Abstract
Phenolic compounds are believed to be promising candidates as complementary therapeutics. Maple syrup, prepared by concentrating the sap from the North American maple tree, is a rich source of natural and process-derived phenolic compounds. In this work, we report the antimicrobial activity of a phenolic-rich maple syrup extract (PRMSE). PRMSE exhibited antimicrobial activity as well as strong synergistic interaction with selected antibiotics against Gram-negative clinical strains of Escherichia coli, Proteus mirabilis, and Pseudomonas aeruginosa. Among the phenolic constituents of PRMSE, catechol exhibited strong synergy with antibiotics as well as with other phenolic components of PRMSE against bacterial growth. At sublethal concentrations, PRMSE and catechol efficiently reduced biofilm formation and increased the susceptibility of bacterial biofilms to antibiotics. In an effort to elucidate the mechanism for the observed synergy with antibiotics, PRMSE was found to increase outer membrane permeability of all bacterial strains and effectively inhibit efflux pump activity. Furthermore, transcriptome analysis revealed that PRMSE significantly repressed multiple-drug resistance genes as well as genes associated with motility, adhesion, biofilm formation, and virulence. Overall, this study provides a proof of concept and starting point for investigating the molecular mechanism of the reported increase in bacterial antibiotic susceptibility in the presence of PRMSE.
INTRODUCTION
Gram-negative and Gram-positive bacteria colonize surfaces in health care settings, on indwelling medical devices, and even on live tissue, leading to infections that often are treated with antibiotic therapy. However, two major factors complicate the effectiveness of antibiotic treatments, namely, (i) the rising number of antibiotic-resistant bacteria and (ii) the formation of biofilms. These complications lead to increased patient morbidity, increased costs of treatment, and higher rates of hospitalization (1, 2). Antibiotic resistance is the inevitable evolutionary survival mechanism of bacteria, and it is aggravated by the overuse of antibiotics in the medical and farming industries. Bacterial biofilms are structured, surface-associated microbial communities, protected by a self-produced matrix of extracellular polymeric substances, and are the most common mode of bacterial growth. Formation of biofilms complicates the treatment of infections, because bacteria in biofilm mode generally are very persistent, requiring considerably higher doses of antibiotics for treatment than planktonic bacteria (3). High antibiotic doses disturb the body's microbiome, putting the patient's health at risk, as well as increasing the potential for development of antibiotic-resistant strains (4). The reduced effectiveness of current therapies and a declining repertoire of clinically useful drugs motivate research for the identification of novel molecules endowed with antimicrobial and/or antibiofilm properties.
Many plants synthesize aromatic substances, most of which are phenols or their oxygen-substituted derivatives (5). These phenolic compounds are believed to be promising candidates as complementary therapeutics (6), since they can modify bacterial behavior by affecting bacterial motility (7, 8, 58), surface adhesion (9), biofilm formation (7, 10), quorum sensing (11), and production of virulence determinants (12, 13). Traditional medicinal approaches owe their significance to the bioactive components that have their origin in plant sources, and many are associated with routine dietary habits. The North American maple tree (genus Acer) plays a central role in Native Americans' traditional medicine (14). The syrup, obtained by concentrating the sap from certain maple species (i.e., the sugar maple, Acer saccharum Marsh, and the red maple, A. rubrum L.), contains a vast number of natural and process-derived phytochemicals, the majority of which are phenolic compounds. Phenolic-rich maple syrup extracts (PRMSE) were obtained by extracting the phenolic compounds of maple syrup with organic solvents. These extracts have been reported to exhibit antiproliferative effects against a panel of human tumor cell lines (15). Although there have been few studies suggesting antimicrobial activity for maple leaf extracts from the sugar and red maple species (16), to date, the potential antimicrobial activity of PRMSE and its phenolic constituents remains relatively unexplored.
In this study, we investigated the antimicrobial activity of PRMSE and its effective phenolic constituents toward selected Gram-negative pathogenic bacteria, namely, Escherichia coli, Pseudomonas aeruginosa, and Proteus mirabilis, in planktonic and biofilm modes. PRMSE was tested for synergistic interactions with antibiotics against the selected bacteria in both planktonic and biofilm mode, and the mechanism(s) of interactions were investigated via quantitative reverse transcription-PCR (qRT-PCR), membrane permeability analysis, and efflux pump assays.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The following organisms were used in this study: E. coli strain CFT073 (ATCC 700928), P. mirabilis HI4320 (17), P. aeruginosa PAO1 (ATCC 15692), and P. aeruginosa PA14 (UCBPP-PA14) (18). Pure stock cultures were maintained at −80°C in 30% (vol/vol) frozen glycerol solution. Starter cultures were prepared by streaking frozen cultures onto LB agar (LB broth contained 10 g liter−1 tryptone, 5 g liter−1 yeast extract, and 5 g liter−1 NaCl, supplemented with 1.5% [wt/vol] agar [Fisher Scientific, ON, Canada]). After overnight incubation at 37°C, a single colony was inoculated into 10 ml of LB broth and the culture was incubated at 37°C on an orbital shaker at 200 rpm for a time length specific to each experiment. LB broth was used for bacterial culture in all experiments unless otherwise specified.
Preparation of PRMSE.
All maple syrup samples (grade D, amber, production year 2013, 66-degrees Brix syrup) were purchased from local markets of Montreal (Quebec, Canada) in August 2013 and stored at −20°C until extraction according to a protocol described by González-Sarrías et al. (19). Briefly, maple syrup was enriched for phenolic content by using Amberlite XAD-16 (Sigma-Aldrich Canada) resin column chromatography (C4919, bed volume of 245 ml; Sigma-Aldrich Canada). The column subsequently was extracted with methanol (Sigma-Aldrich Canada). This extract first was subjected to solvent removal under vacuum and then solubilized in 0.3% (vol/vol) dimethylsulfoxide (DMSO) to yield PRMSE. Extracts were stored at −20°C, thawed at room temperature before each experiment, and filter sterilized using 0.22-μm polyvinylidene difluoride (PVDF) membrane filters (EMD Millipore Millex; Fisher Scientific Canada) before use. PRMSE prepared in this manner was reported to not contain any natural sugar (sucrose, glucose, or fructose) (19).
High-performance liquid chromatography (HPLC) analysis.
The relative level of phenolic compounds in PRMSE was estimated using HPLC, as reported in the literature (20). Samples were analyzed using an Agilent Technologies 1200 series analytical liquid chromatographic system consisting of binary pump LC-20AB, degasser DGU-20A5, column oven CTO-20 AC, autosampler SIL-20 AC, and a UV SPD-M20A detector. Phenolic compounds were separated on a Zorbax SB-C18 column (4.6 by 150 mm, 5 μm; Agilent) at 30°C. Trifluoroacetic acid at 0.2% (phase A) and methanol (phase B) were used as eluents. The elution gradient started with 2% phase B, increased to 50% phase B at 35 min and 80% phase B at 43 min, and held at 80% for 2 min. The injection volume was 10 μl, and the flow rate was 0.5 ml min−1. Data were collected and evaluated by the Analyst 1.4.2 software. The analytes were identified by comparing retention times and UV spectra, recorded in the range of 200 to 400 nm, with the individual chromatogram of each phenolic standard. The following pure phenolic compounds were purchased from Sigma-Aldrich Canada and used as standards: gallic acid, 1,2-dihydroxybenzene (catechol), 3,4-dihydroxybenzaldehyde (catechaldehyde), syringaldehyde, vanillin, and 3-hydroxybenzoic acid.
Determination of MICs.
MICs were determined by preparing 2-fold serial dilutions of PRMSE, pure phenolic compounds, and antibiotics in Mueller-Hinton broth adjusted with Ca2+ and Mg2+ (MHB-II; Oxoid, Fisher Scientific Canada). A range of concentrations of the antibiotics ciprofloxacin (0.0003 to 1.0 μg ml−1) and carbenicillin (0.5 to 512 μg ml−1) was chosen due to their known potency against all four bacterial strains. Dilutions were prepared in flat-bottom, 96-well microtiter plates (Falcon, Corning, Fisher Scientific Canada). Each well of a microtiter plate then was inoculated with the desired bacterial strain (grown in MHB-II and diluted to 106 CFU ml−1), and the plate was incubated at 37°C for 18 h under static conditions. Bacterial growth was assessed by (i) monitoring the optical density of the cell suspension in each well at 600 nm (OD600) and (ii) using the resazurin microtiter plate assay (21). In the resazurin microtiter plate assay, each well of a microtiter plate was supplemented with 20 μM resazurin and incubated in the dark for 20 min at room temperature, followed by fluorescence measurements at excitation and emission wavelengths of 570 and 590 nm, respectively, using a Tecan Infinite M200 Pro microplate reader (Tecan Group Ltd., Switzerland). The lowest concentration of a compound able to prevent an increase in the OD600 and an increase in resazurin fluorescence intensity was recorded as the MIC for that compound.
