SUMMARY
Human centromeres are specified by a stably inherited epigenetic mark that maintains centromere position and function through a two-step mechanism relying on self-templating centromeric chromatin assembled with the histone H3 variant CENP-A, followed by CENP-A-dependent nucleation of kinetochore assembly. Nevertheless, natural human centromeres are positioned within specific megabase chromosomal regions containing α-satellite DNA repeats, which contain binding sites for the DNA sequence specific binding protein CENP-B. We now demonstrate that CENP-B directly binds both CENP-A’s amino-terminal tail and CENP-C, a key nucleator of kinetochore assembly. DNA sequence-dependent binding of CENP-B within α-satellite repeats is required to stabilize optimal centromeric levels of CENP-C. Chromosomes bearing centromeres without bound CENP-B, including the human Y chromosome, are shown to missegregate in cells at rates several fold higher than chromosomes with CENP-B containing centromeres. These data demonstrate a DNA sequence-specific enhancement by CENP-B of the fidelity of epigenetically defined human centromere function.
Keywords: CENP-B, CENP-C, centromere, mitosis, chromosome, α-satellites
INTRODUCTION
The centromere is the fundamental unit for ensuring chromosome inheritance. Correct centromere function is required for preventing errors in chromosome delivery that would lead to genomic instability and aneuploidy, both hallmarks of many human cancers. Although are not conserved across species, centromere regions from fission yeast to man have arrays of repetitive sequences (Fukagawa and Earnshaw, 2014). Human centromeres carry extensive (1,500 to >30,000) copies of imperfectly repeated arrays of a 171 bp element, termed α-satellite DNA. In all but the centromere of the Y chromosome (Earnshaw et al., 1989; 1987; Miga et al., 2014), this array contains a consensus 17 bp motif that is the binding site of CENP-B, the only known mammalian centromeric DNA sequence-specific binding protein (Verdaasdonk and Bloom, 2011).
Nevertheless, human centromere position is epigenetically specified (Ekwall, 2007; Karpen and Allshire, 1997). Among the strongest evidence for an epigenetically defined centromere was the discovery of neocentromeres in humans (Amor et al., 2004; Sart et al., 1997; Ventura, 2004; Warburton, 2004) in which the initial functional centromere has moved from its initial location to a new site in formerly euchromatic DNA without α-satellite repeats or binding of CENP-B (Depinet et al., 1997; Sart et al., 1997; Warburton et al., 1997). Often associated with chromosomal rearrangements and found in some types of cancer, each neocentromere is marked with chromatin stably assembled with CENP-A (Amor et al., 2004), the centromere-specific histone H3 variant (Earnshaw and Rothfield, 1985; Palmer et al., 1987).
CENP-A is essential for centromere identity (Black et al., 2007b). It marks and maintains centromere position (Black et al., 2004; Hori et al., 2013; Mendiburo et al., 2011) and recruits additional centromere and kinetochore components (Carroll et al., 2010; 2009; Foltz et al., 2006; Liu et al., 2006) to both normal and ectopic centromere locations (Barnhart et al., 2011; Guse et al., 2011; Hori et al., 2013; Mendiburo et al., 2011). Indeed, use of gene targeting in both human cells and fission yeast has demonstrated that CENP-A-containing chromatin is the primary epigenetic mark of centromere identity (Fachinetti et al., 2013).
Centromere identity and function is achieved through a two-step mechanism. First, new CENP-A assembly to centromeric chromatin is mediated by its CENP-A Targeting Domain (CATD) (Black et al., 2004; 2007a; Shelby et al., 1997) in conjunction with the CENP-A selective chaperone HJURP (Dunleavy et al., 2009; Foltz et al., 2009; Shuaib et al., 2010) whose activity is tightly controlled across the cell cycle (Müller and Almouzni, 2014). In the second step, either the amino- or carboxy-terminal tails of CENP-A is required for nucleation of assembly of a kinetochore that mediates high fidelity chromosome segregation indefinitely (Fachinetti et al., 2013). Curiously, amino-terminal tail-dependent kinetochore nucleation requires the presence of CENP-B (Fachinetti et al., 2013). Starting from this suggestion of a CENP-B-dependent role in kinetochore function, we now use a combination of gene targeting and replacement in human and mouse cells, coupled with in vitro approaches, to identify a DNA sequence-dependent contribution to fidelity of human centromeric function that is mediated by CENP-B binding to centromeric α-satellite repeats.
RESULTS
CENP-A’s amino-terminal tail directly binds the alphoid DNA binding protein CENP-B
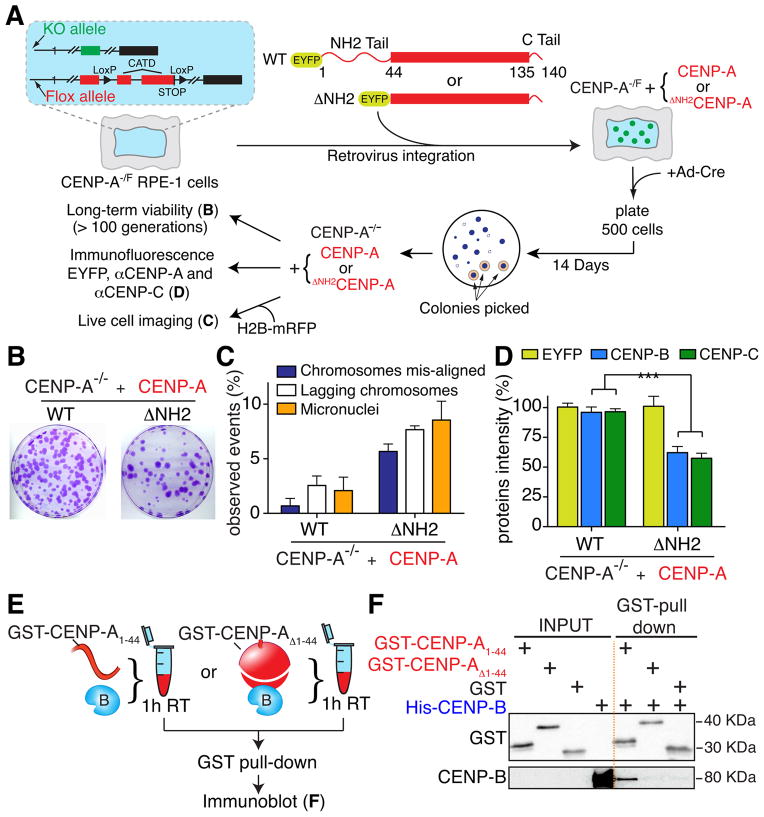
To test the effect that complete loss of the CENP-A amino-terminal tail has on centromere-bound CENP-B and on overall cell viability, we stably expressed (by retroviral integration) a full length CENP-A or a CENP-A variant lacking its amino-terminal tail (ΔNH2CENP-A) in human cells containing one disrupted endogenous CENP-A allele and one floxed allele (CENP-A−/F) (Fig. 1A). After Cre-recombinase mediated inactivation of the floxed allele and subsequent loss of endogenous CENP-A protein (Fig. S1A–B), long-term cell viability was rescued by ΔNH2CENP-A (Fig. 1B), albeit with a 4 fold increase in chromosome missegregation and micronuclei formation (visualized by live cell imaging in cells stably expressing H2B-mRFP to visualize chromosomes) (Fig. 1A, C). Furthermore, loss of the CENP-A amino-terminal tail was accompanied by reduced CENP-B binding at centromeres (Fig. 1A, D), as measured by quantifying centromeric CENP-B intensity by immunofluorescence.
Figure 1. CENP-A amino tail interacts with CENP-B.
(A) Schematic representing the different CENP-A rescue constructs amino terminally tagged with EYFP (enhanced yellow fluorescent protein), the CENP-A−/F cell line and the experiments described in B–D. (B) Images of representative crystal violet–stained colonies from the colony formation assay in A. (C) Frequency of mitotic errors in the indicated cell lines. Bars represent the mean of > 50 cells per condition. Error bars represent the SEM of three independent experiments. (D) Bar graphs showing centromere intensities of EYFP-rescue constructs, CENP-B and CENP-C for the indicated cell lines. Values represent the mean of six independent experiments (> 30 cells per experiment, average of 30 centromeres for cell). Error bars represent the SEM (standard error of the mean). Unpaired t test: *** p < 0.0001. (E) Schematic of the experiment performed in F. (F) GST pull-down and subsequent immunoblot for GST-tagged CENP-A co-incubated with His-tagged CENP-B. See also Figure S1.
