Abstract
As humans contain a currently active L1 (LINE-1) non-LTR retrotransposon family (Ta-1), the human genome database likely provides only a partial picture of Ta-1-generated diversity. We used a non-biased method to clone Ta-1 retrotransposon-containing loci from representatives of four ethnic populations. We obtained 277 distinct Ta-1 loci and identified an additional 67 loci in the human genome database. This collection represents ∼90% of the Ta-1 population in the individuals examined and is thus more representative of the insertional history of Ta-1 than the human genome database, which lacked ∼40% of our cloned Ta-1 elements. As both polymorphic and fixed Ta-1 elements are as abundant in the GC-poor genomic regions as in ancestral L1 elements, the enrichment of L1 elements in GC-poor areas is likely due to insertional bias rather than selection. Although the chromosomal distribution of Ta-1 inserts is generally a function of chromosomal length and gene density, chromosome 4 significantly deviates from this pattern and has been much more hospitable to Ta-1 insertions than any other chromosome. Also, the intra-chromosomal distribution of Ta-1 elements is not uniform. Ta-1 elements tend to cluster, and the maximal gaps between Ta-1 inserts are larger than would be expected from a model of uniform random insertion.
The L1 (LINE-1, long interspersed repeated DNA) family of non-LTR retrotransposons (Fig. 1) is responsible for 27% of the mass of the human genome and has had a major effect on the structure (and presumably function) of modern mammalian genomes (International Human Genome Sequencing Consortium [IHGSC] 2001; Mouse Genome Sequencing Consortium [MGSC] 2002). L1 elements replicate by copying (reverse transcribing) their RNA into genomic DNA (for review, see Furano 2000; Moran and Gilbert 2002). Despite a strong cis preference (Esnault et al. 2000; Wei et al. 2001), the L1 replicative machinery can also copy other RNAs, including Alu (SINE, short interspersed repeated DNA) transcripts (Dewannieux et al. 2003). Thus, L1 retrotransposons likely amplified the one million-member Alu SINE family and numerous processed pseudogenes as well (IHGSC 2001; Buzdin et al. 2003a).
Figure 1.
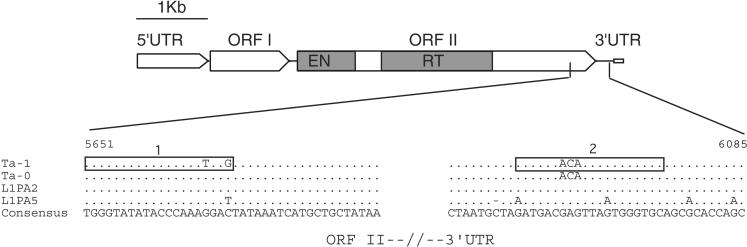
Structure of a typical full-length human L1 element. The 5′ untranslated region (UTR) has a regulatory function; open reading frame 1 (ORF I) encodes an RNA-binding protein; ORF II encodes the L1 replicase and contains an endonuclease (EN) and a reverse transcriptase domain (RT), and the 3′ UTR that contains a conserved G-rich polypurine motif. Genomic copies of L1 usually end in an A-rich stretch (open rectangle, see Moran and Gilbert 2002). The partial alignment of Ta-1, Ta-0, L1PA2, and L1PA5 consensus sequences shows the positions of oligonucleotide 1, which is specific for Ta-1, and of oligonucleotide 2, which includes the ACA trinucleotide, diagnostic of the Ta family (see Methods). The numbers indicate the position of the sequence on alignment ALIGN-000165 at the EMBL_ALIGN database (Boissinot and Furano 2001).
L1 elements have been replicating and evolving in mammalian genomes since before the mammalian radiation, ∼100 million years ago (Mya; Burton et al. 1986; for review, see Smit et al. 1995; Furano 2000). About 100,000 L1 elements have been inserted in the human genome after the mammalian radiation, and all are the products of a single lineage of 16 distinct L1 families (L1PA16–L1PA1) that extends from the radiation to the present (Smit et al. 1995; Boissinot et al. 2001). Only the most recently evolved human-specific L1 family, L1PA1, originally called Ta (Skowronski et al. 1988) and referred to here as such, is currently active, that is, capable of retrotransposition and causing polymorphisms (Boissinot et al. 2000; Sheen et al. 2000; Badge et al. 2003) and genetic defects (Ostertag and Kazazian Jr. 2001).
The persistence of L1 activity without any obvious benefit to its host remains a puzzle. Although recruitment of L1 DNA for a host function or an L1-mediated genetic alteration may be beneficial, such outcomes are too rare to ascribe a beneficial function to L1. In fact, at times L1 activity has been so deleterious that elements capable of replication have been subjected to negative selection (Boissinot et al. 2001). This conclusion was based on the analysis of extinct ancestral L1 families. In contrast, except for the most severely deleterious inserts, the insertional history of the currently replicating Ta family could provide a “real-time” view of L1 amplification. The events necessary for establishing a successful amplification, as well as how these events affect the host, could be revealed by analysis of a currently amplifying family.
The Ta family arose about 5 Mya and ∼2.5 Myr later gave rise to the Ta-1 (sub)family. Ta-1 accounts for almost all L1 replication in humans (Boissinot et al. 2000; Brouha et al. 2003). A number of demonstrably active Ta-1 elements have been isolated from the genome (e.g., see Kimberland et al. 1999; Brouha et al. 2003), and 69% of a small number of Ta-1-containing loci were polymorphic for the presence or absence of the element (Boissinot et al. 2000). Our goal here was to obtain as comprehensive a set as possible of Ta-1 inserts in the human population.
Two approaches were used previously to study Ta elements: extraction of Ta elements from the public databases (Boissinot et al. 2000; Myers et al. 2002) or isolation of polymorphic elements by PCR display (Sheen et al. 2000; Ovchinnikov et al. 2001; Badge et al. 2003; Buzdin et al. 2003b). As public databases would likely be biased in favor of inserts that are found at high frequencies in human populations, the first method will most likely miss a large fraction of the most recent inserts. On the other hand, the second approach is more likely to discriminate against high-frequency polymorphisms (see discussion in Myers et al. 2002). Therefore, we developed a cloning strategy that was free of these biases and applied it successively to the genome of four individuals of different ethnic origins.
We obtained 277 non-redundant Ta-1 inserts to which we added 67 unique inserts from the database (total, 344). For comparison, ∼300 elements belonging to each of the ancestral L1PA2 and L1PA5 families were also collected from the public database. These now-extinct families reached their peak amplification about 9 Mya and 23 Mya, respectively, and essentially all L1PA2 and L1PA5 elements are now fixed in the human genome. We confirmed our earlier finding that the Ta-1 subfamily is currently not as deleterious as were these ancestral L1 families (Boissinot et al. 2001). Also, Ta-1 elements are not randomly distributed in the genome with respect to GC content but recapitulate the distribution of older families. However, the distribution of Ta-1 insertions may not be completely random at either the chromosomal or intra-chromosomal level.
