removed using DpnI (Roche), which cleaves methylated DNA. Amplified plasmids were introduced into the subcloning strain L. lactis MG1363 by electroporation (41) and confirmed by sequencing (primers pNZ-F/pNZ-R) before being moved into L. lactis IL1403 by electroporation (41).
Bacterial growth and stationary-phase mortality experiments.
Bacterial growth was measured at an optical density of 630 nm (OD630; Biotek Synergy 2 spectrophotometer). For each strain tested, eight experimental and three biological replicates were set up in 96-well plates. The growth rate (g) was calculated by manual determination of the slope in the exponential growth phase. In stationary-phase mortality tests, bacteria were grown for 6 h and then sampled at various times (zero hours [t0], t1, t2, t4, and t8). Samples were diluted in cold GM17 medium, spread on GM17 agar medium, and grown overnight prior to counting CFU.
Northern hybridization.
A time course phage infection (noninfected and 2, 10, 20, 30, and 40 min) was carried out as described previously (17). Total RNA first was extracted and purified as described above (5′ RACE PCR). The concentrations of total bacterial RNA extracts were determined using a NanoDrop 2000 and diluted to a concentration of 1 μg/μl. A total of 5 μg of purified RNA was added to formamide loading buffer (98% deionized formamide, 10 mM EDTA, pH 8.0, 0.025% xylene cyanol, and 0.025% bromophenol blue) at a 1:1 ratio and separated on a 10% polyacrylamide–8 M urea denaturing gel. The RNA was electrophoretically transferred to a nylon membrane (Roche) before being fixed by exposure to UV for 2 min (36). A DNA probe complementary to the antiQ antitoxin (GCTCCAATTTTATCAATTCCAACTATGGCTTGGATA) or the abiQ gene (GGGGTATTAATTCGCTGTCAGGAACTGGAATC) was radiolabeled with 32P (Perkin-Elmer) using polynucleotide kinase (Roche) and purified using a Micro Bio-spin P-30 size-exclusion column (Bio-Rad). The radiolabeled probes were diluted to 1 × 106 cpm/ml with a Beckman Coulter LS6500. Hybridization was carried out for 18 h at 42°C, and the filters were washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus SDS 0.05% and revealed by autoradiography using BioMax XAR film (Kodak).
RESULTS
ABIQ cleaves its antitoxin (antiQ) in an adenine-rich region.
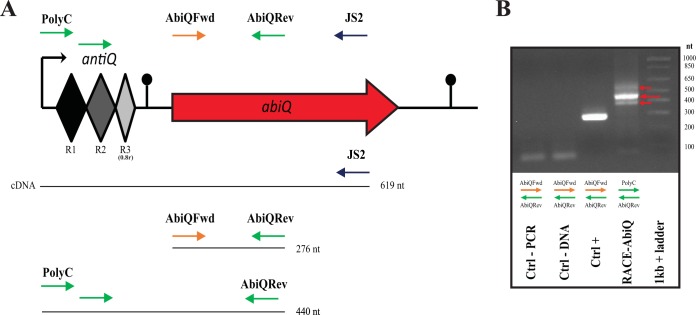
When the noncoding RNA antitoxin, antiQ, is cleaved by its cognate toxin ABIQ, a specific digestion profile is produced (17). To further characterize this RNA maturation process in vivo, we used 5′ RACE PCR to determine the cleavage site and also the transcription initiation site of the AbiQ operon. We extracted total RNA from L. lactis IL1403 AbiQ+ and retrotranscribed it into cDNA before carrying out PCR amplification using the added 5′polyguanine tail as a guide for the primer (Fig. 1). The RACE-AbiQ PCR products included a band at 450 bp surrounded by two bands of lower intensity approximately 50 bp apart. These cDNA fragments corresponded to the expected sizes of digested antiQ RNA fragments (350 to 450 bp) and the addition of 5′ noncomplementary primer nucleotides (49 bp). This PCR extract was cloned into the vector pBS-KS and transferred into E. coli, and the clones were screened for ampicillin resistance and PCR products with the expected insert length (400 to 500 bp). Twelve positive clones were sequenced (Fig. 2).