To determine if this CENP-A-dependent binding of CENP-B at centromeres could result from a direct interaction, recombinant CENP-B was incubated with GST or GST-tagged CENP-A fragments and GST-containing proteins were affinity purified on glutathione-immobilized beads (Fig. 1E, F). CENP-B bound directly to the amino-terminal tail of CENP-A (GST-CENP-A1-44), but not to GST alone or a CENP-A mutant lacking its amino-terminal tail (GST-CENP-AΔ1-44) (Fig. 1F). The first 29 amino acids of the CENP-A tail were sufficient for this interaction (Fig. S1C), in agreement with the observation that the first 29 amino acids of CENP-A’s amino terminal tail stabilize CENP-B binding at centromeres (Fachinetti et al., 2013).
CENP-B supports CENP-C maintenance at centromeres
Deletion of the CENP-A amino-terminal tail not only affected CENP-B binding, but also reduced by half centromere-bound CENP-C (Fig. 1D), a major centromere component required for kinetochore assembly (Fukagawa et al., 1999). In vitro (Carroll et al., 2010; Guse et al., 2011) and in vivo (Fachinetti et al., 2013) findings have reported that the small (6 amino acid) carboxy-terminal tail of CENP-A is one element for CENP-C recruitment to centromeres. Complete loss of the CENP-A carboxy-terminal tail did not, however, abolish centromeric CENP-C binding (Fachinetti et al., 2013), indicating the existence of another pathway for its recruitment.
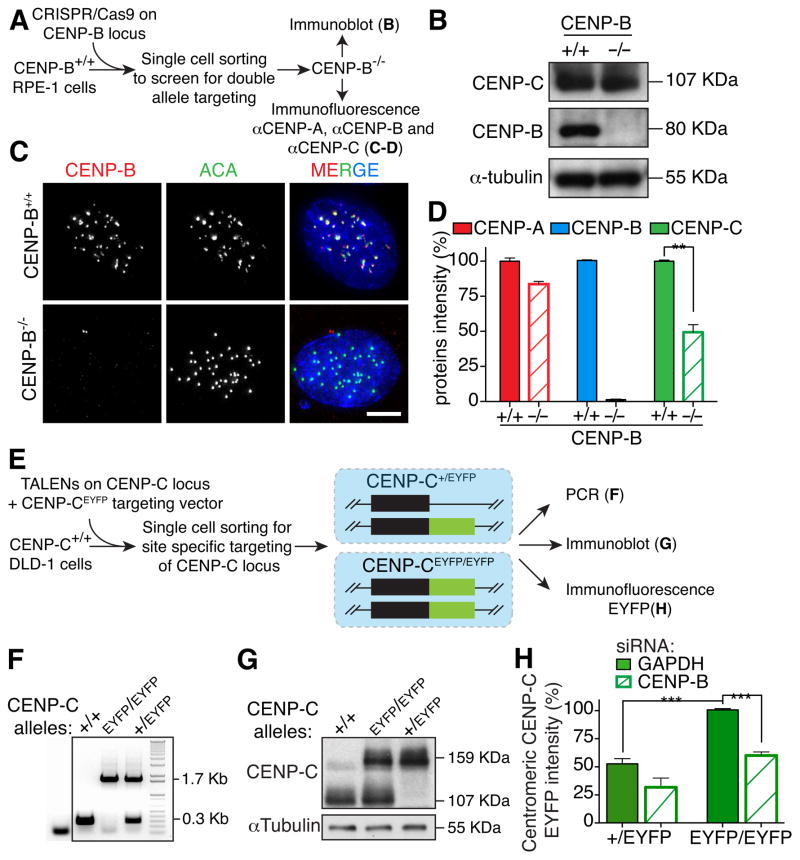
Since the CENP-A amino-terminal tail binds to CENP-B and its deletion reduced both CENP-B and CENP-C bound to centromeres (Fig. 1), we tested if CENP-B was required for the maintenance of a fraction of centromeric CENP-C. Long-term dependency of centromere recruitment of CENP-C on CENP-B was tested by disrupting both CENP-B alleles in human diploid RPE1 cells using a CRISPR/Cas9 nuclease (Fig. 2A and Fig. S2A). Complete loss of CENP-B (Fig. 2B–D) resulted in a 50% reduction of CENP-C at centromeres but not of its total cellular levels (Fig. 2C, D), with only a slight decrease of centromeric CENP-A levels (Fig. 2D), a reduction insufficient to explain the observed CENP-C reduction [CENP-A must be depleted >75% to produce two-fold decrease of centromere-bound CENP-C (Fachinetti et al., 2013)].
Figure 2. CENP-B is required for full CENP-C maintenance at centromeres.
(A) Schematic of the experiments described in B–D with the use of CRISPR/Cas9-mediated gene deletion (B) Immunoblot for accumulated CENP-B and CENP-C after identification of cells with both CENP-B alleles disrupted by action of the CRISPR/Cas9. α-tubulin was used as a loading control. (C) Representative immunofluorescence images of centromere-bound CENP-B following CRISPR-mediated disruption of both CENP-B alleles in RPE1 cells. ACA was used to mark centromere positions. Scale bar = 5 μm. (D) Bar graphs of centromere intensities for CENP-A (red), CENP-B (blue) and CENP-C (green) in the indicated cell lines quantified with specific antibodies as described in A. Bars represent the mean of three independent experiments (> 30 cells per experiment). Error bars represent the SEM. Unpaired t test: ** p < 0.006. (E) Schematic of the experiments described in F–H with the use of TALENs-mediated gene targeting. (F) CENP-C genotypes validated in the indicated cell lines using PCR to distinguish normal (+/+), double tagged (EYFP/EYFP), and single tagged (+/EYFP). (G) Immunoblot for CENP-C to distinguish non-tagged, single or double allele tagged CENP-C with EYFP. (H) Bar graphs represent CENP-C-EYFP centromeres intensity in the indicated cell lines measured by live cell imaging with or without siRNA treatment of CENP-B. Bars represent the mean of three independent experiments (> 30 cells per experiment, average of 30 centromeres for cell). Error bars represent the SEM. Unpaired t test: *** p < 0.0001. See also Figure S2.
To determine if short-term reduction of CENP-B also had consequences on CENP-C maintenance at centromeres, we integrated (at a unique genomic locus using the Flp-In system in DLD-1 cells) an siRNA-resistant, doxycycline-inducible gene encoding CENP-B that was dually tagged with EYFP and AID (an Auxin Inducible Degron), the latter to enable rapid degradation upon addition of the synthetic auxin indole-3-acetic acid (IAA) (Holland et al., 2012). In agreement with our initial gene inactivation approach (Fig. 2D), siRNA-mediated CENP-B depletion led to decrease in centromeric CENP-C by half (Fig. S2B–D). In contrast, doxycycline-induced expression of CENP-BAID-EYFP maintained the initial level of CENP-C at each centromere (Fig. S2B–D), while auxin-induced rapid degradation of CENP-BAID-EYFP was accompanied by reduction (again by half) of CENP-C bound at centromeres (Fig. S2D).
To directly measure the level of CENP-C after siRNA-mediated CENP-B depletion, we EYFP-tagged one or both alleles of the endogenous human CENP-C gene using TALEN-mediated gene targeting (Fig. 2E–G). As expected, quantification of centromere fluorescence intensity of single centromeres in live cells revealed that CENP-C in CENP-C+/EYFP cells was half that of centromeres in which both alleles were targeted (CENP-CEYFP/EYFP) (Fig. 2H). Importantly, in agreement with our indirect immunofluorescence analysis, siRNA-mediated depletion of CENP-B led to loss of half of centromeric EYFP intensity (Fig. 2H).