RESULTS
Collection of Ta-1 Inserts
Table 1 shows the number of Ta-1 containing clones collected from each of the four individuals and the number of different Ta-1 inserts. Of 6751 clones collected, 1256 were sequenced, allowing the identification of 277 different Ta-1 inserts. As expected, the largest number of different inserts was collected from the first individual analyzed (i.e., the Druze), and each subsequent cloning yielded fewer new inserts. Using BLAST, we could unambiguously locate 232 (84%) of the 277 flanking sequences in the human genome database. We could not unambiguously locate 17 flanking sequences because they were very similar to sequences that were repeated in the database. For 28 sequences (10%), no significant match was found. This was expected, as 13% of the nuclear DNA (mostly heterochromatin) is not present in the database. Of the 232 cloned Ta-1 insertions whose sites were identified in the database, 94 (41%) were not occupied by a Ta-1 element. These 94 elements are most likely polymorphic in human populations, and either the elements were absent from the genome of the individuals used by the Human Genome Project or the occupied state was missed during the assembly phase.
Table 1.
Numbers of Ta-1 Inserts Collected and Fractions of Polymorphic Inserts
|
Cloning
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Druze | Biaka | Chinese | Melanesian | Total | Databasea | Totalb | ||
| Clones collected | 1344 | 1576 | 1851 | 1980 | 6751 | — | — | |
| Clones sequenced | 613 | 156 | 253 | 234 | 1256 | — | — | |
| Number of different Ta-1 inserts | 167 | 58 | 35 | 17 | 277 | 67 | 344 | |
| Flanking sequences located in database | 142 | 52 | 26 | 12 | 232 | 67 | 299 | |
| Tested by PCR | 116 | 28 | 21 | 9 | 174 | 43 | 217 | |
| % Polymorphic | 52% | 75% | 71% | 100% | 60% | 72% | 62% | |
| Insertion sites occupied in database | 102 | 19 | 13 | 4 | 138 | 67 | 205 | |
| Tested by PCR | 86 | 14 | 12 | 2 | 114 | 43 | 157 | |
| % Polymorphic | 38% | 57% | 50% | 100% | 43% | 72% | 51% | |
| Insertion sites empty in database | 40 | 33 | 13 | 8 | 94 | — | 94 | |
| Tested by PCR | 29 | 14 | 9 | 7 | 59 | — | 59 | |
| % Polymorphic | 90% | 93% | 100% | 100% | 93% | — | 93% | |
| Flanking sequences not located in database | 25 | 6 | 9 | 5 | 45 | — | 45 | |
| Flanks are repeated DNA | 17 | |||||||
| Flanks not present in db | 28 | |||||||
| Tested by PCRc | 16 | 2 | 4 | 0 | 22 | — | 22 | |
| % Polymorphic | 50% | 0% | 25% | — | 41% | — | 41% | |
Elements from the database that were missed by cloning but retrieved by a BLAST search.
Total number of Ta-1 inserts including cloning and database search.
The polymorphism of these elements was tested with a single PCR using only the flanking primer cognate to the 3′ flanking sequence of the inserts.
BLAST searches also identified 67 Ta-1 elements not represented in our clone collection. Of these, 43 were amenable to PCR and we determined their presence in the four individuals used in the cloning. Ten were absent from the genome of the four individuals and thus could not be cloned. However, 33 inserts did produce a PCR amplification in at least one of the four individuals, and 13 of those appeared fixed in our small panel of humans (see Methods). Therefore, our cloning procedure recovered ∼90% of the Ta-1 inserts present in the genome of the four individuals.
Altogether, we generated a database of 344 Ta-1 inserts, including 139 (40%) elements that were not present in the human genome database. Therefore, the Ta-1 inserts identified in an earlier analysis of the human genome database (Myers et al. 2002) comprise a subset of those reported here. Although additional novel Ta-1 inserts could be collected if more individuals were analyzed, these would be recovered at an ever-decreasing rate. As it is, our collection of Ta-1 elements represents a far more complete census of the Ta-1 subfamily than that in the human genome database.
The chromosomal position of each of the 344 cloned and database Ta-1 elements along with that of their flanking sequences, extent of polymorphism, and the nucleotide sequences of the PCR primers used to detect the inserts are available in the Supplemental material.
Polymorphism of Ta-1 Inserts
The DNA flanks (2 kb each) of the Ta-1 inserts were screened for repeated sequences using RepeatMasker. We could not determine the polymorphism of inserts flanked only by repeated DNA. Otherwise, we designed PCR primers for each flank, and polymorphism was assessed on the panel of eight human DNAs (Methods). PCR was successful for 174 of 232 cloned inserts (Table 1). A Ta-1 insertion was considered polymorphic when we were able to amplify by PCR the empty state (i.e., the absence of insert) in at least one of the eight individuals.
About half (52%) of the Ta-1 elements cloned from the Druze are polymorphic. The fraction of polymorphic elements is higher in the other three individuals (71% to 100%), because most of the fixed Ta-1 inserts should be, and were, present in the Druze collection. In all, 60% of the cloned inserts were polymorphic. As expected, most (93%) of the cloned inserts that correspond to sites that are empty in the database are polymorphic. Also, as expected, 57% of the cloned inserts corresponding to sites that are occupied in the database are fixed. About half (52%) of the polymorphic sites that were empty in the database are found on fewer than four chromosomes in our sample of eight individuals (representing a total of 16 chromosomes), and 21% were detected by PCR only in the individual from whom they were cloned. Therefore, it is not surprising that these seemingly low-frequency inserts were not present in the database.
About 72% of the Ta-1 inserts that were present in the database but not in our cloned collection were polymorphic. This value is similar to the fraction of polymorphic inserts obtained from the pygmy (75%) and Chinese (71%) cloning. Presumably, the Human Genome Project used a different sample of humans from ours and therefore contains the Ta-1 inserts specific to those individuals. Eight of the Ta-1-containing sites that were unique to the database were empty in all 16 chromosomes. Thus, although the database is enriched in high-frequency and fixed Ta-1 inserts, it also contains some low-frequency alleles, unique to those individuals whose DNA was sequenced.
