Abstract
Background
Precise control of developmental and cell-specific expression of the brain-derived neurotrophic factor (BDNF) gene is essential for normal neuronal development and the diverse functions of BDNF in the adult organism. We previously showed that the zebrafish BDNF gene has multiple promoters. The complexity of the promoter structure and the mechanisms that mediate developmental and cell-specific expression are still incompletely understood.
Results
Comparison of pufferfish and zebrafish BDNF gene sequences as well as 5' RACE revealed three additional 5' exons and associated promoters. RT-PCR with exon-specific primers showed differential developmental and organ-specific expression. Two exons were detected in the embryo before transcription starts. Of the adult organs examined, the heart expressed a single 5' exon whereas the brain, liver and eyes expressed four of the seven 5' exons. Three of the seven 5' exons were not detectable by RT-PCR. Injection of promoter/GFP constructs into embryos revealed distinct expression patterns. The 3' flank profoundly affected expression in a position-dependent manner and a highly conserved sequence (HCS1) present in 5' exon 1c in a dehancer-like manner.
Conclusions
The zebrafish BDNF gene is as complex in its promoter structure and patterns of differential promoter expression as is its murine counterpart. The expression of two of the promoters appears to be regulated in a temporally and/or spatially highly circumscribed fashion. The 3' flank has a position-dependent effect on expression, either by affecting transcription termination or post-transcriptional steps. HCS1, a highly conserved sequence in 5' exon 1c, restricts expression to primary sensory neurons. The tools are now available for detailed genetic and molecular analyses of zebrafish BDNF gene expression.
Background
Lasting changes of synaptic transmission are believed to be an essential substrate for the formation and storage of long-term memory. Memory formation begins with sensory input that initially elicits fleeting short-term neuronal signals. These signals eventually result in long-lasting changes of synaptic transmission. There is now considerable evidence that these lasting changes require induction of neuronal gene expression [1]. The genes that are induced earliest, within minutes of the inducing stimulus, are known as immediate early genes (IEGs) [2]. A hallmark of these genes is that their induction does not require protein synthesis. The IEGs are a diverse group, and includes transcription factors, enzymes, structural proteins and growth factors. Their functions are equally diverse. The coordinated actions of these IEG products induce secondary genes and lead to incipient alterations of neuronal structural changes. Further downstream genes are then induced. The IEGs, secondary and downstream genes result in long-lasting changes of neuronal structure and function. The capacity of neurons to undergo these adaptive changes is an important component of neuronal plasticity [3].
A well-studied IEG is BDNF, a member of the neurotrophin family. BDNF has been shown to play important roles in memory, LTP, and neuronal plasticity [4-6]. These functions require tight yet dynamic regulation of neuronal BDNF expression. Regulation has been shown to occur at multiple levels. Neuronal BDNF gene transcription is dynamically regulated by excitatory signals that induce changes in Ca++ flux across the neuronal membrane via L-type VGCC [7-9]. Transcriptional regulation involves multiple alternative promoters whose transcriptional activity is differentially regulated in different neurons. BDNF mRNA is preferentially transported to dendritic spines in response to excitatory signals. BDNF mRNA encodes a BDNF precursor from whose COOH-terminus BDNF is liberated by proteolytic cleavage. The BDNF precursor is N-glycosylated [10,11]. A substantial portion of the precursor is secreted intact. The intact precursor is able to bind to the high affinity BDNF receptor, trkB, and activate intracellular signaling [11] as well as to p75, the low affinity neurotrophin receptor [12]. In the zebrafish only small amounts of mature BDNF are detectable by Western blotting [13]. BDNF can enter either a constitutive or a regulated pathway. In neurons BDNF can be secreted via the regulated pathway [14]. Synaptic secretion is regulated by neuronal activity [15].
The onset and cellular sites of BDNF production during development are determined at the transcriptional level [16,17]. Once induced, BDNF gene transcription is tightly regulated. BDNF production correlates with BDNF mRNA levels, suggesting that translation is not controlled [18]. Kainic acid induces BDNF gene transcription in the hippocampus without changing BDNF mRNA half-life [19]. Thus, the rate of BDNF gene transcription is a major determinant of BDNF production.
To study the transcriptional regulation of BDNF production we have focused on the zebrafish BDNF gene [13,20-22]. We recently showed that the zebrafish gene is more compact than the mammalian gene yet is more complex in its exon organization and primary transcript processing than the mammalian gene[20]. We identified three differentially regulated promoters so far. Transient expression of fusion genes driven by these cloned promoters showed that the 5' flanks are capable of cell-specific expression. We here identified three additional promoters and show that they are transcriptionally active in transiently transfected zebrafish embryos. We also show that not only the 5' flanks, but highly conserved sequences in the 5' non-coding exons and as yet unidentified sequences in the 3' flank control expression. One of the highly conserved sequences, HCS1, acts as a dehancer or enhancer depending on the type of 3' flank that is present. These results show that highly conserved regulatory regions regulate basal zebrafish BDNF gene transcription.
Results
Promoter structure
Identification of potential exons and associated promoters by sequence comparison
The pufferfish and zebrafish are both teleostii and are closely related. We therefore expected that the exon/intron organization of their BDNF genes would be very similar. Exons are generally more conserved than introns. It should therefore be possible to identify potential exons by comparing the nucleotide sequences of the zebrafish and pufferfish BDNF genes. The most highly conserved segments would likely represent exons. Identifying potential exons would allow us to screen a 5' RACE library more specifically and effectively.
A preliminary sequence of the pufferfish genome has been published. The coding exon of the pufferfish BDNF gene had been identified by sequence comparison with the human gene in scaffold_551 of the pufferfish genome sequence (Fugu Ensembl 6.1.1 [23]. The zebrafish genome is also being sequenced. A previously published sequence from the zebrafish genome group at the Sanger Institute, ctg10315, appears to contain the entire zebrafish BDNF transcription unit except the very 5' end.
To identify potential exons, the corresponding sequences in scaffold_551.1 and ctg10315 were compared using the 2-sequence BLAST alignment available at NCBI [24] The alignment revealed 12 segments of significant identity upstream from the coding region. Only 3 of these 12 segments represented known exons. The highly conserved segments HCS1 and HCS2, which are >93% identical in fish and man, were among these three. The remaining 9 segments did not correspond to known exons and therefore were likely to represent as yet undiscovered exons (figure 1).
Figure 1.
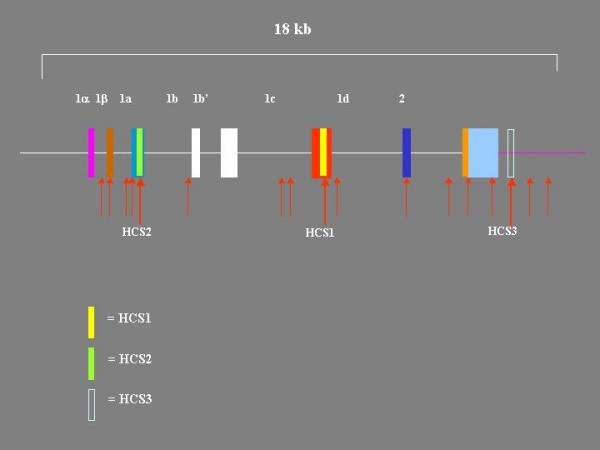
Regions of Zebrafish BDNF gene with high sequence similarity to Pufferfish BDNF gene. Regions of similarity were identified by aligning the zebrafish and pufferfish BDNF gene nucleotide sequences using BLAST. Regions of similarity identified by BLAST are marked with red arrows. Three regions were also highly similar with the mouse BDNF gene. These are marked HCS1, HCS2, and HCS3. The sequences of HCS1, HCS2, and HCS3 are available in reference [20].
5' RACE cloning of exons 1α, 1β, and 1d
To identify these potential exons and their associated promoters we carried out nested 5' RACE using total RNA from 5 and 7 days old larvae. We initially analyzed 47 independent clones from the 5d larvae and 59 from the 7d larvae. The collection of clones was screened for previously cloned exons by PvuII restriction enzyme analysis using size and digestion patterns as criteria. In this fashion 25/47 in the 5d group and 41/59 in the 7d group were identified as 1c.
Five clones from each age group that did not appear to be exon 1c by size and/or restriction enzyme analyses were sequenced. Four clones encoded exon 1c. All four had extensions at the 5' end that turned out to be one or more complete or partial copies of the synthetic oligoribonucleotide that is ligated to the 5' end of the decapped transcripts in the 5' RACE procedure. Interestingly, a BLAST search of the NCBI database with this sequence turned up numerous mRNAs whose 5' ends consist of complete or partial copies of this oligoribonucleotide. The results of one such search are shown in Table 1 [see Additional file 1]. Two clones contained 5' truncated versions of exon 1a. A single clone carried exon 1b/b'. One clone had a tiny insert flanked by the correct internal nested primers. The insert was another corrupted copy of the cloning oligoribonucleotide. Another similar clone had a 110 bp insert entirely unrelated to BDNF or the cloning oligoribonucleotide.
