Abstract
Antibiotic-resistant enterococcal infections are a major concern in hospitals where patients with compromised immunity are readily infected. Enterococcus faecium bacteria are of particular interest as these pathogens account for over 80% of vancomycin-resistant enterococcal infections. Antimicrobial peptides (AMPs) produced at the site of infection by engineered bacteria may offer a potential alternative to traditional antibiotics for the treatment of resistant bacteria such as E. faecium. For this mode of delivery to be effective, it is essential to identify a suitable protein expression system that can be used in the desired delivery bacterium. In this study, we describe a promising chloride-inducible promoter and its application in the bacterial delivery of AMPs from Lactococcus lactis to reduce counts of E. faecium bacteria in vitro. Reporter gene studies show that at chloride concentrations found within the human intestines, the chloride-inducible promoter exhibits high levels of protein expression compared to those of the commonly used nisin-inducible promoter. These results indicate that this system is powerful and would not require the exogenous administration of an inducer molecule. In its application for AMP production against E. faecium in vitro, L. lactis producing AMPs under the chloride promoter rapidly decreased E. faecium counts by nearly 10,000-fold. As an extension of this application, we also demonstrate the potential in using this type of delivery system in combination with traditional antibiotics to slow the development of resistance. Collectively, this study shows the promise of using a chloride-inducible promoter for the bacterial delivery of AMPs in the body for the treatment of vancomycin-resistant enterococci (VRE) and other antibiotic-resistant bacteria.
INTRODUCTION
Enterococcal infections are a rising concern for health care due to the increasing frequency of multidrug-resistant cases. As of 2013, nearly 30% of all reported enterococcal infections were antibiotic resistant (1). This high percentage of resistance is especially disconcerting in hospitals because patients with compromised immune systems or patients who are on antibiotic regimens are particularly susceptible to enterococcal infections (2). Once an infection has occurred, it can be difficult to eradicate not only from the infected patient but also from the entire hospital environment. Antibiotic resistance makes this process even more challenging, and these infections become both more dangerous and costly (3).
Enterococcus faecium and Enterococcus faecalis are the causative pathogens of nearly all vancomycin-resistant enterococcal infections (4). While E. faecalis is more prevalent as an infectious agent, E. faecium is more commonly resistant to antibiotics than E. faecalis and is known for its ability to rapidly transfer antibiotic resistance (2). For example, nearly 81% of E. faecium infections are considered vancomycin resistant (VR), compared to only 5% of E. faecalis infections (4). Additionally, E. faecium carrying resistance to vancomycin is also commonly resistant to many first-line antibiotics, including both β-lactams and aminoglycosides (5). It is thus important to find a means of combating these pathogens as health care providers are often left with limited options for the treatment of infections caused by vancomycin-resistant enterococci (VRE).
Antimicrobial peptides (AMPs) may offer a potential alternative to traditional antibiotics for the treatment of VRE infections. Bacteriocins, a class of AMPs, are short peptides naturally produced by many bacteria as a means of eliminating competing microbes. For example, the bacteriocins enterocin A, enterocin P, and hiracin JM79 are known to be highly active against a wide array of enterococci, including both vancomycin-resistant E. faecalis and E. faecium (6). While bacteriocins are often extremely potent against their target bacteria, their activity is typically species specific, thus making them less destructive to the native microbiota than traditional antibiotics (7). In addition to being less destructive, this narrow-spectrum activity may also avoid unnecessary pressure for resistance development among the unaffected surrounding microbes.
Despite their demonstrated efficacy against many pathogens of interest, AMPs are limited in their application for treatment of internal infections because oral and intravenous delivery are hindered due to the fast degradation of the peptides in the body (8). Because many infections, including those caused by VRE, are often initiated in the gastrointestinal (GI) tract, it is necessary to find a means of delivery of AMPs to the intestines (2).
It may be possible to deliver these peptides to the site of infection using engineered probiotic bacteria. Lactococcus lactis has been previously selected as a delivery vehicle because it has been shown to survive the human gastrointestinal tract and is considered a potentially probiotic organism (9, 10). Indeed, these bacteria have already been successfully used in phase I clinical trials for the delivery of transgenic proteins for the treatment of Crohn's disease in the human intestine (11).
In a previous study, the bacteriocins enterocin A, enterocin P, and hiracin JM79 were successfully produced in L. lactis NZ9000 under the E. faecalis-responsive promoter, PrgX-PrgQ. Borrero and coworkers demonstrated that the expression system was highly effective at both targeting and decreasing E. faecalis populations (6). While the PrgX-PrgQ AMP expression system may be a candidate for the treatment of E. faecalis, E. faecium does not produce the inducer molecule, the sex pheromone peptide cCF10, which is required to activate the PrgX-PrgQ promoter. The use of the PrgX-PrgQ expression system would thus require the exogenous application of cCF10 for treatment of E. faecium or other non-cCF10-producing strains of Enterococcus (6).
The purpose of the current study is to characterize and implement a general AMP expression system against enterococci that can be induced by the conditions found inside the gut. The use of an environmentally inducible promoter is valuable in that it may be used in the treatment of any pathogen. For this study, we have selected the lactococcal chloride-inducible promoter (CIP) previously discovered by Sanders et al. (12) as this promoter has been reported to be active at 0.1 M chloride, which is within the relevant range in the gut (∼0.05 to 0.15 M) (13).
The chloride-inducible promoter is believed to be controlled by the activator protein GadR. GadR's ability to turn on the promoter is thought to be dependent on the environmental chloride concentration though the mechanism behind this interaction remains unclear (12). Due to the reliable presence of chloride in both human and animal guts, this type of expression system could be used for a wide variety of applications employing environment-responsive protein production.
In this study, we first characterized the chloride-inducible promoter and then compared it to the widely used nisin-inducible promoter using reporter protein assays. The chloride-inducible promoter was then used to express the three bacteriocins enterocin A, enterocin P, and hiracin JM79 in L. lactis subsp. cremoris NZ9000. The ability of the chloride-inducible AMP expression system to inhibit a variety of both E. faecium and E. faecalis strains was tested using agar diffusion assays. E. faecium inhibition was further quantified using liquid coculture tests. Last, as an extension of this project, combination treatments using traditional antibiotics and bacterial AMP delivery were explored as a means of reducing the development of AMP resistance.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacteria used in this study are listed in Table 1. L. lactis NZ9000 was cultured at 32°C in M17 broth (Oxoid, Ltd., Basingstoke, United Kingdom) supplemented with 0.5% (wt/vol) glucose (GM17 medium). E. faecium 8-E9 was grown in brain heart infusion (BHI) broth (Oxoid) at 37°C. Escherichia coli MC1061 F′ was grown in LB broth (Fisher Scientific, Fair Lawn, NJ, USA) at 37°C, with shaking. Agar plates were made by the addition of 1.5% (wt/vol) agar (Oxoid) to the liquid medium. When necessary, rifampin (Sigma Chemical Co., St. Louis, MO, USA) was added to the medium at 5 μg/ml for E. faecium, and chloramphenicol (Mediatech Inc., Manassas, VA, USA) was added at 5 μg/ml or 20 μg/ml for L. lactis or E. coli, respectively.