Direct binding of CENP-B to CENP-C
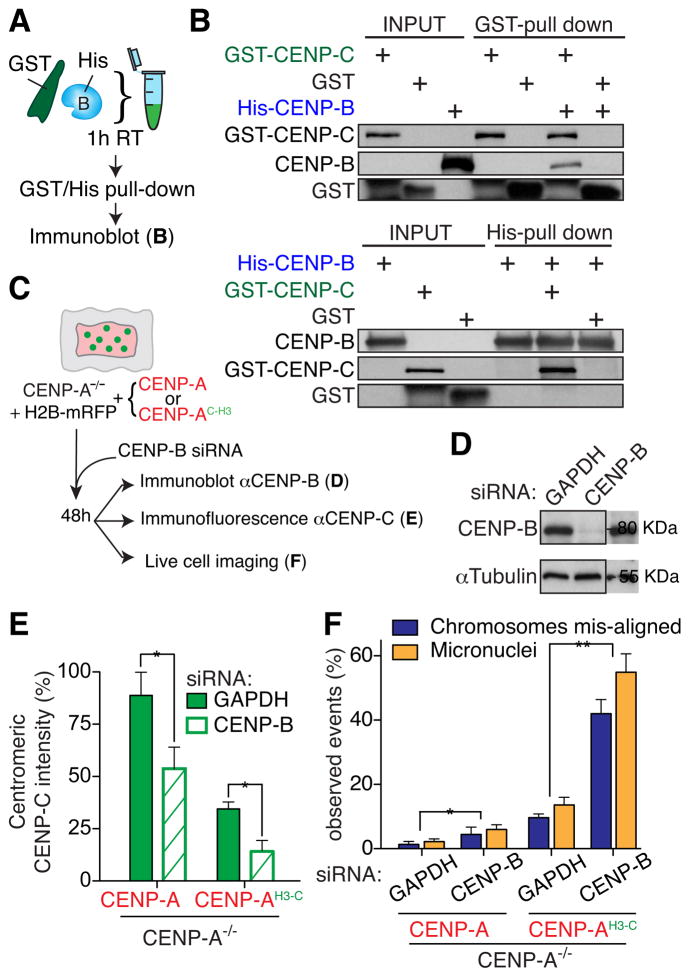
To determine whether the CENP-B influence on the level of centromere-bound CENP-C in cells (Fig. 2) could be mediated by physical interaction between CENP-B and CENP-C, we tested direct binding of the proteins to each other in vitro (Fig. 3A). After co-incubation of GST-CENP-C and His-CENP-B, either protein was affinity purified (Fig. S2E), followed by immunoblot. GST pull-down for CENP-C co-purified a portion of CENP-B and vice versa (Fig. 3B), indicative of direct binding and in agreement with a previous two-hybrid study (Suzuki et al., 2004).
Figure 3. CENP-B supports faithful chromosome segregation via direct CENP-C binding.
(A) Schematic of a GST affinity pull-down assay for testing CENP-B binding to GST-tagged CENP-C. (B) GST (top) or His (bottom)-pull-down using the assay detailed in A. (C) Schematic of the experiments described in D–F. (D) Immunoblot of cell extracts to measure total CENP-B level 48 hours after siRNA treatment. α-tubulin was used as a loading control. (E) Bar graphs of centromere intensities for CENP-C in the indicated cell lines. Bars represent the mean of three independent experiments (> 30 cells per experiment). Error bars represent the SEM (standard error of the mean). Unpaired t test: * p < 0.04 (F) Bar graphs of frequency of micronuclei formation and mis-aligned mitotic chromosomes identified with live cell imaging. Bars represent the mean of > 50 cells per condition. Error bars represent the SEM of three independent experiments. Unpaired t test: * p < 0.04, ** p < 0.006. See also Figure S3.
CENP-B is not required for initial loading of new CENP-C at centromeres
We then tested if CENP-B was also responsible for CENP-C deposition at centromeres beside its role in CENP-C stabilization. To test the loading of CENP-C at centromeres and its dependency on CENP-B, a tetracycline-inducible CENP-C tagged with EYFP and AID was stably integrated at a specific chromosomal locus. After culturing in IAA to induce degradation of the accumulated tagged CENP-C assembled at centromeres, IAA was withdrawn and immunofluorescence was used to monitor CENP-CAID-EYFP re-accumulation and its assembly at centromeres (Fig. S3A, B). Interestingly, siRNA-mediated reduction in CENP-B by more than 90% (Fig. S3C) did not significantly change the percentage of cells that efficiently loaded new CENP-CAID-EYFP (Fig. S3D), but did reduce the intensity of centromeric CENP-CAID-EYFP (Fig. S3E). This result indicates that CENP-B is not required for CENP-C deposition at centromere, but only for its retention.
CENP-B acts to enable highest fidelity of chromosome segregation
To determine the consequence of CENP-B loss on the fidelity of centromere function, we measured chromosome mis-segregation rates using live cell imaging (Fig. 3C) in CENP-A−/− cells deleted in both endogenous CENP-A alleles and with centromere function rescued by stable expression of full length CENP-A or a CENP-A variant (CENP-AH3-C) in which its CENP-C binding site (CENP-A’s carboxy terminus) was replaced with the corresponding region of histone H3 (Fig. 3C, E). In CENP-A−/− cells rescued with full length CENP-A, reduction in CENP-B lowered centromeric CENP-C by 50% and produced a two-fold increase in mitotic errors relative to the siRNA control (Fig. 3C–F). In CENP-A−/− cells rescued by CENP-AH3-C, almost all centromere-bound CENP-C was lost following siRNA-mediated depletion of CENP-B (Fig. 3C, D). This CENP-B-dependent loss of CENP-C at centromeres was accompanied by more than half of mitoses with misaligned chromosomes and micronuclei found within 60% of interphase cells (Fig. 3C, F).
The KMN network (comprised of the Mis12, Knl1, and Ndc80 complexes) is essential for connection between the centromere and spindle microtubules (Hori and Fukagawa, 2012). Recognizing that CENP-C has been proposed to be required for kinetochore function through recruitment of the Mis12 complex (Przewloka et al., 2011; Screpanti et al., 2011), we measured the centromeric levels of Dns1 and Hec1, subunits of the Mis12 and Ndc80 complexes, respectively, on metaphase centromeres in cells with normal or reduced centromere-bound CENP-C (Fig. S3F, G). Reduction of centromeric CENP-C in CENP-B−/− cells or in CENP-A−/− cells rescued by ΔNH2CENP-A lead to correspondingly reduced levels of Dsn1 and slightly reduced Hec1, similar to what was observed in CENP-A−/− cells rescued by CENP-AH3-C (Fig. S3H, I) (Fachinetti et al., 2013). Thus, diminution of centromeric CENP-C drives chromosome mis-segregation at least in part from reduced recruitment of the Mis12 complex.
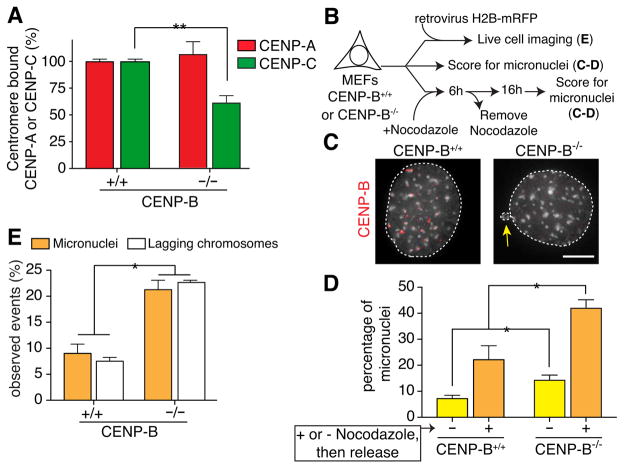
We next tested if a role of CENP-B in supporting centromere function via maintenance of centromeric CENP-C in human cells was conserved in an additional mammalian species. We tested if chromosome mis-segregation rates and centromeric CENP-C loading were affected in immortalized mouse embryonic fibroblasts (MEFs) in which both CENP-B alleles had been inactivated (Kapoor et al., 1998; Okada et al., 2007). As in the human RPE1 or DLD-1 cells following CENP-B reduction, CENP-B null MEFs displayed a 50% reduction in centromeric CENP-C, relative to wild-type MEFs, despite unchanged levels of CENP-A (Fig. 4A). This was accompanied by an increased mitotic error frequency (measured by imaging of fixed cells or by live imaging of cells expressing H2B-mRFP), with accumulation of micronuclei at more than twice the rate seen in CENP-B wild type cells (Fig. 4B–E). Using release from nocodazole arrest to enrich for mitotic errors (Thompson and Compton, 2011), higher error frequencies were again observed in CENP-B deleted MEFs relative to wild type MEFs (Fig. 4B–D).
Figure 4. CENP-B is required for faithful chromosome segregation in mouse cells by supporting CENP-C association with centromeres.