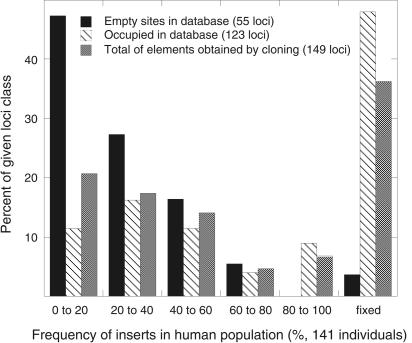
In total, 217 Ta-1 inserts (174 cloned by us and 43 from the human genome database) were analyzed by PCR and 134 of them (62%) were polymorphic (Table 1). Our collection of Ta-1 elements includes a complete range of polymorphism from rare alleles to fixed inserts. Table 1 shows that the extent of polymorphism of the Ta-1-containing sites in the human genome database is significantly lower than the total fraction of polymorphic Ta-1-containg loci (51% vs. 62%; Fisher's exact test, P = 0.006). Therefore, the extent of Ta-1 polymorphism in the database underestimates the level of polymorphism of Ta-1-containing sites in the human population. In a separate study, we determined the frequency of the polymorphic Ta-1 inserts in 190 individuals and we will report these results in full elsewhere. However, Figure 2 shows a subset of these data that compares the frequency distribution in 141 individuals of Ta-1-containing inserts not present in the database with that of those inserts present in the database.
Figure 2.
Frequency distribution of Ta-1-containing alleles. The extent of polymorphism of the indicated Ta-1-containing inserts in 141 individuals was determined as described in the Methods. All DNAs for these studies were obtained from the Coriell Institute for Medical Research and included individuals from the following populations: Chinese, Japanese, Druze, Biaka and Mbuti Pygmy, Melanesian, Atayal, Ami, Caucasian American, and African American.
Characterization of Ta-1 Inserts
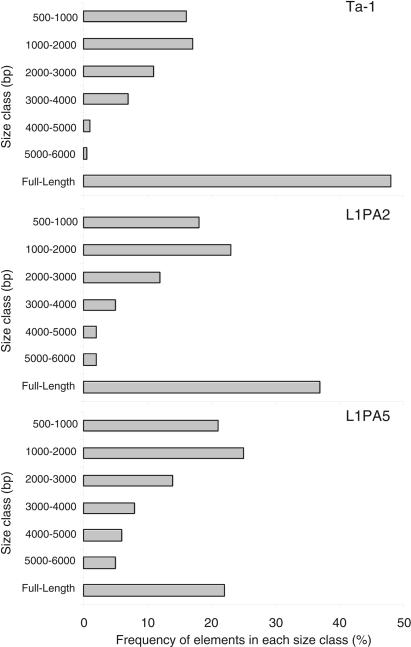
We only analyzed the size distribution of L1 elements on autosomes because we had shown that ancestral full-length (FL) elements were retained at a higher rate on sex chromosomes (Boissinot et al. 2001). About 48% of all autosomal Ta-1 elements are FL (Fig. 3). This is considerably higher than the 35% reported previously (Boissinot et al. 2000), because we only considered elements ≥500 bp. Non-FL elements are either simple truncations (66%) or truncations with inversions of variable length (34%). Both types have been long known (Hutchison III et al. 1989) and will not be discussed further.
Figure 3.
Size distribution of autosomal L1 elements. Elements are grouped in 1000-bp intervals except for the smaller class that shows the frequency of elements 500–1000 bp long. The number of elements analyzed was 183 Ta-1 elements (those whose size was known from the human genome data base), 282 L1PA2 elements, and 305 L1PA5 elements.
The fraction of FL elements is not significantly different between fixed (47%) and polymorphic (45%) Ta-1 elements (Fisher's exact test, P = 0.116). Thus, FL and truncated Ta-1 elements are being fixed in populations at the same rate. Because some polymorphic Ta-1 elements might be close to fixation, we also separately analyzed “low-frequency” Ta-1 elements, that is, those on fewer than four chromosomes (out of 16) by PCR. The fraction of FL elements (50%) of this group was not significantly different than that of all fixed Ta-1 elements (P = 0.117).
In contrast, the percent of autosomal FL L1PA2 (37%) and L1PA5 (22%) elements is significantly lower than that of the Ta-1 family (Fisher's exact test, P = 0.004 and P = 0.000, respectively). The fact that we found fewer FL L1PA5 elements than L1PA2 (P = 0.000) agrees with an earlier study using a smaller sample of these families from chromosomes 21 and 22 (Boissinot et al. 2001). Again, the percentages of FL L1's for each of these families are different from those reported previously because we only examined elements ≥500 bp.
The Genomic Environment of Ta-1 Inserts
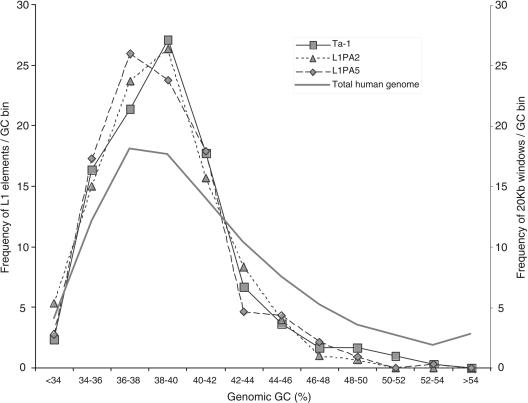
We obtained the GC content of 10 kb, 20 kb, and 100 kb of flanking sequence for 294 Ta-1 inserts. As the results were similar regardless of the window size, only the 20-kb data are shown. Figure 4 shows that Ta-1 elements are not distributed randomly with respect to GC content, being more abundant in regions ≤42% GC than in those ≥42% GC. On average the GC content of Ta-1 flanking sequence is significantly lower than the GC content of the human genome (Student's t-test, P = 0.000). The GC content of the flanking sequences is not significantly different between fixed and polymorphic Ta-1 elements (P = 0.827) or between fixed and low frequency elements (P = 0.153). In contrast with previous reports (Ovchinnikov et al. 2001), Ta-1 elements are found in the same genomic GC compartments as ancestral L1PA2 and L1PA5 subfamilies (Fig. 4); the average GC content of Ta-1–, and L1PA2–, and L1PA5-flanking sequences are not significantly different (P = 0.204 and P = 0.233, respectively).
Figure 4.
Frequency of L1 elements in different GC fractions of the human genome. The % GC was calculated over 20-kb windows. The bins from left to right correspond to an increasing 2% GC fraction. The family-specific L1 curves were built using the DNA flanks of 299 Ta-1 (squares), 300 L1PA2 (triangles), and 324 L1PA5 (diamonds) inserts. The total genome curve (heavy line) was built using Tables “gcPercent” at http://genome.ucsc.edu/ for all chromosomes.
We also determined the repeated DNA content in 100 kb of flanking sequence for 299 Ta-1 inserts (Table 2). Consistent with their presence in regions of relatively low GC content (see Fig. 22 in IHGSC 2001), the flanking sequences of Ta-1 elements are enriched in L1s but deficient in SINEs. The abundance of LTR retrotransposons and all repeats taken together (e.g., SINE, L1, and others) are similar to the genomic average, two features that do not differ much between GC compartments except at a GC content >54%, which represents only a very small fraction of the genome (IHGSC 2001).