TABLE 1.
Bacteria used in this study
| Bacterial strain | Description | Source |
|---|---|---|
| L. lactis NZ9000 | Plasmid-free strain, derivative of MG1363; pepN::nisRK, non-bacteriocin producer | Mobitec |
| E. faecium strains | ||
| 8-E9 | Ampicillin/vancomycin/linezolid resistant | University of Minnesota |
| 6-E6 | Ampicillin/vancomycin/linezolid resistant | University of Minnesota |
| 7A | Ampicillin/vancomycin/linezolid resistant | University of Minnesota |
| 9B | Ampicillin/vancomycin/linezolid resistant | University of Minnesota |
| E. faecalis strains | University of Minnesota | |
| OG1RF | ATCC 47077; plasmid-free, rifampin/fusidic acid-resistant mutant of OG1; common laboratory strain | University of Minnesota |
| V583 | ATCC 700802; first isolated vancomycin-resistant strain and first sequenced E. faecalis genome | University of Minnesota |
| Ch116 | Gentamicin/kanamycin/streptomycin/tetracycline/erythromycin/penicillin-resistant, β-lactamase-producing isolate | University of Minnesota |
| JH2-2 | Rifampin/fusidic acid-resistant mutant; common laboratory strain | University of Minnesota |
| Pan7 | Panose 7; fecal sample from healthy volunteer | University of Minnesota |
| Com1 | Fecal sample from healthy volunteer | University of Minnesota |
| DS5 | ATCC 14508; pAD1, pAMα 1; erythromycin/tetracycline-resistant strain | University of Minnesota |
| E. coli MC1061 F′ | Plasmid-free, recA+, non-amber suppressor strain | Lucigen |
Construction of plasmids.
Plasmids were constructed using standard molecular cloning techniques. All restriction enzymes were purchased from New England BioLabs (Beverly, MA, USA). Fragments obtained and plasmids used are listed in Table 2.
TABLE 2.
Plasmids and DNA fragments used in this study
| Plasmid or DNA fragment | Description | Source or reference |
|---|---|---|
| Plasmids | ||
| Geneart-chloride | Kanr; source of the CIP system | Geneart |
| pNZ8048 | Cmr; inducible expression vector carrying the nisA promoter | 14 |
| pNZC | pNZ8048 derivative containing the CIP | This work |
| pNZ8048L | pNZ8048 derivative containing lacZ under the nisin-inducible promoter | This work |
| pNZCL | pNZC derivative containing lacZ under the CIP | This work |
| pBac | Spcr; source of Bac fragment | 6 |
| pNZCA3 | pNZC derivative containing Bac under the CIP | This work |
| pBK1 | Cmr; source of lacZ | 6 |
| DNA fragments | ||
| CIP systema | 1,317-bp fragment containing the chloride-inducible promoter (Pgad) and the gene encoding the activator protein (gadR) under the control of a constitutive promoter (PgadR) | 15 |
| Bac | 1,610-bp fragment containing the enterocin A structural gene (entA) with its immunity gene (entiA), the enterocin P structural gene (entP) with its immunity gene (entiP), and the hiracin JM79 structural gene (hirJM79) with its immunity gene (hiriJM79) | 6 |
CIP, chloride-inducible promoter.
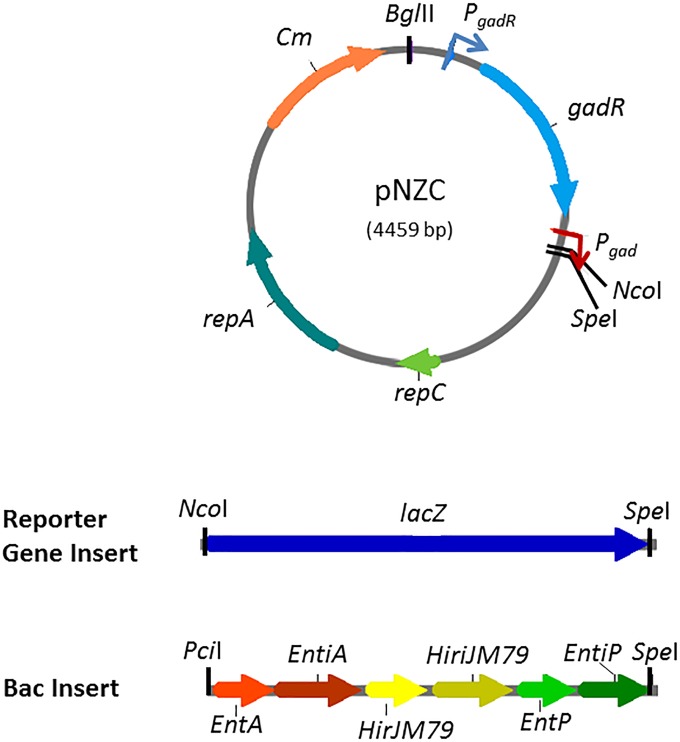
The chloride-inducible promoter (CIP) sequence used in this work was adapted from a CIP sequence of Sanders and coworkers (GenBank accession number AF005098; base pairs 821 to 2,068) (15) and synthesized by Geneart. The sequence was then amplified using primers chloride-F (5′-CGAATTGAAGGAAGGCCG-3′) and chloride-R (5′-GCAGTGAAAGGAAGGCC-3′). The PCR fragment and plasmid pNZ8048 were both digested with restriction enzymes BglII and NcoI (New England BioLabs) and ligated at 16°C for 16 h. The resulting ligation was then transformed into electrocompetent E. coli MC1061 F′ (Lucigen). Successful transformants were identified by colony PCR using the primers pNZ8048-F (5′-GCCCCGTTAGTTGAAGAAGG-3′) and pNZ8048-R (5′-CAATTGAACGTTTCAAGCCTTGG-3′) and further verified by sequencing analysis. The resulting plasmid, pNZC, was isolated from E. coli using a QIAprep Miniprep kit (Qiagen) and then transformed into electrocompetent L. lactis NZ9000 (16).
For β-galactosidase (β-Gal) reporter gene studies, the lacZ reporter gene was inserted downstream of the chloride-inducible promoter. lacZ was amplified using the primers LacZ-F (5′-GCTAAGCCATGGAAG TTACTGACGTAAGATTACGG-3′) and LacZ-R (5′-TCGACTAGTTTATTATTATTTTTGACACCAGACCAACTGG-3′) from pBK1. Both the PCR product obtained and pNZC were then digested with restriction enzymes NcoI and SpeI, ligated, and transformed into E. coli MC1061 F′ and L. lactis as described above. The resulting plasmid is referred to as pNZCL. The plasmid pNZ8048L was created for these studies using an identical procedure to that used to create pNZCL. In this case, however, pNZ8048 rather than pNZC was used for the backbone.