(A) Quantifications of CENP-A and CENP-C protein levels at centromeres in MEFs with or without CENP-B. Bars represent the mean of three independent experiments (> 30 cells per experiment). Error bars represent the SEM (standard error of the mean). Unpaired t test: ** p < 0.0016. (B) Schematic of the experimental design to test for frequency of micronuclei formation in CENP-B+/+ or CENP-B−/− MEFs before and after recovery from nocodazole-induced mitotic arrest or by live cell imaging. (C) CENP-B deletion increases chances of chromosome mis-segregation leading to micronuclei formation. Representative images of nuclei in CENP-B-containing and CENP-B-depleted MEFs quantified in D. Scale bar = 5 μm. An arrow marks a micronucleus in the CENP-B−/− cell. (D) Quantification of micronuclei frequency in CENP-B+/+ and CENP-B−/− MEFs measured as in B. Bars represent the mean of > 100 cells per condition. Error bars represent the SEM of three independent experiments. Unpaired t test: * p < 0.04. (E) Bar graphs quantifying lagging chromosomes in mitosis or micronuclei in interphase in the indicated cell lines. Bars represent the mean of > 50 cells per condition scored by live cell imaging. Error bars represent the SEM of three independent experiments. Unpaired t test: * p < 0.04.
Elevated mis-segregation of a neocentromere without bound CENP-B
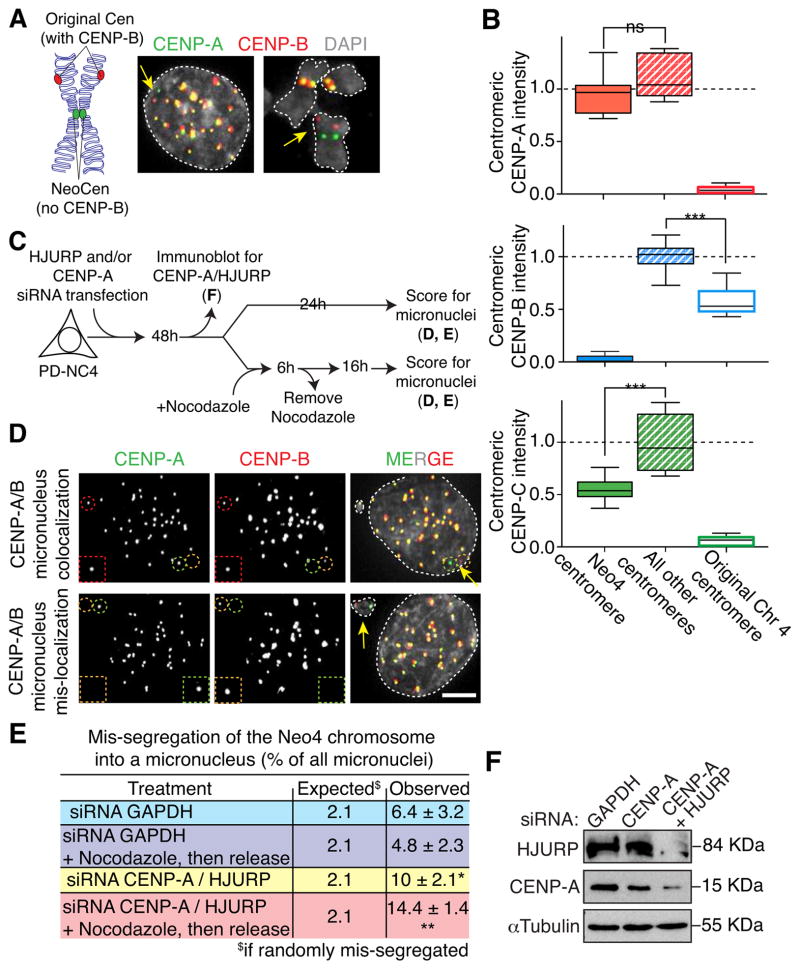
We then tested whether it was the presence of CENP-B per se or the sequence-dependent CENP-B binding at centromeres that was required to reinforce both CENP-C binding at centromeres and fidelity of chromosome segregation. To address this point, we examined a patient-derived cell line (PD-NC4) containing an alphoid DNA-free neocentromere on chromosome 4 (hereafter Neo4) (Amor et al., 2004) (Fig. 5A). The intensities of CENP-A/B/C were measured by immunofluorescence on chromosome spreads. The neocentromere was identified by scoring for a chromosome in which CENP-A (at the Neo4 centromere) and CENP-B (at the inactive, original chromosome 4 centromere DNA locus containing CENP-B boxes in its alphoid DNA repeats) did not colocalized (Fig. 5A). CENP-A level at the Neo4 centromere was nearly normal (83 % of the average level at the other centromeres - Fig. 5B, top), in agreement with previous reports (Bassett et al., 2010; Bodor et al., 2014). In contrast, CENP-C intensity at the CENP-B-free Neo4 was half of that of CENP-B-containing centromeres (49 % reduction; Fig. 5B, bottom). Also, in agreement with a role of CENP-A in stabilizing CENP-B via its amino tail, CENP-B level at the now inactive original centromere on the neocentromere-containing chromosome 4 was also 57 % that of other centromeres (Fig. 5B, middle).
Figure 5. CENP-B is required for faithful chromosome segregation of the neocentromere chromosome.
(A) (Left) Schematic of the neocentromere-containing Neo4 chromosome from PD-NC4 cells. (Right) Representative immunofluorescence images of the neocentromere chromosome in interphase or mitosis (yellow arrows). (B) Box & whisker graphs of CENP-A, CENP-B or CENP-C intensities at the neocentromere (NeoCen), other chromosomes (All other Cens) and the original chromosome 4 measured on metaphase spreads (see Experimental Procedures for details). Error bars represent the SEM. Bar in the box represents the median; the whiskers represent cells distribution. Unpaired t test: *** p < 0.0001. (C) Schematic of the experimental design for testing frequency of micronuclei formation after siRNA treatment to reduce CENP-B or HJURP in PD-NC4 cells with the Neo4 neocentromere, followed by nocodazole-induced mitotic arrest, and subsequent recovery. (D) Chromosome missegregation rates of the Neo4 chromosome or other chromosomes, determined by immunofluorescence. Representative immunofluorescence images of centromere-bound CENP-A and CENP-B after siRNA treatment against GAPDH. Yellow arrows, green and orange circles/squares indicate the neocentromere chromosome, red circles/square a normal chromosome encapsulated in micronuclei. Scale bar = 5 μm. (E) Quantitation of expected and experimentally determined mis-segregation rates from D for the Neo4 chromosome. The observed mis-segregation rate is the frequency with which the Neo4 was found inside micronuclei relative to the total number of micronuclei. The frequency of micronuclei formation was determined by immunofluorescence after the indicated siRNA treatment and, when indicated, following recovery from nocodazole-induced mitotic arrest. Unpaired t test: * p < 0.04, ** p < 0.006. (F) Immunoblot of PD-NC4 cell extracts to measure total CENP-A and HJURP levels after siRNA treatment. α-tubulin was used as a loading control.
We next tested the influence of the presence or absence of cellular CENP-B on CENP-C binding at the Neo4 centromere and the fidelity of segregation of the Neo4 chromosome. To do this, we measured the frequency of Neo4 incorporation into micronuclei by scoring for i) centromeric CENP-A (or CENP-C) without CENP-B (representing the functional Neo4 centromere) and ii) an additional CENP-B spot (representing the inactive original centromere 4). In cells with normal CENP-B levels, Neo4 was found to be encapsulated in a micronucleus with a three-fold higher frequency than other chromosomes. Neo4 was present in 6 % of observed micronuclei, a rate three times higher than the 2.1 % level expected for missegregation by chance, assuming that each chromosome has an equal chance to mis-segregate (Fig. 5C–E).
Since Neo4 centromere function relies only on CENP-A, but not on CENP-B, for CENP-C recruitment and consequently kinetochore nucleation, we then tested whether reduction of CENP-A preferentially increased missegregation of the Neo4 chromosome. We decreased centromere-assembled CENP-A levels (by simultaneous siRNA-mediated reduction of CENP-A and its chaperone HJURP – Fig. 5C, F). Remarkably, this CENP-A reduction selectively and significantly enhanced mis-segregation frequency of the Neo4 containing chromosome, yielding a five- to seven-fold (by release from nocodazole arrest) increased error frequency compared to all other chromosomes (Fig. 5E).