Table 2.
Genomic Environment of Ta-1, L1PA2, and L1PA5 Elements
|
Ta-1
|
||||||
|---|---|---|---|---|---|---|
| Genomic featuresa | Polymorphic | Fixed | Total | L1PA2 | L1PA5 | Genomic average |
| % GC (20 kb) | 39.00 | 38.89 | 38.96 | 38.62 | 38.64 | 41.00b |
| % SINEs (100 kb) | 9.27 | 8.85 | 9.21 | 8.26 | 9.30 | 13.14b |
| % LINEs (100 kb) | 22.35 | 23.20 | 23.12 | 23.59 | 21.97 | 20.42b |
| % LTR-retrotransposons (100 kb) | 8.36 | 8.29 | 8.44 | 8.61 | 8.66 | 8.29b |
| % Repeats (100 kb) | 42.84 | 43.27 | 43.50 | 43.26 | 42.60 | 44.83b |
| Local recombination rate | 1.05 | 1.12 | 1.07 | 1.05 | 1.08 | 1.28c |
| Gene density (5 Mb) | 18.83 | 21.16 | 19.49 | 17.74 | 19.02 | 24.45d |
Numbers in parentheses indicate the window size analyzed.
IHGSC (2001).
Deduced from Web table E of Kong et al. (2002).
Calculated from RefSeq genes table at http://genome.ucsc.edu/.
The flanking sequences of Ta-1 elements are as enriched in L1 DNA and lacking in SINE DNA as the older L1PA2 and L1PA5 elements (Table 2). Ta-1 elements are in genomic regions that are significantly less recombining and have a lower gene density than the genome average (P = 0.000, Student's t-test). This feature is shared between all classes of Ta-1 inserts (fixed, polymorphic, “low”-frequency polymorphism) as well as with the L1PA2 and L1PA5 families. These results are also consistent with the GC-content analysis because GC-rich regions of the genome are known to have a higher recombination rate and gene density.
In conclusion, we found that Ta-1 elements are not randomly distributed in the genome with respect to GC content but are more abundant in genomic compartments with a low GC content. In this regard, there is no difference in distribution between fixed and polymorphic Ta-1 elements and between Ta-1 and the ancestral L1PA2 and L1PA5 families.
The Chromosomal Distribution of Ta-1 Inserts
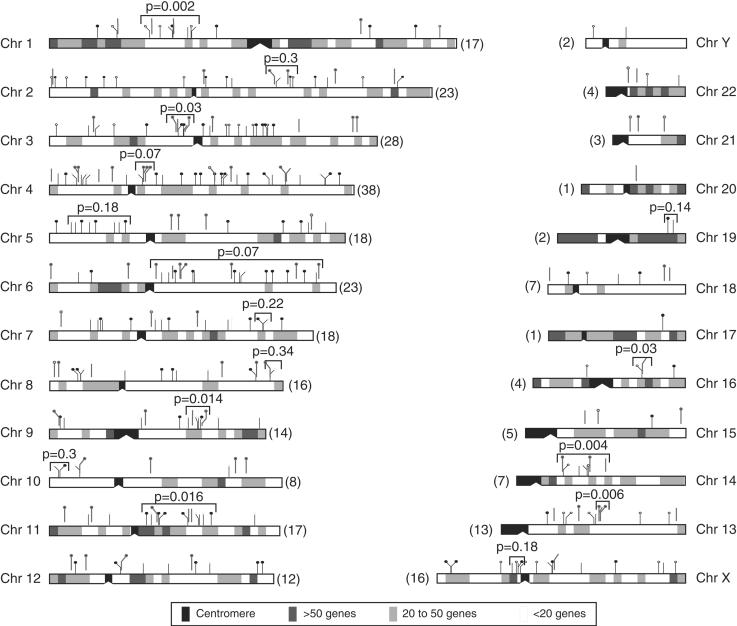
Figure 5 shows the chromosomal distribution of 295 Ta-1 elements. They were found on every chromosome and their number per chromosome ranges from 38 on chromosome 4 to one on chromosomes 17 and 20. Large chromosomes have on average more Ta-1 inserts than smaller ones, and the number of inserts on each chromosome is nearly proportional to the length of the chromosomes (Fig. 6A). Unlike the case for older L1 families (IHGSC 2001), the X and Y chromosomes are not enriched in Ta-1 elements.
Figure 5.
Insertion sites of Ta-1 elements in the human genome. Ta-1 integration sites are shown as tick marks above each chromosome. Tick marks with thinner lines impinging directly on the chromosome diagrams are those identified in the human genome database. Tick marks with heavier lines offset from the chromosomes are the ones that we cloned. Solid circles indicate polymorphic inserts, open circles indicate fixed ones, and tick marks without either are indeterminate (not amenable to PCR, see Methods and Table 1). The number in parentheses indicates the number of Ta-1 insertion sites on each chromosome. Chromosome 15 only shows the positions of 4 of the 5 Ta-1 elements on this chromosome because one of them was located on an unassigned segment. The shaded boxes indicate the number of known genes per 5 Mb segments. The unadjusted k-specific P values for some apparent clusters are given.
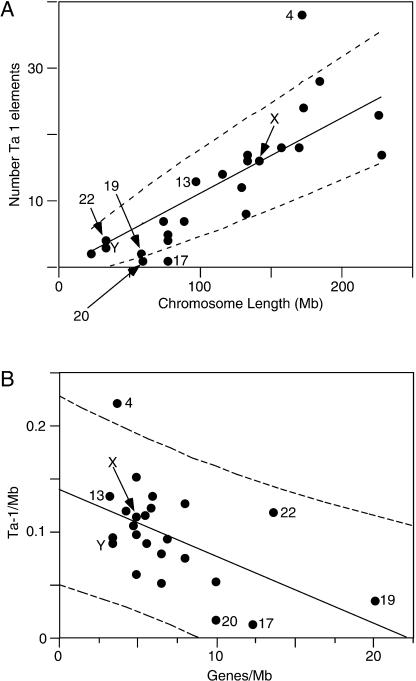
Figure 6.
Chromosomal distribution of Ta-1 elements. (A) The number of Ta-1 elements per chromosome is positively correlated to the chromosome physical length (P < 0.0001) in megabases (Mb, gaps removed). The solid line corresponds to the line expected if Ta-1 elements are distributed proportionally to the length of the chromosomes. The 95% confidence limits (dashed lines) are calculated as ±1.96 times the square root of the predicted number of sites to account for Poisson counting error. (B) The number of Ta-1 elements per chromosome (Ta-1/Mb) is negatively correlated to the gene density (Genes/Mb); R = –0.49, P < 0.004.