Checkerboard microdilution assay.
The checkerboard microdilution assay (22) was used for evaluation of in vitro antimicrobial synergy between two compounds (i.e., antibiotic/PRMSE, antibiotic/pure phenolic compound, and pure phenolic compounds with each other). Twofold serial dilutions were prepared in MHB-II for each of the two compounds under study. The serial dilutions then were loaded into 96-well plates to achieve combinations having different concentrations of each of the two compounds. Each well subsequently was inoculated with 106 CFU ml−1 of the desired bacterial strain and incubated at 37°C for 18 h under static conditions. The fractional inhibitory concentration index (FICI) for each combination was calculated by using the following formulas (22): FICcomponent 1 = MICcomponent 1, in combination/MICcomponent 1, alone and FICI = FICcomponent 1 + FICcomponent 2.
An FICI of ≤0.5 indicated synergy, an FICI of >0.5 and ≤4 indicated no interaction/indifference, and an FICI of >4 indicated antagonism (22).
Biofilm assays.
Biofilm formation was quantified using the standard microtiter plate model (23). Briefly, overnight cultures (LB broth, 37°C, 200 rpm) were diluted 1:100 (vol/vol) into fresh LB broth (with or without PRMSE or catechol) to 106 CFU ml−1. Aliquots (100 μl) of these cultures were transferred into the wells of polystyrene, flat-bottom, nontreated 96-well plates (Falcon, Corning) in triplicate. For all assays, biofilms were allowed to develop for 16 h at 37°C under static conditions, after which OD600 values were recorded, the spent broth was decanted from the wells, and the wells were gently rinsed three times with deionized (DI) water. The washed biofilm was stained with crystal violet (CV). For the CV stain assay, 100 μl of 0.1% (wt/vol) CV was loaded in each well, and the plates were incubated for 15 min under static conditions at room temperature. The wells subsequently were rinsed with DI water to remove excess dye, and the CV adsorbed to the biomass in each well was solubilized in 100 μl of absolute ethanol for 10 min. The solubilized CV then was quantified (at OD570) using a microplate reader. Control experiments were performed with cell-free broth to adjust for background signal.
In vitro assessment of PRMSE for eradication of preformed biofilm on low-surface-energy silicone surfaces (a model biomaterial) was performed using the method described in reference 24, with some modifications. Silicone discs (1-cm diameter) were prepared as described previously (25), placed into wells of a 24-well plate, and incubated overnight at 37°C with 0.5 ml human plasma (P9523; Sigma-Aldrich Canada). After incubation, the plasma solution was removed and replaced with 1 ml of ∼106 CFU ml−1 inoculum of the challenge organism in MHB-II (Oxoid; Fisher Scientific Canada). These plates were incubated further for 24 h at 37°C, after which the discs were washed by gentle shaking at 50 rpm for 30 min in 10 mM sterile phosphate buffer saline solution (PBS; pH 7.0, containing 0.85% NaCl) to remove nonadherent cells. After washing, discs were placed in a fresh 24-well plate containing 1 ml PRMSE solution in each well at different concentrations with and without ciprofloxacin (different concentrations) and incubated at 37°C for 2 h. The discs then were removed and placed in sterile Falcon tubes (centrifuge tube, 15 ml; Fisher Scientific Canada) containing 3 ml of 10 mM sterile PBS solution and sonicated in a bath sonicator (60 Hz and 150 W) for 10 min to disrupt any remaining biofilm. The bacterial cell concentration in the resulting suspension was quantified using standard plate counts. An independently prepared bacterial suspension was subjected to the same sonication conditions to account for any damage to the cells as a result of this treatment.
RNA extraction, cDNA synthesis, and comparative qRT-PCR.
Bacterial cells were grown to an OD600 of 0.5 to 0.8 (16 h, 37°C, 150 rpm) in LB broth with or without different concentrations of PRMSE. Total RNA was extracted using a Direct Zol kit (Zymo Research). RNA concentration was quantified by measuring the absorbance of the sample at 260 and 280 nm, and 300 ng of RNA was used for cDNA synthesis using the high-capacity cDNA reverse transcription kit (Applied Biosystems, Life Technologies Inc., Canada). Expression of target genes was quantified using qRT-PCR with the synthesized cDNA. qRT-PCR was performed with an ABI Prism 7900 HT thermal cycler (Applied Biosystems) using Power SYBR green PCR master mix (Applied Biosystems, Life Technologies Inc., Canada). Conditions for qRT-PCR were the following: 50°C for 2 min, initial denaturation at 95°C for 10 min, and 45 cycles of 15 s at 95°C and 1 min at 60°C. Results were analyzed with SDS software, version 2.2 (Applied Biosystems). Data were normalized to the endogenous reference gene of respective strains. The threshold cycle method (2−ΔΔCT) (26) was used to analyze changes in gene expression in a given sample relative to the control (cells grown under the same conditions without PRMSE). For each sample of cells, qRT-PCR was performed in triplicate and the entire experiment was repeated twice with RNA samples extracted from independent cultures. Oligonucleotide primers (Table 1) were designed using Primer3Plus (27) based on the published genome sequences of CFT073, HI4320, PAO1, and PA14. Moreover, the reported oligonucleotide primer sequences (28–36) used to amplify the gene of interest are listed in Table 1.
TABLE 1.
Primer sequences of the indicated genes used for quantitative RT-PCR
| Organism | Gene | Oligonucleotide sequence (5′–3′) |
Source or reference | |
|---|---|---|---|---|
| Forward | Reverse | |||
| E. coli | gapAa | AAGTTGGTGTTGACGTTGTCGCTG | ATAACCACTTTCTTCGCACCAGCGG | 34 |
| E. coli | fimA | ACTCTGGCAATCGTTGTTCTGTCG | ATCAACAGAGCCTGCATCAACTGC | 28 |
| E. coli | papA2 | ACGGGTGAAATTTGATGGAGCCAC | AATTCGCAACTGCTGAGAAGGCAC | 36 |
| E. coli | flhD | TCCGCTATGTTTCGTCTCGGCATA | ACCAGTTGATTGGTTTCTGCCAGC | 36 |
| E. coli | fliC | ACAGCCTCTCGCTGATCACTCAAA | GCGCTGTTAATACGCAAGCCAGAA | 34 |
| E. coli | acrB | CTGATCATCGTGGTCGGCATGGC | CCAGTCCTTCAAGGAAACGAACGC | 31 |
| E. coli | motB | GCGTTACGTCCACATCTCAA | ATGTCGCGCATATAGGGTTC | This study |
| E. coli | uvrY | GCCAGTTTGTCTGAACGTGA | CTGTTCACCGTTTTCGGACT | This study |
| E. coli | marC | GTCGGAAGAGCTGGAAGATG | GAACTCTGACGCACTGTGGA | This study |
| E. coli | emrA | ACAGGTAGCGCGTTCTCACT | AGCGTGGATAAACCGATACG | This study |
| E. coli | chuA | AGCAAACAACCTGGCTATGG | CTCTTTATCGAAGGCGTTGC | This study |
| E. coli | fimH | GGAACCATTCAGGCAGTGAT | CGGTTTTACAGGCGAATGAC | This study |
| P. mirabilis | rpoAa | GCAAATCTGGCATTGGCCCTGTTA | TAGGGCGCTCATCTTCTTCCGAAT | 35 |
| P. mirabilis | flhD | AAGGCTTCCGCAATGTTTAGAC | GTTGCAAATCATCCACTCTGGA | 29 |
| P. mirabilis | flaA | TGCTGGTGCAACTTCATACG | TTTGTCAGCACCTTCCAGTG | This study |
| P. mirabilis | ureA | GGGGTGCCAGAGATGATAAA | CCGGGGATCATGTTATTACC | This study |
| P. mirabilis | ureD | CCTTACGCACATGCCCTATT | CTTGTGCAACCGTCAATGTC | This study |
| P. mirabilis | atfB | ATTTAGCTGCAGCCGACAGT | GAGTCTGTGCGCCATAATCA | This study |
| P. mirabilis | marC | ATCTCGGCCACAGTGGTATC | AATAAAGCGGGGAGATCAGC | This study |
| P. mirabilis | acrA | GCTGAAATTGCTCGCCTAAC | GCAACAGCTTGAGCGTACTG | This study |
| P. mirabilis | cysJ | CAATGCACGTCGTTTAGCTG | TTCCCCCTGTGTTGAGGTAA | This study |
| P. aeruginosa | rpoDa | GCCGAGATCAAGGAAATCAA | GTGTACTTCTTGGCGATGGAA | 33 |
| P. aeruginosa | lasB | AAGCCATCACCGAAGTCAAG | CGGATCACCAGTTCCACTTT | 33 |
| P. aeruginosa | plcH | TGACTTCGCTGTTCGACTTC | TGGGCTCGTAGGACCAGTAT | 33 |
| P. aeruginosa | phzS | CGTCGGCATCAATATCCAG | ATCGAGTACTGCGGATAGGC | 32 |
| P. aeruginosa | pvdA | GTTCCACCACAGCCAGTACC | CTGTCGTTGAGGTCGATGAA | 32 |
| P. aeruginosa | fliC | AACTTCGACGTAACCGTTGG | TGGTCAGTACACCCTTGTCG | 32 |
| P. aeruginosa | mexA | CGACCAGGCCGTGAGCAAGCAGC | GGAGACCTTCGCCGCGTTGTCGC | 30 |
| P. aeruginosa | mexX | TGAAGGCGGCCCTGGACATCAGC | GATCTGCTCGACGCGGGTCAGCG | 30 |
| P. aeruginosa | oprM | TCAACCTGCGCTACACCA | GCTACCGTCCTCCAGCTTC | This study |
| P. aeruginosa | cupA1 | GCGGCAAACACTATCACATTC | AACAGGGTGGTGAAATGCTC | This study |
| P. aeruginosa | fleQ | GATCAGCTGACCTGCAACAG | GCAGGTACTCGTCCCAACTG | This study |
Housekeeping genes (endogenous control).