Elevated mis-segregation of the CENP-B-free, alphoid DNA-containing Y chromosome
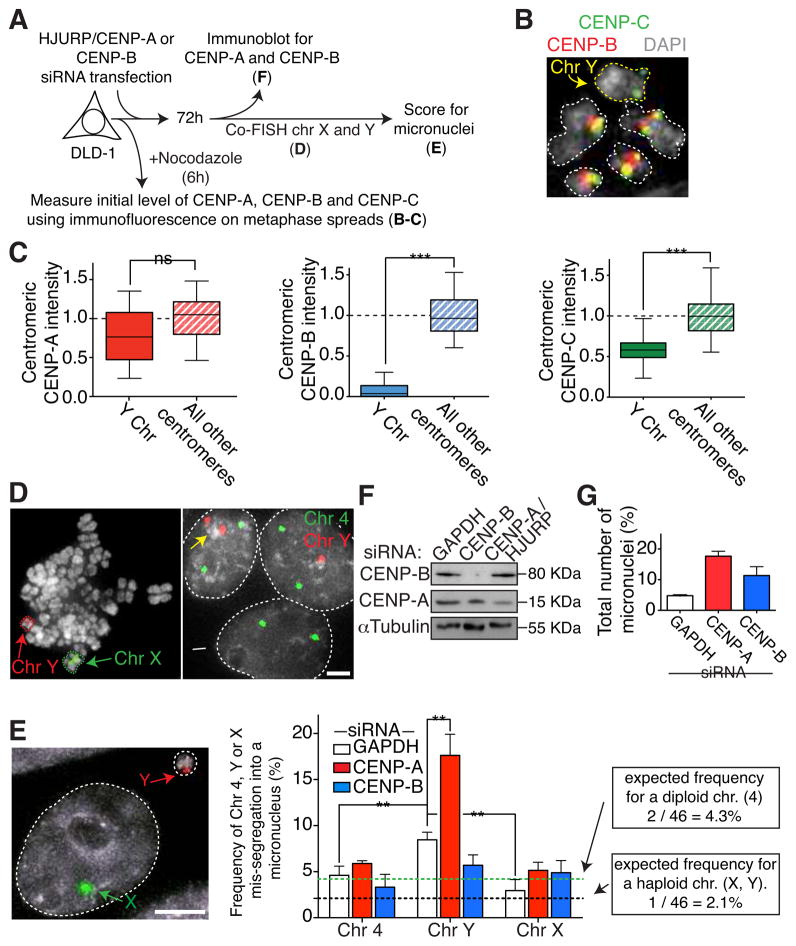
The Y chromosome is known to have repetitive alphoid DNA sequences but lacks functional CENP-B boxes (Earnshaw et al., 1987; 1989; Miga et al., 2014). To determine whether it is binding of CENP-B at each centromere rather than the presence of repetitive alphoid sequences that acts to maintain CENP-C level at centromeres and thereby support fidelity of chromosome segregation, we measured the level of centromeric CENP-C on mitotic chromosomes from a male human cell line (DLD-1) (Fig. 6A, B). CENP-A level on the Y was only slight reduced relative to other centromeres (82 % of the average level on all other centromeres - Fig. 6C, left), in agreement with a previous report (Bodor et al., 2014). However, as was observed for the Neo4 centromere, the intensity of centromeric CENP-C on the CENP-B-free Y centromere (Fig. 6B and C, middle) was only about half (60%) of that of the other chromosomes (Fig. 6B and C, right).
Figure 6. The CENP-B-free alphoid DNA-containing Y chromosome has reduced level of CENP-C and increased rate of chromosome mis-segregation.
(A) Schematic of the experiments shown in B–F. (B) Representative image of Y chromosome in mitosis (yellow arrow) stained with CENP-C (green) and CENP-B (red) antibodies. (C) Box & whisker plots of CENP-A, CENP-B or CENP-C intensities at the centromere of the Y chromosome compared to all other centromeres measured on metaphase spreads (see Experimental Procedures for details). Error bars represent the SEM. Bar in the box represents the median; the whiskers represent points distribution. Unpaired t test: *** p < 0.0001. (D) (left) Representative immunofluorescence image of a FISH experiment on a metaphase spread using centromeric probes for the Y (red) and X (green) chromosomes. (Right) Representative immunofluorescence image of a FISH experiment on interphase cells using centromeric probes for the Y chromosome (red) or chromosome 4 (green). Yellow arrow indicates a nucleus in a cell that has undergone Y chromosome missegregation so as to accumulate two Y chromosomes. White arrow indicates a nucleus in which missegregation has led to loss of the Y chromosome. (E) Chromosome missegregation rates of chromosomes 4, Y and X determined by FISH. (Left) Representative immunofluorescence image of a micronucleus-containing cell after siRNA treatment to lower CENP-A levels. Scale bar = 5 μm. (Right) Bars represent the frequency of micronuclei formation experimentally determined for chromosome 4, Y and X after the indicated siRNA treatment. The observed mis-segregation rate is the frequency with which the chromosome 4, Y or X chromosomes, respectively, were found inside micronuclei relative to the total number of micronuclei. Unpaired t test: ** p < 0.004 (F) Immunoblot of DLD-1 cell extracts to measure total CENP-A and CENP-B levels after siRNA treatment. α-tubulin was used as a loading control. (G) Bars represent the total number of micronuclei after the indicated siRNA treatment. Error bars represent the SEM of three independent experiments.
We next measured the rate of chromosome mis-segregation of the Y chromosome compared to the X chromosome or chromosome 4, the latter two both having alphoid DNA repeats with functional CENP-B boxes. To do this, we used dual fluorescence in situ hybridization (FISH) to concomitantly visualize two chromosomes. We then counted the number of micronuclei containing chromosome 4, X or Y relative to the total number of micronuclei (Fig. 6A, D, E). Remarkably, the CENP-B-free Y chromosome accumulated into micronuclei with four-fold higher frequency (8.3 %) compared to what would be expected from random mis-segregation of haploid chromosomes into micronuclei (2.1 %) or relative to the measured X or 4 mis-segregation rate (2.8 % and 4.8 %, respectively). In agreement with the dependency on CENP-A (but not CENP-B) for chromosome segregation observed for the Neo4 neocentromere-containing chromosome (Fig. 5E), siRNA-mediated reduction of CENP-A by as little as four-fold (Fig. 6F) selectively increased the frequency of Y-positive micronuclei to 18 %, while only slightly increasing mis-segregation of the chromosome X or 4 (Fig 6E). In agreement with what observed before (Fig. 3, 4), an influence of CENP-B on fidelity of chromosome segregation was seen by more than a two-fold elevation in the overall rate of micronuclei formation (Fig. 6G) following CENP-B depletion by siRNA (Fig. 6F). Despite this, reduction in CENP-B did not significantly affect the rate of mis-segregation of the CENP-B-free Y chromosome (Fig. 6E).
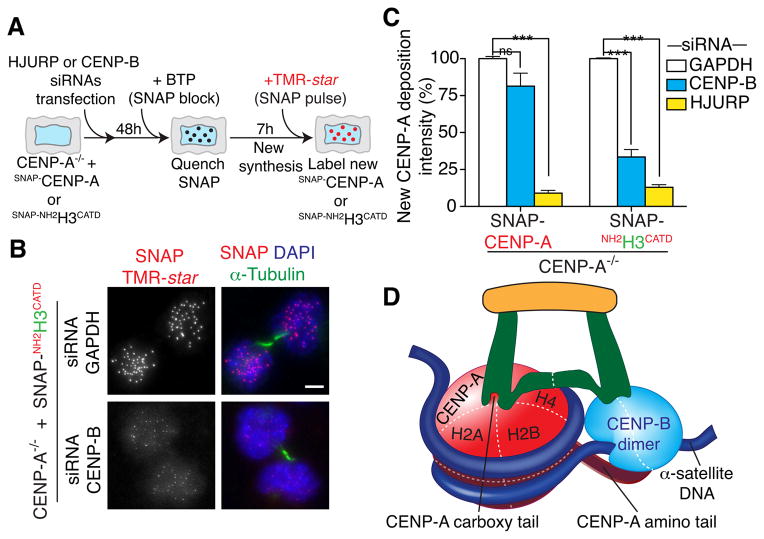
CENP-B contributes to cell cycle-dependent CENP-A loading at centromeres
With our evidence that CENP-B together with CENP-A is required to maintain a normal CENP-C level at each centromere, we tested whether there is a contribution to CENP-A deposition provided by CENP-B. For this test (see schematic in Fig. 7A), we took advantage of the SNAP-tag technique (Jansen et al., 2007) which permits in vivo covalent marking of proteins linked to a 20 kD suicide enzyme (named SNAP). siRNA was used to deplete CENP-B (Fig. S4A) from CENP-A−/− cells in which centromere function was rescued with a stably integrated full-length, SNAP-tagged CENP-A. CENP-A loading at centromeric chromatin was only slightly affected by loss of CENP-B (Fig. 7A, C and Fig. S4A, B). As expected, HJURP depletion, on the other hand, eliminated new CENP-A deposition (Fig. 7C and Fig. S4A, B) (Dunleavy et al., 2009; Foltz et al., 2009). Remarkably, CENP-A−/− cells rescued with a histone H3 variant carrying the CATD and amino terminal tail of CENP-A, but not its carboxy terminal tail (SNAP-NH2H3CATD) showed a marked reduction in new NH2H3CATD deposition following CENP-B depletion (Fig. 7A–C and Supplementary S4B). Accordingly, total centromere-bound CENP-A levels decreased in cells supported by a variant that lacks the CENP-A/CENP-C interaction domain, but not in cells rescued with full-length CENP-A (Fig. S4C).