A single clone had an insert in which a novel 171 bp sequence was located upstream from the coding exon and linked to it at the known splice site. The sequence of the novel 5' fragment was compared with the available sequence of the genomic clone c241 using BESTFIT from the UWGCG sequence analysis software [25]. The comparison showed that the 3' end of the fragment is located 1381 bps upstream from exon 1a and is followed by a classical GTAAG splice donor site (see table 2). These sequence data and comparisons strongly suggest that the novel 5' fragment represents a 5' non-coding exon. This possibility is further strengthened by the presence of TATA-like sequences between -20 and -30 of its 5' end (see table 2). We named this exon 1α. The location of exon 1α in the transcription unit is equivalent to that of rat and mouse exon 1 (see Figure 2). However, sequence comparison by BLAST reveals no significant similarity to the mammalian exon.
Table 2.
5' Flanks, intron-exon and exon-intron junctions of zebrafish BDNF gene. 5' flanks and introns are shown in lower case letters and exons in upper case letters. Splice acceptors are shown underlined and splice donor sites in bold.
| Exon | Sequence |
| 1α | ...tgctctgatggaattttactgataacagaaacggtcgtactGGATTTCGT....ACAGATgtaagta.... |
| 1β | aacttcttaatggtcATTCATT....GGAAAATGgtaagttg.... |
| 1a | ...tatgatttcAGCACTT....TTATAAAGgtgagaa... |
| 1b | ...ccctcccctccGTTCTGA....AGCTTGgtaggtttt... |
| 1b' | ...acagTTTGA....CTTGGgtatgtg... |
| 1c | ...gaggaatgaagccgtcATTTCAA....TAAAGgtaggtt... |
| 1d | ...tttcagttattgccaactCAGTCC....TTGTTgtgctctt |
| 2 | ...tgctgtctccagTTCCAACA... |
Figure 2.
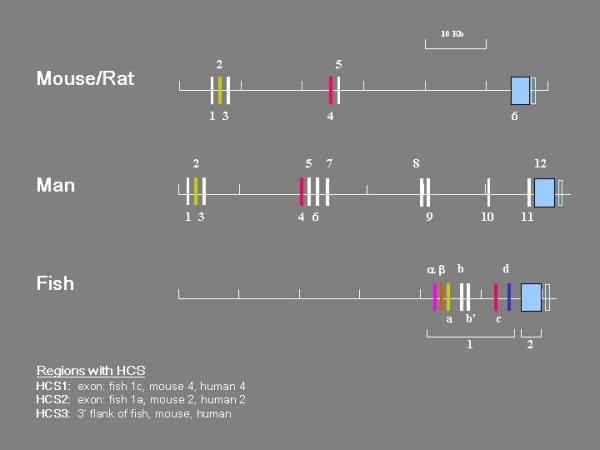
Organization of BDNF gene in mouse, man and fish. The boxes represent exons and the lines introns, 5' flanks and 3' flank. Exons containing highly conserved sequences (HCS) have the same colors. The positions of the mouse BDNF gene exons were taken from sequence posted in the NCBI databank, accession AY057907. The rat exons were published by Timmusk et al. [47] and Bishop et al. [48]. The sequences posted in the NCBI databank (Accession numbers S76799, S76760, S76759, S76758, S76757, X67107, X67106, X67108) were compared with the mouse and found to be >98% identical. Their positions in the rat gene were assumed to be identical to those of the mouse exons. The positions of the human BDNF gene exons were taken from the sequence posted in the NCBI databank, accession number AF411339. The pufferfish BDNF gene sequence was taken from the Ensembl Genome Data Resource of the Wellcome Trust Sanger Centre http://www.sanger.ac.uk/), scaffold_551 and scaffold_1408. Exons 1α, β, and a were mapped by comparison of the 5'RACE clone sequences with the nucleotide sequence of c241 established in this laboratory [20,21]. Exons 1b, b', 1c, and 2 of the zebrafish BDNF gene were mapped on the sequence of clone CH211-251J8, posted in the NCBI databank, accession number AL935207. Zebrafish and pufferfish gene exon sequences were >75% identical including presumptive exon 1d.
The 7d library was screened a second time as described above. A closer analysis of the PvuII digestion patterns indicated that the exon 1c pattern could be mimicked by exon 1b/b' and potentially other exons. The clones with exon 1c patterns were therefore reanalyzed with additional enzymes. These analyses identified a clone with a novel insert. The insert mapped between exons 1α and 1a and was named exon 1β. Exon 1β is spliced directly to the coding exon. In the genomic sequence it is followed by a classical GTAAG splice donor site (table 2). Exon 1β contains two of the 12 upstream segments conserved between zebrafish and pufferfish BDNF genes.
The region between exon 1c and the coding exon contains three conserved segments. One of them is the sixth largest and exhibits the characteristics of an exon (table 2). We named this segment putative exon 1d. Despite analysis of over 300 clones by restriction enzyme digestion and/or nucleotide sequencing we did not find any clones that encoded this putative exon. The 5d and 7d libraries were therefore analyzed by PCR. DNA was extracted from a pool of each library obtained by eluting colonies from plates representing a total of 5000 clones. The primers were positioned at the 5' and 3' ends of the conserved segment. Both libraries contained amplifiable template that yielded product of the expected size. Aliquots of the amplified DNA from each library were cut with HinfI and the digests analyzed by agarose gel electrophoresis. The resulting fragments were of the predicted size. An aliquot of the DNA amplified from the 7d library pool was sequenced. The sequence indicated that the amplified DNA represents exon 1d. We therefore concluded that exon 1d exists. A screen of 1500 clones of the 7d library by colony hybridization has so far been unsuccessful in finding an exon 1d clone. The precise 5' and 3' ends of exon 1d are therefore still unknown.
These results show that the zebrafish BDNF gene has at least seven 5' non-coding exons (Figure 2) that give rise to at least 6 distinct transcripts (Figure 3).
Figure 3.
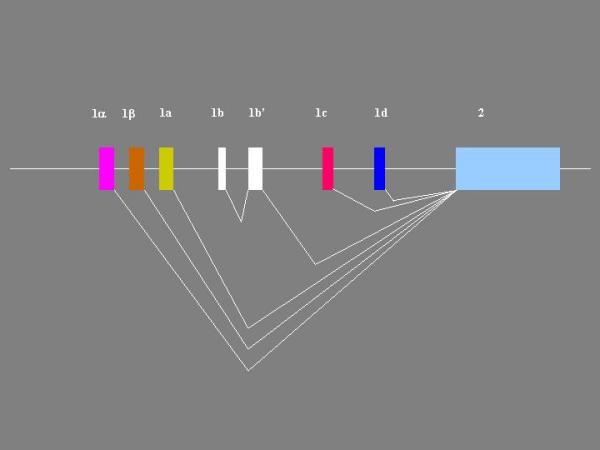
Splicing of Zebrafish BDNF exons into mature transcripts The boxes are exons and the horizontal line represents 5' flanks, introns, and 3' flank. Angled lines to indicate splicing into mature transcripts connect the exons. The depicted combinations were identified by 5' RACE and cDNA cloning. Additional combinations may exist.
Conserved regions in the 3' flank
The BLAST comparison of the zebrafish and pufferfish BDNF genes also revealed four segments of high similarity in the 3' flank. One of these contains HCS3, a segment in which 39 of 41 nucleotides are identical in fish and man.
The sequence similarities abruptly end 1.3 kb downstream from the 3' end of the coding exon. A BLAST search with a 1 kb fragment following the discontinuity was carried out. The search identified 100 distinct zebrafish genomic clones that contain a 280 bp segment that is closely related to a sequence in this 1 kb fragment. Its frequent occurrence and its lack of similarity with the pufferfish genomic sequence suggests it represents a zebrafish-specific repetitive sequence element.
A similar discontinuity between zebrafish and pufferfish sequences was found 2 kb upstream from exon 1α. A BLAST search with a 1.5 kb fragment upstream from the discontinuity revealed a 1.2 kb region within this fragment that is >88% identical with the Tfz.41 transposable element [26]. The orientation of the Tfz.41-like sequence is opposite to that of the BDNF gene. The transposon-like region does not contain any ORFs greater that 20 amino acids.
Developmental and tissue-specific expression of 5' exons
We had previously shown that exon 1c is present in the early embryo before transcription starts at the mid-blastula transition (MBT), indicating that it is part of a maternal transcript. In situ hybridization at subsequent stages revealed that by the larval stage BDNF is expressed in a cell-specific fashion. The common exon was expressed in olfactory epithelium and taste buds, but exon 1c was not. These results suggested that multiple promoters exist and that they are expressed tissue-specifically.
The present as well as previously published results [20-22] show that the zebrafish BDNF gene does indeed possess multiple promoters and 5' exons. Using knowledge of the nucleotide sequences of the 5' exons we performed RT-PCR to study their developmental and organ-specific expression.
Total RNA was extracted from embryos at different developmental stages and various organs of adult females and males. The quantity and quality of the RNA samples were assessed by agarose gel electrophoresis (Figure 4). The RNA was reverse transcribed into cDNA and cDNA quality assessed by PCR using β-actin-specific primers (Figure 5). The cDNA was then examined for reverse copies of transcripts in which any of the 5' exons is spliced to the common coding exon using exon-specific primers in a nested 2-round PCR strategy.
Figure 4.
Agarose Gel Electrophoresis of total RNA from embryos, larvae and adult Zebrafish. Total RNA was extracted as described in methods and subjected to electrophoresis through a native 1% agarose gel. The amplification products were stained with ethidium bromide and photographed under UV light using a digital camera. PMBT = pre-midblastula transition. 80% E = 80% epiboly. 16 Som = 16 somite stage. 5d = 5 days post-dertilization. The two major ribosomal RNA bands are marked in the left hand margin.