Membrane permeabilization and membrane integrity assays.
The outer membrane permeabilization activities of PRMSE and catechol were determined by the 1-N-phenylnapthylamine (NPN; Sigma-Aldrich Canada) assay as described in reference 37, with some modifications. Briefly, overnight bacterial cultures were diluted 1:1 in MHB-II medium to a final volume of 10 ml, with or without sub-MIC supplementation of PRMSE, catechol, or gentamicin (positive control), and grown to an OD600 of 0.5 to 0.6 (37°C, 200 rpm). The cells were harvested, washed with 5 mM HEPES buffer (pH 7.2), and resuspended in the same volume (10 ml) of 5 mM HEPES buffer (pH 7.2) containing 1 mM N-ethylmaleimide (NEM; Sigma-Aldrich Canada). Aliquots (1 ml) were mixed with NPN to a final concentration of 10 μM (in cell suspension), and fluorescence was measured using the microplate reader (excitation, 350 nm; emission, 420 nm).
The BacLight kit (L-13152; Invitrogen, Life Technologies Inc., Canada) was used to assess cell membrane damage (38). Overnight bacterial cultures were diluted 1:40 in fresh MHB-II broth to a final volume of 5 ml, grown to an OD600 of 0.5 to 0.6, washed with filter-sterilized 10 mM PBS (pH 7.0), and resuspended in 1/10 of the original volume. The washed cells then were diluted 1:20 (vol/vol) into PRMSE or catechol at 4× MIC as described in reference 39 or 0.3% (vol/vol) DMSO (control). Cultures were incubated at room temperature (21 ± 2°C) on a tube rocker for 10 min. At the end of the incubation period, an aliquot was taken for CFU counts and the remaining suspension was washed with 10 mM PBS and resuspended to an OD600 of 0.3. An aliquot (100 μl) of each bacterial suspension was removed and added to a 96-well, black, clear-bottom plate (Corning, Fisher Scientific Canada) along with an equal volume of the BacLight reagent (2× stock solution, L13152; Invitrogen, Life Technologies Inc., Canada), and the plates were incubated for 10 min at room temperature in the dark. At the end of the incubation period, fluorescence intensity was recorded for both kit components, SYTO-9 (excitation, 485 nm; emission, 530 nm) and propidium iodide (excitation, 485 nm; emission, 645 nm), using the microplate reader. Fluorescence readings from samples were normalized to the values obtained from the untreated control to determine the ratio of membrane-compromised cells to cells with intact membranes. Cetyltrimethylammonium bromide (CTAB; Sigma-Aldrich Canada), a cationic detergent that is known to cause membrane damage (40), was used at a concentration of 10 μM as a positive control for membrane disruption.
EtBr efflux assay.
To assess the effect of PRMSE and catechol on the inhibition of the proton motive force-driven multidrug efflux pump, an ethidium bromide (EtBr) efflux assay was performed using the method described in reference 41. An overnight-grown culture of each strain was diluted 1:100 using MHB-II broth to a final volume of 10 ml and was grown to an OD600 of 0.8 to 1.0 (at 37°C, 150 rpm). Cells were loaded in polystyrene microcentrifuge tubes (2 ml) and mixed with 5 μM EtBr and PRMSE or catechol at 25% of their MIC or a 100 μM concentration of the proton conductor CCCP (carbonyl cyanide m-chlorophenylhydrazone; Sigma-Aldrich Canada) as a positive control. Replica tubes that did not receive PRMSE, catechol, or proton conductor served as negative controls. The tubes were incubated for 1 h (37°C, 150 rpm). The inoculum then was adjusted to an OD600 of 0.4 with MHB-II broth containing 5 μM EtBr, and 2-ml aliquots of this mixture were pelleted (5,000 × g, 10 min, 4°C). The pellets were incubated on ice immediately, resuspended in 1 ml of MHB-II, and aliquoted (200 μl) into a polystyrene 96-well, black, clear-bottom plate (Corning, Fisher Scientific Canada). EtBr efflux from the cells was monitored at room temperature using the microplate reader (excitation wavelength, 530 nm; emission wavelength, 600 nm). Readings were taken at 5-min intervals for 1 h to monitor efflux pump activity. The background fluorescence of the medium was subtracted from all measurements, and the assay was repeated independently in triplicate.
Statistical analysis.
Where indicated, a two-tailed Student's t test (P < 0.05) was used to determine whether the presence of PRMSE resulted in a significant difference compared to levels for the control. A two-way analysis of variance (ANOVA), followed by Sidak's multiple comparison, was used for biofilm assays to analyze statistical significance of the differences. Throughout the text, all of the changes (increase or decrease) reported were statistically significant. Statistically nonsignificant values were not mentioned in the text.
RESULTS
Characterization of PRMSE.
HPLC chromatograms (see Fig. S1 in the supplemental material) show the presence of six predominant phenolic compounds in PRMSE, gallic acid, 1,2-dihydroxybenzene (catechol), 3,4-dihydroxybenzaldehyde (catechaldehyde), syringaldehyde, vanillin, and 3-hydroxybenzoic acid, which corroborates previous findings on the identified compounds from maple syrup reported in the literature (19, 20). Based on their bioactive properties reported in the literature (42–44), the following four compounds were selected, purchased in pure form, and screened along with PRMSE for growth inhibition and synergy with ciprofloxacin: gallic acid, catechol, catechaldehyde, and syringaldehyde.
Antibacterial activity of PRMSE and phenolic compounds and their interactions with antibiotics.
MICs for PRMSE and ciprofloxacin against the bacterial strains E. coli CFT073, P. mirabilis HI4320, P. aeruginosa PAO1, and P. aeruginosa PA14 are shown in Table 2. To investigate the presence of synergistic interaction between ciprofloxacin (a fluoroquinolone which has biofilm penetration properties) and PRMSE, a checkerboard microdilution analysis was performed. The corresponding functional inhibitory concentration index (FICI) values were <0.5 for all tested strains (Table 2), demonstrating a strong synergistic effect between PRMSE and ciprofloxacin. Because P. aeruginosa is known to be partially resistant to carbenicillin (a β-lactam antibiotic) (45), the effect of PRMSE on the susceptibility of the P. aeruginosa strains to carbenicillin was investigated. The MIC and FICI values in Table S1 in the supplemental material show that PRMSE acted in synergy with carbenicillin against the growth of the two P. aeruginosa strains.
TABLE 2.
Synergistic interactions of ciprofloxacin and PRMSE for growth inhibition
| Bacterial strain | Value for measure of interaction |
|||||
|---|---|---|---|---|---|---|
| Ciprofloxacin |
PRMSE |
FICIb | Synergistic | |||
| MIC (μg ml−1) | FICa | MIC (mg ml−1) | FICa | |||
| E. coli CFT073 | 0.008 | 0.25 | 25 | 0.13 | 0.38 | Yes |
| P. mirabilis HI4320 | 0.016 | 0.02 | 50 | 0.13 | 0.14 | Yes |
| P. aeruginosa PAO1 | 1 | 0.03 | 50 | 0.06 | 0.1 | Yes |
| P. aeruginosa PA14 | 0.06 | 0.03 | 50 | 0.06 | 0.1 | Yes |
FIC, fractional inhibitory concentration. FIC is the MIC of compound 1 in the combination divided by the MIC of compound 1 alone.
FICI, FIC index (FICI = FICcompound 1 + FICcompound 2).
