Abstract
Salmonella remains a leading cause of bacterial food-borne disease, sickening millions each year. Although outbreaks of salmonellosis have traditionally been associated with contaminated meat products, recent years have seen numerous disease cases caused by the consumption of produce. Tomatoes have been specifically implicated, due to the ability of Salmonella spp. to enter the tomato fruit and proliferate within, making the decontamination of the raw product impossible. To investigate the genetic means by which Salmonella is able to survive and proliferate within tomatoes, we conducted a screen for bacterial genes of Salmonella enterica serovar Montevideo specifically induced after inoculation into ripe tomato fruit. Among these genes, we found 17 members of the previously described anaerobic Fur (ferric uptake regulator) regulon. Fur is a transcriptional and posttranscriptional regulator known to sense iron, suggesting the importance of this mineral to Salmonella within tomatoes. To test whether iron acquisition is essential for Salmonella growth in tomatoes, we tested a ΔfepDGC mutant, which lacks the ability to import iron-associated siderophores. This mutant grew significantly more poorly within tomatoes than did the wild type, but the growth defect of the mutant was fully reversed by the addition of exogenous iron, demonstrating the need for bacterial iron scavenging. Further, dependence upon iron was not apparent for Salmonella growing in filtered tomato juice, implicating the cellular fraction of the fruit as an important mediator of iron acquisition by the bacteria.
INTRODUCTION
Salmonellae are ubiquitous pathogens present in a wide variety of warm- and cold-blooded hosts. Nontyphoidal salmonellosis is second to only campylobacterosis in cases of illness—some 1.4 million each year. It is, however, the leading cause of death among food-borne bacterial diseases (1). Because of this association with animals, outbreaks of salmonellosis have traditionally been caused by the consumption of contaminated meat and poultry products. It has become increasingly common, however, for human disease to be caused by Salmonella-contaminated vegetables and produce. In the past decades, tomatoes have been the source of several outbreaks (2–7). The Salmonella serovars implicated in these infections have been varied, including Montevideo, Newport, Javiana, and Braenderup. In some cases, the sources of infection have been found, often being contaminated water sources, while in others, the sources are unknown (4–6).
A mechanism of penetration of Salmonella into the tomato pulp has been proposed: exposure of tomatoes to contaminated water lower in temperature than the tomato itself significantly increased penetration of the fruit (8). In addition, Salmonella has been shown to contaminate tomatoes via soil and from crop debris from previous harvests (9, 10). The bacterium, when either applied to the surface of the fruit or inoculated into the stem scar or the pulp itself, can survive and proliferate (11–15). It can also survive on leaves in the absence of desiccation (16) and proliferates significantly better when coinoculated with a bacterial plant pathogen (14, 17–19). Also important to our work is the finding that Salmonella enterica serovar Montevideo, which has been the cause of human outbreaks and is used in our studies, survives well in ripe tomatoes (12, 13, 16). It is, in fact, a serovar that proliferates highly under experimental conditions (15).
Although the basis of infection of animal hosts by Salmonella has been well defined, much less is known about the genetic determinants of this pathogen required for colonization of produce plants and specifically of tomatoes. A genome-wide screen using Salmonella enterica serovar Typhimurium identified a number of metabolic and regulatory genes induced during growth in tomatoes but did not demonstrate genes required for bacterial survival in this environment (20). Further work in this area showed that the O-antigen portion of bacterial lipopolysaccharide is importance for Salmonella survival in tomatoes (21), as it is in animal hosts.
A method that has been extensively used in the study of bacterium-host interactions is in vivo expression technology (IVET). The basis of the method is the identification of bacterial genes that are selectively expressed when bacteria live in association with the host as a means of both understanding the host environment faced by the pathogen and identifying bacterial genes essential for survival within hosts. This method has been used for a number of bacteria pathogenic in several plant species, including tomato (20, 22–29). Recombinase IVET (RIVET) is a variation on the technique that induces a permanent selectable change in the bacterial phenotype in response to gene expression (30). Although plants can be considered opportunistic hosts for this bacterium, and thus specific infection mechanisms are unlikely to have evolved, it is clear that Salmonella survives and grows for prolonged periods within produce plants, likely in the face of antibacterial responses by the plant (17). It is therefore reasonable to assume that the pathogen invokes metabolic pathways for the utilization of available nutrients and the response to plant defenses. In this work, we describe the use of a RIVET screen to identify Salmonella genes expressed specifically when bacteria live within tomato fruit to gain insight into the requirements for survival and growth of the pathogen in this environment. We demonstrate that components of the bacterial iron acquisition system are selectively expressed, and that this system is required for the prolonged survival within this important produce plant.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Salmonella enterica serovar Montevideo strain ATCC BAA710 and isogenic mutants thereof were used for assays of function and for generation of DNA libraries. A derivate of S. enterica serovar Typhimurium strain ATCC 14028s was used as the vehicle for the RIVET screen, described below. Gene deletions were created using the previously described one-step inactivation method (31). Briefly, PCR products were generated from the chloramphenicol or kanamycin resistance genes of pKD3 or pKD4, respectively, using primers carrying at their 5′ ends 40 bp of homology to the regions flanking the start and stop codons of the gene to be deleted. A derivative of strain BAA710 carrying pKD46, containing the λ Red recombinase for allelic exchange, was transformed with the resultant PCR products. All deletion mutants were checked for the loss of genes by PCR. The recD::lacZ fusion was constructed by first deleting recD and then integrating lacZ at the site of the disruption as previously described (32).
The fepDGC complementing plasmid was made by PCR amplification from BAA710 a 2,898-bp region that includes the fepDGC open reading frames (ORFs) along with 110 bp of 5′ upstream promoter region using the primers 5′-TAAATAGGATCCCTATCGCCGCCCCAGCGGTA-3′ and 5′-TCGGGAAAGCTTTACAACGCCTTAATGCGAGT-3′ and cloning the amplified region into pGB2 cut with HindIII and BamHI (33). The plasmid was maintained by growth in streptomycin (20 μg/ml) and spectinomycin (100 μg/ml).