For AMP production, the genes encoding enterocin A, enterocin P, and hiracin JM79 along with their immunity proteins (fragment Bac from Table 2) were inserted into pNZC. Fragment Bac was amplified using primers AMP-F (5′-CATAACATGTCTACTATGAAAAAAAAGATTATCTC-3′) and AMP-R (5′-CACTAGTTTATCAAAGTCCCGACC-3′), using pBac as the template. pNZC was then digested with NcoI and SpeI while Bac was digested using PciI and SpeI. The digestion products originated were then ligated and transformed into E. coli MC1061 F′. The resulting plasmid, pNZCA3, was then transformed into electrocompetent L. lactis NZ9000.
Beta-galactosidase assays.
L. lactis NZ9000 containing pNZCL or pNZ8048L was grown overnight in GM17 medium. The following day, cells were reinoculated into fresh GM17 medium at an optical density at 600 nm (OD600) of 0.15. Cells were then grown at 37°C to an OD600 of 0.4 to 0.5 at which point they were induced. L. lactis NZ9000/pNZCL was induced by adding NaCl to the medium to obtain final concentrations of 0.01, 0.05, 0.1, 0.3, and 0.5 M Cl−, and L. lactis NZ9000/pNZ8048L was induced by the addition of 5 ng/ml or 40 ng/ml nisin A as previously described (17). After induction, OD600 readings and 1-ml samples were collected each hour. Upon collection, samples were centrifuged for 7 min at 5,600 × g, supernatant was removed, and the pellets were refrigerated until further analysis.
To measure β-Gal activity, we used the traditional Miller assay (18) with some minor modifications. First, pellets were resuspended in 990 μl of Z-buffer (60 mM Na2HPO4·7H2O [Sigma], 40 mM NaH2PO4·H2O [Sigma], 10 mM KCl [Sigma], 1 mM MgSO4·7H2O [Sigma], 50 mM β-mercaptoethanol [Sigma], in deionized [DI] water). A total of 105 μl of each sample was transferred to a polypropylene 96-well plate. Twenty microliters of toluene was then added to each well, and the plate was shaken for ∼30 s. The plate was then covered and incubated for 15 min at 32°C. One hundred microliters of 10 mg/ml o-nitrophenyl-β-d-galactopyranoside (ONPG) (Research Products International Corp., Mt. Prospect, IL, USA) dissolved in Z-buffer was then added to each well. The plate was covered again and then incubated at 32°C for 5 to 15 min until sufficient color had developed. To stop the reaction, 125 μl of 2 M Na2CO3 was then added to the wells. Last, the plate was centrifuged at 4°C for 30 min at 6,130 × g to remove cell debris; the supernatant was transferred to a clean 96-well plate, and the OD420 and OD550 were read using a plate reader (Synergy H1 multimode reader; BioTek, Winooski, VT). The following formula was used to convert cell density to activity units: 1 unit = 1,000(Abs420 − 1.75Abs550)/(incubation time)(0.105 ml)OD600, where incubation time is in minutes, 0.105 ml is the sample volume, and Abs is absorbance.
In this equation, Abs420 is calculated as OD420(sample) × 0.734 + OD420(water) × 0.266 to adjust the final reaction concentration to that of the traditional Miller assay (18).
Supernatant production.
L. lactis containing pNZCA3 was grown overnight in BHI broth. Cells were then reinoculated in fresh BHI broth at an OD600 of 0.07 to 0.1. Because BHI broth contains ∼0.15 M NaCl, no additional salt was added for induction. Cells were then grown for ∼6 h, and then cell-free culture supernatant was obtained by centrifugation of the culture at 12,000 × g at 4°C for 10 min. The supernatant was filtered through 0.2-μm-pore-size filters (Whatman International, Ltd., Maidstone, United Kingdom), and stored at −20°C until use.
Agar diffusion tests.
L. lactis was grown on GM17 plates supplemented with chloramphenicol to produce single colonies while enterococci were grown overnight in BHI broth. The following morning, BHI broth supplemented with agar (0.8%) was inoculated with a 0.005% concentration of the overnight enterococcal culture and poured into a petri dish. Once the plate had solidified, L. lactis colonies were stabbed into the semisolid medium, and the plates were incubated overnight at 37°C. Inhibition was confirmed by the formation of clear zones around the recombinant L. lactis strains.
Coculture and supernatant activity assays. (i) Salt concentration tests.
E. faecium was grown overnight in GM17 medium at 37°C, and L. lactis/pNZCA3 was grown in GM17 medium at 32°C. The following morning, 30 μl of L. lactis and E. faecium overnight cultures was inoculated into 5 ml of fresh GM17 medium supplemented with sodium chloride at the concentrations specified in the figure legends. This resulted in a ∼1:2 ratio of E. faecium to L. lactis bacteria. GM17 medium was used for these experiments because it has a lower salt concentration than BHI broth. Samples of the cultures were then taken at different times, serially diluted, plated (10 μl of each dilution) on GM17 plates containing 5 μg/ml rifampin (GM17-Rif) or 5 μg/ml chloramphenicol (GM17-Cm), and incubated overnight at 37°C for E. faecium and at 32°C for L. lactis. The following day E. faecium or L. lactis CFU were counted on GM17-Rif and GM17-Cm plates, respectively.
(ii) Tests of supernatant combination with rifampin.
E. faecium was grown overnight in BHI broth at 37°C, and L. lactis/pNZCA3 was grown in BHI broth at 32°C. E. faecium was then inoculated at an OD600 of ∼0.06 into 5 ml of fresh BHI broth and allowed to grow to an OD600 of ∼0.1. For antibiotic experiments, 50 μg/ml streptomycin (Chem-Implex International, Inc., Wood Dale, IL, USA), 100 μg/ml ampicillin (Sigma), or 30 μg/ml rifampin (19) was added to the fresh BHI broth. Cultures were then supplemented with 10% of the previously obtained supernatant or 10% of L. lactis/pNZCA3 culture at an OD600 of ∼0.1 (time [t] = 0 h). This resulted in a ∼5:1 ratio of E. faecium to L. lactis bacteria. Samples of the cultures were then taken at different times, serially diluted, plated (10 μl of each dilution) on GM17 plates containing 5 μg/ml rifampin (GM17-Rif) or 5 μg/ml chloramphenicol (GM17-Cm), and incubated overnight at 37°C or 32°C, respectively. The following day E. faecium or L. lactis CFU were counted on GM17-Rif or GM17-Cm plates, respectively.
RESULTS
Reporter gene studies.