FIG 1.
AbiQ operon and PCR products of the 5′RACE assay. (A) The specific position (and orientation) of each primer (arrows) is represented in the schematic form of the operon AbiQ. The PCR products are Ctrl-PCR (AbiQFwd/AbiQRev; water as the template), Ctrl-DNA (AbiQFwd/AbiQRev; RNA without retrotranscription as the template), Ctrl+ (AbiQFwd/AbiQRev; cDNA as the template), and RACE-AbiQ (PolyC/AbiQRev; cDNA as the template). (B) 5′RACE PCR products of the AbiQ operon separated on a 2.0% agarose gel. Red arrows point out the three PCR product bands. The molecular size standard was a 1-kb Plus DNA ladder (Invitrogen).
FIG 2.
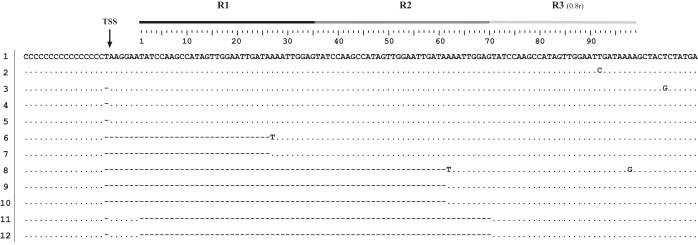
Sequences of the 12 pBS-KS::RACE-AbiQ clones. The repeats (R) and the transcription start site (position −7/−6) are indicated over the first sequence (clone 1), with different shades of gray representing the three repeats. ·, identical sequence; −, the absence of the nucleotide in the sequenced clone.
Point mutations were observed in some of the sequences and are thought to be caused by the error rate of the reverse transcriptase (42) combined with the error rate of the Taq polymerase. In each case, the 5′ end of the sequenced fragment was represented by the polycytosine sequence confirming the clones (Fig. 2). Five clones (numbers 6 to 10) demonstrated the specific ABIQ cleavage site within the antiQ sequence. In all cases, the cleavage site was between adenine 26 and adenine 27 (A/AAA) of the first or second repeats. None of the clones showed cleavage in the last repeat (0.8 repeats), suggesting no cleavage or a lower frequency of cleavage even if the sequence of the last repeat is identical to those of the other two repeats up to nucleotide 29.
Transcription of the AbiQ operon.
Analysis of the sequencing results of clones 1 to 5 (Fig. 2) indicates that transcription of the AbiQ operon starts at one of two adjacent nucleotides: T at the −7 position or A at the −6 position relative to the first nucleotide of the first antiQ repeat. Bioinformatic analysis suggested similar results by predicting the transcription start site (TSS) at the same T with a 99% degree of confidence (Neural Network Promoter Prediction–prokaryote; http://www.fruitfly.org/seq_tools/promoter.html). This also agrees with the −10 and −35 bacterial promoter boxes identified at −8 (TATAAT) and −34 (TTGCAT) (not shown), respectively.
antiQ is cleaved less frequently in the last repeat (0.8 repeat).
The cleavage and transcription start sites described above then were used to associate mature (cleaved) fragments of antiQ to the specific bands previously observed (17) in the in vivo digestion profile as visualized by Northern hybridization (Fig. 3). Of the theoretically possible small RNA fragments, only the 106-nt fragment (transcription start site to last cleavage site [0.8r]) and the 74-nt fragment (first cleavage site to last cleavage site [0.8r]) appeared to be rare or absent, respectively. In both cases, the formation of these fragments is dependent on cleavage by ABIQ within the last repeat, and they support the cloning results described above that suggested a low frequency of cleavage within this 0.8 repeat.