Screen for genes expressed within tomatoes.
We created a library of Salmonella genomic DNA fragments by partial digestion of total genomic DNA from S. enterica serovar Montevideo BAA710, with HaeIII, AluI, or RsaI and then size fractionated the DNA to isolate fragments of 1 to 2 kb. These three libraries were pooled, and fragments were cloned into the PmlI site of the ampicillin-marked plasmid pCA19, placing them upstream of a promoterless derivative of the phage P1 recombinase cre, which lies upstream from a promoterless lacZ (34). A derivative of strain S. Typhimurium 14028s carrying a chloramphenicol resistance marker and a pair of chromosomal loxP sites flanking npt (kanamycin resistance) and sacB (sucrose susceptibility) was P22 transduced with this library, with an initial selection on either M9 minimal medium with 0.2% glycerol or LB agar, supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), chloramphenicol (25 μg/ml), and EGTA (10 mM). S. Typhimurium was used for these experiments as it exhibited a marked sensitivity to sucrose, due to the presence of sacB, which was required for selection.
Approximately 104 independent library transductants were pooled for inoculation into tomatoes. Red-vine tomato fruit from a local grocer was inoculated with 104 to 105 bacteria, representing this pool of 104 clones, in 20 μl of phosphate-buffered saline (PBS). A total of 40 tomatoes were inoculated with 40 independent library pools. For 20 of these tomatoes, the initial selection was on LB agar containing the appropriate antibiotics, and for the remaining 20 tomatoes, selection was on M9 minimal agar with 0.2% glycerol and the appropriate antibiotics. For inoculation, stems were removed, and an 18-gauge blunt needle was used to create four channels into the tomato core tissue located immediately around the stem scar. The 20-μl inoculum was divided evenly among the four channels and to a depth within 1.5 cm of the stem scar. After 48 h, the tomato core tissue segment from 0 to 25 mm below the stem scar was harvested.
The cores were placed in a Whirl-pak filter bag (Nasco) with 1.5 ml LB or M9–0.2% glycerol medium and crushed with a glass bottle, and the liquid was collected through the bag filter. The tomato core liquid was serially diluted and plated onto either LB or M9 agar plates with added chloramphenicol, ampicillin, and 5% sucrose. From each tomato, 10 sucrose-resistant colonies were saved, for a total of 200 clones derived from each medium.
Of the 400 clones, 397 clones were successfully amplified by PCR and Sanger sequenced. The three remaining clones were isolated by preparation of plasmid DNA and sequenced. Inserts were sequenced using a primer from within the cre coding region that allowed for sequencing across the insert junction. The average length of useable sequence reads was 1,077 bp. We confirmed the sizes of all induced fragments to be in the range of 1 to 4.5 kb by PCR amplification using primers homologous to sequence that flanks the inserted fragment. The average insert size was approximately 2 kb. Cloned fragments that induced expression of cre nonspecifically (i.e., while bacteria were growing outside tomatoes) were identified through expression of the fused lacZ by β-galactosidase expression assays in either LB or M9 minimal medium with 0.2% glycerol.
Sequencing.
Sequences were obtained using an Applied Biosystems (Foster City, CA) automated 3730xl DNA analyzer by the Biotechnology Resource Center Genomics Facility at Cornell University. The sequencing primer was 5′-AGGCAAATTTTGGTGTGACC, which is located in cre. The location of each of the cloned fragments was determined by comparison to 50 draft genomes of S. enterica Montevideo (available from the National Center for Biotechnology Information, submitted by the U.S. Food and Drug Administration) and to the sequenced genomes of S. enterica serovar Typhimurium strain LT2 (GenBank accession no. NC_003197) and S. enterica serovar Typhimurium strain SL1344 (GenBank accession no. NC_016810).
Competition assays.
The two strains to be compared were grown separately overnight at 37°C with aeration, subcultured to the exponential growth phase, washed twice with PBS, mixed at a 1:1 ratio, and diluted 1:9 in PBS for inoculation. The number of bacteria in the inoculum was determined by serial dilution and plating onto LB agar supplemented with 80 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and 100 μM ferric chloride (FeCl3). For assays in tomato fruit, red vine tomatoes from a local grocer were inoculated in replicates of five with ∼106 CFU of the mixture of stains in 20 μl of PBS. For inoculation, stems were removed, and an 18-gauge blunt needle was used to create four channels into the tomato core tissue located immediately around the stem scar. The 20-μl inoculum was divided evenly among the four channels and to a depth within 1.5 cm of the stem scar.
Tomatoes were harvested either immediately or after 7 days of incubation at room temperature, and the tomato core tissue segment from 0 to 25 mm below the stem scar was processed. The cores were placed in a Whirl-pak filter bag with 10 ml LB medium and crushed with a glass bottle, and the liquid collected through the bag filter. The tomato core liquid was serially diluted, and aliquots plated onto LB agar supplemented with X-Gal (80 μg/ml) and ferric chloride (100 μM FeCl3). The iron supplement was added to plates to reverse an observed slow-growth phenotype of the ΔfepDGC mutant on LB agar plates without additional iron. Competitive index (CI) values were calculated as the mean ratio of CFU of the two strains, with a CI of >1 indicating the presence of more of the wild-type strain. Growth of strains within tomatoes was determined by comparing the mean colony-forming units per tomato recovered at 7 days to that recovered immediately after inoculation. For the tests of statistical significance, replica plates were averaged, and ratio values were converted logarithmically, while CFU values were divided by 1,000 and converted logarithmically. Data were modeled using analysis of variance (ANOVA), and significance was determined using least square (LS) means with the Tukey honestly significant difference (HSD) test.