The primary motivation for using a chloride-inducible promoter to express antimicrobial peptides is its induction by physiological conditions instead of exogenous induction. It was therefore necessary to characterize the promoter's dependence on chloride and to verify that it would be active within the range of chloride concentrations measured throughout the human GI tract (∼0.05 to 0.15 M) (13). Additionally, it was desirable to compare production under the chloride promoter to that under the widely used nisin-inducible promoter to evaluate the strength of this expression system. To assess these parameters, the lacZ reporter gene was inserted under the control of the chloride-inducible promoter to create the plasmid pNZCL (Fig. 1) or under the control of the nisin-inducible promoter in pNZ8048 to create the plasmid pNZ8048L. L. lactis NZ9000/pNZCL and L. lactis NZ9000/pNZ8048L were then grown to an OD600 of ∼0.45 and induced with chloride (0.01 M, 0.05 M, 0.1 M, 0.3 M, and 0.5 M) or nisin (0 ng/ml, 5 ng/ml, and 40 ng/ml). Beta-galactosidase production under the two promoters was then measured over time.
FIG 1.
Maps of pNZC and inserts used in this study. A chloride-inducible promoter (CIP) was inserted between BglII and NcoI. PgadR is a constitutive promoter controlling the production of the activator protein GadR. lacZ and AMP expression are controlled by the chloride-inducible promoter Pgad (activated by GadR). lacZ and Bac are inserted between cut sites NcoI and SpeI in pNZC to create pNZCL and pNZCA3.
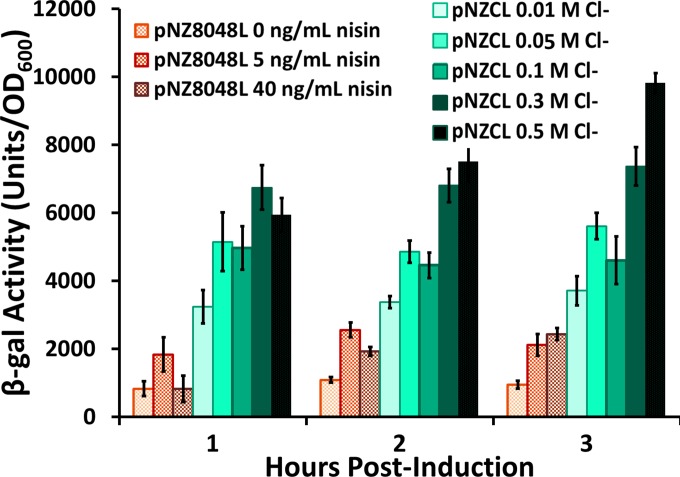
The results shown in Fig. 2 indicate that the chloride-inducible system is highly responsive to the chloride levels in the medium and that, within the range of physiological conditions (0.05 M to 0.1 M), the chloride-inducible system expresses significantly more protein than the fully induced nisin-inducible promoter. It also appears that the production does not vary much within the induction range found in the intestines. The results presented here represent three technical replicates. The experiments used for the data in Fig. 2 were also repeated twice at separate times. Both trials showed that the chloride-inducible system induced with 0.05 M and 0.1 M chloride showed higher β-Gal production than the nisin-inducible promoter induced with either 5 or 40 ng/ml nisin at all time points postinduction (P < 0.005 for t = 1 and 2 h; P < 0.02 for t = 3 h). Experiments comparing β-Gal expression under the chloride-inducible promoter were repeated several times on separate occasions and showed similar trends of increased expression with increasing salt concentrations. These results are promising for the use of this system for the delivery of proteins in the GI tract as they show that high levels of protein expression can be obtained from the promoter under the induction conditions naturally found in the gut.
FIG 2.
Comparison of β-galactosidase production under the nisin-inducible promoter and the chloride-inducible promoter in L. lactis NZ9000. The nisin-inducible promoter was induced with 0 ng/ml, 5 ng/ml, or 40 ng/ml nisin, and the chloride-inducible promoter was induced with 0.01 M, 0.05 M, 0.1 M, 0.3 M, and 0.5 M Cl−. Cells were induced at an OD600 of ∼0.45 and then sampled at the designated time postinduction. Error bars represent ±1 standard deviation of assay triplicates.
It should be noted that significant protein expression occurs even in GM17 medium without any added salt. The chloride concentration in GM17 medium was measured and found to be ∼0.01 M Cl−. On the basis of the trends observed in the data shown in Fig. 2, we may conjecture that the expression level under the chloride-inducible promoter could be further reduced if the chloride concentration was lowered by using a different medium. This approach is used in the agar diffusion tests discussed below. We note, however, that a fully uninduced state (no chloride ions) cannot be obtained because chloride is necessary for bacterial survival.
Delivery of AMPs using the chloride-inducible promoter. (i) Agar diffusion inhibition tests.
To apply the chloride-inducible system for the production of AMPs, the three bacteriocin genes, enterocin A, enterocin P, and hiracin JM79, along with their immunity genes were inserted downstream of the chloride-inducible promoter to create the plasmid pNZCA3 (Fig. 1). L. lactis NZ9000 was then transformed with this plasmid, and its antimicrobial activity was tested using an agar diffusion test using Enterococcus faecium 8-E9 as the indicator strain.
The experiment shown in Fig. 3 demonstrates the inhibition of E. faecium by L. lactis/pNZCA3 at different concentrations of chloride. For these studies, a modified GM17 medium was used which contains half the M17 in traditional GM17 medium. This reduced the basal salt concentration from ∼0.01 M Cl− to ∼0.005 M Cl−, which enabled a more nearly uninduced state. In this experiment, L. lactis/pNZC (chloride-inducible promoter, no AMPs) was used as a negative control. Based on the diameters of the halos produced, it appears that the overall AMP production is salt dependent, as anticipated by the reported gene studies discussed above. Interestingly, there is a significant increase in diameter between cultures treated with 0.005 M and 0.05 M chloride but not between cultures treated with 0.05 M and 0.15 M chloride. The halo sizes observed at 0.05 and 0.15 M in the modified GM17 medium are similar to those observed in traditional GM17 medium (10 to 12 mm). L. lactis and E. faecium growth curves in modified GM17 medium at different salt concentrations can be found in Fig. S1 and S2 in the supplemental material, respectively. It should be noted that at 0.3 M and 0.5 M NaCl, E. faecium growth is significantly slowed (see Fig. S2), which may have contributed to the larger halo diameters observed at the higher salt concentrations.
FIG 3.