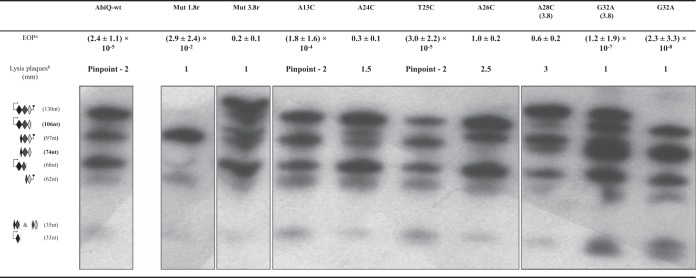
FIG 3.
Efficiency of plaquing (EOP), size of lysis plaques, and digestion profile of antiQ for wild-type and mutated AbiQ operons. One antiQ repeat (GCTCCAATTTTATCAATTCCAACTATGGCTTGGATA) was used as a probe to define the digestion profile for AbiQ mutants in Northern hybridization experiments. Superscript letters: a, EOP and standard deviations were calculated from at least three biological assays; b, P008 infecting L. lactis IL1403 pNZ123 (AbiQ−) produces lysis plaques of 3 to 5 mm.
The number of repeats in antiQ is important to the antiphage activity.
In a previous study, we investigated the effect of point mutations in the gene coding for the toxin protein, ABIQ (17). In the process of generating these mutations, we isolated a few clones that had acquired or lost repeat sequences in the antiQ region (0.8 to 3.8 repeats). These mutants also were detected here, as two clones (11 and 12) lost two complete repeats. Interestingly, one of the clones (11) lost two complete repeats without any AbiQ mutations, suggesting that the variation in the number of repeats is not inevitably associated with mutation in the AbiQ operon (Fig. 2).
We used directed mutagenesis to construct mutants in antiQ that varied in the number of repeats by bringing back the wild-type sequence of abiQ. Mutants containing 1.8 and 3.8 repeats were easily obtained. However, we were unable to isolate clones with only the 0.8 repeat, suggesting that a 0.8 repeat is not enough to avoid the toxic effects of ABIQ. We then compared the antiphage activity of these clones against phage P008 by evaluating the EOP (Fig. 3). Deleting (1.8r) or adding (3.8r) one repeat to the wild-type 2.8 repeats led to a significant reduction (3 to 5 logs) in the phage resistance phenotype. Moreover, in contrast to phage escape mutants obtained on wild-type AbiQ (34), none of the phages from plaques picked on the two mutated antiQ plates had mutations in the orf38 gene (data not shown).
We also performed Northern hybridization experiments using a probe complementary to one repeat of antiQ. As expected, the cleavage pattern of antiQ was affected by the number of repeats, as more RNA fragments were observed with the added repeat while a much simpler profile was obtained with the 1.8 repeats (Fig. 3). However, the expected relative abundance of each fragment was not affected (Fig. 3). We then infected AbiQ-containing strains (Mut 1.8r, Mut 3.8r, and AbiQ-wt) with P008 and performed Northern hybridization experiments targeting antiQ or abiQ. We observed no differences between strains containing variable repeats, demonstrating that the reduction in antiphage activity was not caused by differences at the level of transcription of either antiQ or the toxic gene abiQ (see Fig. S1 in the supplemental material). Taken together, these results suggest that the wild-type length of antiQ (2.8 repeats) is critical for optimal antiphage activity.
More than one mature antiQ fragment plays a role in the regulation of ABIQ toxin.
To further characterize the antiQ gene coding for the antitoxin, we tested the effect of point mutations on AbiQ antiphage and endoribonuclease activities (Fig. 3). The mutations were introduced specifically in the first repeat, leaving a wild-type sequence of 1.8 repeats previously shown to be sufficient for cell survival (described above). The mutations mostly surrounded the cleavage site (A26/A27) in the antiQ repeat (A24C, T25C, A26C, and A28C). The A13C mutation was used as a control, as we observed no differences in phage EOPs and antiQ digestion profile relative to those of wild-type antiQ (Fig. 3). Similar results were obtained for mutation T25C, located only two nucleotides from the cleavage site. This nucleotide likely is not essential for ABIQ recognition and/or cleavage, or it is permissive to pyrimidine bases at this site.