Figure 6A also shows that most small chromosomes are positioned below the line expected if Ta-1 are randomly distributed, whereas most large chromosomes are above the line, indicating that small chromosomes harbor fewer Ta-1 elements than expected given their length. This result may reflect the higher gene density of some of the smallest chromosomes. In general, the chromosomal density of Ta-1 elements is negatively correlated with gene density, P < 0.004 (Fig. 6B).
However, chromosome 4 is a clear exception to these generalizations as it contains significantly more Ta-1 inserts than expected for either its length or gene density. The enrichment of Ta-1 elements on chromosome 4 is not because it is uniquely hospitable to L1 insertions. The chromosomal distribution of L1PA2 and L1PA5 show no such bias towards chromosome 4 (results not shown). Rather, as is the case for L1 DNA in general (IHGSC 2001), the X and the Y chromosomes are enriched in L1PA2 and L1PA5 elements (results not shown).
In addition, examination of Figure 5 suggests that some Ta-1 elements may be clustered on some chromosomes. However, even random insertion of Ta-1 elements could yield apparent clusters of inserts that could be misinterpreted as insertional “hot spots.” Therefore, we assessed the statistical significance of any apparent cluster by testing it against the random (uniform) insertion model. Seemingly over-long gaps (i.e., genomic stretches without insertions) may also be observed, and were also tested for statistical significance. We performed such tests using computer simulations to determine the P value of a run of k consecutive Ta-1 inserts (see Methods for details and definitions). Thus, an apparent cluster is a run that contains an excess of inserts given its span (the distance in bp from the first to the last insert), or equivalently, a run whose span is unusually short given its k.
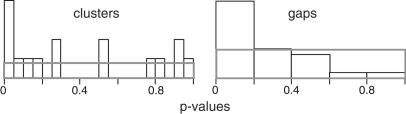
Some of these apparent clusters and their uncorrected P value (k-specific) are indicated on Figure 5. None of the apparent clusters were statistically significant after Bonferroni adjustment. However, as Figure 7 shows, the distribution of k-specific P values (unadjusted) on all chromosomes was skewed towards low values. The results shown are for k = 6, and similar results were found for k = 7 or 8 (results not shown). If the elements were distributed uniformly under a random insertion model, one would expect a rectangular distribution for these P values. This propensity towards small P values can be interpreted as an overall tendency for Ta-1 to cluster but not at high enough density to reach significance in any particular case.
Figure 7.
Distribution of P values. The uncorrected P values for the minimal k-span (k = 6) and maximal gap size for all chromosomes are shown. Similar results were found for k = 7 and 8 (not shown). The rectangles show the expected distribution of P values for a random uniform insertion of inserts.
Thus, although we cannot conclude that any particular cluster is “real” by statistical measures, the results in Figure 7 suggest that Ta-1 insertion is not governed solely by chance. Therefore, we compared the genomic environment of some arbitrarily chosen apparent short Ta-1 clusters with each other and with those apparently not clustered. These groups do not differ significantly from each other with respect to their GC content of their flank (P = 0.443), the local recombination rate (P = 0.826) and the gene density (P = 0.580). They also do not differ in the abundance of L1s in their flanking sequences (P = 0.669), suggesting that the apparent Ta-1 clusters are not genomic compartments where L1 elements accumulate at a higher rate.
Gap lengths (genomic distance between Ta-1 inserts) were measured and the largest for each chromosome was tabulated. Here the most significant large gap (P < 0.007 after Bonferroni correction) fell on Chromosome 19, a chromosome that contained only two inserts. However, one might question the validity of this finding because of the small number of Ta-1 elements. When the 24 P values (unadjusted) were inspected together, they again showed a tendency toward small values, whereas under a random insertion hypothesis, one would expect a uniform, rectangular distribution of values (Fig. 7). This clear tendency for gaps to be larger than expected provides additional evidence against a uniform, random model of Ta-1 insertion.
DISCUSSION
We now have 344 unique Ta-1-containing loci, including 139 that were not present in the human genome database. This represents a near-complete census of Ta-1-containing loci in the four individuals from whom we obtained our clones and in those individuals whose sequence is represented in the human genome database. We previously estimated by quantitative hybridization that an average human haploid genome contains 265 Ta-1 elements (Boissinot et al. 2000). We calculated which fraction of the average haploid number is covered by the present collection as follows: The number of Ta-1 elements at each Ta-1-containing locus for each of the four individuals can be deduced from our PCR data. The 217 loci, which we found amenable to PCR, contribute 255–265 (depending on the individual) Ta-1 elements to the four diploid genomes from which we obtained our clones.
Extrapolation of this value to the total number of Ta-1 inserts collected (344) gives a value of 408 to 422 Ta-1 inserts per individual, which is 204 to 211 Ta-1 elements per haploid genome. These values for haploid Ta-1 content based on cloning are remarkably similar to the 255–265 Ta-1 elements per haploid genome obtained by quantitative hybridization. Therefore, our collection is more representative of the genomic diversity of the active Ta-1 subfamily in humans than deduced from the human genome database. By the same calculation, the Ta-1 elements in the database account for only about one-half of the Ta-1 elements present in any of the four individuals. Thus, not surprisingly, the database provides a relatively incomplete picture of the ongoing Ta-1 amplification event.
The Ta-1 Family Is Not as Deleterious as Older L1 Families
The Ta-1 family contains a significantly larger fraction of autosomal full-length (FL) elements than do the L1PA2 and L1PA5 families (Fig. 3). Furthermore, the fraction of FL Ta-1 elements is the same for both fixed and polymorphic Ta-1 elements. Therefore, FL and truncated Ta-1 elements are (or have been) reaching fixation in humans at the same rate. Thus, FL Ta-1 elements (potentially capable of replication) are not sufficiently deleterious as to be subject to negative selection. These results contrast markedly with the situation for ancestral L1 families. These families suffered a significant loss of their FL L1 elements but not of autosomal L1 elements that are ≤500 bp or SINE elements (Boissinot et al. 2001).
We assessed this loss by using the proportion of FL ancestral L1 elements on the Y chromosome as a proxy for the proportion of FL elements originally present in these families. Unlike autosomes, most of the Y chromosome cannot undergo meiotic recombination with a homolog, a process that facilitates negative selection against deleterious alleles. Thus, the Y chromosome is a repository for deleterious alleles and retrotransposons (see Boissinot et al. 2000, and references therein). For example, the percentage of FL L1PA5 elements on the Y chromosome is ∼7.5 that on autosomes. This corresponds to an 87% loss of autosomal FL L1 elements. We cannot make the Y chromosomal and autosomal comparison for Ta-1 elements because the Y chromosome contains only a few Ta-1 elements.
