Abstract
Lactococcus piscium is a psychrotrophic lactic acid bacterium and is known to be one of the predominant species within spoilage microbial communities in cold-stored packaged foods, particularly in meat products. Its presence in such products has been associated with the formation of buttery and sour off-odors. Nevertheless, the spoilage potential of L. piscium varies dramatically depending on the strain and growth conditions. Additional knowledge about the genome is required to explain such variation, understand its phylogeny, and study gene functions. Here, we present the complete and annotated genomic sequence of L. piscium MKFS47, combined with a time course analysis of the glucose catabolism-based transcriptome. In addition, a comparative analysis of gene contents was done for L. piscium MKFS47 and 29 other lactococci, revealing three distinct clades within the genus. The genome of L. piscium MKFS47 consists of one chromosome, carrying 2,289 genes, and two plasmids. A wide range of carbohydrates was predicted to be fermented, and growth on glycerol was observed. Both carbohydrate and glycerol catabolic pathways were significantly upregulated in the course of time as a result of glucose exhaustion. At the same time, differential expression of the pyruvate utilization pathways, implicated in the formation of spoilage substances, switched the metabolism toward a heterofermentative mode. In agreement with data from previous inoculation studies, L. piscium MKFS47 was identified as an efficient producer of buttery-odor compounds under aerobic conditions. Finally, genes and pathways that may contribute to increased survival in meat environments were considered.
INTRODUCTION
Bacterial spoilage of perishable cold-stored foods is still a serious problem in the modern world. Modified-atmosphere packaging (MAP), based on the inhibitory effect of CO2 on spoilage bacteria, is often used by the food industry to achieve a longer product shelf life. While it eliminates aerobic Gram-negative spoilage bacteria, MAP creates a positive selective pressure for lactic acid bacteria (LAB), which are resistant to high CO2 concentrations (1, 2). Even though spoilage LAB grow slower than aerobic Gram-negative spoilage bacteria, they cause organoleptic and textural spoilage changes, such as discoloration and the formation of slime, gas, biogenic amines, and volatile organic compounds (1), in MAP food products.
The most abundant genera in MAP food spoilage communities are Carnobacterium, Lactobacillus, Lactococcus, Leuconostoc, and Weissella (3). So far, Lactococcus piscium has been the only Lactococcus species with reported food spoilage activity (4, 5). It was first isolated from a diseased salmonid fish (6) but was later detected in MAP meat products, including beef, poultry, pork, and fish, where it frequently belongs to the predominant microbiota at the end of shelf life. L. piscium has been associated with meat spoilage (3, 5, 7–9), and it was recently also shown to be one of the predominating species in chilled packaged vegetable salads (10).
The spoilage potential of L. piscium has been studied by inoculating pork samples (5), raw salmon (11), and bell pepper simulation medium (12). In pork, L. piscium was shown to shorten the sensory shelf life by the formation of buttery and sour off-odors. In the case of raw salmon, L. piscium was described as a light spoiler, and it was associated with buttery off-odors. In bell pepper simulation medium, one L. piscium strain demonstrated a high growth rate and very offensive metabolism, which included the formation of ethanol, acetate, and diacetyl (in the presence of oxygen). That same study also showed a significant variation of spoilage potentials among L. piscium strains, which is in concurrence with the fact that one L. piscium strain, CNCM I-4031, is even considered to be a promising bioprotective organism in seafood products (13, 14).
To date, genome sequences of L. piscium have not been publicly available. In this paper, we present the complete genome sequence of spoilage-related L. piscium strain MKFS47, isolated from MAP broiler filet strips showing early spoilage changes at the end of shelf life (15). The gene content of this species was compared to those of 29 sequenced and annotated Lactococcus species, including L. lactis, L. garvieae, L. raffinolactis, and L. chungangensis. In addition, the time course whole transcriptome, based on glucose metabolism, was generated by capturing the transcriptome at different time points (to determine the changes in the transcriptome over the time period) and analyzed to determine the dynamic regulation of gene expression. Glucose was selected for transcriptome analysis since it is the most abundant and ubiquitous carbohydrate in food products, including meat (16). The main focus of the data analyses was on the genes and pathways involved in carbohydrate metabolism related to food spoilage and especially the production of acetoin and diacetyl.
MATERIALS AND METHODS
Genome sequencing and annotation.
DNA isolation from L. piscium MKFS47 (hereafter called simply L. piscium) grown in de Man-Rogosa-Sharpe (MRS) broth was done by using a method (17) modified from the one described previously by Pitcher et al. (18), and the genomic DNA was mechanically sheared with a needle. The genome sequence was determined by the use of a 454 sequencer with GS Flx chemistry and the Illumina HiScanSQ system. A total of 309,686,454 reads corresponding to 67.4 Mbp were assembled by using Newbler 2.0.00.20 (Roche 454 Life Science), resulting in an average of 27× contig coverage. Genome closure was done by using the Gap4 program in the Staden package (19). The order of contigs was determined by PCR, and Sanger sequencing was used to fill the gaps. Finally, homopolymer errors were corrected by mapping HiScanSQ reads with a length of 27 bp with 1,400× coverage to the genome sequence.
Coding sequences (CDSs) were determined by using the EasyGene (20) and Prodigal (21) programs, followed by a comparison of their outputs and manual resolution of their discrepancies based on the presence of potential ribosomal binding sites, similarity searches, multiple-sequence alignments, and data reported previously. Gene functions were assigned based on a comparison of the outputs of two annotation tools: RAST (22) and PANNZER (23). Predicted functions were manually reviewed by using similarity searches against public databases and bibliomic data. The BAGEL2 (24), CW-PRED (25), and PHAST (26) programs were utilized to determine bacteriocins, LPxTG motif-containing proteins, and prophage regions, respectively. Finally, all genes were classified into functional categories by an RPS-BLAST search against the COG database and checked for the presence of all core functions (27). More details about the bioinformatic genome analysis can be found in the supplemental material.
Ortholog prediction for 30 genomes of the Lactococcus genus.
A comparative analysis of gene contents was performed for 29 lactococci (with sequenced and annotated genomes available by September 2014 [see Table S1 in the supplemental material]) and L. piscium. These lactococci included 5 strains of L. garvieae, 11 strains of L. lactis subsp. lactis, 11 strains of L. lactis subsp. cremoris, and 1 strain each of L. piscium, L. raffinolactis, and L. chungangensis. Their proteomes, except for that of L. piscium, were retrieved from the NCBI and HAMAP public databases. Identification of the orthologous groups of proteins was done by means of OrthoMCL (28). The information about the orthologous groups was used to generate the pangenome matrix of the presence and absence of orthologs in lactococcus genomes (see Data Set S1 in the supplemental material). Based on the pangenome matrix, a pangenome tree was constructed by using the PARS program from the PHYLYP package (29) and visualized by using the iTOL tool (30). Clade-specific genes were determined to be genes that are present in 90% of the genomes in the clade and absent in 90% of the other genomes (see Data Set S2 in the supplemental material). This criterion allows accounting for the missing gene annotations that often occur in draft genomes.
Phenotypic and morphology analyses for functional genomics.
Carbohydrate utilization profiles of 22 L. piscium strains, including that of strain MKFS47, were tested by the API 50CH and API 20 Strep identification systems (bioMérieux, Marcy l'Etoile, France), according to the manufacturer's instructions.
To assess glycerol utilization, L. piscium was precultured in M17 broth with 1% glycerol or 1% glucose aerobically and anaerobically (Anaerogen; Oxoid, United Kingdom) for 24 h, followed by its reinoculation into the same fresh medium as the one used for preculturing. The new cultures were grown for the next 24 h under the same conditions as those used for preculturing, and the final sample optical density at 600 nm (OD600) values (Eppendorf Biophotometer) were measured. The growth of L. piscium in M17 broth without a carbon source was used as the control.
To evaluate the capacities of L. piscium and Leuconostoc gelidum subsp. gasicomitatum LMG 18811T to produce acetoin and/or diacetyl (henceforth “acetoin/diacetyl”), each bacterium was grown aerobically (250 rpm) for 48 h in modified MRS medium without acetate and with 2% glucose (Sigma-Aldrich, USA). Glucose was selected since it is the most abundant carbohydrate in food matrices. Leuc. gelidum subsp. gasicomitatum was used for comparison, because it is a well-known acetoin/diacetyl producer in meat and vegetables packaged under oxygen-containing atmospheres (12, 31). Detection of the total amount of diacetyl and acetoin was performed according to the colorimetric Voges-Proskauer assay (32). The method was modified to enable measurement of the combined amount of diacetyl and acetoin. In order to achieve this, reference curves with known amounts of diacetyl and separately with known amounts of acetoin in the range of 10 to 1,100 μM were constructed. The average absorbance for diacetyl and acetoin at 560 nm (A560) was calculated, and the combined reference curve exhibited linear relationships between the A560 value and the concentration, where 1 A560 unit was equal to 410 μM acetoin/diacetyl (ratio of ∼1:1). The detection limit of the assay was ∼10 μM. Prior to A560 measurements, samples were diluted, and the values obtained were subsequently normalized. In addition, acetoin/diacetyl production from glycerol by L. piscium was evaluated as described above. For this, L. piscium was grown in MRS medium without acetate and with 2% glycerol aerobically for 48 h.