Figure 5.
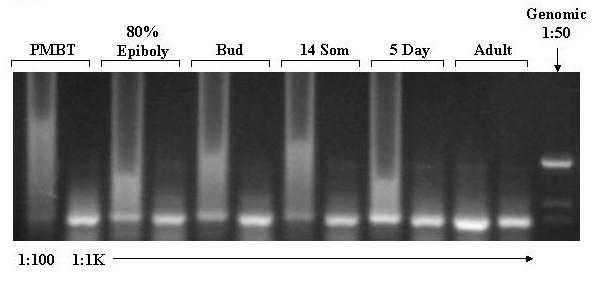
Agarose Gel Electrophoresis of products of amplification of cDNA with β-Actin primers. cDNA was prepared from aliquots of total RNA samples extracted from embryos, larvae and adult zebrafish as described in methods. Aliquots of the cDNA were diluted 1:100 and 1:1000 prior to amplification with β-actin specific primers. The actin primers span three introns. Genomic zebrafish DNA, diluted 1:50 to approximately 100 ng/μl, was also amplified to confirm that the amplification products are derived from cDNA. The primer sequences are provided in the method section. The amplification products were subjected to electrophoresis through a 1.2% agarose gel, stained with ethidium bromide, and photographed under UV light using a digital camera. PMBT = pre-midblastula transition. 16 Som = 16 somite stage. 5d = 5 days post-dertilization. The dilution factors are indicated at the bottom of the panel. The large amount of amplification product in the 1:100 samples led to smearing and "rabbit-earing".
The results in Figure 6 show that, as previously reported, exon 1c is present in embryonic RNA prior to MBT, i.e. before transcription of embryonic genes starts. Exon 1α is also present in the pre-MBT embryo. The other 4 exons are not. By 80% epiboly, exons 1α, β, a, and c are expressed. This expression pattern was seen at all subsequent stages that were examined including 6 somites, 5 days, and the adult (Figure 7). Exons 1b/b' and 1d were not detected at any of the developmental stages that were examined. These results are summarized in table 3.
Figure 6.
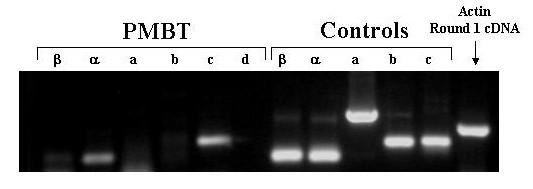
Agarose Gel Electrophoresis of products of amplification of PreMBT cDNA with exon-specific primers. cDNA was prepared from total RNA extracted from pre-MBT embryos. Aliquots of the cDNA samples were ampified in 2 rounds of PCR with the indicated exon-specific nested primers and with β-actin specific primers. Aliquots of 5'RACE clone plasmid DNA were also amplified. The amplification products were subjected to electrophoresis through a 1.5% agarose gel, stained with ethidium bromide, and photographed under UV light using a digital camera. PMBT = pre-midblastula transition.
Figure 7.
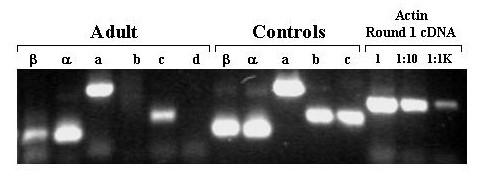
Agarose Gel Electrophoresis of products of amplification of PreMBT cDNA with exon-specific primers. cDNA was prepared from total RNA extracted from adult zebrafish. Aliquots of the cDNA were amplified in 2 rounds of PCR with the indicated exon-specific nested primers and with β-actin specific primers. The cDNA was diluted as indicated prior to amplification with the β-actin primers. Aliquots of 5'RACE clone plasmid DNA corresponding to each exon (except exon 1d for which c241 served as template) were also amplified. The amplification products were subjected to electrophoresis through a 1.5% agarose gel, stained with ethidium bromide, and photographed under UV light using a digital camera.
Table 3.
Expression of 5' non-coding exons at various developmental stages. PMBT = pre-midblastula transition. ND = not done.
| Exon | PMBT | 80% Epiboly | Bud | 14 Somite | 5Day | Adult |
| α | X | X | X | X | X | X |
| β | X | X | X | X | X | |
| a | - | ND | X | X | X | X |
| b/b' | - | - | - | - | - | - |
| c | X | X | X | X | X | X |
| d | - | - | - | - | - | - |
The expression of 5' exons before the onset of transcription was also confirmed using 5' RACE using total RNA from embryos at the 2–8 cell stage. Of 11 5' RACE clones examined by PvuII restriction enzyme analysis 5 had the pattern of exon 1c (Figure 8). The identity of one of these clones as 1c was confirmed by nucleotide sequencing. The remaining clones had fragment patterns consistent with exons 1α.
Figure 8.
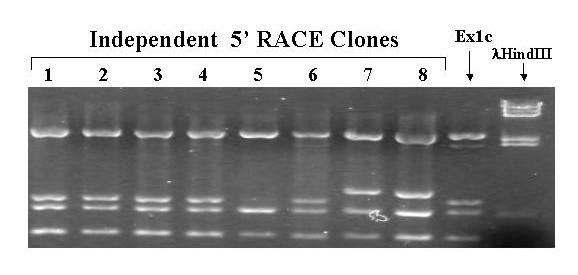
PvuII Restriction Enzyme Digests of 5'RACE Clones From Pre-MBT Embryos. 5' RACE was performed using total RNA extracted from pre-midblastula transition embryos as described in methods. DNA extracted from individual clones was digested with PvuII. A previously identified 5' RACE clone from 10 days old larvae was also digested as a control. Phage λ DNA digested with HindIII was used as a size marker. The digests were subjected to electrophoresis through a 1.5% agarose gel, stained with ethidium bromide, and photographed under UV light using a digital camera.
Of the 4 exons expressed in the adult, the three exons 1α, β, and c were expressed organ-specifically, whereas exon 1a was expressed in all organs examined. Exons 1 b/b' and d were not detected in any of the organs examined including liver, muscle, eye, intestine, heart, brain, and skin. A representative RT-PCR agarose gel electrophoresis is shown in Figure 9. The results for all organs are summarized in table 4.
Figure 9.
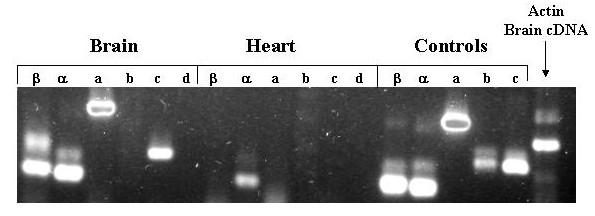
Agarose Gel Electrophoresis of products of amplification of adult organ cDNA with exon-specific primers. Total RNA was extracted from pooled adult zebrafish brains and hearts as described in methods. Aliquots of the RNA were converted to cDNA and then amplified with the indicated exon-specific and β-actin primers. Aliquots of 5'RACE clone plasmid DNA corresponding to each exon (except exon 1d for which c241 served as template) were also amplified. The amplification products were subjected to electrophoresis through a 1.5% agarose gel, stained with ethidium bromide, and photographed under UV light using a digital camera.
Table 4.
Expression of 5' non-coding exons in various organs of adult zebrafish
| Exon | Muscle | Skin | Eyes | Liver | Brain | Heart | Intestine | Gills |
| α | X | X | X | X | X | X | X | X |
| β | X | - | X | X | X | - | - | - |
| a | - | X | X | X | X | - | - | - |
| b/b' | - | - | - | - | - | - | - | - |
| c | - | X | X | X | X | - | X | X |
| d | - | - | - | - | - | - | - | - |
These RT-PCR analyses show that the multiple promoters of the zebrafish BDNF gene are expressed in a developmental and organ-specific fashion.
Promoter activities in transiently transgenic Zebrafish embryos
Promoter 1d
We previously showed that the promoters preceding exons 1a, 1b, and 1c are transcriptionally active in transiently transfected embryos. To determine whether promoter 1d was active, a HindIII fragment of genomic clone c241 that contains exon 1d plus surrounding 5' flank and intron was linked to the EGFP-F reporter. To provide a splice acceptor site, a 300 bp fragment spanning the splice acceptor site preceding the coding exon was placed ahead of the reporter (Figure 10). This construct was called Dex1d. Note that Dex1d contains the 3' UT and polyA site of the SV40 early gene.
Figure 10.
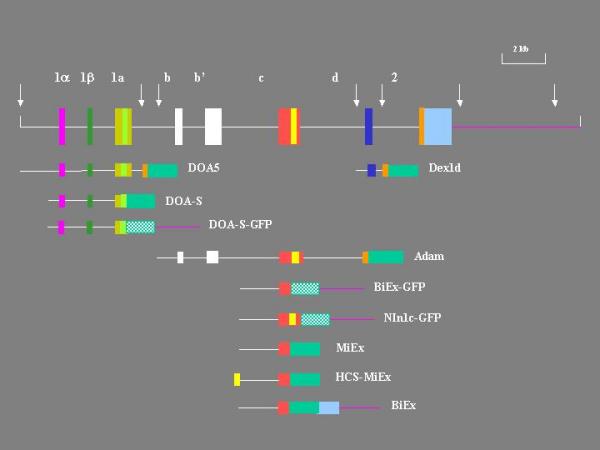
Reporter constructs. The constructs are shown below the gene diagram to indicate their relative positions on the gene. The 5' and 3' flanks are lines and exons and reporters are boxes. The solid green box represents the EGFP-F and the cross-hatched green box the GFP reporter. The yellow bar in exon 1c represents HCS1 and the light green bar in exon 1a HCS2. The white arrows indicate HindIII sites.