Conversely, mutations A24C, A26C, and A28C resulted in significant changes to the digestion profile. The A24C mutation partially reduced the amount of the 97-nt fragment (first cleavage site to terminator) and similarly increased the amount of the 106-nt fragment (transcription start site to last cleavage site [0.8r]). A similar pattern was obtained for mutation A26C, but the transition between these two fragments was more complete. Based on the digestion profile depicted in Fig. 3, these two mutations partially (A24) or completely (A26) prevented cleavage in the first repeat, which likely leads the protein ABIQ to target the 0.8 repeat region. The clone containing the A28C mutation carried a duplication of the mutated repeat. The digestion profile suggested partial inhibition of cleavage for A28C, similar to that for A24C. We performed bioinformatic analysis (RNAfold software) to compare the predicted secondary structures of wild-type and mutated antiQ RNA (see Fig. S2 in the supplemental material). The wild-type antiQ sequence had free access to the first ABIQ cleavage site, unlike the A24 and A28 mutants, where this site likely was sterically hindered. The A26 mutant resulted in no predicted modification to the general structure, suggesting that this nucleotide is essential for recognition and/or cleavage by ABIQ. In all three cases (A24C, A26C, and A28C), the single point mutation led to a phage EOP of almost 1, indicating a significant loss of antiphage activity for these mutated AbiQ systems. These results confirm that the number and relative abundancy of ABIQ-generated antiQ RNA fragments play a role in the control of ABIQ toxicity and antiphage activity.
Antiphage activity of AbiQ can be significantly increased by a point mutation in the putative pseudoknot structure of antiQ.
It has been described for the type III toxin-antitoxin system ToxINPa that specific point mutations in the RNA antitoxin can reduce cell survival, mostly by affecting the pseudoknot secondary structure of RNA (28). Based on the model, the free toxin causes cell death and prevents phage replication. We generated a G32A mutant which would significantly modify the predicted secondary structure (pseudoknot) of the antiQ RNA (see Fig. S3 in the supplemental material). Two different mutants containing G32 mutation were obtained with either 2.8 or 3.8 repeats (duplication of the mutated repeat). Both clones were characterized for their antiphage activity and endoribonuclease digestion profile (Fig. 3).
Interestingly, the antiphage efficiency of ABIQ was significantly increased (3 logs) with a mutated antiQ G32A compared to the efficiency with wild-type antiQ (from an EOP of 10−5 to an EOP of 10−8). Remarkably, the mutated antiQ G32A with 3.8 repeats also increased ABIQ efficacy by 6 logs compared to that of the wild-type antiQ with 3.8 repeats (from an EOP of 10−1 to an EOP of 10−7). No difference in the relative abundance of the RNA fragments was observed between the two clones (other than the extra repeat). However, the abundance of the 97-nt fragment (first cleavage site to terminator) increased relative to that of all other bands in the pattern. It still is not clear if this modification plays a role in AbiQ regulation, but it suggests that the number of inhibitory RNA fragments is important.
To further characterize the mutated antiQ G32A, we analyzed the bacterial fitness of these clones. The comparison between the wild type (2.8r) and mutant (G32A-2.8r) revealed no differences between generation times (60 min; data not shown). Stationary-phase mortality assays also demonstrated no differences between the strains tested (data not shown), strongly suggesting that the antiQ G32A mutation does not affect fitness while increasing antiphage efficiency. Taken together, our results showed the role of the G32A mutation within antiQ for optimization of complete antiphage activity.
DISCUSSION
Described as an efficient abortive infection system against prevalent dairy lactococcal phage groups (936 and c2) (33), the AbiQ mechanism recently has been reported as a type III TA system (17). To understand the molecular basis of the mechanism, two previous studies focused on the role of specific phage proteins in AbiQ antiphage activity (34) and on the specificity of the toxic protein ABIQ within the system (17). Here, we investigated the role of the antitoxin (antiQ, 2.8 repeats of 35 nt), emphasizing the effects of mutations on the antiphage activity.