To test the ability of L. piscium to ferment citrate, a plate screening assay was performed (33). Hydrogen peroxide production was tested as described previously (34), except that M17 broth with 1% glycerol or 1% glucose was used instead of brain heart infusion (BHI) broth. Plates for both assays were incubated for 48 h anaerobically and aerobically.
The twitching motility of L. piscium was tested under different conditions on 1% agar on microscope slides and examined by phase microscopy. The conditions included M17 medium with or without 0.5% glucose or a semidefined medium with or without 0.1% lactose or 0.1% glucose in both aerobic and anaerobic atmospheres for 3 days. For transmission electron microscopy (TEM), 20-μl droplets of a culture of L. piscium grown overnight were inoculated onto M17 plates with 0.5% glucose and 1% agar. Cells from the outermost edge of the inoculum were suspended in a saline solution, stained with 3% uranylacetate, and examined by TEM.
All the tests were done at 25°C in at least three replicates. The average values were calculated across the replicates.
RNA extraction, sequencing, and downstream analysis.
L. piscium was grown in modified MRS broth without acetate at 25°C under microaerobic conditions (50 rpm), with 2% glucose. At the beginning of the experiment, the exponential-phase cell density was ∼106 CFU ml−1. Cells were passed through the medium for at least 10 generations. During 11 h of growth, the OD600 value, the pH (Inolab 720 pH meter; WTW, Weilheim, Germany) of the growth medium, and cell counts (MRS agar without acetate, with incubation for 5 days at 25°C anaerobically) were measured once an hour. Samples were taken at three time points, 3 h, 5 h, and 11 h, in three replicates.
For RNA extraction, samples were treated with a cold 10:1 mixture of ethanol-phenol (Sigma-Aldrich, USA) to inhibit cell metabolism and RNase activity and then centrifuged (5,000 × g for 3 min at 4°C), and the pellets were immediately frozen and stored at −72°C. The cells were disrupted by the use of a mixer mill (FastPrep-24; MP Biomedicals, USA) run at a frequency of 6 Hz twice, for 45 s each time. Finally, RNA was extracted by the use of the RNeasy plant minikit (Qiagen, Germany) with DNase treatment according to the manufacturer's instructions.
Prior to the construction of the RNA sequencing (RNA-seq) libraries, rRNAs were omitted from the total RNA (3 to 5 μg) by using magnetic beads (Ribo-Zero magnetic kit [bacteria], catalog no. MRZB12424; Epicentre). RNA-seq libraries were produced using the Ovation RNA-seq system V2 (part no. 7102; NuGEN). The resulting double-stranded DNA (dsDNA) was sheared by sonication (Bioruptor sonication system; Diagenode) (19 cycles of 30 s on/30 s off). The obtained fragments were polished with T4 DNA polymerase, followed by the ligation of adapters for SOLiD sequencing. Libraries were size selected and thereafter sequenced in five lanes by using SOLiD 5500XL (Life Technologies) to produce 75-bp single-end reads.
Lifescope software (Life Technologies) was used for mapping and counting the RNA-seq reads, including the generation of the normalized read counts in RPKM (reads per kilobase of gene per million mapped reads) (see Data Set S3 in the supplemental material). During the analysis, >90% of the reads in each sample were mapped to the L. piscium MKFS47 genome. In turn, approximately half (45 to 51%) of the mapped reads in each sample belonged to the protein-coding genes, which were used for further analysis. The numbers of CDS mapped reads were in the ranges of 4.1 million to 7.2 million for 3-h replicates, 5.7 million to 7.3 million for 5-h replicates, and 5.9 million to 8.1 million for 11-h replicates. The technical (lanes) and biological replicates were clustered based on their RPKM values by using an R function “heat map.” As a result, all the lanes for the same biological replicate were clustered together, indicating low technical variation in comparison with biological variation. Likewise, biological replicates that belong to the same time point clustered together. This suggests that biological replicates show higher similarity than do samples from different time points. Analysis of differential expression between different time points (5 h over 3 h and 11 h over 5 h) was performed by using two R packages, EBSeq (35) and edgeR (36), which use different inferential approaches. Default parameters were used. The genes were considered to be differentially expressed (DE) if their false discovery rate (FDR) was <0.05 (in the case of edgeR) or if the posterior probability (PP) was >0.95 (in the case of EBSeq) and if the absolute fold change (FC), obtained from the EBSeq output, was >1.5. Absolute fold changes, used to describe DE genes in Results, were calculated based on the ratios between RPKM values (averaged for each time point). Genes that were found to be significantly differentially expressed by at least one program were taken for further analysis.
Accession numbers.
Annotated genomic nucleotide sequences (chromosome, plasmid 1, and plasmid 2) are accessible through the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under accession numbers LN774769, LN774770, and LN774771, respectively. Raw RNA-seq read sequences and raw read counts per gene are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-3245.
RESULTS
General features of the genome and time course transcriptome.
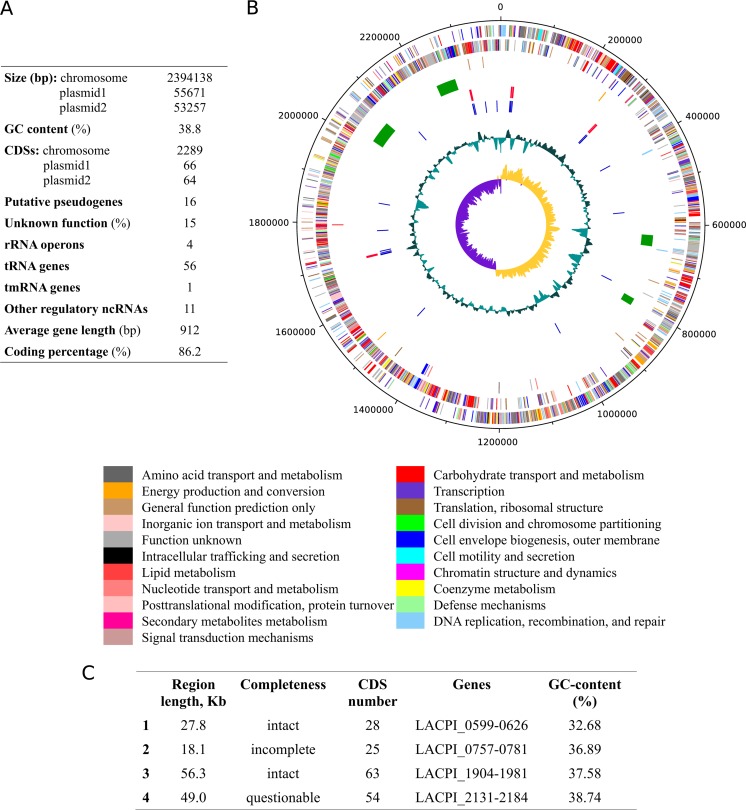
The genome of L. piscium contains one circular chromosome of a moderate size, 2,394,138 bp, and two plasmids (Fig. 1A). The numbers of predicted protein-coding genes are 2,289 on the chromosome and 130 on the plasmids. The chromosomal genes are evenly distributed along both strands, without any major distortions of density (Fig. 1B). The chromosome harbors at least 4 prophage regions (Fig. 1C), constituting ∼6% of the L. piscium genome. In addition to the genes originating from phages, each region contains bacterial genes with a proportion ranging from 21% to 36% for each of the prophage regions. The genome was predicted to have six complete DNA restriction-modification (RM) systems: four on the chromosome (type I, type III, and two type II) and two on the plasmids (both type I). It is noteworthy that the type III DNA methylase gene (LACPI_0416) is a putative pseudogene due to the frameshift, and one of the type II restriction enzymes is disrupted by the transposase gene (LACPI_1775) into two parts: LACPI_1774 and LACPI_1776.
FIG 1.
(A) Main L. piscium genome features. tmRNA, transfer-messenger RNA; ncRNAs, noncoding RNAs. (B) Genome map of L. piscium. Genes are colored according to their COG functional annotations. Moving inwards, the tracks represent the following features: genes on the forward strand, genes on the reverse strand, pseudogenes, prophages (green), rRNA genes (red), tRNA genes (blue), GC plot (cyan), and GC skew (purple and yellow). (C) Prophage regions detected by using PHAST (26).