For assays using tomato juice, tomatoes of the same variety as that used for the fruit assays were cut into quarters, and the sections were placed in the Whirl-pak filter bags and crushed using a glass bottle. We then collected the juice through the bag filters and centrifuged it to remove solid material. Supernatants were recovered, centrifuged again, and then filtered through a 0.45-μm-pore filter. The wild type and the ΔfepDGC mutant were inoculated together into 5 ml of juice at a final concentration of ∼1 × 106 per assay in a tightly capped tube sealed with Parafilm and incubated standing. Bacteria were recovered immediately after inoculation and again after 7 days of incubation at room temperature for enumeration on LB agar plates supplemented with 80 μg/ml X-Gal and 100 μM FeCl3. Assays were performed in replicates of five. All experiments were performed using disposable plastic supplies throughout to reduce the possible contamination of reagents with iron. Iron concentrations were measured using a colorimetric iron assay kit (Sigma-Aldrich) in comparison to a generated standard curve.
RESULTS AND DISCUSSION
Iron-regulated genes are selectively expressed by Salmonella spp. growing in tomatoes.
To identify bacterial functions required for the survival and growth of Salmonella within tomato fruit, we employed a recombinase in vivo expression technology (RIVET) screen. As the Montevideo serovar has frequently been associated with disease outbreaks derived from tomatoes, we introduced a plasmid library of genomic DNA derived from S. enterica serovar Montevideo fused to the recombinase cre into a Salmonella enterica serovar Typhimurium test strain carrying a chromosomal antibiotic resistance cassette flanked by loxP sites (34). After incubation in red ripe tomatoes, bacteria that had undergone recombination of the loxP cassette, indicating selective activation of the cre fusion due to the induction of promoter activity, were obtained by selection on sucrose-containing medium (see Materials and Methods for complete experimental details). This selection yielded S. Montevideo genes encoding numerous predicted functions, including energy metabolism, surface proteins, and gene regulation, that might plausibly be required for survival of the bacterium in this environment.
Conspicuous among the genes identified were those known to be required for the direct acquisition of iron (Table 1). iroC and iroE, located within a single operon, encode proteins utilized for the binding of iron and for the production of the iron siderophore salmochelin, respectively, while sitC encodes an additional iron transporter. In addition, the screen identified several genes with expression known to be controlled by Fur, the ferric uptake regulator. Fur is a transcriptional and posttranscriptional regulator that senses iron concentration and redox state. Recent work with Salmonella Typhimurium has shown it to control expression of genes in classes that include carbohydrate and nucleotide metabolism and bacterial adhesion (35). All of the genes derived from our screen of S. Montevideo in tomatoes that are shown in Table 1 are members of this previously described Fur regulon.
TABLE 1.
Salmonella genes selectively expressed within tomato fruit that are also constituents of the Fur regulon
| Gene | No. of independent isolates from library | Function |
|---|---|---|
| fimA | 5 | Type 1 fimbrial protein |
| STM4305 | 5 | Putative anaerobic dimethyl sulfoxide reductase |
| yjjI | 3 | Putative glycine radical enzyme |
| hrpA | 2 | ATP-dependent RNA helicase |
| tdcE | 2 | Pyruvate formate-lyase |
| bglJ | 1 | DNA-binding transcriptional activator |
| crr | 1 | Glucose-specific phosphotransferase |
| dfp | 1 | Bifunctional phosphopantothenoylcysteine decarboxylase/phosphopantothenate synthase |
| dmsA | 1 | Anaerobic dimethyl sulfoxide reductase |
| eutK | 1 | Ethanolamine utilization protein |
| fmt | 1 | Methionyl-tRNA formyltransferase |
| iroC | 1 | Putative ATP binding cassette (ABC) transporter |
| iroE | 1 | Salmochelin siderophore system protein |
| nrdB | 1 | Ribonucleoside-diphosphate reductase |
| sitC | 1 | Iron/manganese ABC transporter |
| STM3547 | 1 | Putative transcriptional regulator of sugar metabolism |
Interestingly, the Fur regulon was defined in S. Typhimurium using bacteria grown under anaerobic conditions, and this analysis identified regulated genes not known to be controlled by Fur when bacteria were grown in the presence of oxygen (35). That work was conducted specifically to replicate the conditions present in an animal host, where oxygen is limiting. The oxygen tension within tomato fruits under normal storage conditions, however, has been shown to be near that of ambient air (36), suggesting the presence of some additional environmental signal for the induction of the Fur regulon in our experiments. These results thus suggest that within tomato fruit, Salmonella senses and responds to an environment of restricted iron concentration. It is further plausible that the acquisition of exogenous iron in this environment is important for the survival of Salmonella while within tomato fruit.
Genes used for the transport of iron are required for the proliferation of Salmonella in tomatoes.
As iron is a limiting resource in many organisms, and as its acquisition has been shown to be essential for the persistence and virulence of Salmonella in animals (37, 38), we examined further the importance of iron acquisition to bacterial survival in tomatoes. Salmonella uses a pair of siderophores, enterobactin and salmochelin, the C-glycosylated derivative of enterobactin, to scavenge iron from the environment. Enterobactin can be captured by two distinct outer membrane receptors, FepA and IroN, while salmochelin is transported primarily by IroN (39, 40). Both siderophores, however, share a single mechanism of transport through the inner membrane, utilizing the FepDGC transporter (37). To investigate the importance of iron acquisition, we therefore constructed a ΔfepDGC mutant of S. enterica serovar Montevideo, eliminating the ability of this strain to import iron using both enterobactin and salmochelin, and tested its ability to proliferate in tomato fruit.