The MICs for the pure phenolic compounds are presented in Table 3. Among the four selected phenolic constituents of PRMSE, catechol (at 1.25 mg ml−1 for both E. coli CFT073 and P. mirabilis HI4320 and at 2.5 mg ml−1 for P. aeruginosa PAO1 and PA14) and catechaldehyde (at 1.25 mg ml−1 for all four strains) were found to clearly inhibit growth at lower concentrations. Furthermore, the interaction of these four phenolic compounds with ciprofloxacin was examined using a checkerboard microdilution assay. The results presented in Table 3 show that the combination of catechol and ciprofloxacin was the only synergistic combination (FICI of <0.5) for all investigated strains.
TABLE 3.
Interaction of phenolic constituents of PRMSE with ciprofloxacin
| Bacterial strain and constituent | Value for measure of interaction |
|||
|---|---|---|---|---|
| MIC (mg ml−1) | FIC of ciprofloxacin | FIC of compound | FICI | |
| E. coli CFT073 | ||||
| Gallic acid | 5 | 1 | 1 | 2 |
| Catechol | 1.25 | 0.25 | 0.25 | 0.5a |
| Catechaldehyde | 1.25 | 1 | 1 | 2 |
| Syringaldehyde | 5 | 0.5 | 0.25 | 0.75 |
| P. mirabilis HI4320 | ||||
| Gallic acid | 5 | 1 | 1 | 2 |
| Catechol | 1.25 | 0.25 | 0.25 | 0.5a |
| Catechaldehyde | 1.25 | 0.5 | 1 | 1.5 |
| Syringaldehyde | 5 | 0.5 | 0.5 | 1 |
| P. aeruginosa PAO1 | ||||
| Gallic acid | 5 | 1 | 1 | 2 |
| Catechol | 2.5 | 0.25 | 0.25 | 0.5a |
| Catechaldehyde | 1.25 | 0.5 | 0.51 | 1.01 |
| Syringaldehyde | 5 | 1 | 1 | 2 |
| P. aeruginosa PA14 | ||||
| Gallic acid | 5 | 1 | 1 | 2 |
| Catechol | 2.5 | 0.25 | 0.25 | 0.5a |
| Catechaldehyde | 1.25 | 1 | 1 | 2 |
| Syringaldehyde | 5 | 0.5 | 0.25 | 0.75 |
Values represent synergistic interaction.
Synergistic effect among phenolic compounds.
To investigate possible synergistic antimicrobial action among the four phenolic constituents of PRMSE, combinations of gallic acid, catechol, catechaldehyde, and syringaldehyde were tested using the checkerboard microdilution assay; the corresponding FICIs are presented in Table 4. The most potent synergy on growth inhibition was observed for gallic acid-catechol and gallic acid-catechaldehyde pairs, resulting in FICIs of 0.25 to 0.50, whereas all combinations of gallic acid with syringaldehyde led to FICIs ranging between 0.63 and 1.0 (Table 4), indicating no synergistic interaction. Interestingly, catechol exhibited strong synergy for growth inhibition with catechaldehyde against all chosen strains and with syringaldehyde against E. coli CFT073 and P. aeruginosa PAO1, as confirmed by FICIs (Table 4), suggesting that catechol is a potent component of PRMSE and is mainly responsible for its antimicrobial activity.
TABLE 4.
Interaction of individual phenolic constituents of PRMSE
| Bacterial strain and phenolic constituent | FIC |
FICIa |
||||
|---|---|---|---|---|---|---|
| Gallic acid | Catechol | Catechaldehyde | Gallic acid | Catechol | Catechaldehyde | |
| E. coli CFT073 | ||||||
| Catechol | 0.13 | NAb | 0.25* | NA | ||
| Catechaldehyde | 0.25 | 0.25 | NA | 0.5* | 0.5* | NA |
| Syringaldehyde | 0.5 | 0.25 | 0.25 | 0.75 | 0.31* | 0.31* |
| P. mirabilis HI4320 | ||||||
| Catechol | 0.13 | NA | 0.37* | NA | ||
| Catechaldehyde | 0.13 | 0.25 | NA | 0.37* | 0.5* | NA |
| Syringaldehyde | 0.5 | 0.25 | 0.25 | 1 | 0.75 | 0.75 |
| P. aeruginosa PAO1 | ||||||
| Catechol | 0.25 | NA | 0.5* | NA | ||
| Catechaldehyde | 0.13 | 0.25 | NA | 0.37* | 0.37* | NA |
| Syringaldehyde | 0.5 | 0.25 | 0.25 | 1 | 0.5* | 0.5* |
| P. aeruginosa PA14 | ||||||
| Catechol | 0.25 | NA | 0.37* | NA | ||
| Catechaldehyde | 0.25 | 0.25 | NA | 0.5* | 0.5* | NA |
| Syringaldehyde | 0.13 | 0.5 | 0.5 | 0.63 | 1 | 1 |
Asterisks indicate values that represent synergistic interaction.
NA, not applicable.
Inhibitory effect of PRMSE on biofilm formation and biofilm antibiotic susceptibility.
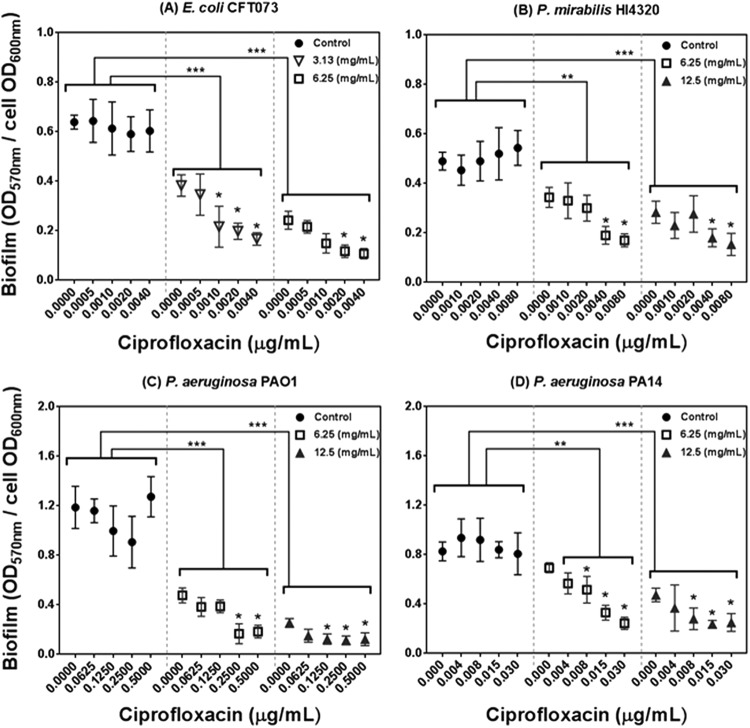
To investigate the potential of PRMSE for biofilm inhibition, biofilms were developed in polystyrene microtiter plates in the presence of different combinations of PRMSE, with and without ciprofloxacin, at sublethal concentrations. These sublethal concentrations of PRMSE (3.13 and 6.25 mg ml−1 for E. coli CFT073 and 6.25 and 12.5 mg ml−1 for P. mirabilis HI4320 and P. aeruginosa PAO1 and PA14) and ciprofloxacin (0.0005 to 0.004 μg ml−1 for E. coli CFT073, 0.001 to 0.008 μg ml−1 for P. mirabilis HI4320, 0.00625 to 0.5 μg ml−1 for P. aeruginosa PAO1, and 0.004 to 0.03 μg ml−1 for P. aeruginosa PA14) were chosen for biofilm studies based on results obtained in the MIC assay. The antibiofilm activity of PRMSE is presented in Fig. 1; the amount of sessile biomass (OD570) was normalized to the level of planktonic growth (OD600) to minimize bias from possible differences in growth levels on biofilm quantification. PRMSE alone showed significant inhibition of monoculture biofilm formation of all tested strains (P < 0.05). PRMSE in combination with ciprofloxacin (at sublethal concentrations) had significant inhibitory effects on biofilm formation (P < 0.05) for E. coli CFT073 (∼70% inhibition at 6.25 mg ml−1), P. mirabilis HI4320 (70% inhibition at 12.5 mg ml−1), P. aeruginosa PAO1 (83% inhibition at 12.5 mg ml−1), and P. aeruginosa PA14 (∼54% inhibition at 12.5 mg ml−1), in a dose-dependent manner. Since catechol was the only compound that exhibited synergy in antimicrobial activity with ciprofloxacin (Table 3), only the combination of catechol and ciprofloxacin was analyzed in this assay. Catechol with and without ciprofloxacin (both at sublethal concentrations) manifested significant inhibitory effects (P < 0.05) on biofilm formation for all four tested strains (see Fig. S2 in the supplemental material). When biofilms formed in the presence of only ciprofloxacin, there was a marginal increase in biofilm formation for P. mirabilis HI4320 and P. aeruginosa PAO1 (Fig. 1B and D). This increase was diminished by the combination of ciprofloxacin with PRMSE or catechol.