The Dex1d construct consistently expressed in a large neuron in the hindbrain situated medially to the otic capsule. The expressing neuron has a large cell body with a small number of large dendrites and a single axon that projects down almost the entire length of the spinal cord in a ventrally located pathway. Along its projection it has numerous small spurs. The location, size, and projection of this neuron suggest it represents a Mauthner cell (Figure 11). The Dex1d construct is also expressed in more rostrally located smaller neurons with a similar pattern of dendrites and projections. Rare expression was also seen in epithelial and notochord cells. These data suggest that the HindIII fragment contains an active promoter.
Figure 11.
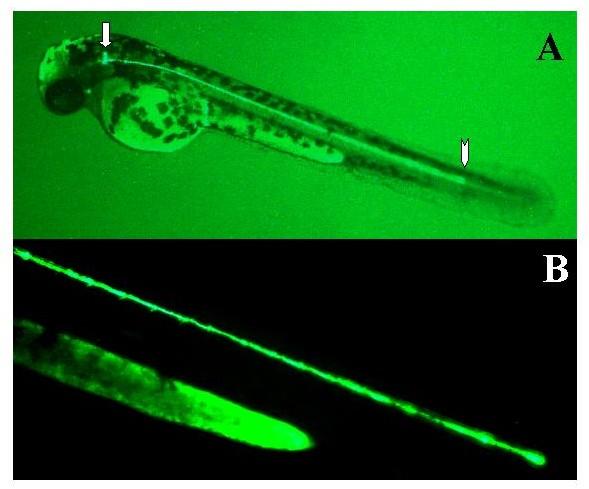
Expression of Dex1d in large hindbrain neuron. Embryos were injected with the Dex1d construct as described in methods. Two days later expression of the reporter was visualized in live embryos using a fluorescent microscope. Expressing embryos were photographed using a digital camera. A single whole embryo is shown in panel A. The expressing neuronal cell body is marked with an arrow with a head. The end of the axon projecting down the spinal cord is marked with a straight arrow. Mag = 2×. Panel B shows an enlargement of the axon. Mag = 10×.
Promoter 1c
We previously reported expression of this promoter [20,21]. The 5' RACE cloning data suggested that this promoter is the most widely expressed and/or transcriptionally most active promoter. In the construct we previously expressed, called MiniExpress (MiEx), exon 1c is directly spliced to the EGFP-F reporter at a BamHI site. The reporter is followed by the 3' UT and polyA site of the SV40 early gene. MiEx contains 1.8 kb of 5' flank (Figure 10). MiEx was expressed in notochord, skeletal and smooth muscle, and epithelial and endothelial cells.
There was no significant expression in neurons. This was surprising because in the mammalian gene this promoter is most actively expressed and regulated in neurons. This situation suggested that MiEx lacked regulatory elements required for neuronal expression, or that the SV40 sequences at the 3' end lead to ectopic expression. Indeed, the 5' end of exon 1c that is contained in MiEx stops short of a highly conserved sequence, which we named HCS1, and whose presence suggested that exon 1c is the ortholog of rat exon 3. To check if this sequence might affect expression, a construct that contains a SapI fragment which extends beyond HCS1 was made, and called Sappie. Sappie was expressed exclusively in Rohon-Beard cells. These are large primary sensory neurons located in the dorsoventral spinal cord. Sappie was missing 730 bps at the 5' end compared with MiEx. To exclude the possibility that this piece contained regulatory sequences required for expression in non-neuronal cells, Sappie was extended at the 5' end to the BamHI site of MiEx to generate DOS. DOS also expressed only in Rohon-Beard cells.
These results suggested that the restricted expression of Sappie and DOS was due to the presence of the HCS1. HCS1 is located in the 5' UT of the fusion gene transcript, and thus could have an effect on RNA stability and/or translation. Alternatively, it could act as a transcriptional regulatory element. To distinguish between these possibilities, HCS1 was placed upstream of the BamHI fragment of MiEx using synthetic oligonucleotides. The resulting construct was called HCS-MiEx (Figure 10). In contrast to its parent, HCS-MiEx was exclusively expressed in Rohon-Beard cells (Figure 12). This result suggested that HCS1 regulates transcription. Moreover, it appeared to have the properties of a dehancer. This possibility was supported further by the fact that HCS1 in the opposite orientation also restricted expression to Rohon-Beard cells.
Figure 12.
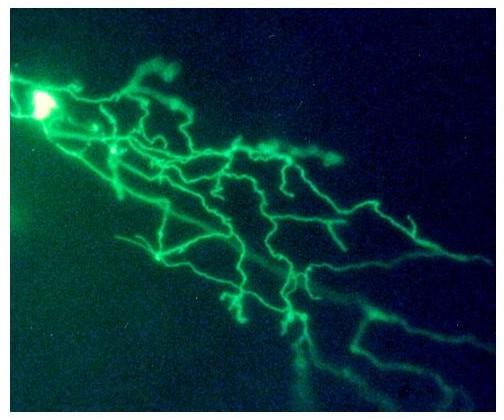
Expression of HCS-MiEx in Rohon-Beard neuron. Embryos were injected with the HCS-MiEx construct as described in methods. Two days later expression of the reporter was visualized in live embryos using a fluorescent microscope. Expressing embryos were photographed using a digital camera. The image shows a Rohon-Beard neuron. The cell body is overexposed to show the widely ramifying peripheral axon. Mag = 30×.
Promoter 1α, 1β and a
We previously tested a construct named DOA5 [20]. DOA5 was constructed from a 5.3 kb HindIII fragment of genomic clone c241 [21]. The present 5' RACE cloning revealed that this fragment contains not only exon 1a but also the newly identified exons 1α and 1β (see Figure 2). In the DOA5 construct there are consequently at least three 5' non-coding exons and their associated promoters. DOA5 was constructed with a splice acceptor site upstream from the reporter. Exon 1a, which is downstream from exon 1 α, is located near the 3' end of the HindIII fragment and is followed by 140 bps of intron. This arrangement of exons and reporter should allow correct splicing of transcripts derived from all three exons. Thus, expression of DOA5 should reflect the activities of all three promoters.
In previous experiments DOA5 exhibited an unusual pattern of expression. The first expression was noted in the pre-somite embryo. The reporter stained primarily yolk platelets rather than cells. No expression was seen in cells up to the 4 days old larval stage [20]. These data suggest that critical regulatory elements are missing from DOA5.
Adam
Our data show that the BDNF gene has multiple distinct promoters. The pattern of expression in the organs we examined suggests that each promoter is independently regulated. However, regulatory elements physically associated with one promoter may need to act on adjacent or even more distantly located promoters. To examine whether there may be such interactions between 1b/b' and 1c we constructed Adam. Adam carries a 9 kb HindIII fragment that contains the known exons 1b, b' and c. Adam uses the EGFP-F reporter and has the SV40 early gene 3' UT and polyA sites.
Adam expressed in a large variety of cells (Figures 13 and 14). We consistently observed expression in primary sensory neurons, spinal cord neurons including motor neurons, epithelial cells, blood cells, skeletal and smooth muscle cells, otic hair cells, otolith, notochord, and as yet unidentified retinal cells.
Figure 13.
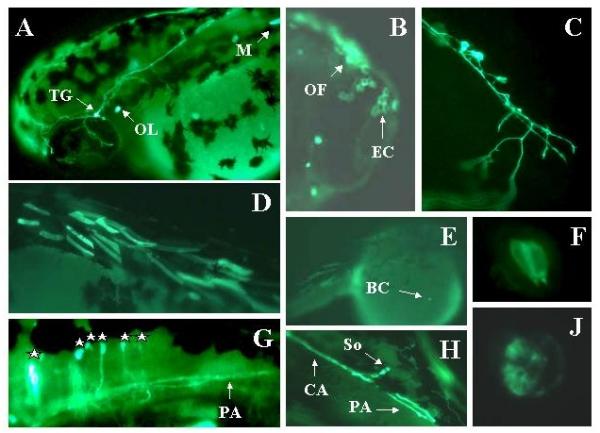
Expression of Adam in live embryos. Embryos were injected with the Adam construct as described in methods. Two days later expression of the reporter was visualized in live embryos using a fluorescent microscope. Expressing embryos were photographed using a digital camera. Panel A: TG = trigeminal. OL = otolith. M = skeletal muscle. Mag = 2×. Panel B: OL = olfactory epithelial cell. EC = epithelial cells. Mag = 30×. Panel C: Rohon-Beard neuron. Mag = 30×. Panel D: skeletal muscle cells. Mag = 2×. Panel E: BC = blood cell temporarily arrested over yolk. Mag = 2×. Panel F: Hair cells in the ear. Note pear-like shape with hair pointing toward lumen of otic capsule (bottom of panel). Mag = 60×. Panel G: Hindbrain neurons. Each cell body is marked with a star. The forebrain is to the left. The most rostral cell body is overexposed. PA = peripheral axon projecting down the spinal cord. Mag = 30×. Panel H: hindbrain neuron with central (CA) and peripheral (PA) axons. Mag = 30×. Panel J: Otolith close-up. Mag = 60×.