We used competitive index (CI) assays to compare the survival of the ΔfepDGC mutant in tomatoes to that of the wild type. Our experiments used as the wild type an isogenic strain carrying a recD::lacZY insertion, which allowed the rapid enumeration of the two strains by color on agar medium containing the chromogenic substrate X-Gal. We had determined previously that the recD::lacZY insertion had no effect on bacterial survival in tomatoes compared to that of the S. Montevideo parent strain (data not shown). We inoculated the ΔfepDGC mutant at 2 × 105 to 3 × 105 CFU per tomato together with a similar number of CFU of the wild type into the stem end of tomato fruit and used competition assays to assess its ability to proliferate over time, assessing bacterial numbers immediately after inoculation and then again 7 days later. As shown in Fig. 1A, our inoculation consisted of slightly more of the ΔfepDGC mutant than the wild type (CI of <1 for day 0, with a CI of 1 signifying equal numbers of both bacterial strains). Conversely, we found that by 1 week postinoculation, there existed a significantly lower number of viable ΔfepDGC mutant bacteria compared to the wild type (median CI of 1.65), indicating a loss of proliferation by the mutant during this period. During these 7 days, both strains grew within tomatoes, increasing their numbers by ∼3 logs. As shown in Fig. 1B, however, the wild-type strain grew significantly better (∼1,300-fold) than did the mutant (∼700-fold), with the difference between the strains corresponding to ∼1 × 108 fewer viable ΔfepDGC mutant bacteria (about 40% reduction) within tomatoes than the isogenic wild-type bacteria.
FIG 1.
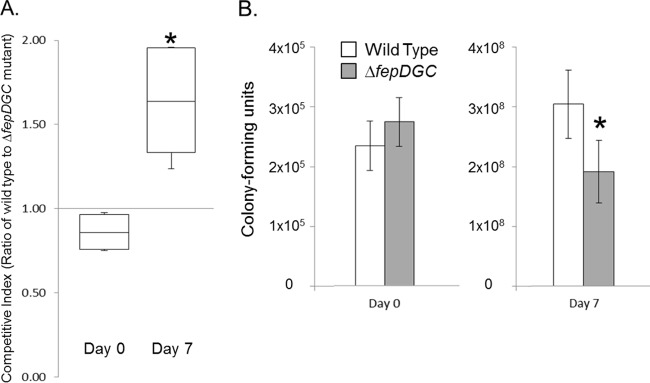
Mutation of fepDGC reduces Salmonella viability in tomato fruit. Wild-type and ΔfepDGC mutant strains were inoculated together in approximately equal numbers into red ripe tomatoes. Bacteria were enumerated by harvesting tomato juice and tissue either immediately after inoculation (day 0) or 1 week later (day 7) with plating onto selective agar. (A) The competitive index (CI) was measured as the ratio of wild-type to ΔfepDGC mutant cells, with a CI of >1 indicating a greater number of the wild-type cells. (B) Numbers of cells of each bacterial strain were determined by colony-forming units per tomato, with white bars indicating the wild type and gray bars the ΔfepDGC mutant. Error bars indicate the standard deviation from the mean, based on five independent biological replicates. Independent axes are used for the two time points for clarity. Significance was determined through ANOVA at P < 0.05. The asterisk indicates a significant difference between cell numbers of the two strains.
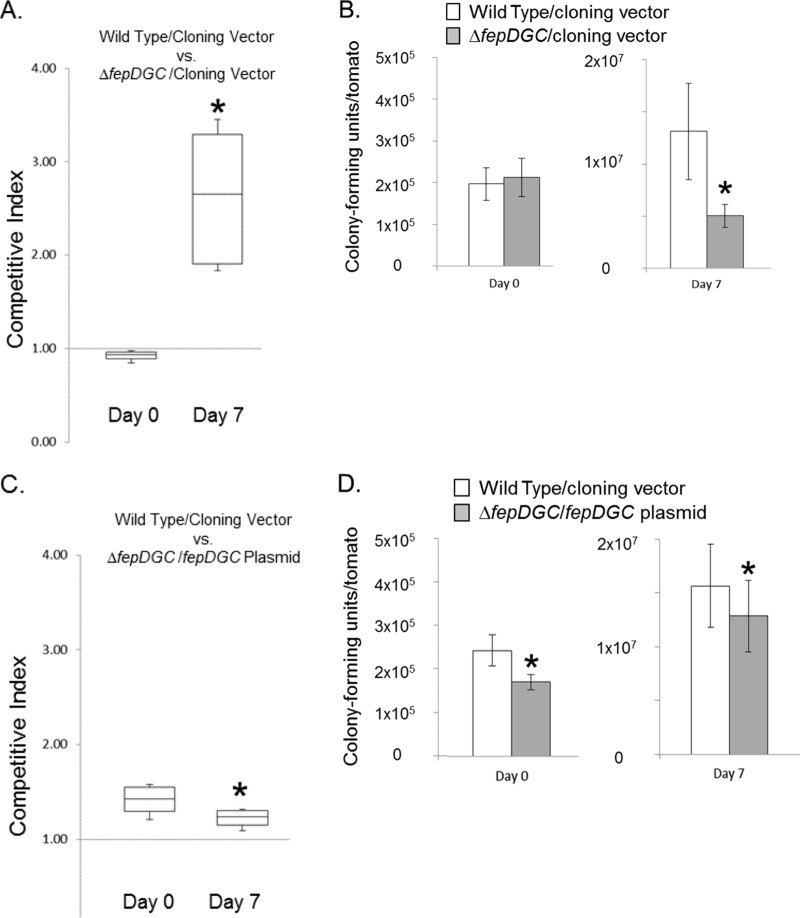
To ensure that the observed phenotype was indeed caused by the loss of fepDGC, we complemented the mutant strain with the fepDGC operon encoded on a low-copy-number plasmid and under the control of its native promoter. As a control, we first compared the survival in tomatoes of the wild type with the ΔfepDGC mutant, both carrying an identical plasmid cloning vector (Fig. 2A). As expected, the presence of this plasmid did not alter the fitness advantage of the wild type, with a median CI of 2.65 for the wild type over the ΔfepDGC mutant achieved by 1 week after bacterial inoculation. In these plasmid experiments, although the amount of the inoculum was similar to that used to assess the ΔfepDGC mutant (Fig. 1B), neither bacterial strain proliferated to as great a degree after 7 days (Fig. 2B and D). Specifically, with the strains containing plasmid cloning vector alone, the wild type increased during this period by only ∼60-fold to 1.3 × 107 and the ΔfepDGC mutant by only ∼20-fold to 5.0 × 106 viable bacteria (Fig. 2B). This general reduction in growth may be due to an added metabolic burden placed on the bacteria by the low-copy-number plasmid within the environment of the tomato fruit. We next compared the ΔfepDGC mutant strain complemented with the fepDGC plasmid to the wild type with the plasmid cloning vector alone. As shown in Fig. 2C, the wild-type strain was inoculated in slightly higher numbers than was the complemented mutant, producing a CI of >1 on day 0. Seven days later, the CI was significantly lower (Fig. 2C), indicating that the plasmid-borne copy of fepDGC could restore the mutant to a growth phenotype superior to that of the wild type. This improved growth of the complemented mutant can be seen in Fig. 2D; although the total number of complemented mutants remained lower than that of the wild type at 7 days, the growth rate of the complemented mutant (75-fold change) surpassed that of the wild type (65-fold change). A subsequent analysis of plasmid retention by selective plating demonstrated no detectable loss of plasmids during the course of these experiments (data not shown), indicating that any fitness burden imparted by the plasmid was not sufficient to select for its loss. Thus, these results demonstrate that the loss of Salmonella proliferation in tomatoes can be attributed solely to the absence of the iron transporter FepDGC. They show also that multicopy expression of FepDGC can enhance Salmonella growth, suggesting that the acquisition of iron is a limiting factor to growth within tomato fruit.