In the time course gene expression study, pure cultures of L. piscium reached the stationary phase after ∼7 h of growth, with average cell counts of 4.6 × 108 to 7.7 × 108 CFU/ml, and the pH of the growth medium decreased from 5.93 to 4.42. According to the mapped RNA-seq data, plasmid 1 is not expressed and was probably lost during cultivation, despite the fact that it contains a toxin-antitoxin system. In turn, genes of plasmid 2 were highly expressed. The total numbers of DE genes, found for the two time intervals, reflect the lengths of the intervals (Table 1): 285 genes in the first, short period (3 to 5 h) compared to 1,359 genes in the second, longer period (5 to 11 h). More than half of the L. piscium genes (56%) changed their expression levels during the second time period, and the proportions of upregulated and downregulated genes were almost equal in both intervals. DE genes were also divided into six groups according to their expression patterns (Table 2). Genes that showed equivalent expression levels in the first period and upregulation (or downregulation) in the second period were considered generally upregulated (or downregulated) during the whole time due to the shortness of the first time interval (3 to 5 h) and the difficulty in detecting DE genes with low FC values. The numbers of the genes in group 1 (mostly upregulated) and group 2 (mostly downregulated) are relatively the same, and they constitute the majority of the DE genes (>90%). As expected, the other four expression patterns (groups) contain small numbers of genes, which, however, could play important roles in the regulation of cellular processes during bacterial growth.
TABLE 1.
Numbers of DE genes in two sequential time intervals
| Time interval (h) | No. (%) of genes |
||
|---|---|---|---|
| Upregulated | Downregulated | Total DE | |
| 3–5 | 136 (48) | 149 (52) | 285 |
| 5–11 | 707 (52) | 652 (48) | 1,359 |
TABLE 2.
Groups of DE genes with different expression patterns
| Group | Direction of change in expression levela |
No. of genes | Gene(s) and/or pathway(s) | |
|---|---|---|---|---|
| 3–5 h | 5–11 h | |||
| 1 | → or ↑ | ↑ | 677 | General stress and oxidative stress protection; catabolism of carbohydrates (other than glucose), glycerol, and agmatine |
| 2 | → or ↓ | ↓ | 621 | Biosynthesis of the cell wall and nucleotides; branched-chain-amino-acid biosynthesis and uptake; oligopeptide uptake |
| 3 | ↑ | → | 37 | Acetolactate synthase alsS |
| 4 | ↓ | → | 41 | Sucrose phosphorylase (LACPI_1289) |
| 5 | ↑ | ↓ | 54 | Ribulose-phosphate 3-epimerase; acetolactate decarboxylase; translation initiation factor IF-2 |
| 6 | ↓ | ↑ | 39 | Glutamate-pyruvate aminotransferase; 1-phosphofructokinase; malolactic enzyme and malate/Na+ symporter |
↑ indicates upregulation, ↓ indicates downregulation, and → indicates equivalent expression.
Comparative genomics.
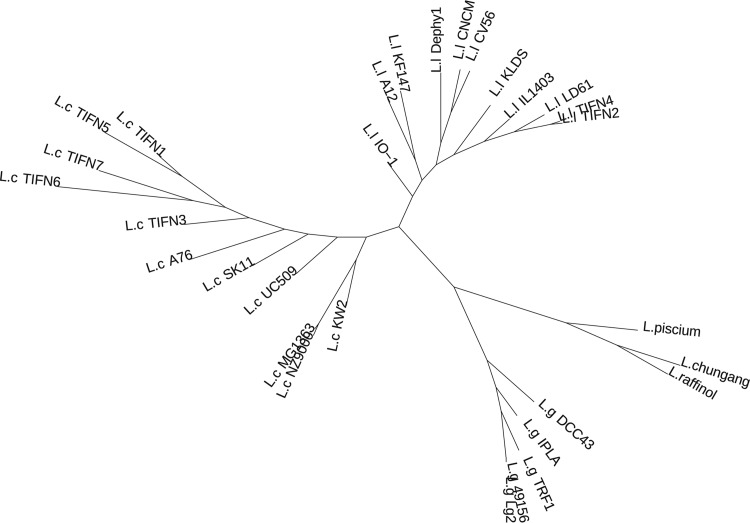
The number of orthologous groups of genes determined for 30 lactococcus genomes was 4,938 (excluding the groups containing only paralogs from the same genome). Among them, 337 groups (11 to 19% of lactococcus genomes) are present in all analyzed genomes and represent the core genes. Sixteen groups out of the core genes have completely unknown functions (see Table S2 in the supplemental material). By using information about the presence or absence of orthologs in the genomes, an unrooted pangenome tree of the Lactococcus genus was constructed (Fig. 2). It has three major clades: the first (“L. garvieae” clade) includes species of animal and environmental origins, including L. piscium, L. raffinolactis, L. chungangensis, and L. garvieae strains, and the second and third clades consist of all L. lactis subsp. lactis and L. lactis subsp. cremoris genomes, respectively. L. piscium, L. raffinolactis, and L. chungangensis form a separate branch within the clade, as opposed to L. garvieae species.
FIG 2.
Pangenome tree of the Lactococcus genus, constructed based on information on the presence or absence of orthologs. Abbreviations: L.g, L. garvieae; L.l, L. lactis subsp. lactis; L.c, L. lactis subsp. cremoris; L. chungang, L. chungangensis; L. raffinol, L. raffinolactis.
Based on the gene collections, the “L. garvieae,” “L. lactis subsp. lactis,” and “L. lactis subsp. cremoris” clades have 33, 54, and 67 clade-specific genes, of which proteins with completely unknown function constitute 18, 42, and 52%, respectively (see Data Set S2 in the supplemental material). Interesting examples of clade-specific genes include the phosphatidylglycerol lysyltransferase gene mprF in the L. garvieae clade, which is known to protect bacteria from cationic antimicrobial peptides (37); the phosphoketolase gene ptk in the L. lactis subsp. lactis clade; and the nitroreductase gene in the L. lactis subsp. cremoris clade.
A detailed comparison of the most phylogenetically close species, L. raffinolactis and L. chungangensis, showed that L. piscium shares more orthologous groups with L. raffinolactis than with L. chungangensis (Fig. 3), while L. raffinolactis shares almost the same number of genes with both species. Their core genomes consist of 1,278 genes, which is more than three times higher than the number of genes in the core genomes of 30 analyzed lactococci. An analysis of the genes shared by only two species (see Data Set S4 in the supplemental material) showed that (i) in comparison with L. piscium, two other species possess pyruvate and lactate oxidase genes and lack the citrate catabolic operon; (ii) L. piscium shares the agmatine, glycerol, sorbitol, and xylose catabolic genes and the malolactic operon with L. raffinolactis; and (iii) common genes for L. piscium and L. chungangensis include the LPxTG-like pilus genes and the riboflavin biosynthesis operon. Interestingly, L. piscium and L. raffinolactis have different galactose degradation pathways, the Leloir pathway and the tagatose-6-phosphate pathway, respectively, while L. chungangensis has neither of them.
FIG 3.
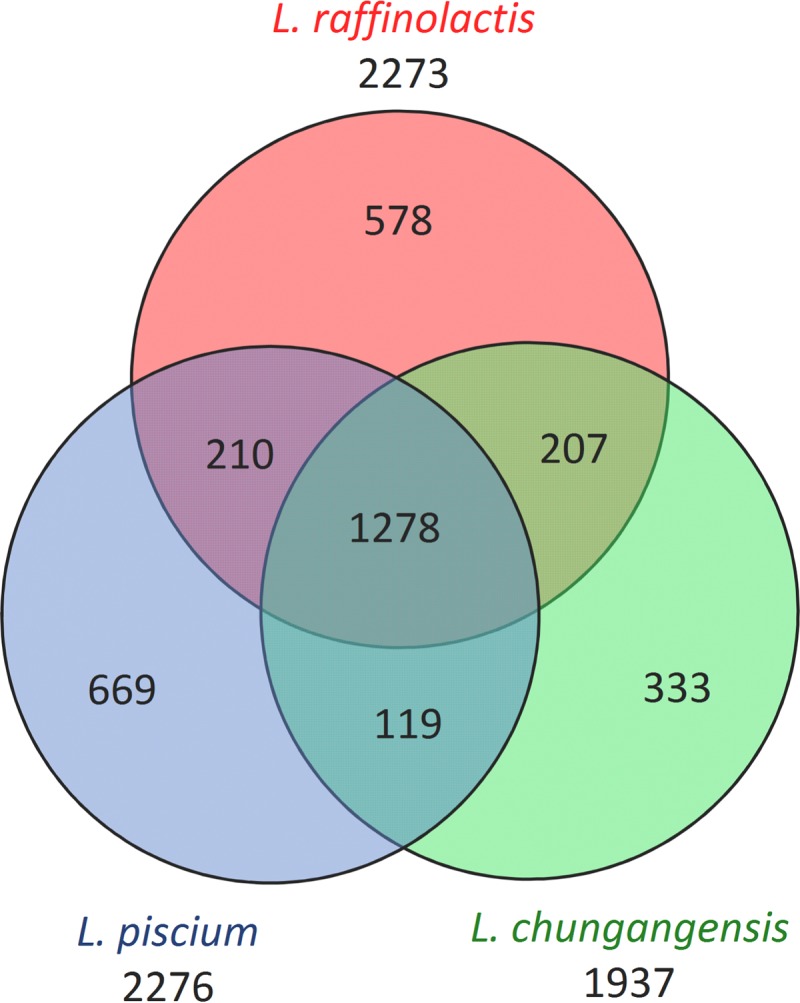
Venn diagram of the distribution of the orthologous gene groups among the three closely related species L. piscium, L. raffinolactis, and L. chungangensis.