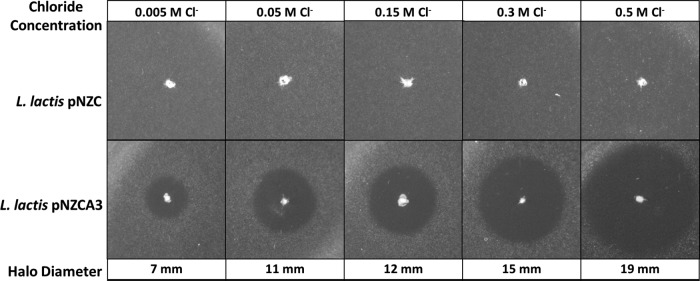
Agar diffusion inhibition test of L. lactis NZ9000 producing enterocin A, enterocin P, and hiracin JM79 under the CIP (pNZCA3) or with no AMPs under the CIP (pNZC) at different chloride concentrations. E. faecium 8-E9 was used as the indicator strain. Modified GM17 medium was used to obtain a lower basal level chloride concentration (0.005 M).
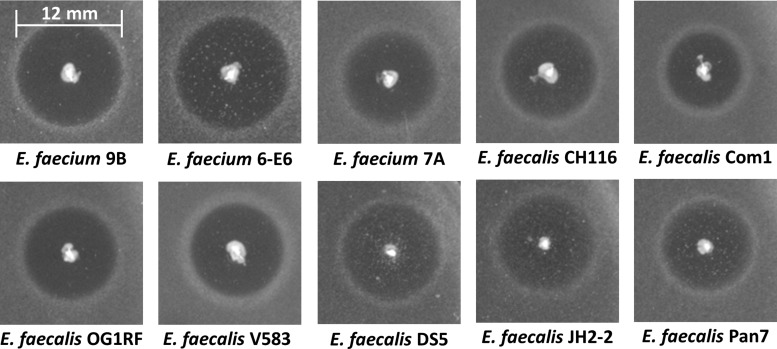
An important benefit of using an environmentally inducible promoter for the delivery of AMPs is that, in principle, it can be used against any type of pathogen. To demonstrate that the chloride-inducible promoter expressing the three AMPs, enterocin A, enterocin P, and hiracin JM79, can be used to target a broad range of enterococci, halo tests were performed against several pathogenic, antibiotic-resistant strains of both E. faecium and E. faecalis. Figure 4 shows the results of agar diffusion tests of L. lactis/pNZCA3 against 10 strains of enterococci (data not shown for E. faecium 8-E9). In all cases we observed clear halos of ∼8 mm to 13 mm in diameter. All halo tests shown in Fig. 4 were done on BHI broth-agar (∼0.15 M Cl−). Activity was also tested against two E. faecalis isolates from healthy patients (isolates Com1 and Pan7). The observed activity against these strains indicates that this system could potentially impact commensal enterococcal species.
FIG 4.
Agar diffusion inhibition test of L. lactis NZ9000 producing enterocin A, enterocin P, and hiracin JM79 under the CIP (pNZCA3) against 10 strains of Enterococcus. The indicator strain is shown below each halo. See Table 1 for descriptions of the strains. Halo diameters range from ∼8 to 13 mm. Tests were done in BHI medium-agar (∼0.15 M chloride).
(ii) Liquid coculture inhibition tests.
To further quantify the effect of the chloride-inducible AMP expression cassette on E. faecium growth, coculture inhibition tests were done using E. faecium 8-E9 and L. lactis expressing enterocin A, enterocin P, and hiracin JM79 under the chloride-inducible promoter. Figure 5A shows the counts of viable E. faecium 8-E9 bacteria at different time points. Results for E. faecium grown alone and for E. faecium treated with AMP-producing L. lactis in GM17 medium with 0.01 M Cl−, 0.1 M Cl−, or 0.3 M Cl− are shown. E. faecium growth was also tested in the presence of L. lactis producing no AMPs (pNZC) and was found to be nearly identical to that of normal E. faecium growth (data not shown). Figure 5B shows the corresponding L. lactis counts in each culture. Interestingly, the highest inhibition of E. faecium by L. lactis/pNZCA3 coculture was that of the 0.01 M culture. Similar results were observed in three individual experiments, conducted at separate times, in which L. lactis induced with a higher concentration of chloride showed reduced inhibition of E. faecium. This is likely due to the faster growth of L. lactis at lower NaCl concentrations, as seen in the data shown in Fig. 5B. At 4 h postinduction, the counts of viable L. lactis bacteria were approximately 2- and 4-fold higher in the 0.01 M cultures than in the cultures supplemented with 0.1 and 0.3 M NaCl, respectively. The difference in growth at increasing salt concentrations is significantly more pronounced than that observed when L. lactis is grown alone rather than in coculture with E. faecium (see Fig. S1 in the supplemental material).
FIG 5.
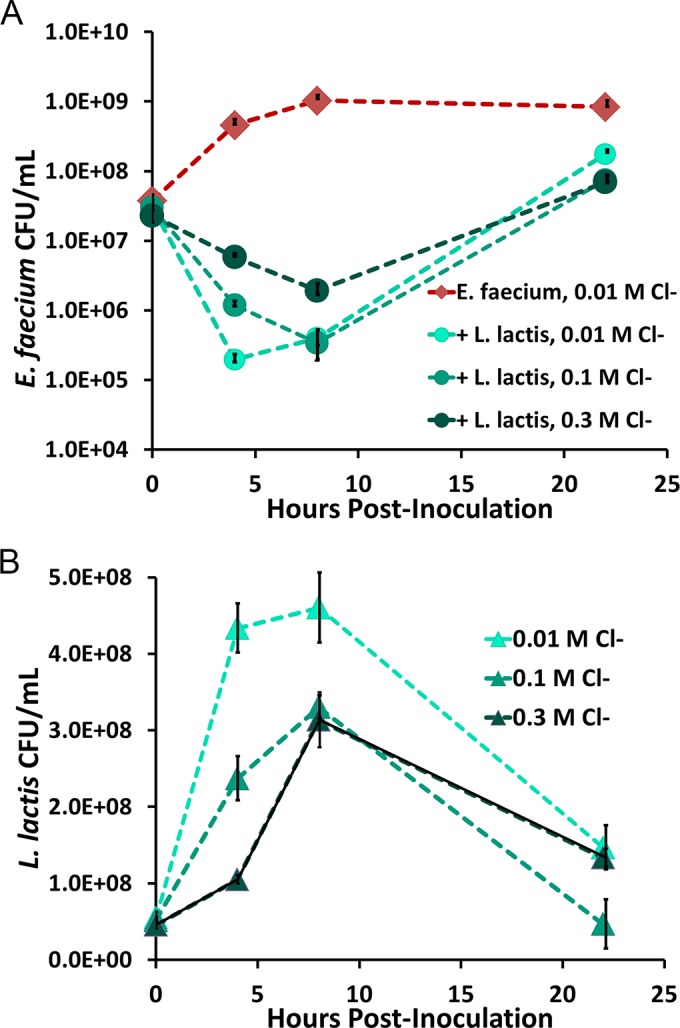
(A) Growth of E. faecium alone or with L. lactis producing enterocin A, enterocin P, and hiracin JM79 under the chloride-inducible promoter at 0.01 M, 0.1 M, and 0.3 M chloride. Data points represent the averages of three technical replicates. Error bars represent ±1 standard deviation calculated from the sample triplicates. (B) L. lactis/pNZCA3 counts from the coculture test shown in panel A. Note the reduced growth at higher salt concentrations. Data points represent the averages of three technical replicates. Error bars represent ±1 standard deviation calculated from the sample triplicates.