Figure 7. CENP-B supports centromeric chromatin replication and function.
(A) Schematic of SNAP-tagging experiment to measure new CENP-A deposition at centromeres after siRNA-mediated reduction of CENP-B or HJURP. (B) Representative immunofluorescence images show localization and intensity of SNAP-NH2H3CATD after siRNA of GAPDH or CENP-B. α-tubulin staining was used to identify telophase/early G1 cells. Scale bar = 5 μm. (C) Quantitation of level of centromeric CENP-A deposition after depletion of CENP-B or HJURP. Bars represent the mean of four independent experiments (> 30 cells per experiment). Error bars represent the SEM (standard error of the mean). Unpaired t test: *** p < 0.0001. (D) Model of CENP-B enhanced centromere function through its interaction with CENP-C. Centromere function is supported by a dual mechanism for CENP-C recruitment and subsequent CENP-C-dependent nucleation of kinetochore assembly, mediated both by CENP-A and by CENP-B. See also Figure S4.
CENP-B depletion also reduced M18BP1 (a subunit of the Mis18 complex) binding at centromeres assembled with a CENP-C binding-deficient CENP-A variant (Fig. S4D), in agreement with a previously reported CENP-C/Mis18 interaction (Dambacher et al., 2012; Moree et al., 2011) and consistent with the CENP-B-dependent portion of CENP-A loading at centromeres mediated in part through the Mis18 complex.
DISCUSSION
CENP-A-containing chromatin has previously been demonstrated to be the epigenetic mark that is sufficient for long-term maintenance of human centromere positioning and identity and for nucleation of a functional kinetochore (Fachinetti et al., 2013). Adding to this, our current evidence has identified a parallel DNA sequence-specific role for direct stabilization of CENP-C and recruitment of CENP-A that is provided by CENP-B, the sole sequence-specific mammalian centromeric DNA binding protein identified so far (Muro et al., 1992; Stimpson and Sullivan, 2010). This discovery offers an explanation for the existence and retention of CENP-B boxes and the high degree of CENP-B conservation in divergent mammals (Sullivan and Glass, 1991).
An initial suggestion of a functional role at centromeres for CENP-B emerged from Masumoto and colleagues’ demonstration that CENP-B strongly enhanced de novo centromere formation in artificial chromosomes (Ikeno et al., 1998; Ohzeki, 2002; Okada et al., 2007; Okamoto et al., 2007), possibly by altering modifications to centromere-associated histone H3 (Okada et al., 2007), thereby affecting HJURP and CENP-A recognition/deposition (Bergmann et al., 2010). To these earlier proposals, our effort has established that repetitive alphoid DNA-containing centromeric DNA sequences to which CENP-B binds do provide a contribution for binding of centromeric CENP-A and CENP-C on native centromeres. Indeed, we have now shown that a CENP-A variant without its CENP-C binding domain (NH2H3CATD), that fully supports long-term centromere function despite inability to directly recruit CENP-C (Fachinetti et al., 2013), supports incorporation or stabilization of new CENP-A into centromeric chromatin in a CENP-B-dependent manner (Fig. 7C). This CENP-B-dependent loading/stabilization of CENP-A occurs via the recruitment of CENP-C to centromeres in a parallel pathway to direct CENP-C recruitment by the CENP-A carboxy-terminal tail (Carroll et al., 2010; Guse et al., 2011; Fachinetti et al., 2013; Kato et al., 2013). Our findings on this mutual dependency of CENP-A and CENP-C are in agreement with the demonstration that targeting of CENP-C to an ectopic chromosomal site is sufficient for CENP-A recruitment following removal of the endogenous centromere on that chromosome (Hori et al., 2013) and with the CENP-C-dependent recruitment of the methyl-transferase DNMT3B (Gopalakrishnan et al., 2009; Kim et al., 2012) or the Mis18 complex (Dambacher et al., 2012; Hori et al., 2013; Moree et al., 2011) which in turn is recognized by the CENP-A chaperone HJURP (Barnhart et al., 2011).
It is important to note that this CENP-B-dependent stabilization of centromeric CENP-A and CENP-C requires sequence-dependent DNA binding, as it is found only on centromeres carrying CENP-B boxes and indeed is absent from the Y centromere and neocentromeres. In addition, our findings that CENP-B does not play a role in initial CENP-C recruitment (Fig. S3) but stabilizes it once centromere bound reinforces the importance of a crucial epigenetically defined contribution to centromere function. CENP-A-containing chromatin is essential for the mutual reinforcement of binding of CENP/A/B/C binding at centromeres. Without it, the silenced centromere on the Neo4 chromosome cannot stabilize CENP-C despite a direct affinity for CENP-B bound to the CENP-B boxes in its α-satellite repeats.
The idea that CENP-B might play an important role in centromere function had previously been dismissed by demonstration that CENP-B null mice are viable (Hudson et al., 1998; Kapoor et al., 1998; Perez-Castro et al., 1998). In addition, the existence of human neocentromeres and the Y chromosome lacking CENP-B boxes had established that CENP-B is dispensable for human centromere function.
Nevertheless, we have shown here in both mouse and human cells that depletion of CENP-B yields a higher rate of chromosome mis-segregation for chromosomes with centromeres to which CENP-B had initially been bound (Fig. 3, 4). This observation offers at least a partial explanation for the reproductive and developmental dysfunctions reported in CENP-B null mice due to disruption in the normal morphogenesis of the highly mitotically active uterine epithelial tissue (Fowler et al., 2000; 2004). When combined with evidence that widespread somatic aneuploidy is surprisingly well tolerated in mice [e.g., in mice with reduced levels of the centromeric motor protein CENP-E (Weaver et al., 2007), the mitotic checkpoint protein Mad2 (Michel et al., 2001), or mitotic checkpoint kinase Bub1 (Jeganathan et al., 2007)], we predict that mice deficient in CENP-B develop similar somatic aneuploidy. While this was not tested by any of the three groups that had produced CENP-B null mice (none of which have been maintained) (Hudson et al., 1998; Kapoor et al., 1998; Perez-Castro et al., 1998), we have demonstrated that fibroblasts from those mice (Kapoor et al., 1998) are chromosomally unstable with a chronically elevated rate of mis-segregation (Fig. 4).
We have also demonstrated that two human chromosomes that lack DNA-containing CENP-B binding sites (the Y chromosome and the neocentromere) mis-segregate at higher frequency than do chromosomes with the alphoid DNA-containing, CENP-B-bound centromeres (Fig. 5, 6). [Note that an additional contributor for neocentromere mis-segregation may be inefficient Aurora B function, as previously proposed (Bassett et al., 2010)]. The reduced segregation fidelity, which had previously been reported for the Y chromosome to be associated with shorter survival and high risk of blood cancer in an age- and smoke-dependent manner (Dumanski et al., 2015; Forsberg et al., 2014; Nath et al., 1995), correlates with reduced CENP-C binding, which we argue is the probable cause of reduced centromere function. While our evidence has only tested CENP-B influence on mitotic chromosome segregation, if a similar contribution is also present in meiosis the selective pressure for maintaining males will act to maintain the Y chromosome despite increased mis-segregation frequency. Therefore, our findings strongly implicate CENP-B as a key contributor to centromere strength and the rate of faithful chromosome segregation through stabilization of binding of other centromere/kinetochore proteins.