FIG 1.
Effect of PRMSE with and without ciprofloxacin on biofilm formation of E. coli CFT073 (A), P. mirabilis HI4320 (B), P. aeruginosa PAO1 (C), and P. aeruginosa PA14 (D). The graph presents normalized biofilm levels (OD570/OD600) versus different subinhibitory concentrations of ciprofloxacin for each strain grown in LB medium (control) or in LB medium amended with subinhibitory concentrations of PRMSE (3.13, 6.25, and 12.5 mg ml−1). Error bars show the standard deviations from values obtained from three replications. Statistically significant differences are indicated for each sample treated with PRMSE and ciprofloxacin compared to the control (sample treated with the corresponding concentration of ciprofloxacin only) (**, P < 0.01; ***, P < 0.001) and also for samples treated with PRMSE plus ciprofloxacin compared to sample treated with the same concentration of PRMSE without ciprofloxacin (*, P < 0.05). The legend in each graph shows the concentration of PRMSE.
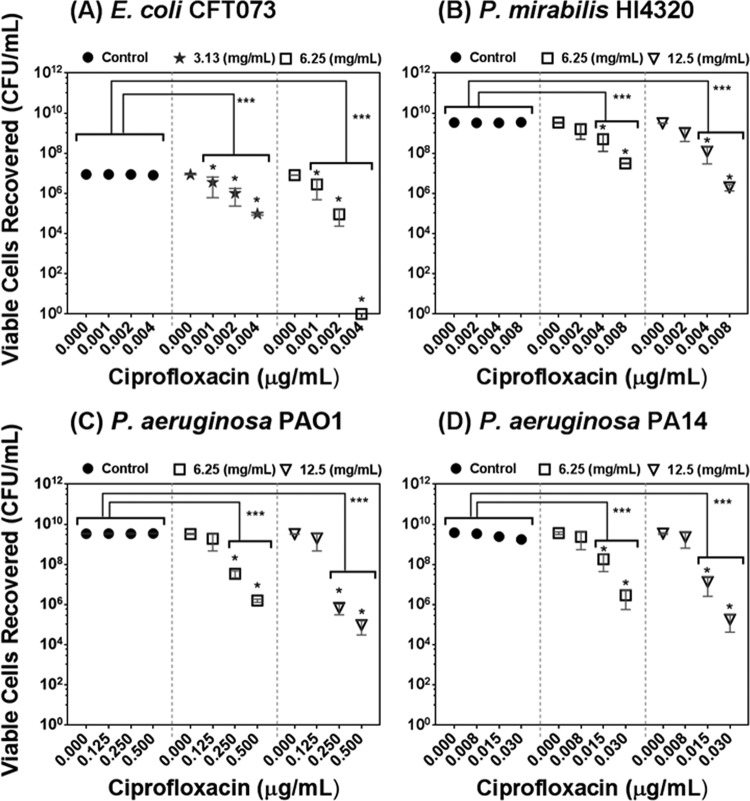
To analyze the applicability of PRMSE to eradicate biofilm from biomaterial surfaces, an in vitro biofilm assay was performed using silicone discs. The data for the biofilm eradication property of PRMSE in combination with ciprofloxacin are presented in Fig. 2; viable cell recoveries from the silicone discs after exposure to different combinations of PRMSE and ciprofloxacin was evaluated. PRMSE alone did not show significant eradication of monoculture biofilm on silicone discs for any of the tested strains, while PRMSE in combination with a sublethal concentration of ciprofloxacin had a significant biofilm eradication effect, with synergistic interaction at higher concentrations of ciprofloxacin (P < 0.05). These results are correlated with the above-mentioned antibiofilm activity of PRMSE.
FIG 2.
Effect of PRMSE, with and without ciprofloxacin, on eradication of monoculture biofilms on silicone discs of E. coli CFT073 (A), P. mirabilis HI4320 (B), P. aeruginosa PAO1 (C), and P. aeruginosa PA14 (D). The graph presents recovered bacterial cells from biofilm on silicone discs versus different subinhibitory concentrations of ciprofloxacin for each strain exposed to PBS (control) or in PBS amended with subinhibitory concentrations of PRMSE (3.13, 6.25, and 12.5 mg ml−1). Error bars show the standard deviations of values obtained from three replicates. Statistically significant differences are indicated for each sample treated with PRMSE plus ciprofloxacin compared to the control (sample treated with the corresponding concentration of ciprofloxacin only) (**, P < 0.01; ***, P < 0.001) and also for samples treated with PRMSE plus ciprofloxacin compared to sample treated with the same concentration of PRMSE without ciprofloxacin (*, P < 0.05). The legend in each graph shows the concentration of PRMSE.
Effect of PRMSE on bacterial membrane integrity and efflux pump inhibition.
In an effort to elucidate the mechanism(s) for the observed synergistic interactions, we quantified the change in bacterial outer membrane permeability using 1-N-phenylnapthylamine (NPN) as an indicator. The results show that both PRMSE and catechol, at sublethal concentrations, increased membrane permeability in all tested strains (Table 5; also see Fig. S3 in the supplemental material). These effects were similar to those of the known outer membrane-permeabilizing effect of gentamicin (see Fig. S3).
TABLE 5.
Biological effects of PRMSE and catecholg
| Strain | Effect based on bioassays | Value for: |
Value for positive controle |
||||
|---|---|---|---|---|---|---|---|
| Untreated cells | Phenolic extract (PRMSE) | Pure compound (catechol) | Gentamicin (μg ml−1) | CCCP | CTAB | ||
| E. coli CFT073 | Minimum permeabilization concna (mg ml−1) (NPN assay) | 1.6 | 0.06 | 0.006 | NAf | NA | |
| % reduction in fluorescence intensity due to efflux of EtBrb | 36.9 | 22.7 | 34.3 | NA | 15.8 | NA | |
| % of cells with uncompromised membranec (BacLight assay) | 100 | 88.7 | 68 | NA | NA | 0 | |
| Change in bacterial colony-forming abilityd (log CFU ml−1) | 0 | 0.8 | 2.3 | NA | NA | 5.7 | |
| P. mirabilis HI4320 | Minimum permeabilization concn (mg ml−1) | 1.6 | 1 | 0.013 | NA | NA | |
| % reduction in fluorescence intensity due to efflux of EtBr | 35.9 | 23.4 | 33.9 | NA | 14.5 | NA | |
| % of cells with uncompromised membrane | 100 | 72.6 | 50.3 | NA | NA | 0 | |
| Change in bacterial colony-forming ability (log CFU ml−1) | 0 | 1.2 | 3.2 | NA | NA | 4.5 | |
| P. aeruginosa PAO1 | Minimum permeabilization concn (mg ml−1) | 0.8 | 0.5 | 1 | NA | NA | |
| % reduction in fluorescence intensity due to efflux of EtBr | 37.6 | 22.3 | 35.8 | NA | 19.3 | NA | |
| % cells with uncompromised membrane | 100 | 87.9 | 80.4 | NA | NA | 0 | |
| Change in bacterial colony-forming ability (log CFU ml−1) | 0 | 0.9 | 1.5 | NA | NA | 5.1 | |
| P. aeruginosa PA14 | Minimum permeabilization concn (mg ml−1) | 0.8 | 0.5 | 1 | NA | NA | |
| % reduction in fluorescence intensity due to efflux of EtBr | 30.9 | 19 | 29.9 | NA | 17 | NA | |
| % of cells with uncompromised membrane | 100 | 89.1 | 78.6 | NA | NA | 0 | |
| Change in bacterial colony-forming ability (log CFU ml−1) | 0 | 0.9 | 1.9 | NA | NA | 4.9 | |
Assay was performed at concentrations below the MICs of extract and pure compound compared to the MIC of the reference antibiotic (gentamicin at above and below the MICs). These concentrations led to a maximal increase in NPN (1-N-phenylnapthylamine) uptake based on fluorescence intensity recorded. A 2.5% membrane permeabilization was considered the baseline value for the determination of minimum permeabilization concentrations. Raw data are provided in Fig. S3 in the supplemental material.
The ratio of green to red fluorescence was normalized to that of the untreated control and expressed as a percentage of the control. Cells were treated at 4× MIC of pure compound and extract for 10 min.
Reduction in fluorescence intensity as a percentage of that at the first time point of recording. Cells were treated at 0.25 times the MIC of pure compound and extract overnight.
Bacterial colonies were counted from the LB agar plate, and the log decrease in CFU/ml compared to that of the untreated control was calculated.
CCCP, carbonyl cyanide m-chlorophenylhydrazone (100 μM); CTAB, cetyltrimethylammonium bromide (10 μM).