It is evident from the results shown in Fig. 5A that even under nonoptimal growth conditions, L. lactis is able to inhibit E. faecium immediately upon treatment. It is clear, however, that within 10 h the pathogen begins to regrow. To test whether this was truly due to the appearance of E. faecium with stable resistance to the peptides or due simply to decreased AMP concentrations over time, E. faecium bacteria that arose from the culture treated with AMPs were regrown in fresh GM17 medium for ∼10 generations. Growth of the original culture and of the regrown resistant E. faecium was then monitored in GM17 medium with or without 10% L. lactis supernatant containing AMPs to determine if resistance was still present. Figure 6 shows the growth curves of both the original E. faecium culture and the supposed resistant culture with and without AMPs. Even after growth for 10 generations in the absence of AMPs, the resistant culture was only mildly impacted by the AMPs. These results indicate that the surviving E. faecium bacteria from the coculture experiments are, in fact, stably resistant to the AMPs for at least 10 generations. The resistance observed in these experiments was somewhat expected as the development of resistance to class IIa bacteriocins has been previously reported and studied in multiple bacterial species (20). The proposed mechanisms of this resistance are further discussed below.
FIG 6.
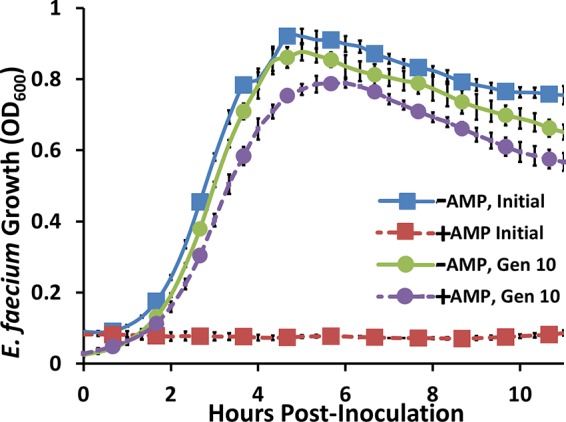
Stability of E. faecium resistance to AMPs after 10 generations of regrowth. The blue curve represents wild-type E. faecium growth in the absence of AMPs, and the red curve represents wild-type E. faecium growth in the presence of AMPs. After 15 h, E. faecium bacteria grown in the presence of AMPs (red) were inoculated into GM17 medium with no AMPs and grown for 10 generations (Gen 10). The green curve shows the growth of the resulting E. faecium grown in the absence of AMPs, and the purple curve shows the growth of this same E. faecium grown in the presence of AMPs. Results indicate that resistance is maintained for at least 10 generations. Error bars represent ±1 standard deviation of biological triplicates.
Resistance prevention using combined treatment with class IIa bacteriocins and rifampin.
Though L. lactis producing enterocin A, enterocin P, and hiracin JM79 under the chloride-inducible promoter offers promise in temporarily decreasing E. faecium populations in coculture, the rise in resistant mutants was concerning. To further demonstrate the potential of the AMP delivery system, we aimed to identify a means of combating this resistance. It is well-known that the combination of multiple antibiotics can help to postpone the development of antibiotic resistance (21). Additionally, it has been observed in some cases that antibiotics as well as AMPs can act synergistically against the target pathogen (22, 23). One can thus imagine the potential in either producing additional AMPs from our currently engineered L. lactis or combining bacterial AMP delivery with traditional antibiotic treatments.
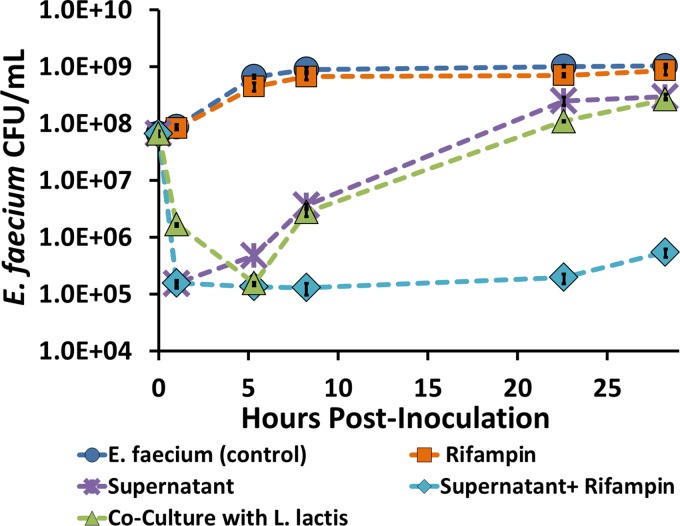
In an attempt to eliminate regrowth of E. faecium, several common antibiotics, including streptomycin, ampicillin, and rifampin, were tested in combination with the three AMPs used in this study. Alone, none of these drugs showed significant activity against vancomycin-resistant E. faecium at clinically relevant concentrations (19) (Fig. 7; also data not shown). It was found, however, that treating E. faecium with a combination of rifampin and the supernatant containing the three AMPs both decreased E. faecium counts by nearly four orders of magnitude and prevented regrowth of the pathogen for over 24 h (Fig. 7). Data in Fig. 7 are technical triplicates obtained from one of three individual tests. Similar decreases in E. faecium numbers were observed in all three tests and were consistent with values from several other colony-counting experiments performed with this pathogen. The effects of rifampin and of rifampin with AMPs on E. faecium are also representative of all three trials. Synergy of the AMPs with streptomycin and ampicillin was also tested and was found to be minor with these antibiotics (data not shown).
FIG 7.
E. faecium inhibition by addition of the following treatments: 30 μg/ml rifampin; supernatant from L. lactis producing enterocin A, enterocin P, and hiracin JM79 under the CIP (L. lactis containing pNZCA3); supernatant and rifampin; or culture containing L. lactis/pNZCA3 induced with 0.15 M chloride. Data points represent the averages of three technical replicates. Error bars represent ±1 standard deviation calculated from the sample triplicates.
Because the L. lactis bacteria used in these studies were not rifampin resistant, supernatant rather than coculture was used with the antibiotic. A comparison between inhibition by 10% cell-free supernatant and that by 10% L. lactis/pNZCA3 demonstrates that similar stable inhibition might be obtained if rifampin-resistant L. lactis/pNZCA3 rather than supernatant was used in combination with the antibiotic. In practice, these rifampin-resistant L. lactis bacteria would be used in conjunction with the antibiotic.