The number of groups of unique genes/paralogs for L. piscium not present in any of the analyzed genomes of the Lactococcus genus is 395 (16% of the genome). These unique genes include, for instance, the pilBTCA cluster, encoding type IV pilus proteins; the ula operon for l-ascorbate transport and degradation; and three putative bacteriocins (see Data Set S5 in the supplemental material). On the other hand, the L. piscium genome lacks (i) gluconate kinase, (ii) cytochrome d ubiquinol oxidase/transporter and menaquinone biosynthesis, and (iii) arginine deiminase genes, while these genes are present in 29, 25, and 24 other lactococcus genomes, respectively.
General metabolism.
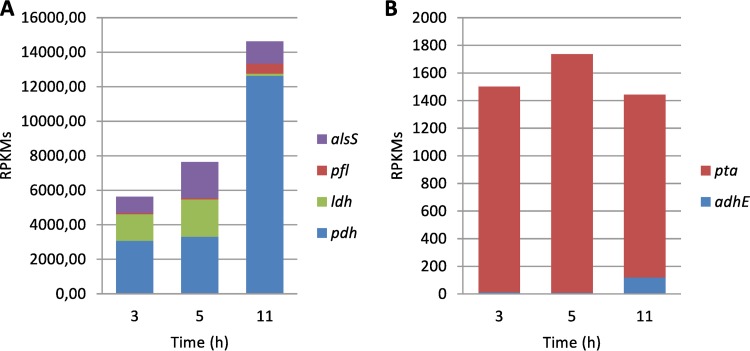
The primary catabolic pathway for glucose (and other hexoses) in L. piscium is predicted to be glycolysis, since all the required glycolytic enzymes are present in the genome as well as in the genomes of all other analyzed lactococci. On the other hand, L. piscium is missing the phosphoketolase gene, the key enzyme for the phosphoketolase pathway that is present in the 12 other lactococcal genomes. The above-described findings allow L. piscium to be classified as a homofermentative lactic acid bacterium (38). In addition, L. piscium possesses a partial pentose phosphate pathway that allows the catabolism of pentoses such as arabinose and xylose. Four pathways for pyruvate utilization have been predicted, including acetoin/diacetyl, pyruvate dehydrogenase, l-lactate dehydrogenase, and pyruvate-formate lyase pathways. Significantly, many spoilage substances, such as acetoin/diacetyl and acetate, are produced as a result of these pathways (Table 3). All the genes involved in the pyruvate utilization routes show differential expression during the time course (Fig. 4A). The pyruvate dehydrogenase complex pdhABCD (LACPI_0822 to LACPI_0825 [LACPI_0822-0825]) has the highest level of expression (averaged for all subunits) across all time points, compared to the other three enzymes. Another observation is that the overall expression level of the pyruvate-dissipating enzymes increased during this time. The pyruvate dehydrogenase complex and the pyruvate-formate lyase gene pfl (LACPI_1736) were highly upregulated after 5 h, with FC values of ∼4 and 8, respectively. In turn, the lactate dehydrogenase gene ldh (LACPI_1375) was highly downregulated (FC, ∼16) at the same time. Both the pyruvate dehydrogenase and pyruvate-formate lyase pathways produce acetyl coenzyme A (acetyl-CoA), which can be reduced to ethanol by the aldehyde-alcohol dehydrogenase AdhE (LACPI_2207) or converted to acetyl-phosphate by the phosphate acetyltransferase Pta (LACPI_0981). It can be seen that the level of pta RNA expression was much higher than that of adhE at any time. However, adhE was significantly upregulated (FC, ∼13) in the late hours, while the RNA expression changes of pta were not found to be significant (Fig. 4B). Acetoin/diacetyl production-associated genes showed very different expression patterns. The acetolactate synthase gene alsS (LACPI_1143) was found to be upregulated only in the early hours; the expression of the acetolactate decarboxylase gene aldB (LACPI_1151) first increased and then decreased in the second period; and diacetyl reductases, encoded by the chromosomal gene budC (LACPI_0294) and the plasmid gene butA (LACPI_2399), were both upregulated during the whole time. However, the expression level of budC was much lower (34 to 72 times) than that of butA, indicating that butA is the main diacetyl-reducing enzyme in L. piscium.
TABLE 3.
Enzymes and their encoding genes associated with the production of spoilage substances in L. piscium
| Spoilage compound | Enzyme | Gene(s) | Locus tag(s) |
|---|---|---|---|
| Acetate | Acetate kinase | ackA | LACPI_1846 |
| N-Acetylglucosamine-6-phosphate deacetylase | nagA | LACPI_1268 | |
| Citrate lyase complex | citCDEF | LACPI_1076-1079 | |
| CO2 | Oxaloacetate decarboxylase | citM | LACPI_1074 |
| Acetolactate synthase | alsS | LACPI_1143 | |
| Alpha-acetolactate decarboxylase | aldB | LACPI_1151 | |
| Pyruvate dehydrogenase complex | pdhABCD | LACPI_0822-0825 | |
| Phosphogluconate dehydrogenase | gnd | LACPI_2003 | |
| Acetoin/diacetyl | Acetolactate synthase | alsS | LACPI_1143 |
| Alpha-acetolactate decarboxylase | aldB | LACPI_1151 | |
| Diacetyl reductase [(S)-acetoin forming] | budC | LACPI_0294 | |
| Diacetyl reductase [(S)-acetoin forming] | butA | LACPI_2399 |
FIG 4.
Expression levels of the four pyruvate-dissipating enzymes (A) and two acetyl-CoA-dissipating enzymes (B). RPKM, reads per kilobase of a gene per million mapped reads, averaged across all replicates. Designations: pdh, pyruvate dehydrogenase complex; ldh, lactate dehydrogenase; pfl, pyruvate-formate lyase; alsS, acetolactate synthase; adhE, aldehyde-alcohol dehydrogenase; pta, phosphate acetyltransferase.
Despite living in a protein-rich environment, L. piscium has genes for the biosynthesis of all amino acids, except for phenylalanine. In addition, the genome contains a full set of genes for the de novo biosynthesis of purines/pyrimidines and several cofactors/vitamins such as riboflavin, folate, CoA, NAD, lipoate, and polyprenyls. Since the L. piscium genome carries a glycerol transporter and a set of glycerol degradation genes, we tested its growth with glycerol as the sole carbon source. Growth occurred only in the presence of oxygen, and the final OD600 (∼4), after 24 h of growth in M17 medium, was approximately the same as that after growth with glucose as the carbon source. Glycerol catabolism genes were well expressed and mainly upregulated (Fig. 5), especially the phosphotransferase system (PTS)-dependent dihydroxyacetone (DHA) kinase genes dhaMLK (LACPI_1470-1472), the expression of which increased dramatically during the second period, with FC values of ∼70 to 250 for the different subunits. These were the highest FCs among all L. piscium DE genes. Peculiarly, we did not find glycerol dehydrogenase in the genome, which would oxidize glycerol to DHA that can be subsequently phosphorylated by DhaMLK. We hypothesize that DHA could be directly taken up from the extracellular medium by the glycerol facilitator GlpF (LACPI_1162), which has been shown in Escherichia coli to transport a variety of neutral straight-chain compounds such as DHA (39).
FIG 5.
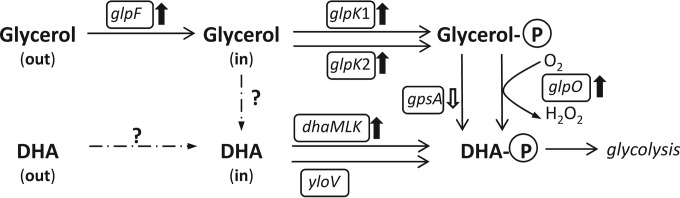
Differential expression of glycerol catabolism genes. Designations: glpF, glycerol permease; glpK, glycerol kinase; glpO, alpha-glycerophosphate oxidase; gpsA, glycerol-3-phosphate dehydrogenase; dhaMLK, PTS-dependent dihydroxyacetone kinase; yloV, putative dihydroxyacetone kinase. Abbreviations: DHA, dihydroxyacetone; out, outside the cell; in, inside the cell. The direction of the arrows adjacent to the enzyme names indicates up- or downregulation.
Carbohydrate metabolism.