As there is no easy way to excise interspersed L1 elements we proposed that the loss of FL autosomal elements from the ancestral L1PA2-L1PA5 families was due to negative selection, that is, the loss of FL L1-containing alleles from the population (Boissinot et al. 2001). Because only FL elements are capable of retrotransposition, presumably some aspect of L1 activity was sufficiently deleterious to have elicited negative selection. However, Ovchinnikov et al. (2001) proposed two other explanations for the different proportion of FL elements in the Ta and ancestral L1 families: more proficient production of FL elements by the Ta family than by ancestral families; and continual loss of FL elements from ancestral L1 families. However, the former explanation would not account for the difference between the Y chromosomal and autosomal content of FL L1 elements of the ancestral L1 families (Boissinot et al. 2000). The latter explanation is inconsistent with the fact that all ancestral L1 elements (FL and truncated) are now fixed in humans. Because selection acts on genetic variation, none of them could be cleared from the population by this means.
FL Ta-1 elements may not be as deleterious as were FL L1PA2 or L1PA5 elements for several reasons. As the Ta-1 family just emerged and is only about 5–10% the size of the ancestral families, it might not generate enough L1 products (e.g., L1 RNA or L1 proteins) that could be toxic to its host. Perhaps Ta-1 elements are inherently less active or the host has evolved ways to repress their activity. Interestingly, the fraction of FL L1PA2 is intermediate between that of the Ta-1 and L1PA5 families (Fig. 3 and Boissinot et al. 2001). Also, the copy number of L1PA2 is about one-half that of L1PA5 (∼5000 vs. ∼10,000, based on RepeatMasker output). Therefore, the activity of L1 in the human lineage has apparently decreased from L1PA5 to L1PA2 and then to Ta (also called L1PA1, IHGSC 2001), and along with it, the intensity of negative selection against it.
Biased Distribution of Ta-1 Inserts in GC-Poor Genomic Regions
On average, Ta-1 elements (fixed and polymorphic) are most abundant in GC-poor regions of the genome, and their distribution mimics the distribution of ancestral L1PA2 and L1PA5 families (Fig. 4). This observation agrees with the findings on whole-genome analysis (IHGSC 2001) but contradicts results that showed that, unlike older L1 families, Ta elements are randomly distributed with regard to GC content (Ovchinnikov et al. 2001). However, this latter analysis was based on a much smaller number of elements than ours (24 polymorphic Ta and 57 ancestral elements), and the method used to collect Ta elements may have been biased in favor of Ta elements located in GC-rich regions. Ovchinnikov et al. (2001) amplified Ta elements from the genome using 10-mer PCR primers of arbitrary sequence. This size primer would generally favor the generation of PCR products originating from GC-rich primer annealing sites (because of increasing stability of the primer template pairs with increasing GC content).
There are two possible explanations for the insertional bias of Ta-1 elements in GC-poor regions. First, Ta-1 elements could insert preferentially in AT-rich regions because such regions are statistically more likely to contain the target site recognized by the L1 endonuclease whose consensus is TTAAAA (Cost and Boeke 1998). However, the L1 endonuclease is not very specific and the majority of L1 elements have inserted into sites that differ from this consensus (Jurka 1997; Cost and Boeke 1998). Alternatively, Ta-1 elements may not be excluded from GC-rich regions but are selected against because they are deleterious. As GC-rich regions generally are gene rich, L1 insertions could be deleterious in any number of ways including insertional gene inactivation; alteration of gene transcriptional activity; alteration of transcript processing, for example, by the introduction of alternate splice sites or transcriptional termination signals (see Ostertag et al. 2002).
We found Ta-1 elements inserted into the introns of 46 known genes and they were twice as often oriented in the antisense direction with respect to the gene (67%) as compared with the sense direction (33%). There was no difference with respect to FL or truncated elements in this regard and the ancestral families showed the same trend. If the bias in orientation is due to negative selection, it is presumably based on the deleterious effect of either sense L1 DNA sequences or of the sense L1 transcripts themselves. An example of the former effect would be the introduction of the L1 polyadenylation site into an intron leading to premature transcriptional termination of the gene (Harendza and Johnson 1990; Ostertag and Kazazian Jr. 2001; Perepelitsa-Belancio and Deininger 2003). Another would be the introduction of signals that interfere with RNA processing (Lindtner et al. 2002; Floyd et al. 2003). The orientation bias could also have been imposed during insertion, say by interaction of the L1 retrotransposition apparatus with the transcriptional or transcript processing machinery. The fact that that snRNA U6–L1 and U4–L1 hybrids have been retroposed continually during the course of mammalian evolution may imply some intimacy between L1 retrotransposition and the splicing machinery (Buzdin et al. 2002, 2003a).
Both polymorphic (i.e., recently inserted) and fixed Ta-1 inserts show the same distribution with respect to GC-rich regions. Thus, if the relative lack of Ta-1 elements in GC-rich regions were due to negative selection then it must be relatively strong, as even putative recent inserts here would have already been lost from the population. Perhaps, inserts here are so deleterious that they are embryonic lethal and therefore would never be present in the population.
Chromosomal Distribution of Ta-1 Inserts
Ta-1 elements are not distributed randomly between chromosomes. Although there is a general tendency for the number of Ta-1 insertions to be directly related to chromosomal size and inversely related to gene content, chromosome 4 is an obvious exception. As there is no such bias of the ancestral L1PA2 and L1PA5 families towards chromosome 4, the biased distribution of Ta-1 on this chromosome is not because it is a haven for L1 insertions. In fact, the chromosomal distribution of ancestral L1PA2 and L1PA5 families is biased towards the sex chromosomes as is typical of L1 DNA in general (IHGSC 2001). In contrast, we observed no such bias of the Ta-1 family for the sex chromosomes. This distribution may merely reflect the fact that the Ta-1 amplification is still in its infancy. On the other hand, if accumulation on the sex chromosomes is a proxy for the deleterious effect of an L1 family (see Results and Boissinot et al. 2001), then the lack of sex chromosome bias of the Ta-1 family may be additional evidence that the Ta-1 is less deleterious than the ancestral L1 families.
The distribution of some of the Ta-1 elements within chromosomes also seems not to be uniform. Although our simulation studies showed that although any particular apparent Ta-1 cluster could not be supported statistically, Figure 7 shows that the distribution of P values for both clusters and gaps skewed towards low values greater than would be expected by a completely random uniform model of Ta-1 insertion. Although verifying the clustering of L1 inserts requires additional studies, previous evidence indicated that some genomic regions apparently experience a higher rate of L1 insertions than others. For instance, of 14 L1 insertions in known genes, four are in dystrophin and three are in the factor VIII gene (Ostertag and Kazazian Jr. 2001). In addition, two L1 elements inserted recently and independently in the same 1 kb region of gorilla and humans (DeBerardinis and Kazazian Jr. 1998). Given the size of primate genomes, it seems unlikely that these two independent insertions occurred by chance.