FIG 2.
Complementation of fepDGC restores Salmonella viability in tomato fruit. (A and B) The wild-type and ΔfepDGC mutant strains carrying the identical cloning vector were inoculated together into red ripe tomatoes, with bacterial numbers determined immediately after inoculation (day 0) and 1 week later (day 7). The competitive index (CI) was measured as the ratio of wild-type to ΔfepDGC mutant cells, with a CI of >1 indicating a greater number of the wild-type cells. The numbers of cells of each bacterial strain were determined by colony-forming units per tomato, with white bars indicating the wild type and gray bars the ΔfepDGC mutant, both with the cloning vector. In a separate experiment (C and D), the wild type carrying the cloning vector was inoculated with the ΔfepDGC mutant carrying a fepDGC complementing plasmid and similarly treated. In both cases, a CI of >1 indicates a greater number of the wild-type than the ΔfepDGC mutant cells. Error bars indicate standard deviation from the mean, based on five independent biological replicates. Independent axes are used for the two time points for clarity. For each experiment, significance was determined through ANOVA at P < 0.05. The asterisk indicates a significant difference between cell numbers of the two strains.
Iron is required for the proliferation of Salmonella in tomatoes.
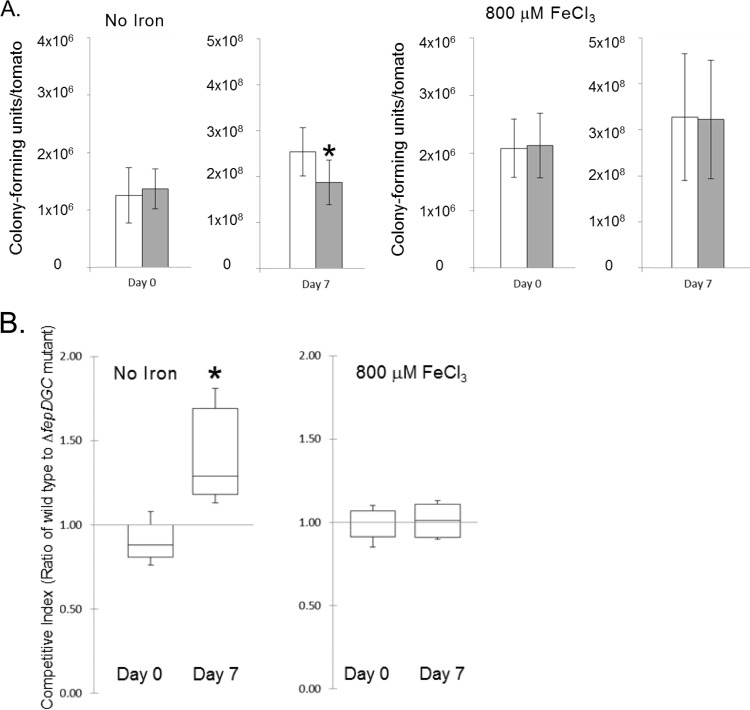
Although FepDGC is known to be a transporter of iron, it remained possible that other, uncharacterized functions of this transporter might make it important for the survival or growth of Salmonella in tomatoes. To ensure that the observed survival defect of this mutant was attributable specifically to the failure of Salmonella to acquire iron within tomato fruit, we next tested the phenotypic complementation of the mutation by the addition of exogenous iron. We again performed competition assays using the wild-type and ΔfepDGC mutant strains, but added to the inoculum 800 μM FeCl3, a concentration that we had previously determined not to inhibit the growth or affect recovery of either strain on laboratory media (data not shown). We found that this addition of iron did not appreciably change the ability of the wild-type strain to proliferate within tomatoes; the numbers of cells of this strain harvested after 7 days were not significantly different with or without added iron. Specifically, the wild type grew by ∼200-fold in the absence of exogenous iron and by ∼160-fold with iron added (Fig. 3A). This result indicates that the iron concentration naturally present in tomatoes does not limit the growth of wild-type Salmonella and therefore provides sufficient iron in the presence of intact bacterial iron-scavenging systems. Importantly, however, inclusion of FeCl3 in the inoculum did eliminate the growth defect of the ΔfepDGC mutant, restoring numbers of this mutant to those of the wild type (Fig. 3A). Without iron added, the ΔfepDGC mutant demonstrated the expected growth defect in tomatoes, with a CI at 7 days significantly higher than that at the day of inoculation (Fig. 3B). With added iron, however, the wild type and the ΔfepDGC mutant strain had CIs near 1 at both the time of inoculation and 7 days later, with the CIs at the two times not significantly different from each other (Fig. 3B). These results thus demonstrate that the known function of FepDGC, transport of iron into the bacterium, is that which is required for the normal growth of Salmonella within the tomato. Furthermore, our data show that iron acquisition is required for the growth of the pathogen in this fruit, as it is known to be in animal tissues. Our results also support previous work in plants that demonstrated siderophores and iron to be important for Salmonella colonization of alfalfa roots (41).