Within the AbiQ system, the toxic protein (ABIQ) is a sequence-dependent endoribonuclease that specifically cleaves its cognate noncoding RNA antitoxin (17). Our 5′ RACE experiments showed that the cleavage site was located within the repeat between adenine 26 and adenine 27 in an adenine-rich region (A/AAA), one nucleotide away from a predicted in silico ABIQ-antiQ cleavage site (/AAAA) (43). Interestingly, the toxin of the two other type III TA systems also cleaved RNA fragments in adenine-rich regions (ToxINPa, AA/AU; ToxINBt, A/AAAA) (32). In antiQ, this polyadenine sequence is found in the complete repeats (R1 and R2) and also in the last 0.8 repeat (R3). Our data showed that ABIQ seems to cleave at the same frequency between the first and the second repeats. However, cleavage in the last repeat is either rare or absent. This may be explained by the presence of a stem-loop structure (transcriptional terminator) next to the last repeat that could sequester the cleavage site. Moreover, secondary structure prediction of antiQ RNA suggests that the polyadenine region interacts with polyuracil residues of the terminator sequence, increasing the total length of the stem-loop structure. In a few cases, this extension of the stem-loop structure (rho-independent terminator) can lead to reduced termination efficiency (44). It also is possible that nucleotides 30 to 35 (absent from the last repeat) are important for recognition and/or cleavage by ABIQ.
Using 5′ RACE PCR, the transcription start site of abiQ was determined to be 6 or 7 nucleotides upstream of the start of the first repetition, more specifically at a thymine or adenine residue, respectively. This double transcription site phenomenon has been described in the characterization of transcription start sites in E. coli (45). Interestingly, thymine and adenine were identified as transcription start sites in 35% (7 nt upstream) and 31% (6 nt upstream) of cases. We also investigated the role of specific modifications within the antiQ region. Our data showed that 1.8 repeats (antiQ) are enough to enable cell survival (and neutralize ABIQ) because clones were obtained, which was not possible with only a 0.8 repeat. In comparison, ToxINPa needs 2.5 repeats (out of 5.5 repeats) to prevent ToxN toxicity in vivo (46). However, when the protein (ToxN) is expressed in trans, only 1.5 repeats are necessary (25). Presumably, the key would be to have at least one complete mature repeat fragment (from cleavage site to cleavage site), with this fragment playing a critical role in toxin regulation within the type III TA systems (28, 32).
Two clones with either Mut 1.8 or Mut 3.8 repeats were used to see the effect on antiphage activity of ABIQ. In both cases, the EOP of phage P008 was reduced by only 1 or 2 logs compared to the phage titer on the sensitive strain without AbiQ. This value is far from the EOP of 10−5 obtained with the wild-type AbiQ system. Interestingly, phages replicating on the mutated strains (Mut 1.8r and Mut 3.8r) do not have mutations in orf38, in contrast to AbiQ (2.8r), where escaping P008 phages were shown to be mutated in that gene (34). This indicates that modification in the number of repeats (antiQ) leads to reduced efficiency of AbiQ that enables the wild-type phage to directly bypass the system rather than selecting a phage-escaping mutant. It is plausible that one additional repeat (Mut 3.8r) leads to an increased number of regulatory RNA fragments, thereby regulating the toxin ABIQ, increasing cell survival, and decreasing antiphage activity. Conversely, it is unclear how Mut 1.8r reduces the antiphage activity, since the deletion did not cause a change at the level of antiQ or abiQ transcription.
Characterization of antiQ point mutants also gives key information on AbiQ mechanism. Our data showed the importance of specific nucleotides in the cleavage of antiQ and suggested that the secondary structure of RNA also is important in the cleavage. Three mutations (A24C, A26C, and A28C) led to significant reductions in antiphage activity and were associated with changes in the profile of antiQ digestion by ABIQ. Blocking cleavage in the first repeat brought ABIQ to cleave in the last repeat and changed the ratio of specific fragments, increasing the 106-nt fragment (transcription start site to last cleavage site [0.8r]) and lowering the 97-nt fragment (first cleavage site to terminator). Experiments assessing the EOPs of phage P008 suggest that this 106-nt fragment is implicated in the regulation of ABIQ toxin, preventing its release, favoring cell survival, and leading to normal replication of the phage. This fragment could be another inhibitory RNA, like the mature repeat (cleaved RNA, 36 nt) identified for the type III TA ToxINPa (28) that probably also is involved in regulation in the AbiQ system. On the other hand, it also is possible that the 97-nt fragment (first cleavage site to terminator) acts as an anti-inhibitory fragment and that reduction of this fragment leads to this phenomenon.