As Ta-1 insertion sites are not particularly enriched in old L1 elements, they apparently do not correspond to preferred sites for L1 insertion. Actually, preliminary analysis also indicates an apparent clustering of ancestral L1PA2 and L1PA5 inserts. Additionally, some of these apparent clusters do not overlap (A.V. Furano, L. Young, P.J. Munson, and S. Boissinot, unpubl.). The pattern of L1 insertions presumably reflects some aspect of the retrotransposition mechanism itself or the consequences of the inserts on genome function or integrity, or both. Furthermore, the possibility that the insertion patterns differ for L1 families of different evolutionary age suggests that evolutionary changes in the host may play a role in determining the insertional pattern. These evolutionary changes by the host may even in part be a response to L1 activity.
Conclusion
A major premise that motivated this work was that as the human Ta-1 family is currently undergoing amplification, the human genome database would provide only an incomplete census of Ta-1-containing loci in the human population. This proved to be the case; our polling of a single representative of just four ethnic populations identified 139 new Ta-1-containing loci as compared with the 205 identified in the human genome database (Table 1, column 8). As our cloning technique recovered ∼90% of Ta-1 elements from the examined individuals, we are confident that the current collection of 344 Ta-1-containing loci provides a less-biased “real time” view of this ongoing L1 amplification then that provided by the human genome database.
METHODS
Cloning of Ta-1 Inserts and Their Flanking Sequences
The 3′ termini and the 3′ flanking sequences of Ta-1 inserts were cloned from the DNA of four males from different ethnic origin: a Druze (Coriell Institute ID number: NA11522); a Biaka pygmy (NA10469); a Chinese (NA11321); and a Melanesian (NA10540). To minimize bias in collecting Ta-1-containing loci, we used the following anchor-PCR cloning strategy: DNA samples were sheared physically by nebulization and then ∼2 μg was treated for 30 min at 12°C with 6 units of T4 DNA polymerase in a total volume of 40 μL (50 mM Tris-HCl at pH7.5, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, 75 μg/mL BSA, 0.1 mM dNTP) to create blunt-ended fragments. After inactivation of the T4 DNA polymerase (10 min at 75°C), the 5′ hydroxyl termini were phosphorylated by incubating the reaction at 37°C for 30 min with 10 units of T4 polynucleotide kinase (with 5% polyethylene glycol 8000). After inactivation of the kinase (20 min at 75°C), the DNA fragments were ligated to a double-stranded DNA anchor (5′-TAGCTACAGCTGTAGCTGACAT-3′) with 400 units of T4 DNA ligase and 10μM of the double strand anchor. After three hours at room temperature the unligated anchors were removed by chromatography on a 1ml Sepharose CL-4B column. Ta-1 containing fragments were then amplified from ∼50 ng of the anchor ligated DNA in four 25 μl PCR reactions using an Idaho Technology Air-Thermo Cycler with 1 μM Ta-1-specific biotinylated primer (Primer 1 on Fig. 1) and 0.2μM of the anchor primer in 50 mM Tris-HCl (pH8.3), 2 mM MgCl2, 0.2 mM of each dNTP, 250 μg/ml BSA, 2% sucrose, 0.1 mM Cresol Red using the following conditions: denaturation, 94°C, 0”; annealing, 50°C, 0”; extension, 72°C, 60”, 35 cycles. The PCR products were then chromatographed on a Sepharose CL-4B column, ethanol precipitated, dissolved in 40μl TE (pH 8.0), and captured on streptavidin coated magnetic beads (Dynabeads M-280) following the procedure recommended by the company (DYNAL). The beads were then resuspended in 20μl of TE (pH8.0) and 1 μl was used in a PCR reaction as above but using 0.5 μM each of the non-biotinylated Ta-1-specific and anchor primers. The PCR products were purified through Sepharose CL-4B, ethanol precipitated, and cloned into the pGEM-T vector (Promega). Bacterial colonies were lifted from the plates onto Hybond-N membranes (Amersham) and processed as described in (Buluwela et al. 1989). The filters were hybridized overnight at 43°C to an oligonucleotide probe (Primer 2 on Fig. 1) cognate to the ACA trinucleotide diagnostic of the Ta family (Boissinot et al. 2000). Positive clones were grown overnight in 100 μL of LB media. We determined the orientation and the size of each cloned insert by PCR using the Ta-1 specific primer and primers located in the plasmid. These PCRs were carried out in 96-well plates in an MJ Research PTC 100 Thermocycler under the following conditions: denaturation, 94°C, 1 h; annealing, 48°C, 1 h; extension, 72°C, 1 h. Because the Ta-1 specific primer is located 484 bp from the 3′ end of an L1 element, only inserts that were at least 700 bp long were sequenced (starting at the anchor end) to ensure the isolation of flanking genomic sequence. This procedure was followed for the first individual analyzed, the Druze, but for the three next individuals (Pygmy, then Chinese, and finally Melanesian) an additional step was added to avoid the sequencing of inserts already cloned from the preceding individuals. Dot blots of clones longer than 700 bp were hybridized to pools of the PCR primers cognate to the 3′ flanks of the Ta-1 elements collected from the individuals analyzed previously. Only clones that did not hybridize to primer pools were sequenced.
PCR Analysis of Ta-1-Containing Loci
The polymorphism of the Ta-1 inserts was determined using two PCRs as described before (Boissinot et al. 2000). PCRs were performed on a panel of eight individuals of different ethnic origins: Biaka Pygmy (NA10469); Druze (NA11522); Chinese (NA11321); Maya (NA10975); Mbuti Pygmy (NA10492); Melanesian (NA10540); Cambodian (NA11373); and Karitiana (NA10965). All DNAs were purchased from the Coriell Institute for Medical Research.
Bioinformatics
For each Ta-1 insert we determined six parameters.
Chromosomal location. We identified the genomic location of the cloned 3′-flanking sequences in the public data bases using both the BLAST (Altschul et al. 1990) and BLAT search programs (http://genome.ucsc.edu). Two identifiers were associated with each flanking sequence: the GenBank accession number of the sequence with the highest similarity to our cloned sequence and the coordinates of the cloned sequence in the December 2001 assembly as given by the human genome browser at UCSC (the format of the coordinates is ChrN:start-end). To be certain that our database of Ta-1 elements was complete, we used the sequences of primers 1 and 2 (Fig. 1) in a BLAST search of the database to collect all of the intact Ta-1 elements that might have been missed by our cloning.