Carbohydrate utilization by L. piscium MKFS47 and the predicted carbohydrate catabolic pathways/transporters are presented in Table 4. It can be easily perceived that L. piscium is capable of consuming a wide range of mono- and disaccharides as carbon and energy sources. Moreover, L. piscium is well equipped for the catabolism of higher-order oligosaccharides and even polysaccharides of different types (see Table S3 in the supplemental material). In particular, the genome has a metabolic cluster for glycogen. The cluster consists of the glycogen biosynthesis genes glgBCDA (LACPI_0555-0558) and the glycogen phosphorylase gene glgP (LACPI_0559) for glycogen breakdown. Nevertheless, all L. piscium isolates tested negative for glycogen utilization; also, no extracellular amylase was found in the genome. L. piscium also lacks a ribose pyranase gene, encoding the first enzyme in the ribose degradation pathway, therefore indicating the inability of this strain to utilize ribose. This prediction is in accordance with the finding that most L. piscium isolates do not utilize ribose (5). Many carbohydrate catabolic pathways and transporters were found to be significantly upregulated (FC, >4) during the whole time or the second period. These carbohydrates include fructose, mannose, mannitol, xylose, lactose/galactose, maltose, beta-glucosides, and glycogen (the whole biosynthesis/degradation operon glg, with FC values of ∼7 to 17). Interestingly, the universal ABC transporter ATP-binding protein-encoding gene msmK (LACPI_0501), which energizes multiple carbohydrate ABC transporters (40), was also highly upregulated (FC, ∼10) in the second time period. Accordingly, the glucose uptake family protein (LACPI_2218) became highly downregulated.
TABLE 4.
Predicted carbohydrate degradation pathways, transporters, and acid production from carbohydrates in L. piscium MKFS47 based on the API 50CH and API 20 Strep tests
| Degradation enzyme/pathway | Transporter(s)b | Acid productiona |
|---|---|---|
| Monosaccharides | ||
| d-Glucose | LACPI_2218, manXYZ | + |
| d-Fructose | levDEFG, fruABC, LACPI_0871-0873 | + |
| d-Mannose | manXYZ, ptnDCAB | + |
| d-Galactose | ? | +/− |
| N-Acetylglucosamine | manXYZ | ? |
| d-Mannitol | mtlFA | + |
| d-Sorbitol | srlAEB | +/− |
| d-Glucuronate | ? | ? |
| l-Arabinose | ? | + |
| d-Xylose | xylHGF | +/− |
| Disaccharides | ||
| Sucrose | scrA | + |
| Maltose | mdxEFG | + |
| Lactose | lacEF1F2 | +/− |
| Trehalose | treP | + |
| Cellobiose | ∼8–10 PTS beta-glucoside transporters | + |
| Melibiose | melB, msmG1F1E1 | +/− |
| Polysaccharides | ||
| Inulin | msmG2F2E2 | −c |
+, positive reaction; +/−, weak or variable reaction; −, negative reaction; ?, not tested.
Unknown transporters are indicated by “?.”
However, six strains of L. piscium were positive for inulin according to the API 20 Strep test.
Aerobic acetoin/diacetyl production from glucose and associated pathways.
Under specified conditions (see Materials and Methods), L. piscium generated a significant amount of acetoin/diacetyl, reaching a final concentration of 8.5 mM after 48 h of growth (OD600 of ∼2.2 in MRS medium), whereas Leuc. gelidum subsp. gasicomitatum did not produce any detectable amounts of these substances. All the necessary genes for acetoin/diacetyl production from pyruvate are present in the L. piscium genome (Table 3). In addition to the catabolic acetolactate synthase gene alsS (LACPI_1143), the genome also contains the anabolic acetolactate synthase genes ilvBH (LACPI_0280-0281), usually involved in branched-chain amino acid biosynthesis, which were shown previously to increase the level of acetoin/diacetyl production in the case of plasmid-directed expression (41). Factors such as the catabolism of citrate, aspartate, or alanine as well as the presence of oxygen are known to enhance acetoin/diacetyl production in LAB (38, 42).
L. piscium harbors an operon for citrate catabolism, citMCDEFXG (LACPI_1074 and LACPI_1076-1080), and the citrate/malate transporter gene cimH (LACPI_1075). Besides L. piscium, this operon is present in only four strains of L. lactis subsp. lactis: L. lactis subsp. lactis strain IL1403 and L. lactis subsp. lactis bv. diacetylactis strains LD61, TIFN2, and TIFN4. Citrate utilization assays showed no citrate fermentation by L. piscium during 48 h, both aerobically and anaerobically. In addition, RNA expression levels were found to be very low for the citrate catabolic cluster (RPKM ranges of ∼0.15 to 2.62 for citCDEFXG and 1.99 to 22.83 for citM), although citrate was present in the growth medium.
The first step in the catabolism of aspartate and alanine to pyruvate requires (i) aspartate (LACPI_0652) and alanine (LACPI_0317) transaminases and (ii) alpha-ketoglutarate, which can be produced from glutamate by glutamate dehydrogenase (LACPI_1533). Besides L. piscium, glutamate dehydrogenase is present only in L. raffinolactis and L. chungangensis. While alanine is directly converted to pyruvate, aspartate is first deaminated to oxaloacetate, which can be converted to pyruvate by the oxaloacetate decarboxylase CitM (LACPI_1074).
The L. piscium genome harbors two NADH oxidase genes, noxE (LACPI_1670) and noxC (LACPI_1783 [putative]), that are assumed to mediate the role of oxygen as an electron acceptor. Oxygen-dependent glycerol catabolism (Fig. 5) can also contribute to pyruvate excess and, hence, increased production of aroma compounds (43). The Voges-Proskauer test showed that L. piscium produced 1.1 mM acetoin/diacetyl from glycerol under aerobic conditions after 48 h of growth (OD600 of ∼1.0 in MRS medium).
Predicted factors contributing to increased survival and predominance within the microbial meat spoilage community.
The L. piscium genome possesses all the genes required for purine/pyrimidine (deoxyribo)nucleoside transport and catabolism. These genes include the nucleoside ABC transporter genes rnsDCAB (LACPI_0560-0563), the nucleoside phosphorylase genes punA (LACPI_1420) and pdp (LACPI_1499), the phosphopentomutase gene deoB (LACPI_1421), and the deoxyribose-phosphate aldolase gene deoC (LACPI_1498). Nucleosides can be used as alternative carbon and energy sources.
The genome also contains an operon, aguCADBR (LACPI_2110-2114), allowing the catabolism of agmatine with the production of putrescine, ammonia, and ATP. Therefore, agmatine can be used as an alternative energy source. According to the RNA-seq data, the agmatine catabolic operon was significantly upregulated during the second period (FC, ∼2 to 6). Since the growth medium includes meat extract, it could contain a trace amount of agmatine, which is, however, assumed to be insignificant.
Adhesion is another important survival factor. Since sortase-dependent proteins are often involved in adhesion in LAB (44), we scanned the genome for the presence of proteins with a C-terminal LPxTG-like motif and found three such proteins on the chromosome: the collagen adhesion protein YbeF (LACPI_1319) (encoded by a putative pseudogene due to the frameshift) and two pilus proteins (LACPI_0532 and LACPI_0533), which show homology to the adhesive pilus proteins SpaCED in Lactobacillus rhamnosus. Interestingly, LACPI_0532 seems to be a fusion between two proteins that correspond to SpaC and SpaE. Surprisingly, the genome possesses an operon for type IV pili, pilBTCA (LACPI_0070-0073), which were shown to be involved in twitching motility and adhesion (45). However, when twitching motility was tested under different conditions on 1% agar on microscope slides and examined by phase microscopy, no motility was observed. In addition, TEM was performed to examine if pili were present on the surface of L. piscium MKFS47 cells, but no pili were observed. According to RNA-seq data, the pilBTCA genes are expressed at very low levels (RPKM range, ∼0.84 to 9.69). Finally, the L. piscium genome has a fibronectin-binding protein (LACPI_1373) and two proteins with mucus-binding domains (LACPI_1655 and LACPI_1656).
As putative antimicrobial factors, L. piscium harbors genes for the biosynthesis of three putative bacteriocins (LACPI_0296, LACPI_0897-0901, and LACPI_2233-2235). We also found several enzymes that could be involved in hydrogen peroxide production: the superoxide dismutase SodA (LACPI_0465), the alpha-glycerophosphate oxidase GlpO (LACPI_1161), and two NADPH-dependent flavin mononucleotide (FMN) reductases (LACPI_0686 and LACPI_0687) showing homology to the corresponding enzymes from L. johnsonii involved in hydrogen peroxide production (46). According to data from the hydrogen peroxide production assay, L. piscium produced hydrogen peroxide only from glycerol under aerobic conditions but not from glucose both aerobically and anaerobically.
DISCUSSION
L. piscium is a psychrotrophic food spoilage LAB, the numbers of which may be underestimated in microbial analyses of cold-stored MAP food products if mesophilic bacterial enumeration techniques utilizing incubation temperatures close to or above 30°C are applied (10). Leuc. gelidum subsp. gelidum, Leuc. gelidum subsp. gasicomitatum, and L. piscium have often been reported to be the most abundant species in cold-stored MAP food independently of the composition of food matrices (3, 10). The annotated genomes of the first two species have been available for some time (47, 48), while the present study delivers genome and time course glucose catabolism-based transcriptome information for L. piscium.