DISCUSSION
We characterized and implemented a chloride-inducible expression system for the bacterial delivery of antimicrobial peptides. We demonstrated the efficacy of a previously discovered chloride-inducible promoter as a potentially useful expression system for the delivery of antimicrobial peptides by L. lactis. In this paper we have focused on developing a system to eliminate antibiotic-resistant E. faecium; however, this type of expression system can easily be expanded with different AMPs to target a wide variety of pathogens.
The first section of this study focuses on characterizing protein expression under the chloride-inducible promoter using reporter gene assays. By comparing β-Gal production levels of the chloride-inducible promoter and the commonly used nisin-inducible promoter, we found the chloride-inducible promoter to be a powerful expression system which is strongly activated at typical chloride concentrations found inside the human GI tract. These results were promising because they indicated that this promoter could be used to express AMPs (or other proteins) in the GI tract without additional induction. The reporter studies also showed that significant promoter activity was observed in the lowest-attained induction state (0.01 M Cl−). The high expression level observed in GM17 medium could be somewhat problematic in the production of more toxic proteins, but this issue could likely be diminished by further optimizing the medium to reduce the chloride concentration.
In the future it may be of interest to further explore the responsiveness of the chloride-inducible promoter to other environmental signals. For example, there is evidence that the promoter activity is also impacted by the glutamate concentration and pH of the culture (15). Glutamate availability, pH, or other environmental conditions could thus play an important role in the delivery of AMPs in the body and should be examined in the future. These variables can also provide additional parameters for improving promoter control for manufacturing and growth purposes.
The second portion of this study was to implement the chloride-inducible promoter for the production of AMPs against VRE. In a recent study from our lab, the E. faecalis-responsive PrgX-PrgQ promoter was used to express enterocin A, enterocin P, and hiracin JM79 to eliminate E. faecalis. While the PrgX-PrgQ system shows promise in targeting E. faecalis, it is not ideal for the treatment of E. faecium or other pathogens lacking the inducer pheromone cCF10 as these bacteria would not induce AMP production from the PrgX-PrgQ system. We thus sought an environmentally inducible system that could be used for general AMP production against any pathogen.
To test the utility of this new expression system, the chloride-inducible promoter was used to express the same three AMPs (enterocin A, enterocin P, and hiracin JM79) for the elimination of our indicator pathogen, E. faecium 8-E9, as well as a variety of other enterococcal strains at chloride concentrations obtainable in the intestines. It was shown that all 11 enterococcal strains tested were significantly inhibited by the lactococcal AMP delivery system. The wide-spectrum activity that can be obtained with this system demonstrates the potential benefit in using the chloride-inducible promoter for AMP delivery. Though this type of system will likely be effective in eliminating pathogenic enterococci, it was also observed that the two commensal E. faecalis strains were also inhibited by our AMPs. In the future, selection of AMPs with minimal impacts on the native gut microbiota will be especially important when the chloride-inducible promoter is used since promoter activity will not be localized to the pathogen.
In liquid coculture tests, the system proved to be extremely effective in reducing E. faecium counts. Interestingly, it was seen that increasing the salt concentration did not result in increased killing of the pathogen. Based on the colony counts of the L. lactis bacteria from the cultures with 0.01 M, 0.1 M, and 0.3 M chloride concentrations, the ability to kill the pathogen was likely impacted by reduced growth (and productivity) of L. lactis early in the coculture. These results contrasted with those observed in the agar diffusion tests, which showed increased halo diameters at increased salt concentrations.
It should be noted that the impact on the growth of L. lactis cultures of chloride concentrations of 0.01 M and 0.1 M was far more pronounced in coculture than in L. lactis cultures grown alone. These differences may be due to the competition for nutrients between L. lactis and E. faecium. We recognize that the nutrient availability and environmental stresses found within the GI tract are significantly different from those found in vitro and that only in vivo tests can tell the true utility of this system. Based on the studies performed thus far, however, high AMP production under the chloride-inducible promoter appears robust to growth and induction conditions, which is invaluable for the proposed application.
The liquid coculture tests also revealed that while the AMPs produced under the chloride-inducible expression system were initially very effective against E. faecium, resistance began to overtake the culture within 10 h. Resistance was verified by monitoring the growth of surviving bacteria in the presence and absence of AMPs. These results were not surprising as resistance to bacteriocins by L. lactis, E. faecalis, and Listeria monocytogenes has been previously reported (20). Several mechanisms of resistance for class IIa bacteriocins have been hypothesized and explored. Most of these proposed mechanisms involve the mannose-phosphotransferase system (Man-PTS), which is believed to be the receptor of these class IIa bacteriocins. The Man-PTS is a major sugar uptake system found in many bacteria. Class IIa bacteriocins are thought to change the conformation of the Man-PTS in such a way that allows a free flow of ions across the membrane, which ultimately leads to cell death (20). Some of the major proposed resistance mechanisms include downregulation of the Man-PTS, alterations to membrane composition and charge, and random mutations in the Man-PTS locus (20, 24, 25). These mechanisms have been found to differ among species as well as among mutants of the same species found to have various levels of resistance (24).
Due to the rapid development of resistance to all three class IIa bacteriocins and the high fraction of resistant mutants in the unexposed culture (∼1 mutant/50,000 bacteria), it is tempting to propose that the primary mode of resistance observed in this study relies on the downregulation of the Man-PTS. As previously discussed by Kjos and coworkers, it is possible that this downregulation could be the result of randomness in E. faecium's metabolic gene regulation, a survival tactic referred to as metabolic variability (20). This hypothesis is further supported by a transcriptome analysis of pediocin-resistant E. faecalis mutants that found that mutants had altered transcription of approximately 200 genes, most of which related to metabolism and transport (24). It is possible that E. faecium bacteria have a subpopulation that has switched their metabolisms to paths not requiring the Man-PTS, relying on alternative carbon sources. At this point, this is only speculation, and further, more extensive studies will be needed to determine the true cause of resistance.
As an extension of the delivery system developed in this study, we explored the combination of bacterial AMP delivery with traditional antibiotic therapies to help improve our current system by reducing the rise of resistant mutants (21). This type of combination therapy is commonly used to avoid antibiotic resistance when traditional antibiotics are used. Additionally, this type of combination therapy is conceptually similar to our future goal of adding alternative AMPs with orthogonal targets to those of the bacteriocins currently in our system. These combination studies successfully showed that the application of 30 μg/ml of rifampin held off resistant mutants for over 24 h when it was combined with the three AMPs. These results are interesting because VRE are often considered inherently resistant to rifampin. It is possible that the AMPs help permeabilize the cell membrane as previous studies have found that cell membrane permeability likely plays a major role in bacterial susceptibility to rifampin (26). The reduction of bacterial resistance is essential and must be carefully considered for both traditional and new antibiotic technologies.
Concluding remarks.