NA, not applicable.
Values are compared to those of reference antibacterial agents with known mechanisms of action for the ability to permeabilize the outer membrane, inhibit efflux pumps, and damage membrane integrity.
Bacterial cell membrane damage was further examined using the BacLight assay. As shown in Table 5, catechol and PRMSE exhibited moderate membrane-disruptive effects. The cells exposed to PRMSE or catechol also were tested for their ability to form colonies on solid agar medium. Catechol reduced the number of CFU compared to those of the negative control (reductions of ≥1.5 log CFU ml−1 for E. coli CFT073, P. mirabilis HI4320, and P. aeruginosa PA14) during the 10-min exposure period, whereas PRMSE had a weak effect on the colony-forming ability of chosen strains. The CFU measurement correlates well with the BacLight results; however, because bacterial membrane damage is not necessarily a lethal event, the severity of the effect reflected by each assay is different.
We further investigated the effect of PRMSE and catechol on inhibition of the bacterial drug resistance efflux pump using the ethidium bromide (EtBr) efflux pump assay. PRMSE exhibited a significant inhibitory effect on the efflux pump for all bacterial strains with values comparable to that for the positive control, CCCP, which is an established proton motive force modulator (Table 5). There was weak reduction in fluorescence intensity for cells treated with PRMSE, indicating accumulation of EtBr in the cell as a result of efflux pump inhibition. Cells treated with catechol also exhibited a reduction in EtBr efflux but to a lesser extent than cells treated with PRMSE (Table 5).
Effect of PRMSE on gene transcription.
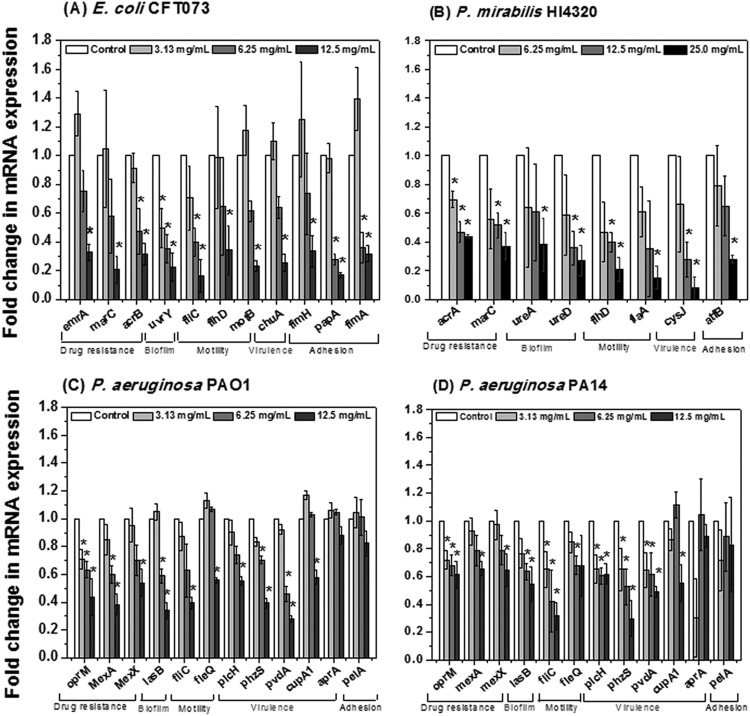
To explore the genetic basis for the synergy in antimicrobial activity observed between PRMSE and antibiotics, as well as the effect of PRMSE on bacterial biofilms, transcriptional analysis was performed using qRT-PCR to observe the differential expression of genes associated with bacterial motility, virulence, drug resistance, adhesion, and biofilm formation for each of the four bacterial strains. The results in Fig. 3 indicate that PRMSE, at sublethal concentrations, repressed the expression of genes associated with multiple drug resistance (emrA, acrB, and marC in CFT073; acrA and marC in HI4320; oprM, mexA, and mexX in PAO1 and PA14), motility (fliC, flhD, motB, fimH, fimA, and papA2 in CFT073; flaA and flhD in HI4320; fliC and fleQ in PAO1 and PA14), virulence determinants (chuA in CFT073; cysJ in HI4320; plcH, phzS, and pvdA in PAO1 and PA14), adhesion (fimH, fimA, and papA2 in CFT073; atfB in HI4320; cupA1 and pelA in PAO1 and PA14), and biofilm formation (uvrY in CFT073; ureD in HI4320; lasB in PAO1 and PA14). Transcriptional analysis confirms the trends observed with the biofilm assay (i.e., biofilm inhibition in the presence of PRMSE correlates with downregulation of biofilm-associated genes) and antibiotic synergy tests (i.e., downregulation of multiple drug resistance genes correlates with increased antibiotic susceptibility in the presence of PRMSE). PRMSE was used at sublethal concentrations for this experiment and does not affect the bacterial growth of any of the four strains (see Fig. S4 in the supplemental material). This is important, because conditions that affect growth can inhibit gene expression. Based on these results, it can be concluded that the observed differences in gene expression were due to the presence of PRMSE and not due to growth inhibition by this extract.
FIG 3.
Effect of PRMSE on expression of virulence genes for E. coli CFT073 (A), P. mirabilis HI4320 (B), P. aeruginosa PAO1 (C), and P. aeruginosa PA14 (D). Error bars show the standard deviations of values obtained from three replications. All cases of gene expression were normalized to levels for the corresponding housekeeping gene (gapA of E. coli CFT073, rpoA of P. mirabilis HI4320, and rpoD of P. aeruginosa PAO1 and PA14) and then were related to the normalized expression level of the same gene in the control. An asterisk indicates a statistically significant difference in measured values compared to the control (P < 0.05 by Student's t test).
DISCUSSION
In this work, we demonstrated the antimicrobial and antibiofilm effects of a cocktail of phenolic compounds extracted from maple syrup (PRMSE) against a range of pathogenic bacteria. We further showed that PRMSE can potentiate antibiotic susceptibility in both planktonic and biofilm modes of growth, which could be due partially to its ability to permeabilize the bacterial membrane, inhibit multidrug resistance efflux pumps, and downregulate genes associated with multidrug resistance. Among the four tested phenolic constituents of PRMSE, our results indicated that catechol plays a key role in the synergistic activity of PRMSE with ciprofloxacin.
Derivatives of A. saccharum Marsh (sugar maple) are used in traditional medicinal treatments for various health conditions, such as sores, cough, diarrhea, cataracts, and shortness of breath (14). Maple syrup is obtained by boiling and concentrating maple sap (46). During this intensive heating process, a complex cocktail of native phenolics (originally present in the xylem sap) and process-derived compounds (formed through chemical reactions during processing) is formed (19, 46). In our work, the MIC values of PRMSE were higher than those reported in the literature for ethanol extracts of maple leaf (16). The MIC values indicate that PRMSE is a mild antibacterial agent that is better suited to combinatorial therapy. Our results further show that PRMSE synergistically interacts with conventional antibiotics at sublethal concentrations to inhibit the growth of the chosen bacterial strains (Table 2; also see Table S1 in the supplemental material). This synergism-based antibiotic treatment is a promising approach for expanding the antimicrobial spectrum of treatments, preventing the emergence of resistant mutant strains during antibiotic treatment, and minimizing potential cytotoxicity due to high antibiotic doses.
Phenolic extracts of maple syrup have been reported to exhibit antioxidant and anticancer activity linked with the presence of diverse phytochemical constituents, including phenylpropanoids (19). Of the 51 known metabolites in maple syrup (from A. saccharum) (19), four compounds were investigated in more detail in the present study. The combination of catechol with ciprofloxacin, gallic acid, catechaldehyde, or syringaldehyde and the combination of gallic acid with catechaldehyde exhibited a synergistic antimicrobial effect against all chosen strains in vitro (Tables 3 and 4). Catechol has been reported previously to interact synergistically with other phenolic compounds in terms of antioxidant capacity (47). Moreover, catechol is made synthetically and can be readily available for potential use as a disinfectant. Phenolic compounds from other plant extracts or essential oils also have been reported previously for synergistic antimicrobial activity with synthetic drugs, such as erythromycin or vancomycin and antibiotics against Gram-negative bacteria (48). The mechanism(s) of this antimicrobial synergy remains a topic for investigation.