Figure 14.
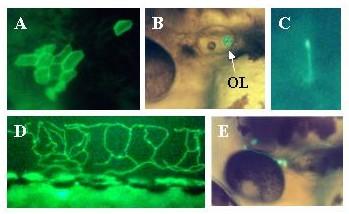
Expression of Adam in live embryos. Embryos were injected with the Adam construct as described in methods. Two days later expression of the reporter was visualized in live embryos using a fluorescent microscope. Expressing embryos were photographed using a digital camera. Panel A: Epithelial cells. Mag = 10×. Panel B: Otolith. Mag = 10×. Panel C: Motor neuron. Mag = 30×. Panel D: Rohon-Beard neuron arborizing among epithelial cells of dorsal fin. Mag = 10×. Panel E: TG = Trigeminal neuron. Mag = 10×.
We had previously examined expression of promoter 1b separately. Promoter 1b was exclusively expressed in trunk skeletal myocytes [20]. The spectrum of cells that express Adam is thus much larger than the sum of the sets in which promoters 1b and 1c express when the two promoters are not physically linked.
Effect of 3' flank on promoter 1c activity
Transgenic mouse experiments reported by Timmusk et al [27] showed that not only the 5' flanks, but also the 3' flanks were required for cell-specific and regulated expression. Moreover, the 3' UT and flank are highly conserved. HCS3, a 41 nts segment that is >95% identical in fish and man, is located in the 5' portion of the 3' UT, 63 nts downstream from the translation stop codon [20]. Three additional segments that are highly conserved between zebrafish and pufferfish lie in the next 740 nts. The high degree of conservation together with the experimental evidence reported by Timmusk et al. suggests that the 3' UT and flank have important regulatory functions.
Since MiEx had a repertoire of expression that was somewhat unexpected, and this may have been in part due to the presence of SV40 sequences at the 3' end of the reporter, these sequences were replaced with a 1.3 kb SmaI/HindIII fragment of c241 that begins in the propeptide region of the coding exon and extends 800 bps beyond the stop codon. This includes the entire 3' UT and 600 bps of 3' flank. This construct was called BiEx. Expression was seen exclusively in Rohon-Beard cells. Thus, the 3' flank has a profound effect on expression. To determine if this effect is position-dependent the same SmaI/HindIII fragment was placed downstream from the 3' flank of MiEx to generate ME41-9. The expression of ME41-9 was indistinguishable from that of MiEx. Thus, its effect is position-dependent. Although it does not prove it, position-dependence suggests it acts at the level of transcription termination and/or mRNA stability.
Since BiEx contained a significant fragment of the coding exon, and extended only 600 bps into the 3' flank, we made a derivative of MiEx in which the reporter is followed by a 2 kb fragment of 3' flank that begins at the stop codon of the coding exon. In addition the EGFP-F reporter was replaced with hrGFP (Stratagene, La Jolla, CA, USA), a potentially less cytotoxic but also less sensitive reporter. This construct was named BiEx-GFP. We could not detect any expression of this construct. A trivial explanation was the lower sensitivity of the reporter. Another possibility was the absence of HCS1. To test this possibility a derivative, called NIn1c-GFP was constructed, in which exon 1c extends further 3' and thus includes HCS1. NIn1c-GFP was predominantly expressed in primary sensory neurons, i.e. trigeminal and Rohon-Beard cells (Figure 15). These results indicate that the hrGFP reporter is adequately sensitive and that HCS1 acts as an enhancer in the context of the larger 3' flank. Interestingly, NIn1c-GFP expressed in a larger set of cells than BiEx.
Figure 15.
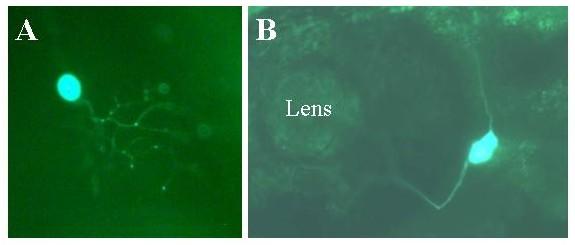
Expression of NIn1c-GFP in live embryos. Embryos were injected with the Nin1c-GFP construct as described in methods. Two days later expression of the reporter was visualized in live embryos using a fluorescent microscope. Expressing embryos were photographed using a digital camera. Panel A: Rohon-Beard neuron. Note that the axons are only weakly fluorescent with the GFP reporter compared with the EGFP-F reporter (for example figures 10 and 11). Mag = 30×. Panel B: Trigeminal neuron. Mag = 30×.
Discussion
Our results reveal a high degree of structural and regulatory complexity of the zebrafish BDNF gene. We have so far identified five distinct promoters by cloning, and a sixth promoter by sequence similarity and transient expression. We also cloned a mature BDNF message with three exons indicating that the primary transcript of the zebrafish BDNF gene undergoes more complex splicing than its mammalian counterpart. The sites of expression and the functional significance of this tripartite transcript remain to be determined.
Transient expression in embryos and larvae showed that a highly conserved sequence, HCS1, in the 5' non-coding exon 1c has a profound effect on expression. The specific effect appears to be dependent on the context. In particular, in the MiEx context HCS1 restricted expression to Rohon-Beard cells. MiEx expressed in a variety of non-neuronal cell types without HCS1. In the MiEx context the restricting effect was independent of the position and orientation of HCS1. By these criteria HCS1 qualifies as a dehancer.
On the other hand, BiEx-GFP expression was below the level of detection until HCS1 was added to create NIn1c-GFP (Figure 10). Nin1c was consistently expressed in trigeminal neurons and Rohon-Beard cells. Thus, in this context HCS1 appears to have an enhancing effect. BiEx-GFP and MiEx differ in the reporter and the 3' UT and flank. It is unlikely that the difference in the reporter accounts for the difference in the effect of HCS1. A more likely explanation is the difference in 3' UTs and flanks. If this were so it would suggest that HCS1 and sequences in the 3' UTs and flank interact. Interestingly, when the SV40-derived 3'UT of MiEx was replaced with the corresponding BDNF region to create BiEx expression was diminished. This effect was position-dependent. It remains to be seen whether the modification of the effect of HCS1 by the 3' UT and flank depends on position or not. We also do not yet know whether the enhancing effect of HCS1 in the BiEx-GFP context, is independent of position and/or orientation, i.e. enhancer-like, as it is in the MiEx context.
We expressed HCS1 in a third context. Adam contains exon 1c in its entirety including HCS1. In contrast to HCS-MiEx, BiEx and BiEx-GFP, Adam also contains 2.8 kb of downstream intron (Figure 10). Adam also contains 6.3 kb of 5' flank of exon 1c, and, embedded in the flank, exons 1b and b'. The presence of a splice acceptor in front of the EGFP-F reporter in Adam presumably allows splicing of these exons to the reporter. Note that Adam has the same 3' UT and flank as HCS-MiEx and MiEx. Adam was expressed in a wide range of cells. Clearly, in this context HCS1 did not restrict expression to Rohon-Beard cells. Obviously Adam also contains promoter 1b. Promoter 1b with 1.8 kb of 5' flank was strictly expressed in skeletal muscle. Even if myocyte expression is attributed to promoter 1b, the remaining set of cells is diverse and far greater that the MiEx and BiEx sets. This fact clearly indicates that HCS1 in the Adam context does not have a restrictive effect.
The set of cells that express Adam is not merely the sum of the cells expressing promoters 1b and 1c. Several explanations are possible. Perhaps the presence of a splice acceptor leads to more efficient expression in cells that would otherwise express below the level of detection. Another possibility is that the region downstream from exon 1c, i.e. intron 1c, contains regulatory elements that are responsible for the expansion of the observed expression. These elements must be downstream from exon 1c because a construct in which the 5' flank of MiEx was extended to the same 5' point as it extends in Adam, a construct we called MaxiEx, was expressed identically to MiEx. Some of these elements may be located in a 43 nts stretch 147 nts downstream from exon 1c that is conserved between zebrafish and pufferfish (see Figure 1). The next conserved region is exon 1d. Thus, the third explanation, namely that the intron following exon 1c contains another promoter and its associated 5' exon, although possible is quite unlikely.
In any event, in the MiEx context HCS1 does act as a dehancer. This raises the question whether HCS1 might bind transcription factors. A sequence analysis using TESS [28] suggests that in fact there are several potential transcription factor binding sites in HCS1 (table 5). Potential binding sites for Pbx1, Cdx, CHOP, Gata, HSF and AML-1a were found. AML-1, also known as Runx1, contains a runt type of DNA binding domain [29]. Runx1 must form a dimer with CBFβ to recognize and stably bind to its DNA binding site. A runx1 gene was cloned from the Zebrafish [30]. It is expressed in the developing blood system and, most interestingly, in a subset of Rohon-Beard cells [31,32]. Antagonizing runx1 mRNA with morpholino antisense oligonucleotides disturbs the orderly alignment of Rohon-Beard cells along the dorsolateral spinal cord and causes them to position themselves at ectopic sites. Their central axon projections along the dorsolateral fascicle are also stunted. These observations suggest that Runx1 plays a role in Rohon-Beard cell positioning and axon growth and possibly target finding. Runx1 may also play a role in directing BDNF gene expression to Rohon-Beard cells.