To assess gene content-based similarity among different lactococcus genomes, we exploited pangenome tree construction. As opposed to phylogenetic trees, pangenome trees reflect not only the phylogeny of a species (vertical gene transfer) but also the horizontal gene transfer between species and their phenotypic differences (49). The analysis of the tree revealed three major clades within the Lactococcus genus, where L. piscium clustered together with L. garvieae strains, L. raffinolactis, and L. chungangensis. The clustering observed probably reflects niche adaptation, since most of the species in this clade were isolated from animal bodies (50, 51) or considered to be environmental (7, 10, 52, 53). While L. garvieae strains appear to be more functionally related to L. piscium than to L. lactis strains based on the gene content, a previous phylogenetic analysis showed that L. garvieae is evolutionarily closer to L. lactis strains than to L. piscium or L. raffinolactis (5). As expected, based on the gene composition, the closest species to L. piscium are L. raffinolactis and L. chungangensis, which were also phylogenetically the closest species to L. piscium (5, 52). Unlike the majority of Lactococcus species, L. piscium is predicted to be unable to degrade gluconate and arginine as well as to respire aerobically. The latter conclusion is in agreement with the finding that the growth of L. piscium in the presence of oxygen and heme is not stimulated (5).
L. piscium has many catabolic pathways for plant-specific carbohydrates, including arabinose, xylose, fructose, cellobiose, melibiose, raffinose, arabinogalactan type I oligosaccharides, and inulin, which might suggest a plant origin of L. piscium. The genome is particularly enriched in PTS transporters (containing 8 to 10) and phospho-glucosidases (containing 13) for beta-glucosides, which are also well catabolized by L. piscium according to the API 50CH test. The genome possesses the glycogen biosynthesis/degradation operon. Glycogen is abundant in meat, and its catabolism would be beneficial in a meat environment. The inability to grow on glycogen and the absence of extracellular amylase in the genome suggests that the glg operon is meant for intracellular glycogen production and degradation. The upregulation of the carbohydrate catabolic pathways, including the glycogen metabolic cluster (54), during the time course can be explained by the relief of carbon catabolite repression (CCR) (55), which occurs when the preferred carbon source (glucose for L. piscium) is exhausted and the bacteria are ready to utilize alternative carbon sources. This also explains the decreased expression level of the glucose uptake protein. Other upregulated pathways that are subject to CCR include the agmatine (56) and glycerol (57) catabolic operons. The increase in CCR-controlled gene expression occurs in the absence of alternative carbon sources in the medium, suggesting that the low glucose concentration is itself enough to turn on the expression of CCR-regulated genes.
The presence of several pyruvate utilization pathways in L. piscium increases its adaptive flexibility under various conditions (38) and, more importantly, becomes a basis for the formation of spoilage substances. According to the DE gene analysis, there is a switch in pyruvate utilization from the lactate dehydrogenase pathway to the acetyl-CoA generation pathways in the late hours of growth. This makes bacterial metabolism more flexible and efficient, since acetyl-CoA can be used not only as an electron acceptor but also as a precursor for amino acid biosynthesis and ATP production. Consistent with the downregulation of lactate dehydrogenase, the expression of aldehyde-alcohol dehydrogenase increases during the second period, taking over the role of NADH reoxidation. Along with it, elevated acetyl-CoA production increases acetate and ethanol formation during the late period. Relating to acetoin/diacetyl formation, the upregulation of both diacetyl reductases during the whole time leads to an increased conversion of diacetyl to acetoin, where the latter compound is preferable, since it has a significantly less powerful aroma than does diacetyl (58).
According to Rahkila et al. (5), meat inoculated with L. piscium strains had an odor described as buttery and sour after 14 to 16 days. The buttery aroma is caused mainly by the presence of diacetyl and, to a much lesser extent, by acetoin (58). After 48 h of aerobic growth with glucose, the concentration of acetoin/diacetyl produced by L. piscium was 8.5 mM, which is comparable to the amounts of acetoin/diacetyl produced by L. lactis subsp. lactis bv. diacetylactis strains after 25 h of aerobic growth (59). L. lactis subsp. lactis bv. diacetylactis strains are used in cheese production to generate aroma compounds such as diacetyl and acetoin, which are desirable in cheese but not in meat. Leuc. gelidum subsp. gasicomitatum did not produce any acetoin/diacetyl in the presence of oxygen and glucose in the medium. The same was observed for anaerobic growth with glucose (60). However, it has been shown that heme-based respiration (31) and catabolism of pentoses (60) induce the accumulation of significant amounts of these substances in Leuc. gelidum subsp. gasicomitatum cultures. Heme, glucose, and pentoses are particularly abundant in meat; thus, the production capabilities for acetoin/diacetyl in L. piscium and Leuc. gelidum subsp. gasicomitatum in meat are complementary.
Efficient acetoin/diacetyl production occurs when there is excess pyruvate, which is accomplished in two ways: by the production of additional pyruvate without the generation of reduced NADH (e.g., catabolism of citrate [38], aspartate, or alanine [42]) or by reducing the conversion of pyruvate into lactate in the presence of additional electron acceptors (e.g., oxygen [38]). The absence of citrate fermentation and the very low RNA expression levels of citrate catabolic genes while citrate was present in the growth medium indicate that citrate does not contribute to acetoin/diacetyl production in L. piscium. Relating to aspartate/alanine catabolism, the presence of the required transaminases and glutamate dehydrogenase suggests that L. piscium (as well as L. raffinolactis and L. chungangensis) is a potential good producer of acetoin/diacetyl from these amino acids, which, different from other lactococci (58), does not require exogenous alpha-ketoglutarate to be present in the medium. Besides the formation of acetoin/diacetyl, pyruvate surplus favors its conversion into acetate with the concurrent formation of ATP (38). Hence, aspartate and alanine may play the role of an alternative energy source in a meat environment abundant in amino acids. The presence of oxygen was shown previously to increase acetoin/diacetyl production by L. lactis (59, 61, 62). We observed the same for L. piscium (data not shown). Out of two NADH oxidases found in the genome, noxE is assumed to play the major role in oxygen-dependent NAD regeneration (63, 64). The role of noxC remains to be determined. In L. piscium, oxygen also acts as an electron acceptor during glycerol catabolism mediated by the glpFKO operon. We showed that L. piscium can grow on glycerol as the sole carbon source and that the growth efficiency is comparable to or lower than that on glucose, depending on the type of growth medium used. While utilizing glycerol, L. piscium produced acetoin/diacetyl, the amount of which, however, was approximately eight times lower than that with glucose. This is partly explained by the lower (∼2 times) efficiency of glycerol-based growth than of glucose-based growth when an MRS medium is used. Taking into account that glycerol is present in meat (16) as a product of the degradation of triacylglycerols (which are abundant in meat), it can be speculated that glycerol might be used by L. piscium as an alternative carbon source in meat, leading to the formation of buttery off-odors.
In addition, the presence of several other pathways/gene sets in the genome was predicted to give a competitive advantage to L. piscium in a meat environment, explaining its predominance. The L. piscium genome is well equipped for the catabolism of nucleosides, which are abundant in meat and are thought to be used as alternative carbon and energy sources by other meat-inhabiting bacteria (48, 65) when glucose is depleted. Due to the absence of arginine-degrading genes, this species cannot gain energy directly from this amino acid; however, the genome possesses an operon for the energetic catabolism of agmatine, which is produced from arginine by arginine decarboxylase-containing bacteria (66) and naturally present in meat (67, 68). In addition to ATP, the agmatine degradation pathway generates putrescine, a biogenic amine, which is very undesirable in meat due to its repellent smell. Another important survival factor is the ability to persist in the food processing environment and quickly colonize food surfaces, which can be achieved through adhesion. Several putative adhesins were found in the genome, including two types of pili: LPxTG-like and type IV. Nevertheless, the absence of twitching motility and pilus structures on the L. piscium cell surface as well as the low gene RPKM values indicate that both types of pili are not expressed under the examined conditions. Finally, the presence of putative bacteriocin-coding genes in the genome and hydrogen peroxide production from glycerol might contribute to the development of antimicrobial properties. The formation of hydrogen peroxide can also be associated with meat discoloration, which was recently attributed to L. piscium-mediated veal meat spoilage (C. Denis, S. La Carbona, A. Hanin, S. Chaillou, M. Zagorec, and M.-C. Champomier-Vergés, presented at Food Micro 2014, 24th International ICFMH Conference, Nantes, France, 1 to 4 September 2014).
In conclusion, the present study expands the knowledge of the genomics and transcriptomics of the core psychrotrophic food spoilage LAB, creating a base for future studies of food spoilage microbial communities, such as exploration of the interspecies interactions and mechanisms involved in the establishment of a stable population structure. As soon as the genomic sequences of other L. piscium strains are available, a comparative analysis can be done to identify the genetic determinants underlying the strain-dependent variations in spoilage potential. Regarding this, the L. piscium MKFS47 strain described here was identified as an efficient producer of buttery-odor compounds, which are known to be detrimental to meat quality.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by the Academy of Finland (grant no. 267755 and the Center of Excellence Program 2008–2013 in Microbial Food Safety Research), the Finnish Funding Agency for Technology and Innovation (TEKES) (grant no. 40046/11), and a joint ELVIRA program of the Academy of Finland and TEKES.