Finally, our evidence for a direct contribution of CENP-B to CENP-C stabilization at centromeres together with the discovery of a direct interaction between CENP-A and CENP-B (via the CENP-A amino terminal tail) demonstrates a redundant, mutual dependency among CENP-A, CENP-B, and CENP-C for centromere association and function. Unresolved is whether individual CENP-C dimers bind simultaneously to both CENP-A and CENP-B. Thus, while human centromeres are specified primarily by an epigenetic mechanism mediated through CENP-A-containing chromatin, we have identified a sequence-dependent component to centromere function provided by the centromeric DNA binding protein CENP-B. We now propose that CENP-B does this by supporting a parallel pathway for CENP-A stabilization and kinetochore formation via CENP-C recruitment. This CENP-B-mediated enhancement in CENP-C binding at centromeres acts as a backup mechanism to ensure chromosome segregation when CENP-A levels are compromised (Fig. 7D). This might explain the observation that centromere function is still partially retained even in cells with as little as 1% of the original CENP-A level (Fachinetti et al., 2013).
Although other natural metoazoan centromeres had been identified to be without repetitive sequences including horses (Wade et al., 2009), chicken (Shang et al., 2010), and orangutans (Locke et al., 2011), the overall evidence implies that repetitive alphoid sequences can provide increased fidelity to human centromere function, consistent with the suggestion that the observed human neocentromeres are likely to be centromeres “in progress” (Marshall et al., 2008). Incorporation of repetitive sequences during evolution is therefore likely to have enhanced full centromere maturation and fixation, as proposed in primates (Ventura et al., 2007). Additionally, it is tantalizing to speculate that α-satellites and CENP-B play a role in the meiotic drive for egg specification (the preferential segregation of one chromosome over another) (Dawe and Henikoff, 2006; Henikoff et al., 2001), not only by supporting CENP-A binding (Chmátal et al., 2014; Marshall and Choo, 2012), but also by actively reinforcing centromere function.
EXPERIMENTAL PROCEDURES
siRNA, SNAP-tagging, clonogenic colony assay and Adeno-Cre treatment
siRNAs were introduced using Lipofectamine RNAiMax (Invitrogen). A pool of four siRNAs directed against CENP-C, CENP-B, HJURP, GAPDH (Fachinetti et al., 2013), CENP-A (GAGCACACACCUCUUGAUA, UUACAUGCAGGCCGAGUUA, AGACAAGGUUGGCUAAAGG, UAAAUUCACUCGUGGUGUG), were purchased from Dharmacon. SNAP labeling was conducted as described previously (Jansen et al., 2007). Clonogenic colony assays and Adeno-Cre treatment were done as described (Fachinetti et al., 2013).
CENP-B gene deletion by the CRISPR/Cas9 system
An expression vector (Addgene plasmid 42230) expressing cas9 and sgRNA (Cong et al., 2013) was digested with BbsI and the linearized vector was gel purified. A pair of complementary oligos to the targeting site (5′ GAAGAACAAGCGCGCCATCC 3′) were annealed and ligated into the vector. RPE-1 CENP-Af/− cells were co-transfected with the sgRNA expression vector and a GFP expressing vector (to identify transfected cells) by nucleofection (Lonza) using program T-020 and nucleofector solution V (Lonza). 48 hours post transfection, high GFP-expressing cells were single cell sorted into 96 well plates using a FACSAria II. Genomic DNA sequencing, immunostaining and immunoblotting were used to identify cells with disruption of both targeted alleles.
CENP-C gene targeting with TALENs
TALENs were assembled using the Golden Gate cloning strategy and library as described previously (Cermak et al., 2011) and cloned into a modified version of pcDNA3.1 (Invitrogen) containing also the Fok I endonuclease domain as previously described (Miller et al., 2011). TALENs were designed to the C-terminal region of CENP-C gene: GAGGAAAGTGTTCTTC and GGTTGATCTTTCATC. DLD-1 cells were co-transfected with the TALEN expression vectors and the donor cassette (containing the two homology arms for CENP-C C-terminal region and the AID and EYFP gene) by nucleofection (Lonza) using program T-020 and positive clones were selected by FACS.
Protein purification
GST tagged human CENP-A (1-29 or 1-44 aa) and 6xHis tagged CENP-B variants were expressed in Rosetta E. coli after induction with IPTG. GST-CENP-C variants were over-expressed in Sf9/High-Five insect cells using the Bac-to-Bac expression system (Invitrogen). Briefly, genes encoding GST tagged human CENP-C variants were cloned into pFastBac1. Bacmids were obtained from DH10Bac E.coli after transformation with the pFastBac1 containing CENP-C gene and used to transfect Sf9 cells (72 hr) to produce baculovirus. Hi-Five insect cells were infected by CENP-C baculovirus (1:10 dilution) for 48 hr, collected, and frozen until required. Cells were resuspended in PBS containing protease inhibitors and lysed by sonication. Soluble lysate was recovered after centrifugation and proteins were affinity purified over nickel-nitrilotriacetic acid sepharose beads (Qiagen) or glutathione sepharose beads (GE Healthcare Life Sciences).
In vitro binding assay
In vitro binding assays were conducted in 50 mM Tris-HCl pH 7.7, 100 mM KCl, 0.1% Triton X-100, 10% glycerol. For the GST pull-down assay, glutathione sepharose-bound GST-tagged protein and other recombinant proteins were combined and incubated at room temperature for 1 h. Bound protein complexes were washed four times with 20 volumes of the binding buffer, eluted from the beads in 15 mM glutathione buffer. For the His pull-down assay, nickel sepharose-bound 6xHis-tagged protein and other recombinant proteins were combined and incubated in the presence of 20 mM imidazole at 4°C for 1 h. Bound protein complexes were washed four times with 20 volumes of the binding buffer supplemented with 20 mM imidazole, and eluted from the beads in 200 mM imidazole buffer. Eluted proteins were analyzed by immunoblotting.
FISH experiment
Cells were fixed in Carnoy’s Fixative (Meth-Acetic Acid 3:1) for 15 minutes at room temperature, rinsed in 80% ethanol and air dried for 5 minutes. Probe mixtures (MetaSystems) were applied and sealed with a coverslip. Slides were denatured at 75°C for 2 minutes and incubated at 37°C overnight in a humidified chamber. Slides were washed with 0.4xSCC at 72°C for 2 minutes, 4xSCC, 0.1% Tween-20 at room temperature for 1 minute, and rinsed with ddH2O. Slides were incubated with DAPI solution for 10 minutes before mounting in anti-fade reagent.
For information regarding DNA constructs, cell culture conditions, generation of stable cell lines, the surveyor nuclease assay, antibodies and experimental conditions for immunoblotting and immunofluorescence, live-cell microscopy and procedures for centromere quantifications see Supplemental Experimental Procedures
Supplementary Material
Acknowledgments
The authors would like to thank P. Kalitsis (Murdoch Childrens Research Institute Victoria, Australia), A. Desai, R. Khaliullin, Neil Hattersley and C. Bartocci (Ludwig, La Jolla), F. Dibazar, Y. Arbely, B. Vitre and others members of the Cleveland lab for helpful suggestions. P. Maddox (University of North Carolina), I. Cheeseman (MIT, Boston), A. Desai (Ludwig, La Jolla), P. Kalitsis (Murdoch Children’s Research Institute Victoria, Australia), H. Masumoto (Kazusa DNA Research Institute, Japan) and B.E. Black (University of Pennsylvania, Philadelphia) for providing reagents. We also thank the Neuroscience Microscopy Shared Facility (P30 NS047101, University of California, San Diego) and the FACS facility in the Sanford Consortium for Regenerative Medicine, La Jolla, CA. This work was supported by a grant (R01-GM 074150) from the National Institutes of Health to D.W.C., who receives salary support from the Ludwig Institute for Cancer Research.