In persistent infections, bacterial biofilms commonly are more resistant to antibiotics and antimicrobial agents than bacteria in planktonic mode (1). We found that PRMSE and catechol exhibited antibiofilm activity against the four pathogenic strains examined. Genes associated with biofilm formation (uvrY, fimH, fimA, and papA2 of E. coli, ureD, ureA, and atfB of P. mirabilis, and lasB and cupA1 of P. aeruginosa [49–51]) were downregulated upon supplementation with PRMSE. In addition, PRMSE and catechol synergized with ciprofloxacin at sublethal concentrations to further inhibit the formation of biofilms (Fig. 1; also see Fig. S2 in the supplemental material). Ciprofloxacin has good penetration properties in the biofilm matrix; nevertheless, resistance has been reported in P. aeruginosa and E. coli biofilms (52). This suggests that the mechanism of resistance of these biofilms to ciprofloxacin goes beyond poor drug penetration and may be due to the presence of persister or resistant cells. Moreover, PRMSE exhibits synergy with ciprofloxacin for eradication of monoculture biofilm from silicone substrates, suggesting the applicability of PRMSE as a biofilm eradication strategy along with antibiotics. These observations led us to perform a more detailed investigation of the effect of PRMSE on antibiotic penetration into the bacterial cell.
The outer membrane is an advanced barrier shielding the bacterial cell against external antimicrobial compounds (53). To achieve the target cytoplasmic membrane, antimicrobial agents must overcome the barrier of the outer membrane in Gram-negative bacteria (54). This involves displacement of divalent cations from their binding sites on cell wall lipopolysaccharides (LPS) and the consequent permeabilization of the outer membrane (37). Our results show that PRMSE, at sublethal concentrations, can increase the uptake of NPN into the cell outer membrane, indicating its ability to permeabilize the membrane (Table 5). Our results also indicate that outer membrane permeabilization by PRMSE was achieved without altering the cell membrane integrity. Altered membrane permeability did not necessarily have a lethal effect (as indicated by the CFU counts); however, it may have contributed to the observed antimicrobial synergy with antibiotics by facilitating drug penetration. Interestingly, pure catechol exhibited a weaker effect on both outer membrane permeabilization and overall membrane integrity.
Multidrug efflux pumps of Gram-negative bacteria that traverse both the outer and inner membranes make a key contribution to intrinsic antimicrobial resistance (27). PRMSE showed significant inhibition of the EtBr transport across the cell envelope, which indicates a decreased activity of bacterial multidrug resistance efflux pumps (Table 5) and reduction in expression of genes associated with multidrug resistance (Fig. 3). Active efflux involves multidrug resistance efflux pump assemblies in the bacterial cell membranes that transport structurally unrelated compounds, including different classes of antibiotics, antiseptics, and cationic dyes, such as ethidium bromide and acriflavin (55). Thus, the observed inhibition of efflux pumps is of interest, as it potentially can result in (i) increased intracellular drug concentration, (ii) restoration of drug activity against resistant strains, (iii) minimization of further development of resistance (56), and (iv) reduced biofilm formation (57). In our study, the antimicrobial synergy of PRMSE with both ciprofloxacin (a fluoroquinolone) and carbenicillin (a β-lactam) suggests that it interacts with multiple membrane transporters.
In summary, we showed that PRMSE can increase the potency of conventional antibiotics against planktonic and biofilm cells of four pathogenic bacterial strains through a strong synergistic effect. Membrane permeabilization and efflux pump inactivation contribute to this antibacterial and antibiofilm synergy. In addition, this study shows that among the defined phenolic constituents of PRMSE, catechol was the major contributor to the antibiofilm and antibacterial synergy effects. Our results suggest the potential for the combined use of PRMSE (or its active component, catechol) with antibiotics to target bacterial biofilms. By combining antibiotics with mild antimicrobial agents (such as PRMSE) that exhibit synergistic interaction, it may be possible to decrease the dosage of antibiotics used to eradicate bacterial biofilms. Further studies are needed to evaluate the antimicrobial efficacy of PRMSE in vivo.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the financial support of the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canada Research Chairs (CRC) Program.
We thank K. Gibbs (Harvard University) for kindly providing P. mirabilis HI4320.
We have no conflict of interest to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00239-15.
REFERENCES
- 1.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saint S, Chenoweth CE. 2003. Biofilms and catheter-associated urinary tract infections. Infect Dis Clin N Am 17:411–432. doi: 10.1016/S0891-5520(03)00011-4. [DOI] [PubMed] [Google Scholar]
- 4.Antunes LC, Han J, Ferreira RB, Lolic P, Borchers CH, Finlay BB. 2011. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother 55:1494–1503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geissman TA. 1963. Flavonoid compounds, tannins, lignins and related compounds, p 265 In Florkin M, Stotz EH (ed), Pyrrole pigments, isoprenoid compounds and phenolic plant constituents. Elsevier, New York, NY. [Google Scholar]
- 6.Haslam E. 1996. Natural polyphenols (vegetable tannins) as drugs: possible modes of action. J Nat Prod 59:205–215. doi: 10.1021/np960040+. [DOI] [PubMed] [Google Scholar]
- 7.Borges A, Saavedra MJ, Simoes M. 2012. The activity of ferulic and gallic acids in biofilm prevention and control of pathogenic bacteria. Biofouling 28:755–767. doi: 10.1080/08927014.2012.706751. [DOI] [PubMed] [Google Scholar]
- 8.O'May C, Tufenkji N. 2011. The swarming motility of Pseudomonas aeruginosa is blocked by cranberry proanthocyanidins and other tannin-containing materials. Appl Environ Microbiol 77:3061–3067. doi: 10.1128/AEM.02677-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eydelnant IA, Tufenkji N. 2008. Cranberry derived proanthocyanidins reduce bacterial adhesion to selected biomaterials. Langmuir 24:10273–10281. doi: 10.1021/la801525d. [DOI] [PubMed] [Google Scholar]
- 10.Lee HI, Lee JH, Park KH, Sangurdekar D, Chang WS. 2012. Effect of soybean coumestrol on Bradyrhizobium japonicum nodulation ability, biofilm formation, and transcriptional profile. Appl Environ Microbiol 78:2896–2903. doi: 10.1128/AEM.07336-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burt SA, Ojo-Fakunle VT, Woertman J, Veldhuizen EJ. 2014. The natural antimicrobial carvacrol inhibits quorum sensing in Chromobacterium violaceum and reduces bacterial biofilm formation at sub-lethal concentrations. PLoS One 9:e93414. doi: 10.1371/journal.pone.0093414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, Zhou XD, Wu CD. 2011. The tea catechin epigallocatechin gallate suppresses cariogenic virulence factors of Streptococcus mutans. Antimicrob Agents Chemother 55:1229–1236. doi: 10.1128/AAC.01016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khokhani D, Zhang C, Li Y, Wang Q, Zeng Q, Yamazaki A, Hutchins W, Zhou SS, Chen X, Yang CH. 2013. Discovery of plant phenolic compounds that act as type III secretion system inhibitors or inducers of the fire blight pathogen, Erwinia amylovora. Appl Environ Microbiol 79:5424–5436. doi: 10.1128/AEM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnason T, Hebda RJ, Johns T. 1981. Use of plants for food and medicine by native peoples of eastern Canada. Can J Bot 59:2189–2325. doi: 10.1139/b81-287. [DOI] [Google Scholar]
- 15.Gonzalez-Sarrias A, Li L, Seeram NP. 2012. Effects of maple (Acer) plant part extracts on proliferation, apoptosis and cell cycle arrest of human tumorigenic and non-tumorigenic colon cells. Phytother Res 26:995–1002. doi: 10.1002/ptr.3677. [DOI] [PubMed] [Google Scholar]
- 16.Wu D, Wu XD, You XF, Ma XF, Tian WX. 2010. Inhibitory effects on bacterial growth and beta-ketoacyl-ACP reductase by different species of maple leaf extracts and tannic acid. Phytother Res 24:S35–S41. doi: 10.1002/ptr.2873. [DOI] [PubMed] [Google Scholar]
- 17.Mobley HL, Warren JW. 1987. Urease-positive bacteriuria and obstruction of long-term urinary catheters. J Clin Microbiol 25:2216–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 19.González-Sarrías A, Li L, Seeram NP. 2012. Anticancer effects of maple syrup phenolics and extracts on proliferation, apoptosis, and cell cycle arrest of human colon cells. J Funct Foods 4:185–196. doi: 10.1016/j.jff.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Sadiki M, Martin N. 2013. Solid-phase extraction and procedure for determination of phenolic compounds in maple syrup. Food Anal Methods 6:737–744. doi: 10.1007/s12161-012-9474-7. [DOI] [Google Scholar]