It has been shown that the pseudoknot structure of type III antitoxin RNA is essential for preventing toxicity of the cognate toxin (28). However, it had not been shown that mutations affecting this structure could increase antiphage activity. Our data showed that a single specific mutation (G32A) led to a significant increase (3 logs) in antiphage activity. Even the G32A-3.8r mutant led to a remarkable 6-log EOP reduction compared to the level for the 3.8r mutant. Surprisingly, this G32A mutation also increased AbiQ antiphage activity against the previously isolated (34) AbiQ-resistant phage P008-Q12 (EOP of 10−4; data not shown). We propose that the G32A mutation inactivated the inhibitory fragments, thereby preventing phage replication. Interestingly, the G32A mutants did not reduce the fitness of AbiQ (G32A)-containing bacteria. Taken together, a single specific mutation in antiQ can increase the antiphage activity of AbiQ.
In summary, AbiQ is a complex molecular system that provides phage resistance. Our results suggest that different inhibitory RNA fragments regulate ABIQ. Under stress conditions (phage infection), a specific phage protein can destabilize the regulatory complex by interacting with the antitoxin or by changing ABIQ activity. Freeing the toxin would enable it to cleave phage and cellular mRNA, cause growth arrest, and prevent phage replication. Also, our data showed that we can optimize the AbiQ system through a single mutation in the pseudoknot structure. Better comprehension of antiphage systems is the key to limiting rapid phage evolution, where phages often find a strategy to avoid the bacterial barrier (47).
Supplementary Material
ACKNOWLEDGMENTS
We thank Barbara-Ann Conway for editorial assistance and Julie Samson and Alex Hynes for discussion.
This work was supported through a strategic grant from the Natural Sciences and Engineering Research Council of Canada. M.B. is the recipient of a scholarship from Quebec Protein Structure, Function and Engineering Research Network (PROTEO). S.M. holds a Tier 1 Canada Research Chair in Bacteriophages.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00572-15.
REFERENCES
- 1.Rousseau GM, Moineau S. 2009. Evolution of Lactococcus lactis phages within a cheese factory. Appl Environ Microbiol 75:5336–5344. doi: 10.1128/AEM.00761-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 3.Chopin M-C, Chopin A, Bidnenko E. 2005. Phage abortive infection in lactococci: variations on a theme. Curr Opin Microbiol 8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Durmaz E, Klaenhammer TR. 2007. Abortive phage resistance mechanism AbiZ speeds the lysis clock to cause premature lysis of phage-infected Lactococcus lactis. J Bacteriol 189:1417–1425. doi: 10.1128/JB.00904-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holubova J, Josephsen J. 2007. Potential of AbiS as defence mechanism determined by conductivity measurement. J Appl Microbiol 103:2382–2391. doi: 10.1111/j.1365-2672.2007.03507.x. [DOI] [PubMed] [Google Scholar]
- 6.Haaber J, Moineau S, Fortier L-C, Hammer K. 2008. AbiV, a novel antiphage abortive infection mechanism on the chromosome of Lactococcus lactis subsp. cremoris MG1363. Appl Environ Microbiol 74:6528–6537. doi: 10.1128/AEM.00780-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bidnenko E, Chopin MC, Ehrlich SD, Anba J. 2002. Lactococcus lactis AbiD1 abortive infection efficiency is drastically increased by a phage protein. FEMS Microbiol Lett 214:283–287. doi: 10.1111/j.1574-6968.2002.tb11360.x. [DOI] [PubMed] [Google Scholar]
- 8.Bidnenko E, Ehrlich D, Chopin M-C. 1995. Phage operon involved in sensitivity to the Lactococcus lactis abortive infection mechanism AbiD1. J Bacteriol 177:3824–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bidnenko E, Ehrlich SD, Chopin M-C. 1998. Lactococcus lactis phage operon coding for an endonuclease homologous to RuvC. Mol Microbiol 28:823–834. [DOI] [PubMed] [Google Scholar]
- 10.Curtis FA, Reed P, Sharples GJ. 2005. Evolution of a phage RuvC endonuclease for resolution of both Holliday and branched DNA junctions. Mol Microbiol 55:1332–1345. doi: 10.1111/j.1365-2958.2004.04476.x. [DOI] [PubMed] [Google Scholar]