FIG 3.
Exogenous iron suppresses the growth defect of the Salmonella ΔfepDGC mutant in tomato fruit. The wild-type and ΔfepDGC mutant strains were inoculated together in approximately equal cell numbers into red ripe tomatoes, without added iron or with the addition of 800 μM FeCl3 at the time of inoculation. (A) The numbers of cells of each bacterial strain were determined by colony-forming units per tomato, with white bars indicating the wild type and gray bars the ΔfepDGC mutant. Error bars indicate the standard deviation from the mean, based on five independent biological replicates. Independent axes are used for the two time points for clarity. (B) The competitive index (CI) was measured as the ratio of wild-type to ΔfepDGC mutant cells, with a CI of >1 indicating a greater number of the wild-type cells. Significance was determined through ANOVA at P < 0.05. The asterisk indicates a significant difference between cell numbers of the two strains.
Proliferation of Salmonella in tomato juice is not affected by the ability to acquire iron.
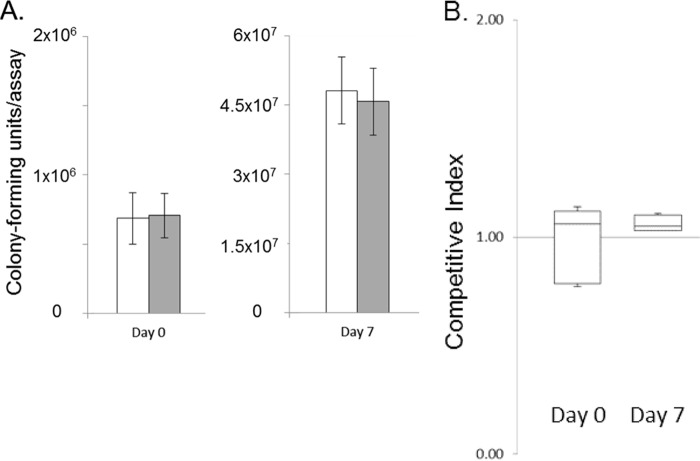
The internal environment of the tomato fruit is comprised of a complex milieu of liquid and cellular material. To determine whether iron concentration in the liquid fraction of this environment alone dictated the survival of Salmonella, we further tested bacterial survival in tomato juice. Filtered juice extracts obtained from the same variety of tomato used in the fruit survival assays were employed in competition assays identical to those described above. We found that the proliferation of Salmonella in tomato juice was similar to that in intact tomatoes (Fig. 4A). The inoculation of ∼7 × 105 wild-type bacteria yielded 70-fold bacterial growth after 7 days of incubation. Importantly, the ΔfepDGC mutant survived and grew 65-fold, not significantly different from the wild type, demonstrating no loss of viability, in contrast to that observed in whole fruit (Fig. 4A). The CI calculation further demonstrated this parity of growth between the two strains; there was no significant difference in the ratios of the wild type and the ΔfepDGC mutant at inoculation and 7 days afterwards (Fig. 4B). To further investigate the discrepancy in iron-dependent growth between tomato fruit and juice, we measured the concentration of iron in filtered juice extracts. The iron concentration varied within the single micromolar concentration range among juice samples. Although this is far less than the concentration we used to supplement iron in tomatoes, it greatly exceeds the concentration known to be required to support the growth of Salmonella. In defined laboratory medium, wild-type Salmonella has been shown to grow in concentrations of iron as low as 0.1 μM (42), thus indicating that the iron concentration present in our juice samples was sufficiently great to permit bacterial growth even by the ΔfepDGC mutant, with its diminished capacity to acquire iron.
FIG 4.
Iron acquisition provides no advantage to Salmonella during growth in tomato juice. The wild-type and ΔfepDGC mutant strains were inoculated together into filtered tomato juice. Bacteria were enumerated from the juice either immediately after inoculation (day 0) or 1 week later (day 7) with plating onto selective agar. (A) The numbers of cells of each bacterial strain were determined by colony-forming units per 5-ml juice assay, with white bars indicating the wild type and gray bars the ΔfepDGC mutant. Error bars indicate the standard deviation from the mean, based on five independent biological replicates. Independent axes are used for the two time points for clarity. (B) The competitive index (CI) was measured as the ratio of wild-type to ΔfepDGC mutant cells, with a CI of >1 indicating a greater number of the wild-type cells. Significance was determined through ANOVA at P < 0.05.
These findings have important implications for understanding the interaction of Salmonella with tomatoes. They demonstrate that the acquisition of iron is essential for the growth of this pathogen within the tomato fruit; a ΔfepDGC mutant, unable to efficiently scavenge iron, grew significantly less well than did the isogenic wild type strain. Importantly, we also found that the concentration of iron in extracted juice was sufficient to support the growth of both strains competent for iron acquisition and those deficient, demonstrating that the ability to salvage iron from within the fluid milieu does not alone dictate the survival and growth of this pathogen within tomatoes. These results, taken together, thus suggest that some factor or factors present within the tomato fruit reduce the ability of the pathogen to acquire the iron that should be available to it in this environment. We are tempted to speculate that the fruit itself harbors means to chelate iron, thus sequestering it from the bacteria. It has long been known that animal tissues readily capture free iron and that Salmonella must compete for this nutrient, through the production of siderophores, to establish infections in animal hosts (37). Some plants are similarly known to chelate iron through the production of phytosiderophores, molecules secreted into the rhizosphere through the roots by graminaceous plants as a means to acquire environmental iron (43). Although the existence of such iron chelators in tomatoes is thus plausible, we know of no similar example in any fruit. Alternatively, it remains possible that the environment within the tomato represses Salmonella functions essential for the acquisition or use of iron. We think this explanation unlikely, however, as our work stems from the identification of components of the iron regulon specifically induced within the tomato fruit (Table 1), implicating an active role in iron acquisition by the bacterium. Thus, the data presented here, taken together, show that Salmonella uses the iron stores that exist in the tomato fruit to survive and proliferate within this important produce crop and suggest that the tomato itself acts to limit the availability of iron to the pathogen.