Table 5.
Potential transcription factor binding sites in HCS1 The sequence was searched for potential transcription factor binding sites with TESS [28]. The URL is http://www.cbil.upenn.edu/tess/index.html.
| 1 ATTACCTCAA CCATGCAATT TCCACCATCA ATAATTTAA | entry | score | |
| ---------> | M00096 | Pbx-1 | 90.2 |
| <----- | M00271 | AML-1a | 88.7 |
| <------- | M00101 | CdxA | 88.6 |
| -------> | M00101 | CdxA | 88.6 |
| <------- | M00101 | CdxA | 88.6 |
| <----- | M00029 | HSF | 86.9 |
| <----- | M00028 | HSF | 85.9 |
| <------- | M00100 | CdxA | 85.9 |
| -------------> | M00249 | CHOP-C | 85.5 |
| <---------- | M00075 | GATA-1 | 85.3 |
Gata-1 is the prototype of the Gata family [33]. All family members have four zinc-fingers. Gata-1 plays a major role in the development of the hematopoetic system [34]. Two family members, Gata-2 and Gata-3 also have functions in the nervous system [35]. Gata-2 was shown to be involved in the specification of V2 motor neurons [36]. Gata-3 is expressed in rhombomere 4 of the mouse embryo hindbrain in neurons that project to the cochlea [37]. In Gata-3 knockout mice these projections become aberrant. Gata-3 is also expressed in the mesenchymal cells surrounding the otocyst and may have a function in target finding of efferent neurons [38]. Interestingly, Adam, which contains HCS1 and its Gata-1 binding site, is expressed in blood cells and neurons. The mammalian BDNF gene is known to be expressed in macrophages.
The results of the RT-PCR experiments presented here as well as previous results of in situ hybridization and immunocytochemistry indicate that BDNF is a maternal message in the early embryo. The present results independently confirm the earlier RT-PCR results by 5' RACE cloning. Our present data show that a second 5' exon in addition to exon 1c, namely exon 1β, is present in the pre-transcriptional embryo. Exon 1β contains a 170 nt segment that is highly similar to human BDNF gene exon 1 and a region of the pufferfish BDNF gene, suggesting they may be subject to similar regulatory mechanisms and functions. BDNF is secreted by cumulus granulosa cells of the mammalian ovary [39] and has been shown to participate in mammalian oocyte maturation [40]. It is not known which of the multiple exons of the mammalian BDNF gene are expressed in the ovary. The function of the maternal transcripts in zebrafish oocyte development and the pre-transcriptional embryo are unknown. A potential high affinity receptor for BDNF in the zebrafish is TrkB, a transmembrane protein tyrosine kinase [41]. The mammalian counterpart, TrkB, is the preferred high affinity receptor for mammalian BDNF [42]. We previously showed that a trkB transcript that lacks a kinase domain, is present in the pre-transcriptional zebrafish embryo [13]. One function of BDNF in early embryo development could thus be paracrine, or in cell adhesion, movement, or guidance.
RT-PCR analysis of adult organs revealed an organ-specific pattern of expression of the 5' non-coding exons. Every organ we examined expressed at least one of the seven exons. The heart expressed only exon 1α. This exon was also expressed in all other organs. The widespread expression suggests expression in a cell-type present in all organs such as endothelial cells. Exon-specific in situ hybridization will be necessary to address this possibility. Interestingly, although exon 1α is widely expressed in the adult zebrafish, suggesting an important function, its sequence is not at all similar to any of the mammalian exons. Indeed, a BLAST search against all currently known sequences revealed no significant similarity to any of them. Exon 1 α thus appears to be quite unique to the zebrafish.
Despite a thorough search for exons 1b/b' and d we could not detect them with RT-PCR during development in whole specimens or in organs of the adult. The successful cloning by RT-PCR from 7 days and 10 days old larvae of exons 1d and 1b/b', respectively, indicates that they exist. Perhaps these exons are expressed in transient and/or very small cell populations and this may account for our failure to detect them by RT-PCR. The highly specific expression of the Dex1d construct in Mauthner cells is consistent with this explanation. It is possible that future in situ hybridization or microchip analyses will detect transcripts containing exons 1b/b' and 1d.
Conclusions
The data presented here show that the structure and organization of the zebrafish BDNF gene rivals in complexity that of its mammalian counterpart. The existence of two additional promoters was proven by 5'RACE cloning and a third promoter was identified by sequence comparison and transient expression. The zebrafish BDNF gene thus has at least five promoters and possibly as many as six. Two of the seven 5' non-coding exons associated with promoters were found by RT-PCR in the embryo before transcription starts and therefore must be parts of maternally transmitted transcripts. Their existence in the pre-transcriptional embryo was independently confirmed by 5'RACE. RT-PCR showed that by 80% epiboly four of the six were expressed and remained active at all stages of development into adulthood. The other two, promoters 1b and 1d, could not be detected at any stage or in any adult organ. The four active promoters were differentially expressed in adult zebrafish organs. Transient expression of fusion genes driven by various promoters also revealed distinct patterns of cellular expression. A highly conserved sequence, HCS1, in the 5' exon associated with promoter 1c has properties of a dehancer and, depending on the sequence context, an enhancer. The 3' UT and flank exert a position-dependent restrictive effect on promoter 1c and switch HCS1 from dehancer to enhancer.
Methods
5' RACE
Amplified cDNA was generated using the Invitrogen GeneRacer kit according to the manufacturer's directions [43]. The template for cDNA preparation was total RNA extracted from 2–4 cell stage embryos, 10 days old larvae, pooled organs from adult fish, and whole adult fish using the guanidine thiocyanate method of Chomczynski [44]. cDNA was generated using random hexamer oligonucleotides. The cDNA was amplified with the GeneRacer primer and a BDNF antisense primer, P1, complementary to 18 nucleotides near the 5' end of the coding exon. Nested PCR was then performed using the internal 5' GeneRacer primer and a second BDNF-specific primer, P2 consisting of the 25 nucleotides immediately upstream of P1.
The amplified cDNA was cloned using the Invitrogen TOPO Cloning kit. DNA was extracted from individual clones. The extracted DNA was analyzed by restriction enzyme digestion followed by agarose gel electrophoresis and nucleotide sequencing at a commercial sequencing facility (Davis Sequencing, LLC. Davis, CA 95616. [45]).
RT-PCR
Total RNA was extracted from 2–4 cell stage embryos, 10 days old larvae, pooled organs from adult fish, and whole adult fish using the guanidine thiocyanate method of Chomczynski [44]. The integrity of the RNA was checked by agarose gel electrophoresis. Approximately 1 μg of total RNA was reverse transcribed using random hexamers for priming of murine Maloney virus reverse transcriptase. Aliquots of the samples were amplified with nested BDNF exon-specific primers in two rounds of amplification. The antisense primers in all cases were P1 and P2, P2 being situated upstream from P1. 5' RACE cDNA clones were used as positive control templates and size markers. β-actin primers were used as internal controls for the quality of cDNA synthesis. The amplified DNA was subjected to agarose gel electrophoresis and exon-specific bands identified by size.
Oligonucleotide Primer Sequences
P1 (outside antisense): ATAGTAACGAACAGGATG
P2 (inside antisense): GTCATCACTCTTCTAACCTGTTG
Exon 1α nested
Outside sense: ggaattaaccctcactaaagggaCGCCAAGGTGTAAGAGAG
Inside sense: CAAGATAAAGGCGTACGG
Exon 1β nested
Outside sense: ggaattaaccctcactaaagggaGGATTTCGTGACTTCCA
Inside sense: CATAAGTCACCCAAGTGG
Exon 1a nested
Outside sense: GCACTTTGGACAGAGGCA
Inside sense: GGAGTCGTTGAACGCTGC
Exon 1b nested
Outside sense: ACATGGACTGAGGAGTAG
Inside sense: TCAATAACAAATCTGAGC
Exon 1c nested
Outside sense: CTCAATGCGCACTAC
Inside sense: GCTCAGTCATGGGAGTCC
Exon 1d nested
Outside sense: ggaattaaccctcactaaagggaTCCCAGAGCAATAAAGTG
Inside sense: CGTCTAGATTACGTTTTA
β-actin: CAACGGCTCCGGCATGTG/TGCCAGGGTACATGGTGG
Construction of Fusion Genes
Starting materials were the previously cloned genomic clone c241, various subclones of c241, and plasmids pEGFP and pEGFP-F from Clontech, Palo Alto, CA http://www.clontech.com. EGFP-F is a derivative of GFP that exhibits enhanced fluorescence and possesses a farnesylation signal at the COOH-terminal [46]. The farnesylation signal is derived from the src protein. Once farnesylated EGFP-F becomes anchored in the cell membrane [46]. The reporter encoded by the plasmid phrGFP-1 from Stratagene, La Jolla, CA http://www.stratagene.com was used for the construction of fusion genes containing the hrGFP reporter.
Standard methods of restriction enzyme digestion and ligation of selected fragments were applied to generate the various expression constructs. The junctions of heterologous fragments were sequenced for all constructs to confirm correct order and orientation of the heterologous fragments in each fusion gene.