- 11.Fortier L-C, Bouchard JD, Moineau S. 2005. Expression and site-directed mutagenesis of the lactococcal abortive phage infection protein AbiK. J Bacteriol 187:3721–3730. doi: 10.1128/JB.187.11.3721-3730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C, Villion M, Semper C, Coros C, Moineau S, Zimmerly S. 2011. A reverse transcriptase-related protein mediates phage resistance and polymerizes untemplated DNA in vitro. Nucleic Acids Res 39:7620–7629. doi: 10.1093/nar/gkr397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domingues S, Chopin A, Ehrlich SD, Chopin M-C. 2004. The lactococcal abortive phage infection system AbiP prevents both phage DNA replication and temporal transcription switch. J Bacteriol 186:713–721. doi: 10.1128/JB.186.3.713-721.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domingues S, McGovern S, Plochocka D, Santos MA, Ehrlich SD, Polard P, Chopin M-C. 2008. The lactococcal abortive infection protein AbiP is membrane-anchored and binds nucleic acids. Virology 373:14–24. doi: 10.1016/j.virol.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Haaber J, Samson JE, Labrie SJ, Campanacci V, Cambillau C, Moineau S, Hammer K. 2010. Lactococcal abortive infection protein AbiV interacts directly with the phage protein SaV and prevents translation of phage proteins. Appl Environ Microbiol 76:7085–7092. doi: 10.1128/AEM.00093-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haaber J, Rousseau GM, Hammer K, Moineau S. 2009. Identification and characterization of the phage gene sav, involved in sensitivity to the lactococcal abortive infection mechanism AbiV. Appl Environ Microbiol 75:2484–2494. doi: 10.1128/AEM.02093-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samson JE, Spinelli S, Cambillau C, Moineau S. 2013. Structure and activity of AbiQ, a lactococcal endoribonuclease belonging to the type III toxin-antitoxin system. Mol Microbiol 87:756–768. doi: 10.1111/mmi.12129. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi Y, Park JH, Inouye M. 2011. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet 45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 19.Ogura T, Hiraga S. 1983. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A 80:4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unterholzner SJ, Poppenberger B, Rozhon W. 2013. Toxin-antitoxin systems: biology, identification, and application. Mob Genet Elements 3:e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnuson RD. 2007. Hypothetical functions of toxin-antitoxin systems. J Bacteriol 189:6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 23.Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A 108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Wang X, Wood TK. 2011. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol 77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A 106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fozo EM, Hemm MR, Storz G. 2008. Small toxic proteins and the antisense RNAs that repress them. Microbiol Mol Biol Rev 72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerdes K, Christensen SK, Løbner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol 3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 28.Blower TR, Pei XY, Short FL, Fineran PC, Humphreys DP, Luisi BF, Salmond GP. 2011. A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat Struct Mol Biol 18:185–190. doi: 10.1038/nsmb.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masuda H, Tan Q, Awano N, Yamaguchi Y, Inouye M. 2012. A novel membrane-bound toxin for cell division, CptA (YgfX), inhibits polymerization of cytoskeleton proteins, FtsZ and MreB, in Escherichia coli. FEMS Microbiol Lett 328:174–181. doi: 10.1111/j.1574-6968.2012.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda H, Tan Q, Awano N, Wu KP, Inouye M. 2012. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol Microbiol 84:979–989. doi: 10.1111/j.1365-2958.2012.08068.x. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Lord DM, Cheng HY, Osbourne DO, Hong SH, Sanchez-Torres V, Quiroga C, Zheng K, Herrmann T, Peti W, Benedik MJ, Page R, Wood TK. 2012. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol 8:855–861. doi: 10.1038/nchembio.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Short FL, Pei XY, Blower TR, Ong SL, Fineran PC, Luisi BF, Salmond GP. 2013. Selectivity and self-assembly in the control of a bacterial toxin by an antitoxic noncoding RNA pseudoknot. Proc Natl Acad Sci U S A 110:E241–E249. doi: 10.1073/pnas.1216039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emond E, Dion E, Walker SA, Vedamuthu ER, Kondo JK, Moineau S. 1998. AbiQ, an abortive infection mechanism from Lactococcus lactis. Appl Environ Microbiol 64:4748–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samson JE, Bélanger M, Moineau S. 2013. Effect of the abortive infection mechanism and type III toxin/antitoxin system AbiQ on the lytic cycle of Lactococcus lactis phages. J Bacteriol 195:3947–3956. doi: 10.1128/JB.00296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. 2009. Enumeration of bacteriophages by double agar overlay plaque assay, p 69–76. In Clokie MRJ, Kropinski A (ed), Bacteriophages: methods and protocols, volume 1: isolation, characterization, and interactions. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Russel DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 37.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 38.Bonfield JK, Whitwham A. 2010. Gap5–editing the billion fragment sequence assembly. Bioinformatics 26:1699–1703. doi: 10.1093/bioinformatics/btq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ullmann A, Jacob F, Monod J. 1967. Characterization by in vitro complementation of a peptide corresponding to an operator-proximal segment of the β-galactosidase structural gene of Escherichia coli. J Mol Biol 24:339–343. doi: 10.1016/0022-2836(67)90341-5. [DOI] [PubMed] [Google Scholar]
- 41.Holo H, Nes IF. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55:3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Potter J, Zhang W, Lee J. 2003. Thermal stability and cDNA synthesis capability of SuperScript reverse transcriptase. Focus 25.1:19–24. [Google Scholar]
- 43.Blower TR, Short FL, Rao F, Mizuguchi K, Pei XY, Fineran PC, Luisi BF, Salmond GP. 2012. Identification and classification of bacterial type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res 40:6158–6173. doi: 10.1093/nar/gks231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson KS, von Hippel PH. 1995. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc Natl Acad Sci U S A 92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendoza-Vargas A, Olvera L, Olvera M, Grande R, Vega-Alvarado L, Taboada B, Jimenez-Jacinto V, Salgado H, Juárez K, Contreras-Moreira B, Huerta AM, Collado-Vides J, Morett E. 2009. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One 4:e7526. doi: 10.1371/journal.pone.0007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blower TR, Fineran PC, Johnson MJ, Toth IK, Humphreys DP, Salmond GP. 2009. Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J Bacteriol 191:6029–6039. doi: 10.1128/JB.00720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samson JE, Magadán AH, Sabri M, Moineau S. 2013. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 48.Chopin A, Chopin M-C, Moillo-Batt A, Langella P. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260–263. doi: 10.1016/0147-619X(84)90033-7. [DOI] [PubMed] [Google Scholar]
- 49.Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP, van Sinderen D, Kok J. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 189:3256–3270. doi: 10.1128/JB.01768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 51.de Vos WM. 1987. Gene cloning and expression in lactic streptococci. FEMS Microbiol Rev 46:281–295. [Google Scholar]
- 52.Mahony J, Deveau H, Mc Grath S, Ventura M, Canchaya C, Moineau S, Fitzgerald GF, van Sinderen D. 2006. Sequence and comparative genomic analysis of lactococcal bacteriophages jj50, 712 and P008: evolutionary insights into the 936 phage species. FEMS Microbiol Lett 261:253–261. doi: 10.1111/j.1574-6968.2006.00372.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.