ACKNOWLEDGMENTS
This work was supported by grant no. 2011-67017-3023 from the USDA National Institute of Food and Agriculture Program in Food Safety, Nutrition, and Health: Food-Borne Plant-Pathogen Interactions.
We thank Francoise Vermeylen and Erika Louise Mudrak of the Cornell Statistical Consulting Unit for assistance with statistical analysis.
REFERENCES
- 1.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States. Emerg Infect Dis 5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2007. Multistate outbreaks of Salmonella infections associated with raw tomatoes eaten in restaurants—United States, 2005–2006. MMWR Morb Mortal Wkly Rep 56:909–911. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2005. Outbreaks of Salmonella infections associated with eating Roma tomatoes—United States and Canada, 2004. MMWR Morb Mortal Wkly Rep 54:325–328. [PubMed] [Google Scholar]
- 4.Cummings K, Barrett E, Mohle-Boetani JC, Brooks JT, Farrar J, Hunt T, Fiore A, Komatsu K, Werner SB, Slutsker L. 2001. A multistate outbreak of Salmonella enterica serotype Baildon associated with domestic raw tomatoes. Emerg Infect Dis 7:1046–1048. doi: 10.3201/eid0706.010625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greene SK, Daly ER, Talbot EA, Demma LJ, Holzbauer S, Patel NJ, Hill TA, Walderhaug MO, Hoekstra RM, Lynch MF, Painter JA. 2008. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol Infect 136:157–165. doi: 10.1017/S095026880700859X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta SK, Nalluswami K, Snider C, Perch M, Balasegaram M, Burmeister D, Lockett J, Sandt C, Hoekstra RM, Montgomery S. 2007. Outbreak of Salmonella Braenderup infections associated with Roma tomatoes, northeastern United States, 2004: a useful method for subtyping exposures in field investigations. Epidemiol Infect 135:1165–1173. doi: 10.1017/S0950268807007911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedberg CW, Angulo FJ, White KE, Langkop CW, Schell WL, Stobierski MG, Schuchat A, Besser JM, Dietrich S, Helsel L, Griffin PM, McFarland JW, Osterholm MT, The Investigation Team . 1999. Outbreaks of salmonellosis associated with eating uncooked tomatoes: implications for public health. Epidemiol Infect 122:385–393. doi: 10.1017/S0950268899002393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhuang RY, Beuchat LR, Angulo FJ. 1995. Fate of Salmonella montevideo on and in raw tomatoes as affected by temperature and treatment with chlorine. Appl Environ Microbiol 61:2127–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barak JD, Liang A, Narm KE. 2008. Differential attachment to and subsequent contamination of agricultural crops by Salmonella enterica. Appl Environ Microbiol 74:5568–5570. doi: 10.1128/AEM.01077-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barak JD, Liang AS. 2008. Role of soil, crop debris, and a plant pathogen in Salmonella enterica contamination of tomato plants. PLoS One 3:e1657. doi: 10.1371/journal.pone.0001657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beuchat LR, Mann DA. 2008. Survival and growth of acid-adapted and unadapted Salmonella in and on raw tomatoes as affected by variety, stage of ripeness, and storage temperature. J Food Prot 71:1572–1579. [DOI] [PubMed] [Google Scholar]
- 12.Iturriaga MH, Escartin EF, Beuchat LR, Martinez-Peniche R. 2003. Effect of inoculum size, relative humidity, storage temperature, and ripening stage on the attachment of Salmonella Montevideo to tomatoes and tomatillos. J Food Prot 66:1756–1761. [DOI] [PubMed] [Google Scholar]
- 13.Iturriaga MH, Tamplin ML, Escartin EF. 2007. Colonization of tomatoes by Salmonella Montevideo is affected by relative humidity and storage temperature. J Food Prot 70:30–34. [DOI] [PubMed] [Google Scholar]
- 14.Noel JT, Joy J, Smith JN, Fatica M, Schneider KR, Ahmer BM, Teplitski M. 2010. Salmonella SdiA recognizes N-acyl homoserine lactone signals from Pectobacterium carotovorum in vitro, but not in a bacterial soft rot. Mol Plant Microbe Interact 23:273–282. doi: 10.1094/MPMI-23-3-0273. [DOI] [PubMed] [Google Scholar]
- 15.Shi X, Namvar A, Kostrzynska M, Hora R, Warriner K. 2007. Persistence and growth of different Salmonella serovars on pre- and postharvest tomatoes. J Food Prot 70:2725–2731. [DOI] [PubMed] [Google Scholar]
- 16.Rathinasabapathi B. 2004. Survival of Salmonella Montevideo on tomato leaves and mature green tomatoes. J Food Prot 67:2277–2279. [DOI] [PubMed] [Google Scholar]
- 17.Meng F, Altier C, Martin GB. 2013. Salmonella colonization activates the plant immune system and benefits from association with plant pathogenic bacteria. Environ Microbiol 15:2418–2430. doi: 10.1111/1462-2920.12113. [DOI] [PubMed] [Google Scholar]
- 18.Pollard S, Barak J, Boyer R, Reiter M, Gu G, Rideout S. 2014. Potential interactions between Salmonella enterica and Ralstonia solanacearum in tomato plants. J Food Prot 77:320–324. doi: 10.4315/0362-028X.JFP-13-209. [DOI] [PubMed] [Google Scholar]