We thank Erja Merivirta, Henna Niinivirta, Eeva-Marja Turkki, Kirsi Lipponen, Harri Kangas, and Päivi Laamanen for their excellent technical assistance. We also appreciate the help of Kui Qian with the sequencing data analysis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00320-15.
REFERENCES
- 1.Borch E, Kant-Muermans ML, Blixt Y. 1996. Bacterial spoilage of meat and cured meat products. Int J Food Microbiol 33:103–120. doi: 10.1016/0168-1605(96)01135-X. [DOI] [PubMed] [Google Scholar]
- 2.Gram L, Ravn L, Rasch M, Bruhn JB, Christensen AB, Givskov M. 2002. Food spoilage—interactions between food spoilage bacteria. Int J Food Microbiol 78:79–97. doi: 10.1016/S0168-1605(02)00233-7. [DOI] [PubMed] [Google Scholar]
- 3.Nieminen TT, Vihavainen E, Paloranta A, Lehto J, Paulin L, Auvinen P, Solismaa M, Björkroth KJ. 2011. Characterization of psychrotrophic bacterial communities in modified atmosphere-packed meat with terminal restriction fragment length polymorphism. Int J Food Microbiol 144:360–366. doi: 10.1016/j.ijfoodmicro.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Pothakos V. 2014. Psychrotrophic lactic acid bacteria (LAB) as a source of fast spoilage occurring on packaged and cold-stored food products. Ph.D. thesis Ghent University, Ghent, Belgium. [Google Scholar]
- 5.Rahkila R, Nieminen T, Johansson P, Säde E, Björkroth J. 2012. Characterization and evaluation of the spoilage potential of Lactococcus piscium isolates from modified atmosphere packaged meat. Int J Food Microbiol 156:50–59. doi: 10.1016/j.ijfoodmicro.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 6.Williams AM, Fryer JL, Collins MD. 1990. Lactococcus piscium sp. nov. a new Lactococcus species from salmonid fish. FEMS Microbiol Lett 68:109–113. doi: 10.1111/j.1574-6968.1990.tb04132.x. [DOI] [PubMed] [Google Scholar]
- 7.Sakala RM, Hayashidani H, Kato Y, Kaneuchi C, Ogawa M. 2002. Isolation and characterization of Lactococcus piscium strains from vacuum-packaged refrigerated beef. J Appl Microbiol 92:173–179. doi: 10.1046/j.1365-2672.2002.01513.x. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y, Gao F, Xu XL, Su Y, Ye KP, Zhou GH. 2010. Changes in the bacterial communities of vacuum-packaged pork during chilled storage analyzed by PCR-DGGE. Meat Sci 86:889–895. doi: 10.1016/j.meatsci.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 9.Macé S, Cornet J, Chevalier F, Cardinal M, Pilet M-F, Dousset X, Joffraud J-J. 2012. Characterisation of the spoilage microbiota in raw salmon (Salmo salar) steaks stored under vacuum or modified atmosphere packaging combining conventional methods and PCR-TTGE. Food Microbiol 30:164–172. doi: 10.1016/j.fm.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Pothakos V, Snauwaert C, De Vos P, Huys G, Devlieghere F. 2014. Psychrotrophic members of Leuconostoc gasicomitatum, Leuconostoc gelidum and Lactococcus piscium dominate at the end of shelf-life in packaged and chilled-stored food products in Belgium. Food Microbiol 39:61–67. doi: 10.1016/j.fm.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Macé S, Joffraud J-J, Cardinal M, Malcheva M, Cornet J, Lalanne V, Chevalier F, Sérot T, Pilet M-F, Dousset X. 2013. Evaluation of the spoilage potential of bacteria isolated from spoiled raw salmon (Salmo salar) fillets stored under modified atmosphere packaging. Int J Food Microbiol 160:227–238. doi: 10.1016/j.ijfoodmicro.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Pothakos V, Nyambi C, Zhang B-Y, Papastergiadis A, De Meulenaer B, Devlieghere F. 2014. Spoilage potential of psychrotrophic lactic acid bacteria (LAB) species: Leuconostoc gelidum subsp. gasicomitatum and Lactococcus piscium, on sweet bell pepper (SBP) simulation medium under different gas compositions. Int J Food Microbiol 178:120–129. doi: 10.1016/j.ijfoodmicro.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Matamoros S, Pilet MF, Gigout F, Prévost H, Leroi F. 2009. Selection and evaluation of seafood-borne psychrotrophic lactic acid bacteria as inhibitors of pathogenic and spoilage bacteria. Food Microbiol 26:638–644. doi: 10.1016/j.fm.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Fall PA, Leroi F, Cardinal M, Chevalier F, Pilet MF. 2010. Inhibition of Brochothrix thermosphacta and sensory improvement of tropical peeled cooked shrimp by Lactococcus piscium CNCM I-4031. Lett Appl Microbiol 50:357–361. doi: 10.1111/j.1472-765X.2010.02801.x. [DOI] [PubMed] [Google Scholar]
- 15.Vihavainen EJ, Björkroth KJ. 2007. Spoilage of value-added, high-oxygen modified-atmosphere packaged raw beef steaks by Leuconostoc gasicomitatum and Leuconostoc gelidum. Int J Food Microbiol 119:340–345. doi: 10.1016/j.ijfoodmicro.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 16.Casaburi A, Piombino P, Nychas G-J, Villani F, Ercolini D. 2015. Bacterial populations and the volatilome associated to meat spoilage. Food Microbiol 45:83–102. doi: 10.1016/j.fm.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Björkroth J, Korkeala H. 1996. rRNA gene restriction patterns as a characterization tool for Lactobacillus sake strains producing ropy slime. Int J Food Microbiol 30:293–302. doi: 10.1016/0168-1605(96)00955-5. [DOI] [PubMed] [Google Scholar]
- 18.Pitcher DG, Saunders NA, Owen RJ. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol 8:151–156. doi: 10.1111/j.1472-765X.1989.tb00262.x. [DOI] [Google Scholar]
- 19.Staden R, Beal KF, Bonfield JK. 2000. The Staden package, 1998. Methods Mol Biol 132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 20.Larsen TS, Krogh A. 2003. EasyGene—a prokaryotic gene finder that ranks ORFs by statistical significance. BMC Bioinformatics 4:21. doi: 10.1186/1471-2105-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koskinen P, Törönen P, Nokso-Koivisto J, Holm L. 8 January 2015. PANNZER: high-throughput functional annotation of uncharacterized proteins in an error-prone environment. Bioinformatics doi: 10.1093/bioinformatics/btu851. [DOI] [PubMed] [Google Scholar]
- 24.De Jong A, van Heel AJ, Kok J, Kuipers OP. 2010. BAGEL2: mining for bacteriocins in genomic data. Nucleic Acids Res 38:W647–W651. doi: 10.1093/nar/gkq365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Litou ZI, Bagos PG, Tsirigos KD, Liakopoulos TD, Hamodrakas SJ. 2008. Prediction of cell wall sorting signals in gram-positive bacteria with a hidden Markov model: application to complete genomes. J Bioinform Comput Biol 6:387–401. doi: 10.1142/S0219720008003382. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klimke W, O'Donovan C, White O, Brister JR, Clark K, Fedorov B, Mizrachi I, Pruitt KD, Tatusova T. 2011. Solving the problem: genome annotation standards before the data deluge. Stand Genomic Sci 5:168–193. doi: 10.4056/sigs.2084864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166. [Google Scholar]
- 30.Letunic I, Bork P. 2011. Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jääskeläinen E, Johansson P, Kostiainen O, Nieminen T, Schmidt G, Somervuo P, Mohsina M, Vanninen P, Auvinen P, Björkroth J. 2013. Significance of heme-based respiration in meat spoilage caused by Leuconostoc gasicomitatum. Appl Environ Microbiol 79:1078–1085. doi: 10.1128/AEM.02943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholson WL. 2008. The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Appl Environ Microbiol 74:6832–6838. doi: 10.1128/AEM.00881-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kempler GM, McKay LL. 1980. Improved medium for detection of citrate-fermenting Streptococcus lactis subsp. diacetylactis. Appl Environ Microbiol 39:926–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito M, Seki M, Iida K, Nakayama H, Yoshida S. 2007. A novel agar medium to detect hydrogen peroxide-producing bacteria based on the Prussian blue-forming reaction. Microbiol Immunol 51:889–892. doi: 10.1111/j.1348-0421.2007.tb03971.x. [DOI] [PubMed] [Google Scholar]
- 35.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BMG, Haag JD, Gould MN, Stewart RM, Kendziorski C. 2013. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hachmann A-B, Angert ER, Helmann JD. 2009. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob Agents Chemother 53:1598–1609. doi: 10.1128/AAC.01329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Axelsson L. 2004. Lactic acid bacteria: classification and physiology, p 1–66. In Salminen S, von Wright A, Ouwehand A (ed), Lactic acid bacteria. Microbiological and functional aspects, 3rd ed Marcel Dekker, Inc, New York, NY. [Google Scholar]