- 21.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. 2002. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 23.Merritt JH, Kadouri DE, O'Toole GA. 2005. Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1:Unit 1B.1. doi: 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenblatt J, Reitzel R, Dvorak T, Jiang Y, Hachem RY, Raad II. 2013. Glyceryl trinitrate complements citrate and ethanol in a novel antimicrobial catheter lock solution to eradicate biofilm organisms. Antimicrob Agents Chemother 57:3555–3560. doi: 10.1128/AAC.00229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan M, Hidalgo G, Asadishad B, Almeida S, Muja N, Mohammadi MS, Nazhat SN, Tufenkji N. 2013. Inhibition of bacterial motility and spreading via release of cranberry derived materials from silicone substrates. Colloids Surf B Biointerfaces 110:275–280. doi: 10.1016/j.colsurfb.2013.03.047. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper LA, Simmons LA, Mobley HL. 2012. Involvement of mismatch repair in the reciprocal control of motility and adherence of uropathogenic Escherichia coli. Infect Immun 80:1969–1979. doi: 10.1128/IAI.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cusick K, Lee YY, Youchak B, Belas R. 2012. Perturbation of FliL interferes with Proteus mirabilis swarmer cell gene expression and differentiation. J Bacteriol 194:437–447. doi: 10.1128/JB.05998-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dumas JL, van Delden C, Perron K, Kohler T. 2006. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol Lett 254:217–225. doi: 10.1111/j.1574-6968.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- 31.Elkins CA, Mullis LB, Lacher DW, Jung CM. 2010. Single nucleotide polymorphism analysis of the major tripartite multidrug efflux pump of Escherichia coli: functional conservation in disparate animal reservoirs despite exposure to antimicrobial chemotherapy. Antimicrob Agents Chemother 54:1007–1015. doi: 10.1128/AAC.01126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosseinidoust Z, Tufenkji N, van de Ven TG. 2013. Predation in homogeneous and heterogeneous phage environments affects virulence determinants of Pseudomonas aeruginosa. Appl Environ Microbiol 79:2862–2871. doi: 10.1128/AEM.03817-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosseinidoust Z, van de Ven TG, Tufenkji N. 2013. Evolution of Pseudomonas aeruginosa virulence as a result of phage predation. Appl Environ Microbiol 79:6110–6116. doi: 10.1128/AEM.01421-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane MC, Alteri CJ, Smith SN, Mobley HL. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104:16669–16674. doi: 10.1073/pnas.0607898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson MM, Rasko DA, Smith SN, Mobley HL. 2010. Transcriptome of swarming Proteus mirabilis. Infect Immun 78:2834–2845. doi: 10.1128/IAI.01222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simms AN, Mobley HL. 2008. PapX, a P fimbrial operon-encoded inhibitor of motility in uropathogenic Escherichia coli. Infect Immun 76:4833–4841. doi: 10.1128/IAI.00630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falla TJ, Karunaratne DN, Hancock RE. 1996. Mode of action of the antimicrobial peptide indolicidin. J Biol Chem 271:19298–19303. doi: 10.1074/jbc.271.32.19298. [DOI] [PubMed] [Google Scholar]
- 38.Boulos L, Prevost M, Barbeau B, Coallier J, Desjardins R. 1999. LIVE/DEAD BacLight: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J Microbiol Methods 37:77–86. doi: 10.1016/S0167-7012(99)00048-2. [DOI] [PubMed] [Google Scholar]
- 39.Hilliard JJ, Goldschmidt RM, Licata L, Baum EZ, Bush K. 1999. Multiple mechanisms of action for inhibitors of histidine protein kinases from bacterial two-component systems. Antimicrob Agents Chemother 43:1693–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Neill AJ, Miller K, Oliva B, Chopra I. 2004. Comparison of assays for detection of agents causing membrane damage in Staphylococcus aureus. J Antimicrob Chemother 54:1127–1129. doi: 10.1093/jac/dkh476. [DOI] [PubMed] [Google Scholar]
- 41.Kaatz GW, Seo SM, O'Brien L, Wahiduzzaman M, Foster TJ. 2000. Evidence for the existence of a multidrug efflux transporter distinct from NorA in Staphylococcus aureus. Antimicrob Agents Chemother 44:1404–1406. doi: 10.1128/AAC.44.5.1404-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chanwitheesuk A, Teerawutgulrag A, Kilburn JD, Rakariyatham N. 2007. Antimicrobial gallic acid from Caesalpinia mimosoides Lamk. Food Chem 100:1044–1048. doi: 10.1016/j.foodchem.2005.11.008. [DOI] [Google Scholar]
- 43.Jeong E-Y, Jeon J-H, Lee C-H, Lee H-S. 2009. Antimicrobial activity of catechol isolated from Diospyros kaki Thunb. roots and its derivatives toward intestinal bacteria. Food Chem 115:1006–1010. doi: 10.1016/j.foodchem.2009.01.021. [DOI] [Google Scholar]
- 44.Mellegard H, Stalheim T, Hormazabal V, Granum PE, Hardy SP. 2009. Antibacterial activity of sphagnum acid and other phenolic compounds found in Sphagnum papillosum against food-borne bacteria. Lett Appl Microbiol 49:85–90. doi: 10.1111/j.1472-765X.2009.02622.x. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez-Tebar A, Rojo F, Damaso D, Vazquez D. 1982. Carbenicillin resistance of Pseudomonas aeruginosa. Antimicrob Agents Chemother 22:255–261. doi: 10.1128/AAC.22.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L, Seeram NP. 2011. Quebecol, a novel phenolic compound isolated from Canadian maple syrup. J Funct Foods 3:125–128. doi: 10.1016/j.jff.2011.02.004. [DOI] [Google Scholar]
- 47.Freeman BL, Eggett DL, Parker TL. 2010. Synergistic and antagonistic interactions of phenolic compounds found in navel oranges. J Food Sci 75:C570–C576. doi: 10.1111/j.1750-3841.2010.01717.x. [DOI] [PubMed] [Google Scholar]
- 48.Hemaiswarya S, Kruthiventi AK, Doble M. 2008. Synergism between natural products and antibiotics against infectious diseases. Phytomedicine 15:639–652. doi: 10.1016/j.phymed.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Holling N, Lednor D, Tsang S, Bissell A, Campbell L, Nzakizwanayo J, Dedi C, Hawthorne JA, Hanlon G, Ogilvie LA, Salvage JP, Patel BA, Barnes LM, Jones BV. 2014. Elucidating the genetic basis of crystalline biofilm formation in Proteus mirabilis. Infect Immun 82:1616–1626. doi: 10.1128/IAI.01652-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitra A, Palaniyandi S, Herren CD, Zhu X, Mukhopadhyay S. 2013. Pleiotropic roles of uvrY on biofilm formation, motility and virulence in uropathogenic Escherichia coli CFT073. PLoS One 8:e55492. doi: 10.1371/journal.pone.0055492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qaisar U, Luo L, Haley CL, Brady SF, Carty NL, Colmer-Hamood JA, Hamood AN. 2013. The pvc operon regulates the expression of the Pseudomonas aeruginosa fimbrial chaperone/usher pathway (cup) genes. PLoS One 8:e62735. doi: 10.1371/journal.pone.0062735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans DJ, Allison DG, Brown MR, Gilbert P. 1991. Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: effect of specific growth rate. J Antimicrob Chemother 27:177–184. doi: 10.1093/jac/27.2.177. [DOI] [PubMed] [Google Scholar]
- 53.Bolla JM, Alibert-Franco S, Handzlik J, Chevalier J, Mahamoud A, Boyer G, Kiec-Kononowicz K, Pages JM. 2011. Strategies for bypassing the membrane barrier in multidrug resistant Gram-negative bacteria. FEBS Lett 585:1682–1690. doi: 10.1016/j.febslet.2011.04.054. [DOI] [PubMed] [Google Scholar]
- 54.Martinez de Tejada G, Sanchez-Gomez S, Razquin-Olazaran I, Kowalski I, Kaconis Y, Heinbockel L, Andra J, Schurholz T, Hornef M, Dupont A, Garidel P, Lohner K, Gutsmann T, David SA, Brandenburg K. 2012. Bacterial cell wall compounds as promising targets of antimicrobial agents. I. Antimicrobial peptides and lipopolyamines. Curr Drug Targets 13:1121–1130. doi: 10.2174/138945012802002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raherison S, Gonzalez P, Renaudin H, Charron A, Bebear C, Bebear CM. 2002. Evidence of active efflux in resistance to ciprofloxacin and to ethidium bromide by Mycoplasma hominis. Antimicrob Agents Chemother 46:672–679. doi: 10.1128/AAC.46.3.672-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aparna V, Dineshkumar K, Mohanalakshmi N, Velmurugan D, Hopper W. 2014. Identification of natural compound inhibitors for multidrug efflux pumps of Escherichia coli and Pseudomonas aeruginosa using in silico high-throughput virtual screening and in vitro validation. PLoS One 9:e101840. doi: 10.1371/journal.pone.0101840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kvist M, Hancock V, Klemm P. 2008. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl Environ Microbiol 74:7376–7382. doi: 10.1128/AEM.01310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hidalgo G, Chan M, Tufenkji N. 2011. Inhibition of Escherichia coli CFT073 fliC expression and motility by cranberry materials. Appl Environ Microbiol 77:6852–6857. doi: 10.1128/AEM.05561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.