- 19.Potnis N, Soto-Arias JP, Cowles KN, van Bruggen AH, Jones JB, Barak JD. 2014. Xanthomonas perforans colonization influences Salmonella enterica in the tomato phyllosphere. Appl Environ Microbiol 80:3173–3180. doi: 10.1128/AEM.00345-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noel JT, Arrach N, Alagely A, McClelland M, Teplitski M. 2010. Specific responses of Salmonella enterica to tomato varieties and fruit ripeness identified by in vivo expression technology. PLoS One 5:e12406. doi: 10.1371/journal.pone.0012406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marvasi M, Cox CE, Xu Y, Noel JT, Giovannoni JJ, Teplitski M. 2013. Differential regulation of Salmonella typhimurium genes involved in O-antigen capsule production and their role in persistence within tomato fruit. Mol Plant Microbe Interact 26:793–800. doi: 10.1094/MPMI-09-12-0208-R. [DOI] [PubMed] [Google Scholar]
- 22.Barr M, East AK, Leonard M, Mauchline TH, Poole PS. 2008. In vivo expression technology (IVET) selection of genes of Rhizobium leguminosarum biovar viciae A34 expressed in the rhizosphere. FEMS Microbiol Lett 282:219–227. doi: 10.1111/j.1574-6968.2008.01131.x. [DOI] [PubMed] [Google Scholar]
- 23.Boch J, Joardar V, Gao L, Robertson TL, Lim M, Kunkel BN. 2002. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol Microbiol 44:73–88. doi: 10.1046/j.1365-2958.2002.02877.x. [DOI] [PubMed] [Google Scholar]
- 24.Brown DG, Allen C. 2004. Ralstonia solanacearum genes induced during growth in tomato: an inside view of bacterial wilt. Mol Microbiol 53:1641–1660. doi: 10.1111/j.1365-2958.2004.04237.x. [DOI] [PubMed] [Google Scholar]
- 25.Czelleng A, Bozso Z, Ott PG, Besenyei E, Varga GJ, Szatmari A, Kiraly L, Klement Z. 2006. Identification of virulence-associated genes of Pseudomonas viridiflava activated during infection by use of a novel IVET promoter probing plasmid. Curr Microbiol 52:282–286. doi: 10.1007/s00284-005-0208-6. [DOI] [PubMed] [Google Scholar]
- 26.Giddens SR, Jackson RW, Moon CD, Jacobs MA, Zhang XX, Gehrig SM, Rainey PB. 2007. Mutational activation of niche-specific genes provides insight into regulatory networks and bacterial function in a complex environment. Proc Natl Acad Sci U S A 104:18247–18252. doi: 10.1073/pnas.0706739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preston GM, Bertrand N, Rainey PB. 2001. Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Mol Microbiol 41:999–1014. DOI: 10.1046/j.1365-2958.2001.02560.x. [DOI] [PubMed] [Google Scholar]
- 28.Silby MW, Rainey PB, Levy SB. 2004. IVET experiments in Pseudomonas fluorescens reveal cryptic promoters at loci associated with recognizable overlapping genes. Microbiology 150:518–520. doi: 10.1099/mic.0.26871-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XX, Lilley AK, Bailey MJ, Rainey PB. 2004. The indigenous Pseudomonas plasmid pQBR103 encodes plant-inducible genes, including three putative helicases. FEMS Microbiol Ecol 51:9–17. doi: 10.1016/j.femsec.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Slauch JM, Camilli A. 2000. IVET and RIVET: use of gene fusions to identify bacterial virulence factors specifically induced in host tissues. Methods Enzymol 326:73–96. doi: 10.1016/S0076-6879(00)26047-3. [DOI] [PubMed] [Google Scholar]
- 31.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellermeier CD, Janakiraman A, Slauch JM. 2002. Construction of targeted single copy lac fusions using λ Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161. doi: 10.1016/S0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 33.Churchward G, Belin D, Nagamine Y. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 34.Altier C, Suyemoto M. 1999. A recombinase-based selection of differentially expressed bacterial genes. Gene 240:99–106. doi: 10.1016/S0378-1119(99)00427-8. [DOI] [PubMed] [Google Scholar]
- 35.Troxell B, Fink RC, Porwollik S, McClelland M, Hassan HM. 2011. The Fur regulon in anaerobically grown Salmonella enterica sv. Typhimurium: identification of new Fur targets. BMC Microbiol 11:236. doi: 10.1186/1471-2180-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berry AD, Sargent SA. 2009. Real-time microsensor measurement of internal oxygen partial pressure in tomato fruit under hypoxic conditions. Postharvest Biol Technol 52:240–242. doi: 10.1016/j.postharvbio.2009.01.007. [DOI] [Google Scholar]
- 37.Crouch ML, Castor M, Karlinsey JE, Kalhorn T, Fang FC. 2008. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol Microbiol 67:971–983. doi: 10.1111/j.1365-2958.2007.06089.x. [DOI] [PubMed] [Google Scholar]
- 38.Kirby MF, Devoy B, Law RJ, Ward A, Aldridge J. 2008. The use of a bioassay based approach to the hazard/risk assessment of cargo derived toxicity during shipping accidents: a case study—the MSC Napoli. Mar Pollut Bull 56:781–786. doi: 10.1016/j.marpolbul.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Hantke K, Nicholson G, Rabsch W, Winkelmann G. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci U S A 100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabsch W, Voigt W, Reissbrodt R, Tsolis RM, Baumler AJ. 1999. Salmonella typhimurium IroN and FepA proteins mediate uptake of enterobactin but differ in their specificity for other siderophores. J Bacteriol 181:3610–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hao LY, Willis DK, Andrews-Polymenis H, McClelland M, Barak JD. 2012. Requirement of siderophore biosynthesis for plant colonization by Salmonella enterica. Appl Environ Microbiol 78:4561–4570. doi: 10.1128/AEM.07867-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi T, Nakanishi H, Nishizawa NK. 2010. Recent insights into iron homeostasis and their application in graminaceous crops. Proc Jpn Acad Ser B Phys Biol Sci 86:900–913. doi: 10.2183/pjab.86.900. [DOI] [PMC free article] [PubMed] [Google Scholar]