- 39.Heller KB, Lin ECC, Wilson TH. 1980. Substrate specificity and transport properties of the glycerol facilitator of Escherichia coli. J Bacteriol 144:274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marion C, Aten AE, Woodiga SA, King SJ. 2011. Identification of an ATPase, MsmK, which energizes multiple carbohydrate ABC transporters in Streptococcus pneumoniae. Infect Immun 79:4193–4200. doi: 10.1128/IAI.05290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benson KH, Godon J-J, Renault P, Griffin HG, Gasson MJ. 1996. Effect of ilvBN-encoded α-acetolactate synthase expression on diacetyl production in Lactococcus lactis. Appl Microbiol Biotechnol 45:107–111. doi: 10.1007/s002530050656. [DOI] [Google Scholar]
- 42.Kieronczyk A, Skeie S, Langsrud T, Le Bars D, Yvon M. 2004. The nature of aroma compounds produced in a cheese model by glutamate dehydrogenase positive Lactobacillus INF15D depends on its relative aminotransferase activities towards the different amino acids. Int Dairy J 14:227–235. doi: 10.1016/j.idairyj.2003.07.001. [DOI] [Google Scholar]
- 43.Alvarez MF, Medina R, Pasteris SE, Strasser de Saad AM, Sesma F. 2004. Glycerol metabolism of Lactobacillus rhamnosus ATCC 7469: cloning and expression of two glycerol kinase genes. J Mol Microbiol Biotechnol 7:170–181. doi: 10.1159/000079826. [DOI] [PubMed] [Google Scholar]
- 44.Vélez MP, De Keersmaecker SCJ, Vanderleyden J. 2007. Adherence factors of Lactobacillus in the human gastrointestinal tract. FEMS Microbiol Lett 276:140–148. doi: 10.1111/j.1574-6968.2007.00908.x. [DOI] [PubMed] [Google Scholar]
- 45.Giltner CL, Nguyen Y, Burrows LL. 2012. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev 76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hertzberger R, Arents J, Dekker HL, Pridmore RD, Gysler C, Kleerebezem M, Teixeira de Mattos MJ. 2014. H2O2 production in species of the Lactobacillus acidophilus group: a central role for a novel NADH-dependent flavin reductase. Appl Environ Microbiol 80:2229–2239. doi: 10.1128/AEM.04272-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung JY, Lee SH, Jeon CO. 2012. Complete genome sequence of Leuconostoc gelidum strain JB7, isolated from kimchi. J Bacteriol 194:6665. doi: 10.1128/JB.01806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johansson P, Paulin L, Säde E, Salovuori N, Alatalo ER, Björkroth KJ, Auvinen P. 2011. Genome sequence of a food spoilage lactic acid bacterium, Leuconostoc gasicomitatum LMG 18811T, in association with specific spoilage reactions. Appl Environ Microbiol 77:4344–4351. doi: 10.1128/AEM.00102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snipen L, Ussery DW. 2010. Standard operating procedure for computing pangenome trees. Stand Genomic Sci 2:135–141. doi: 10.4056/sigs.38923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morita H, Toh H, Oshima K, Yoshizaki M, Kawanishi M, Nakaya K, Suzuki T, Miyauchi E, Ishii Y, Tanabe S, Murakami M, Hattori M. 2011. Complete genome sequence and comparative analysis of the fish pathogen Lactococcus garvieae. PLoS One 6:e23184. doi: 10.1371/journal.pone.0023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gabrielsen C, Brede DA, Hernández PE, Nes IF, Diep DB. 2012. Genome sequence of the bacteriocin-producing strain Lactococcus garvieae DCC43. J Bacteriol 194:6976–6977. doi: 10.1128/JB.01864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho S-L, Nam S-W, Yoon J-H, Lee J-S, Sukhoom A, Kim W. 2008. Lactococcus chungangensis sp. nov., a lactic acid bacterium isolated from activated sludge foam. Int J Syst Evol Microbiol 58:1844–1849. doi: 10.1099/ijs.0.65527-0. [DOI] [PubMed] [Google Scholar]
- 53.Meslier V, Loux V, Renault P. 2012. Genome sequence of Lactococcus raffinolactis strain 4877, isolated from natural dairy starter culture. J Bacteriol 194:6364. doi: 10.1128/JB.01697-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goh YJ, Klaenhammer TR. 2013. A functional glycogen biosynthesis pathway in Lactobacillus acidophilus: expression and analysis of the glg operon. Mol Microbiol 89:1187–1200. doi: 10.1111/mmi.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stülke J, Hillen W. 1999. Carbon catabolite repression in bacteria. Curr Opin Microbiol 2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- 56.Linares DM, del Río B, Ladero V, Redruello B, Martín MC, Fernández M, Alvarez MA. 2013. The putrescine biosynthesis pathway in Lactococcus lactis is transcriptionally regulated by carbon catabolic repression, mediated by CcpA. Int J Food Microbiol 165:43–50. doi: 10.1016/j.ijfoodmicro.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 57.Hindle Z, Smith CP. 1994. Substrate induction and catabolite repression of the Streptomyces coelicolor glycerol operon are mediated through the GylR protein. Mol Microbiol 12:737–745. doi: 10.1111/j.1365-2958.1994.tb01061.x. [DOI] [PubMed] [Google Scholar]
- 58.Le Bars D, Yvon M. 2008. Formation of diacetyl and acetoin by Lactococcus lactis via aspartate catabolism. J Appl Microbiol 104:171–177. doi: 10.1111/j.1365-2672.2007.03539.x. [DOI] [PubMed] [Google Scholar]
- 59.Bassit N, Boquien C, Picque D. 1993. Effect of initial oxygen concentration on diacetyl and acetoin production by Lactococcus lactis subsp. lactis biovar diacetylactis. Appl Environ Microbiol 59:1893–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jääskeläinen E, Vesterinen S, Parshintsev J, Johansson P, Riekkola M-L, Björkroth J. 2015. Production of buttery-odor compounds and transcriptome response in Leuconostoc gelidum subsp. gasicomitatum LMG18811T during growth on various carbon sources. Appl Environ Microbiol 81:1902–1908. doi: 10.1128/AEM.03705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cogan JF, Walsh D, Condon S. 1989. Impact of aeration on the metabolic end-products formed from glucose and galactose by Streptococcus lactis. J Appl Bacteriol 66:77–84. doi: 10.1111/j.1365-2672.1989.tb02457.x. [DOI] [Google Scholar]
- 62.De Felipe FL, Starrenburg M, Hugenholtz J. 1997. The role of NADH-oxidation in acetoin and diacetyl production from glucose in Lactococcus lactis subsp. lactis MG1363. FEMS Microbiol Lett 156:15–19. [Google Scholar]
- 63.Neves AR, Ramos A, Costa H, van Swam II, Hugenholtz J, Kleerebezem M, de Vos W, Santos H. 2002. Effect of different NADH oxidase levels on glucose metabolism by Lactococcus lactis: kinetics of intracellular metabolite pools determined by in vivo nuclear magnetic resonance. Appl Environ Microbiol 68:6332–6342. doi: 10.1128/AEM.68.12.6332-6342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tachon S, Chambellon E, Yvon M. 2011. Identification of a conserved sequence in flavoproteins essential for the correct conformation and activity of the NADH oxidase NoxE of Lactococcus lactis. J Bacteriol 193:3000–3008. doi: 10.1128/JB.01466-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaillou S, Champomier-Vergès M-C, Cornet M, Crutz-Le Coq A-M, Dudez A-M, Martin V, Beaufils S, Darbon-Rongère E, Bossy R, Loux V, Zagorec M. 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat Biotechnol 23:1527–1533. doi: 10.1038/nbt1160. [DOI] [PubMed] [Google Scholar]
- 66.Curiel JA, Ruiz-Capillas C, de Las Rivas B, Carrascosa AV, Jiménez-Colmenero F, Muñoz R. 2011. Production of biogenic amines by lactic acid bacteria and enterobacteria isolated from fresh pork sausages packaged in different atmospheres and kept under refrigeration. Meat Sci 88:368–373. doi: 10.1016/j.meatsci.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 67.Hernandez-Jover T, Izquierdo-Pulido M, Veciana-Nogues MT, Vidal-Carou MC. 1996. Ion-pair high-performance liquid chromatographic determination of biogenic amines in meat and meat products. J Agric Food Chem 44:2710–2715. doi: 10.1021/jf9506803. [DOI] [Google Scholar]
- 68.Ruiz-Capillas C, Jimenez-Colmenero F. 2004. Biogenic amine content in Spanish retail market meat products treated with protective atmosphere and high pressure. Eur Food Res Technol 218:237–241. doi: 10.1007/s00217-003-0848-3. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.