The fusion genes were constructed as follows:
1. EGFP-FM. Cut pEGFP-F with BamHI, blunt-end, and religate. Eliminates multicloning sites at 3' end and creates ClaI site.
2. Mi-Ex. Insert BamHI fragment of genomic clone c241 containing promoter 1c into BamHI site of pEGFP to create subclone 25. The insert is in the sense orientation. To replace EGFP with EGFP-F reporter in subclone 25 remove small AgeI/StuI fragment from subclone 25 and replace with small AgeI/StuI fragment of pEGFP-F. Mi-Ex has multiple cloning sites at the 5' and 3' ends.
3. MJ. Remove small AgeI/StuI fragment from subclone 25 and replace with small AgeI/StuI fragment of pEGFP-FM. This construct is identical to Mi-Ex but lacks the multicloning sites at the 3' end of the EGFP-F reporter.
4. Greenie. Cut MJ with BamHI and religate to remove BamHI insert. In Greenie the EGFP-F reporter is under control of the LacZ promoter allowing green/white screening of bacterial colonies for inserts in the multicloning site.
5. Bi-Ex. SmaI fragment of subclone 41 of genomic clone c241 into blunt-ended EcoRI site 3' to reporter of Mi-Ex. Subclone 41 carries a HindIII fragment of genomic clone c241 in pEGFP. The HindIII fragment contains the coding exon and 800 bps of 3' flank. The HindIII fragment is in the sense orientation in subclone 41.
6. HCS-MiEx: Amplify HCS1 with primers containing SalI sites, cut with SalI, insert into SalI site of MJ. Both orientations were obtained.
7. Maxi-Express: Insert into MJ both a HindIII/BamHI fragment containing exons 1b and 1b' and a BamHI fragment containing exon 1c. Both fragments are from genomic clone c241.
8. Sappie: Insert blunt-ended SapI fragment of genomic clone c241 containing exon 1c into SmaI site of MJ.
9. DOS: Cut Sappie with BamHI, insert BamHI fragment of genomic clone c241 containing exon 1c.
10. ME41-9: SmaI fragment of subclone 41 into StuI site of Mi-Ex.
11. Clay: Amplify 300 bp fragment spanning 5' end of coding exon and 3' end of preceding intron with primers containing BamHI and AgeI sites. Clone into BamHI/AgeI site of Greenie.
12. Adam: Large HindIII fragment of genomic clone c241 containing exons 1b, 1b' and 1c into HindIII site of Clay.
13. phrGFP-CMV: Cut phrGFP-1 with NsiI/SacI, blunt-end, religate to eliminate CMV promoter.
14. LJ: Amplify 2 kb of 3' flank with primers containing EcoRI and MluI sites. Cut phrGFP-CMV with EcoRI/MluI, remove smaller piece. Ligate amplified, EcoRI/MluI digested 3' flank into larger piece.
15. LJB: Partially cut LJ with BamHI, blunt-end, religate. Identify clones with inactive BamHI site in 3' flank.
16. BiEx-GFP: BamHI fragment of genomic clone c241 containing exon 1c into BamHI site of LJB.
17. Dex1a-GFP: BglII fragment of genomic clone c241 that contains exons 1α,1β, and part of 1a into BamHI site of LJB.
18. Dex1d: HindIII frgament of genomic clone c241 that contains exon 1d into HindIII site of Clay.
19. DOA5: HindIII fragment of genomic clone c241 that contains exons 1α, 1β, and 1a plus intron downstream from exon 1a into HindIII site of Clay.
20. Nin1c-GFP: Amplify a fragment containing 2 kb of 5' flank upstream from exon 1c and all of exon 1c including HCS1. The primers contain NotI and XmaI sites. Digest amplified DNA with NotI/XmaI. Ligate into LJ cut with NotI/XmaI.
Preparation of DNA and microinjection of embryos
Plasmids were digested with restriction enzymes to remove SV40 enhancers and other vector sequences. The desired restriction fragments were purified by agarose gel electrophoresis. DNA was dissolved in 100 mM KCl at a concentration of 10–50 μg/ml. Phenol red was added to visualize injected DNA solution. DNA was injected into a blastomere or into the cytoplasmic stream below the blastomere(s) at the 1–8 cell stages. Embryos were enzymatically dechorionated with Pronase and extensively washed in embryo medium prior to injection. Embryo medium = 13 mM NaCl; 4.2 mM NaHCO3; 0.54 mM KCl; 0.025 mM Na2HPO4; 0.044 mM KH2PO4; 1.3 mM CaCl2; 1 mM MgSO4). Embryos were injected and maintained for 14 hours post fertilization on a bed of 0.7% agarose in embryo medium. Subsequently they were placed into deionized water in clear plastic dishes for observation with a Zeiss AxioVert 200 M fluorescent microscope or further growth. Pictures were taken with a digital camera and acquired with the AxioVision software package, version 3.0.6. The images were further processed with Microsoft PhotoEditor, Microsoft PowerPoint and Adobe PhotoDeluxe Business Edition.
Supplementary Material
Identification of GeneRacer oligoribonucleotide at 5' ends of sequences deposited in data banks. The GeneRacer kit from Invitrogen is a kit for full-length RNA ligase-mediated rapid amplification of 5' and 3' cDNA ends (RLM-RACE). The procedure includes the ligation of a synthetic oligoribonucleotide to the 5' end of decapped mRNAs. The oligoribonucleotide serves as a universal primer site for subsequent amplification of cDNAs. We used the kit for 5' RACE of zebrafish transcripts derived from the gene encoding brain-derived neurotrophic factor. When we searched the databanks for sequences related to our 5' RACE clones using BLAST we noticed that the GeneRacer oligoribonucleotide sequence was present in single or multiple copies at the 5' ends of many unrelated sequences. This suggests that its presence was overlooked by investigators before they deposited their sequences and care should be taken to remove it prior to future depositions. The sequence of the oligoribonucleotide is CGA CUG GAG CAC GAG GAC ACU GAC AUG GAC UGA AGG AGU AGA AA. The result of a BLAST search for the GeneRacer oligoribonucleotide sequence in the sequence databases that are searched by BLAST is shown in table 1 of additional files. The file is in Microsoft Word document format. The left hand column lists the sequence identifier for each sequence that contained one or more copies of the GeneRacer oligoribonucleotide. The sequence identifiers are hyperlinked to the corresponding sequences in the databases. The right hand column gives the number of identities between the target sequence and the GeneRacer oligoribonucleotide. Since the oligoribonucleotide has 44 nts, 88 identities indicates the presence of two copies of the GeneRacer oligoribonucleotide. The hyperlinks associated with the entries in the right hand column are inactive since they refer to a specific search.
Acknowledgments
Acknowledgements
This work was supported by a Merit Review Grant from the Veterans Administration. We thank Shan Siva for excellent technical help.
Contributor Information
Gerhard Heinrich, Email: gheinrich@ucdavis.ed.
Carl John Pagtakhan, Email: mvmntalum809@hotmail.com.