Footnotes
AUTHOR CONTRIBUTION
J. S. H. performed in vitro protein purification. D.F. and J.H. performed the binding assay. D.F. and M.A.M. performed gene-targeting of CENP-B and CENP-C. D.F. and P.L. performed FISH experiments. D.F., A.A. and A.J.W. performed all the remaining experiments. D.F. and D.W.C. conceived the experimental design, analyzed the data, and wrote the manuscript.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amor DJ, Bentley K, Ryan J, Perry J, Slater H, Choo KHA. Human centromere repositioning “in progress”. PNAS. 2004:6542–7. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PHJL, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. The Journal of Cell Biology. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, Wood S, Salimian KJ, Ajith S, Foltz DR, Black BE. Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. The Journal of Cell Biology. 2010;190:177–185. doi: 10.1083/jcb.201001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, guez MGOMRI, Martins NMC, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LET, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. The EMBO Journal. 2010;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Brock MA, Bédard S, Woods VL, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. PNAS. 2007a;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LET, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere Identity Maintained by Nucleosomes Assembled with Histone H3 Containing the CENP-A Targeting Domain. Molecular Cell. 2007b;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE. The quantitative architecture of centromeric chromatin. eLife. 2014;3:e02137. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. The Journal of Cell Biology. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Silva MCC, Godek KM, Jansen LET, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82–e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 2014;24:2295–2300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher S, Deng W, Hahn M, Sadic D, Fröhlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 2012;3:101–110. doi: 10.4161/nucl.18955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe RK, Henikoff S. Centromeres put epigenetics in the driver’s seat. Trends in Biochemical Sciences. 2006;31:662–669. doi: 10.1016/j.tibs.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Depinet TW, Zackowski JL, Earnshaw WC, Kaffe S, Sekhon GS, Stallard R, Sullivan BA, Vance GH, Van Dyke DL, Willard HF, et al. Characterization of Neo-Centromeres in Marker Chromosomes Lacking Detectable Alpha-satellite DNA. Human Molecular Genetics. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- Dumanski JP, Rasi C, Lönn M, Davies H, Ingelsson M, Giedraitis V, Lannfelt L, Magnusson PKE, Lindgren CM, Morris AP, et al. Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science. 2015;347:81–83. doi: 10.1126/science.1262092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP Is a Cell-Cycle-Dependent Maintenance and Deposition Factor of CENP-A at Centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie H, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Sullivan KF, Machlin PS, Cooke CA, Kaiser DA, Pollard TD, Rothfield NF, Cleveland DW. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. The Journal of Cell Biology. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K. Epigenetic control of centromere behavior. Annu Rev Genet. 2007;41:63–81. doi: 10.1146/annurev.genet.41.110306.130127. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LET, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 2013;15:1–13. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Bailey AO, Yates JR, III, Bassett EA, Wood S, Black Ben E, Cleveland DW. Centromere-Specific Assembly of CENP-A Nucleosomes Is Mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Black BE, Bailey AO, Yates JR, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Forsberg LA, Rasi C, Malmqvist N, Davies H, Pasupulati S, Pakalapati G, Sandgren J, Diaz de Ståhl T, Zaghlool A, Giedraitis V, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet. 2014;46:624–628. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KJ, Hudson DF, Salamonsen LA, Edmondson SR, Earle E, Sibson MC, Choo KH. Uterine dysfunction and genetic modifiers in centromere protein B-deficient mice. Genome Research. 2000;10:30–41. [PMC free article] [PubMed] [Google Scholar]
- Fowler KJ, Wong LH, Griffiths BK, Sibson MC, Reed S, Choo KHA. Centromere protein b-null mice display decreasing reproductive performance through successive generations of breeding due to diminishing endometrial glands. Reproduction. 2004;127:367–377. doi: 10.1530/rep.1.00102. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Earnshaw WC. The centromere: chromatin foundation for the kinetochore machinery. Developmental Cell. 2014;30:496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Pendon C, Morris J, Brown W. CENP-C is necessary but not sufficient to induce formation of a functional centromere. The EMBO Journal. 1999;18:4196–4209. doi: 10.1093/emboj/18.15.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Sullivan BA, Trazzi S, Valle, Della G, Robertson KD. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Human Molecular Genetics. 2009;18:3178–3193. doi: 10.1093/hmg/ddp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The Centromere Paradox: Stable Inheritance with Rapidly Evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Han JS, Cleveland DW. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. PNAS. 2012;109(49):E3350–E3357. doi: 10.1073/pnas.1216880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang WH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. The Journal of Cell Biology. 2013;200:45–60. doi: 10.1083/jcb.201210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Fukagawa T. Establishment of the vertebrate kinetochores. Chromosome Res. 2012;20:547–561. doi: 10.1007/s10577-012-9289-9. [DOI] [PubMed] [Google Scholar]
- Hudson DF, Fowler KJ, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford NG, de Kretser DM, Cancilla MR, et al. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. The Journal of Cell Biology. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. The Journal of Cell Biology. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. The Journal of Cell Biology. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, de Oca Luna RM, Liu G, Lozano G, Cummings C, Mancini M, Ouspenski I, Brinkley BR, May GS. The cenpB gene is not essential in mice. Chromosoma. 1998;107:570–576. doi: 10.1007/s004120050343. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends in Genetics. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A Conserved Mechanism for Centromeric Nucleosome Recognition by Centromere Protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IS, Lee M, Park KC, Jeon Y, Park JH, Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH, et al. Roles of Mis18α in epigenetic regulation of centromeric chromatin and CENP-A loading. Molecular Cell. 2012;46:260–273. doi: 10.1016/j.molcel.2012.03.021. [DOI] [PubMed] [Google Scholar]
- Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. The Journal of Cell Biology. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Choo KHA. Putative CENP-B paralogues are not present at mammalian centromeres. Chromosoma. 2012;121:169–179. doi: 10.1007/s00412-011-0348-3. [DOI] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KHA. Neocentromeres: New Insights into Centromere Structure, Disease Development, and Karyotype Evolution. The American Journal of Human Genetics. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P. Drosophila CENH3 Is Sufficient for Centromere Formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VVVS, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Research. 2014;24:697–707. doi: 10.1101/gr.159624.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. The Journal of Cell Biology. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T. Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. The Journal of Cell Biology. 1992;116:585–596. doi: 10.1083/jcb.116.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Almouzni G. A network of players in H3 histone variant deposition and maintenance at centromeres. Biochim Biophys Acta. 2014;1839:241–250. doi: 10.1016/j.bbagrm.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Nath J, Tucker JD, Hando JC. Y Chromosome aneuploidy, micronuclei, kinetochores and aging in men. Chromosoma. 1995;103:725–731. doi: 10.1007/BF00344234. [DOI] [PubMed] [Google Scholar]
- Ohzeki JI. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. The Journal of Cell Biology. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Ohzeki JI, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B Controls Centromere Formation Depending on the Chromatin Context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- Okamoto Y, Nakano M, Ohzeki JI, Larionov V, Masumoto H. A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. The EMBO Journal. 2007;26:1279–1291. doi: 10.1038/sj.emboj.7601584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. The Journal of Cell Biology. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Castro AV, Shamanski FL, Meneses JJ, Lovato TL, Vogel KG, Moyzis RK, Pedersen R. Centromeric protein B null mice are viable with no apparent abnormalities. Dev Biol. 1998;201:135–143. doi: 10.1006/dbio.1998.9005. [DOI] [PubMed] [Google Scholar]
- Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Sart du D, Cancilla MR, Earle E, Mao JI, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KH. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. Direct Binding of Cenp-C to the Mis12 Complex Joins the Inner and Outer Kinetochore. Current Biology. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang WH, Hori T, Toyoda A, Kato J, Popendorf K, Sakakibara Y, Fujiyama A, Fukagawa T. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Research. 2010;20:1219–1228. doi: 10.1101/gr.106245.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into Centromeric Chromatin Requires a Cooperative Array of Nucleosomal DNA Contact Sites. The Journal of Cell Biology. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. PNAS. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Sullivan BA. Epigenomics of centromere assembly and function. Current Opinion in Cell Biology. 2010;22:772–780. doi: 10.1016/j.ceb.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Sullivan KF, Glass CA. CENP-B is a highly conserved mammalian centromere protein with homology to the helix-loop-helix family of proteins. Chromosoma. 1991;100:360–370. doi: 10.1007/BF00337514. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Nakano M, Nozaki N, Egashira S-I, Okazaki T, Masumoto H. CENP-B interacts with CENP-C domains containing Mif2 regions responsible for centromere localization. J Biol Chem. 2004;279:5934–5946. doi: 10.1074/jbc.M306477200. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. PNAS. 2011;108:17974–17978. doi: 10.1073/pnas.1109720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M. Recurrent Sites for New Centromere Seeding. Genome Research. 2004;14:1696–1703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Antonacci F, Cardone MF, Stanyon R, D’Addabbo P, Cellamare A, Sprague LJ, Eichler EE, Archidiacono N, Rocchi M. Evolutionary formation of new centromeres in macaque. Science. 2007;316:243–246. doi: 10.1126/science.1140615. [DOI] [PubMed] [Google Scholar]
- Verdaasdonk JS, Bloom K. Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol. 2011;12:320–332. doi: 10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326:865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Current Biology. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.