With this study, we have identified and implemented a chloride-inducible promoter for the production of AMPs. This expression system shows promise for the production of a broad range of AMPs in GI tract environments without the need for added induction molecules. As an example of the application of the chloride-inducible promoter, we showed that the expression of AMPs under the new expression system drastically decreases counts of E. faecium bacteria. Furthermore, we showed that by combining the antibiotic rifampin with three AMPs produced from this system, the inhibition of E. faecium is longer lasting, with limited regrowth of resistant mutants. This study gives promise that the chloride-inducible promoter can be used as a general expression system for the delivery of a wide array of AMPs targeting different pathogens.
In the future, we will explore the mechanisms of resistance to the AMPs enterocin A, enterocin P, and hiracin JM79. Additionally we will test the survivability of the engineered L. lactis bacteria under more physiologically relevant conditions to further evaluate their effectiveness in this application. The need for new antimicrobial therapies is becoming increasingly urgent, and the in vivo production of AMPs may offer a new tool against even the most resistant pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gary Dunny and Patricia Ferrieri of the University of Minnesota Medical School for supplying isolates of E. faecium and E. faecalis with varied spectra of antibiotic resistance for these studies.
This work was supported by grants from the National Institutes of Health (GM111358) and by a grant from the National Science Foundation (CBET-1412283). Support from the University of Minnesota Biotechnology Institute is gratefully acknowledged. K.G. was also supported by a National Science Foundation Graduate Research Fellowship (00039202).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00227-15.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf. [Google Scholar]
- 2.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neely AN, Maley MP. 2000. Survival of enterococci and staphylococci on hospital fabrics and plastic. J Clin Microbiol 38:724–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias CA, Mendes RE, Stilwell MG, Jones RN, Murray BE. 2012. Unmet needs and prospects for oritavancin in the management of vancomycin-resistant enterococcal infections. Clin Infect Dis 54(Suppl 3):S233–S238. doi: 10.1093/cid/cir924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marothi YA, Agnihotri H, Dubey D. 2005. Enterococcal resistance—an overview. Indian J Med Microbiol 23:214–219. [PubMed] [Google Scholar]
- 6.Borrero J, Chen Y, Dunny GM, Kaznessis YN. 2015. Modified lactic acid bacteria detect and inhibit multiresistant enterococci. ACS Synth Biol 4:299–306. doi: 10.1021/sb500090b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kjos M, Borrero J, Opsata M, Birri DJ, Holo H, Cintas LM, Snipen L, Hernández PE, Nes IF, Diep DB. 2011. Target recognition, resistance, immunity and genome mining of class II bacteriocins from Gram-positive bacteria. Microbiology 157:3256–3267. doi: 10.1099/mic.0.052571-0. [DOI] [PubMed] [Google Scholar]
- 8.Park S-C, Park Y, Hahm K-S. 2011. The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. Int J Mol Sci 12:5971–5992. doi: 10.3390/ijms12095971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klijn N, Weerkamp AH, de Vos WM. 1995. Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl Environ Microbiol 61:2771–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drouault S, Juste C, Marteau P, Renault P, Corthier G. 2002. Oral treatment with Lactococcus lactis expressing Staphylococcus hyicus lipase enhances lipid digestion in pigs with induced pancreatic insufficiency. Appl Environ Microbiol 68:3166–3168. doi: 10.1128/AEM.68.6.3166-3168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braat H, Rottiers P, Hommes DW, Huyghebaert N, Remaut E, Remon J-P, van Deventer SJH, Neirynck S, Peppelenbosch MP, Steidler L. 2006. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn's disease. Clin Gastroenterol Hepatol 4:754–759. doi: 10.1016/j.cgh.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 12.Sanders JW, Venema G, Kok J. 1997. A chloride-inducible gene expression cassette and its use in induced lysis of Lactococcus lactis. Appl Environ Microbiol 63:4877–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fordtran JS, Locklear TW. 1966. Ionic constituents and osmolality of gastric and small-intestinal fluids after eating. Am J Dig Dis 11:503–521. doi: 10.1007/BF02233563. [DOI] [PubMed] [Google Scholar]
- 14.Kuipers OP, de Ruyter PGG, Kleerebezem M, de Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol 64:15–21. doi: 10.1016/S0168-1656(98)00100-X. [DOI] [Google Scholar]
- 15.Sanders JW, Leenhouts K, Burghoorn J, Brands JR, Venema G, Kok J. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol 27:299–310. doi: 10.1046/j.1365-2958.1998.00676.x. [DOI] [PubMed] [Google Scholar]
- 16.Holo H, Nes IF. 1995. Transformation of Lactococcus by electroporation. Methods Mol Biol 47:195–199. [DOI] [PubMed] [Google Scholar]
- 17.Volzing K, Borrero J, Sadowsky MJ, Kaznessis YN. 2013. Antimicrobial peptides targeting Gram-negative pathogens, produced and delivered by lactic acid bacteria. ACS Synth Biol 2:643–650. doi: 10.1021/sb4000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozlowicz BK, Bae T, Dunny GM. 2004. Enterococcus faecalis pheromone-responsive protein PrgX: genetic separation of positive autoregulatory functions from those involved in negative regulation of conjugative plasmid transfer. Mol Microbiol 54:520–532. doi: 10.1111/j.1365-2958.2004.04286.x. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert D, Moellering R, Eliopoulos G, Chambers HF, Saag MS (ed). 2013. The Sanford guide to antimicrobial therapy 2013, 43rd ed. Antimicrobial Therapy, Inc, Sperryville, VA. [Google Scholar]
- 20.Kjos M, Nes IF, Diep DB. 2011. Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Appl Environ Microbiol 77:3335–3342. doi: 10.1128/AEM.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis K. 2013. Platforms for antibiotic discovery. Nat Rev Drug Discov 12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 22.Rand KH, Houck H. 2004. Daptomycin synergy with rifampicin and ampicillin against vancomycin-resistant enterococci. J Antimicrob Chemother 53:530–532. doi: 10.1093/jac/dkh104. [DOI] [PubMed] [Google Scholar]
- 23.Vaara M, Porro M. 1996. Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob Agents Chemother 40:1801–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Opsata M, Nes IF, Holo H. 2010. Class IIa bacteriocin resistance in Enterococcus faecalis V583: the mannose PTS operon mediates global transcriptional responses. BMC Microbiol 10:224. doi: 10.1186/1471-2180-10-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vadyvaloo V, Hastings JW, van der Merwe MJ, Rautenbach M. 2002. Membranes of class IIa bacteriocin-resistant Listeria monocytogenes cells contain increased levels of desaturated and short-acyl-chain phosphatidylglycerols. Appl Environ Microbiol 68:5223–5230. doi: 10.1128/AEM.68.11.5223-5230.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abadi FJ, Carter PE, Cash P, Pennington TH. 1996. Rifampin resistance in Neisseria meningitidis due to alterations in membrane permeability. Antimicrob Agents Chemother 40:646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.