References
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kaczmarek L. Expression of c-fos and other genes encoding transcription factors in long-term potentiation. Behav Neural Biol. 1992;57:263–266. doi: 10.1016/0163-1047(92)90276-a. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Sandstrom DJ, Hoeffer CA, Ramaswami M. AP-1 functions upstream of CREB to control synaptic plasticity in Drosophila. Nature. 2002;416:870–874. doi: 10.1038/416870a. [DOI] [PubMed] [Google Scholar]
- Messaoudi E, Ying SW, Kanhema T, Croll SD, Bramham CR. Brain-derived neurotrophic factor triggers transcription-dependent, late phase long-term potentiation in vivo. J Neurosci. 2002;22:7453–7461. doi: 10.1523/JNEUROSCI.22-17-07453.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes PE, Alexi T, Walton M, Williams CE, Dragunow M, Clark RG, Gluckman PD. Activity and injury-dependent expression of inducible transcription factors, growth factors and apoptosis-related genes within the central nervous system. Prog Neurobiol. 1999;57:421–450. doi: 10.1016/S0301-0082(98)00057-4. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Southard T, Sarvey JM, Herkenham M, Brady LS. Unilateral LTP triggers bilateral increases in hippocampal neurotrophin and trk receptor mRNA expression in behaving rats: evidence for interhemispheric communication. J Comp Neurol. 1996;368:371–382. doi: 10.1002/(SICI)1096-9861(19960506)368:3<371::AID-CNE4>3.3.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Finkbeiner S. Calcium regulation of the brain-derived neurotrophic factor gene. Cell Mol Life Sci. 2000;57:394–401. doi: 10.1007/PL00000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh PB, Ghosh A. Molecular mechanisms underlying activity-dependent regulation of BDNF expression. J Neurobiol. 1999;41:127–134. doi: 10.1002/(SICI)1097-4695(199910)41:1<127::AID-NEU16>3.3.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/S0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Heymach J. V., Jr., Kruttgen A, Suter U, Shooter EM. The regulated secretion and vectorial targeting of neurotrophins in neuroendocrine and epithelial cells. J Biol Chem. 1996;271:25430–25437. doi: 10.1074/jbc.271.41.25430. [DOI] [PubMed] [Google Scholar]
- Mowla SJ, Farhadi HF, Pareek S, Atwal JK, Morris SJ, Seidah NG, Murphy RA. Biosynthesis and post-translational processing of the precursor to brain-derived neurotrophic factor. J Biol Chem. 2001;276:12660–12666. doi: 10.1074/jbc.M008104200. [DOI] [PubMed] [Google Scholar]
- Lee R, Kermani P, Teng KK, Hempstead BL. Regulation of cell survival by secreted proneurotrophins. Science. 2001;294:1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- Lum T, Huynh G, Heinrich G. Brain-derived neurotrophic factor and TrkB tyrosine kinase receptor gene expression in zebrafish embryo and larva. Int J Dev Neurosci. 2001;19:569–587. doi: 10.1016/S0736-5748(01)00041-7. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Narz F, Heumann R, Lessmann V. BDNF-GFP containing secretory granules are localized in the vicinity of synaptic junctions of cultured cortical neurons. J Cell Sci. 1998;111 ( Pt 11):1483–1493. doi: 10.1242/jcs.111.11.1483. [DOI] [PubMed] [Google Scholar]
- Canossa M, Gartner A, Campana G, Inagaki N, Thoenen H. Regulated secretion of neurotrophins by metabotropic glutamate group I (mGluRI) and Trk receptor activation is mediated via phospholipase C signalling pathways. Embo J. 2001;20:1640–1650. doi: 10.1093/emboj/20.7.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Meyer M, Matsuoka I, Wetmore C, Olson L, Thoenen H. Enhanced synthesis of brain-derived neurotrophic factor in the lesioned peripheral nerve: different mechanisms are responsible for the regulation of BDNF and NGF mRNA. J Cell Biol. 1992;119:45–54. doi: 10.1083/jcb.119.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castren E, Berninger B, Leingartner A, Lindholm D. Regulation of brain-derived neurotrophic factor mRNA levels in hippocampus by neuronal activity. Prog Brain Res. 1998;117:57–64. doi: 10.1016/s0079-6123(08)64007-8. [DOI] [PubMed] [Google Scholar]
- Heinrich G. A novel BDNF gene promoter directs expression to skeletal muscle. BMC Neurosci. 2003;4:11. doi: 10.1186/1471-2202-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh G, Heinrich G. Brain-derived neurotrophic factor gene organization and transcription in the zebrafish embryo. Int J Dev Neurosci. 2001;19:663–673. doi: 10.1016/S0736-5748(01)00046-6. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Heinrich G. Brain-derived neurotrophic factor gene expression in the developing zebrafish. Int J Dev Neurosci. 1997;15:983–997. doi: 10.1016/S0736-5748(97)00017-8. [DOI] [PubMed] [Google Scholar]
- FuguEnsemble http://www.fugu-sg.org/
- NCBI http://www.ncbi.nlm.nih.gov/
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam WL, Lee TS, Gilbert W. Active transposition in zebrafish. Proc Natl Acad Sci U S A. 1996;93:10870–10875. doi: 10.1073/pnas.93.20.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk T, Lendahl U, Funakoshi H, Arenas E, Persson H, Metsis M. Identification of brain-derived neurotrophic factor promoter regions mediating tissue-specific, axotomy-, and neuronal activity-induced expression in transgenic mice. J Cell Biol. 1995;128:185–199. doi: 10.1083/jcb.128.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J, Overton GC. Modeling transcription factor binding sites with Gibbs Sampling and Minimum Description Length encoding. Proc Int Conf Intell Syst Mol Biol. 1997;5:268–271. [PubMed] [Google Scholar]
- Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- Kataoka H, Ochi M, Enomoto K, Yamaguchi A. Cloning and embryonic expression patterns of the zebrafish Runt domain genes, runxa and runxb. Mech Dev. 2000;98:139–143. doi: 10.1016/S0925-4773(00)00445-7. [DOI] [PubMed] [Google Scholar]
- Kalev-Zylinska ML, Horsfield JA, Flores MV, Postlethwait JH, Vitas MR, Baas AM, Crosier PS, Crosier KE. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129:2015–2030. doi: 10.1242/dev.129.8.2015. [DOI] [PubMed] [Google Scholar]
- Crosier PS, Kalev-Zylinska ML, Hall CJ, Flores MV, Horsfield JA, Crosier KE. Pathways in blood and vessel development revealed through zebrafish genetics. Int J Dev Biol. 2002;46:493–502. [PubMed] [Google Scholar]
- Watt P, Lamb P, Squire L, Proudfoot N. A factor binding GATAAG confers tissue specificity on the promoter of the human zeta-globin gene. Nucleic Acids Res. 1990;18:1339–1350. doi: 10.1093/nar/18.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw E, Tsai SF, Hogben P, Orkin SH. Regulated expression of globin chains and the erythroid transcription factor GATA-1 during erythropoiesis in the developing mouse. Mol Cell Biol. 1990;10:6596–6606. doi: 10.1128/mcb.10.12.6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardelli J, Thiesson D, Fujiwara Y, Tsai FY, Orkin SH. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev Biol. 1999;210:305–321. doi: 10.1006/dbio.1999.9278. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yamamoto M, Engel JD. GATA2 is required for the generation of V2 interneurons. Development. 2000;127:3829–3838. doi: 10.1242/dev.127.17.3829. [DOI] [PubMed] [Google Scholar]
- Pata I, Studer M, van Doorninck JH, Briscoe J, Kuuse S, Engel JD, Grosveld F, Karis A. The transcription factor GATA3 is a downstream effector of Hoxb1 specification in rhombomere 4. Development. 1999;126:5523–5531. doi: 10.1242/dev.126.23.5523. [DOI] [PubMed] [Google Scholar]
- Pandolfi PP, Roth ME, Karis A, Leonard MW, Dzierzak E, Grosveld FG, Engel JD, Lindenbaum MH. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet. 1995;11:40–44. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- Seifer DB, Feng B, Shelden RM, Chen S, Dreyfus CF. Brain-derived neurotrophic factor: a novel human ovarian follicular protein. J Clin Endocrinol Metab. 2002;87:655–659. doi: 10.1210/jc.87.2.655. [DOI] [PubMed] [Google Scholar]
- Ojeda SR, Romero C, Tapia V, Dissen GA. Neurotrophic and cell-cell dependent control of early follicular development. Mol Cell Endocrinol. 2000;163:67–71. doi: 10.1016/S0303-7207(99)00242-7. [DOI] [PubMed] [Google Scholar]
- Martin SC, Marazzi G, Sandell JH, Heinrich G. Five Trk receptors in the zebrafish. Dev Biol. 1995;169:745–758. doi: 10.1006/dbio.1995.1184. [DOI] [PubMed] [Google Scholar]
- Klein R, Nanduri V, Jing SA, Lamballe F, Tapley P, Bryant S, Cordon-Cardo C, Jones KR, Reichardt LF, Barbacid M. The trkB tyrosine protein kinase is a receptor for brain-derived neurotrophic factor and neurotrophin-3. Cell. 1991;66:395–403. doi: 10.1016/0092-8674(91)90628-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Invitrogen http://www.invitrogen.com.
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Davis-Sequencing http://www.davissequencing.com.
- Harvey KJ, Lukovic D, Ucker DS. Membrane-targeted green fluorescent protein reliably and uniquely marks cells through apoptotic death. Cytometry. 2001;43:273–278. doi: 10.1002/1097-0320(20010401)43:4<273::AID-CYTO1059>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Bishop JF, Mueller GP, Mouradian MM. Alternate 5' exons in the rat brain-derived neurotrophic factor gene: differential patterns of expression across brain regions. Brain Res Mol Brain Res. 1994;26:225–232. doi: 10.1016/0169-328X(94)90094-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of GeneRacer oligoribonucleotide at 5' ends of sequences deposited in data banks. The GeneRacer kit from Invitrogen is a kit for full-length RNA ligase-mediated rapid amplification of 5' and 3' cDNA ends (RLM-RACE). The procedure includes the ligation of a synthetic oligoribonucleotide to the 5' end of decapped mRNAs. The oligoribonucleotide serves as a universal primer site for subsequent amplification of cDNAs. We used the kit for 5' RACE of zebrafish transcripts derived from the gene encoding brain-derived neurotrophic factor. When we searched the databanks for sequences related to our 5' RACE clones using BLAST we noticed that the GeneRacer oligoribonucleotide sequence was present in single or multiple copies at the 5' ends of many unrelated sequences. This suggests that its presence was overlooked by investigators before they deposited their sequences and care should be taken to remove it prior to future depositions. The sequence of the oligoribonucleotide is CGA CUG GAG CAC GAG GAC ACU GAC AUG GAC UGA AGG AGU AGA AA. The result of a BLAST search for the GeneRacer oligoribonucleotide sequence in the sequence databases that are searched by BLAST is shown in table 1 of additional files. The file is in Microsoft Word document format. The left hand column lists the sequence identifier for each sequence that contained one or more copies of the GeneRacer oligoribonucleotide. The sequence identifiers are hyperlinked to the corresponding sequences in the databases. The right hand column gives the number of identities between the target sequence and the GeneRacer oligoribonucleotide. Since the oligoribonucleotide has 44 nts, 88 identities indicates the presence of two copies of the GeneRacer oligoribonucleotide. The hyperlinks associated with the entries in the right hand column are inactive since they refer to a specific search.