Size and structure. These were obtained from the human genome sequence when the insertion site was occupied by a Ta-1 element in the public database. Many cloned Ta-1-containing loci lacked an insert in the database. We determined the size of these inserts by PCR.
GC content of the insertion site. The GC content of 10 kb (5 kb on each side of the Ta-1 inserts), 20 kb, and 100 kb of flanking DNA was determined using the RepeatMasker program (A.F.A. Smit and P. Green, http://ftp.genome.washington.edu/RM/RepeatMasker.html). As suggested by Ovchinnikov et al. (2001), the immediate 150 bp adjacent to the elements were excluded because of the A-rich tail that flanks L1 elements at their 3′ end. The RepeatMasker server is at www.repeatmasker.org.
Abundance of other repeated DNAs. The numbers of SINEs, LINEs, and LTR-containing retrotransposons as well as the total abundance of repeats in 100 kb of flanking DNA were also determined using the RepeatMasker program.
Local recombination rate. Recombination rates were obtained from Web Table E of Kong et al. (2002). The recombination rate of each Ta-1-containing genomic region was scored as the recombination rate calculated for the closest marker analyzed by Kong et al. (2002) within a window of 1 Mb.
Gene density. The number of known genes found in the same 5-Mb window as each Ta-1 element was obtained from the RefSeq Genes table available at http://genome.ucsc.edu/. The function and orientation of all genes located within 200 kb of a Ta-1 insertion site were also obtained from the human genome browser at UCSC and from the SOURCE Web page at http://genome-www5.stanford.edu/.
For comparison, we also collected elements belonging to the older L1PA2 and L1PA5 families (Smit et al. 1995; Boissinot et al. 2001) that are no longer active. These were randomly selected from table chrN_rmsk at http://genome.ucsc.edu/. So that they were comparable to the cloned Ta-1 inserts, only elements that were at least 500 bp long were analyzed. For each of these elements we collected the same information as for the Ta-1 elements.
Statistical Tests for the Distribution of Ta-1 Inserts Between Chromosomes
Proportionality of the number of insertions to the length of a chromosome was tested with simple linear regression and was highly significant (data not shown). Least-squares linear regression constrained through the origin was used to estimate the proportionality factor. The 95% confidence limits for an individual observation were calculated assuming only the Poisson counting variability. These confidence limits were determined by adding and subtracting 1.96 × the square root of the predicted value to the regression predicted value. Gene density and Ta-1 insertion (hereafter referred to as insert) density were determined by dividing the total number of genes or inserts by the length of the chromosome after removal of the nonsequenced regions.
Statistical Tests for Distributions of Ta-1 Inserts Within Each Chromosome
Apparent Clusters
We refer to any group of k consecutive inserts as a run, which is characterized by its span, that is, the distance (in bp) from the first to the last insert. To search for clusters we first determined the runs of all possible k (i.e., k = 2, 3...n, where n is the number of inserts on the chromosome) by grouping neighboring inserts. Initially, all overlapping runs of k = 2 were identified, then all overlapping runs of k = 3, and so on up to k = n. For example, we grouped inserts 1 and 2, then 2 and 3, then 3 and 4, then 1, 2, and 3, then 2, 3, and 4, etc. Next, the number of bases spanned by each run was calculated; this distance is the k-span. Finally, we determined the shortest span over all runs of a given k (minimal k-span) and checked whether the minimal k-span would qualify as a cluster by comparing it with the minimal k-span expected in the “null” distribution of elements.
We determined the “null” distribution of the minimal k-span for a given chromosome by simulating a random insertion process for the same number of inserts into a sequence of the same length. The minimal k-span was determined for 10,000 simulations to yield 10,000 minimal k-spans that were then sorted so that the quantiles of the null distribution could be determined. For example, for each k the 1 percentile is the minimal span that is greater than or equal to 1% of the 10,000 random minimal k-span values. The statistical significance of an apparent cluster in the original data (for a prespecified k) may now be checked by reference to this null distribution. If the length of an apparent cluster falls below the 1 percentile length, then we would consider the putative cluster significant at P = 0.01. We refer to the significance level for this k-specific test as the “k-specific P-value.”
Because we could not a priori specify the number of inserts (k) that would be found in an apparent cluster, the k-specific test was determined for all values of k, from two to n (the number of elements on a chromosome) and selected the apparent cluster with the minimal k-specific P value. However, testing for significance at all values of k involves multiple tests, and as each test has some probability of error, the P values need to be adjusted for this effect. We did this by performing a second simulation to find the null distribution of the “most significant minimal k-span”, that is, that run of k sites associating with the smallest quantile of the k-specific null distribution. Here, the minimal k-spans for a simulated, randomly inserted sequence was determined for all possible k values, along with their corresponding quantiles from the k-specific null distribution. The minimal quantile was recorded, and the process repeated 10,000 times. A statistical test, adjusting for the fact that k is not specified (i.e., all k's are considered), of the significance of the smallest quantile obtained in the actual data can easily be made by comparison to this second null distribution of random minimal quantiles. P values from this test are referred to here as “k-unspecific P values.”
Apparent Gaps
Gaps are defined as the number of bases between adjacent inserts on a chromosome. The distribution for the largest gap in a chromosome can be found in the same manner as above under the random insertion model, and the quantiles of the null distribution can be determined. An observed large gap is tested for significance by comparison with the appropriate quantile of the null distribution. Significance is inferred when the observed gap exceeds the (1-p) quantile, where p is the desired significance level.
Polymorphic Clusters Near Fixed Elements
A special test of the potential clustering of polymorphic inserts near fixed ones was performed as follows. Instead of considering the span or length of any run of k inserts, only those runs containing at least one fixed and one polymorphic insert were considered. The random uniform insertion model was modified to allow for random insertion of only the polymorphic inserts. Other details were as for the test for apparent clusters.
Multiple Comparisons Adjustment
Each of these statistical tests was computed separately for each of the 24 chromosomes. To adjust for the fact that 24 independent tests were performed, we applied a Bonferroni adjustment by multiplying the P values for the tests given above by 24. To avoid confusion, we refer to the original P values as “unadjusted” and to the P values adjusted by the Bonferroni correction for multiple chromosomes as “corrected” or “adjusted” P values.
Acknowledgments
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2326704. Article published online before print in June 2004.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Badge, R.M., Alisch, R.S., and Moran, J.V. 2003. ATLAS: A system to selectively identify human-specific L1 insertions. Am. J. Hum. Genet. 72: 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot, S. and Furano, A.V. 2001. Adaptive evolution in LINE-1 retrotransposons. Mol. Biol. Evol 18: 2186–2194. [DOI] [PubMed] [Google Scholar]
- Boissinot, S., Chevret, P., and Furano, A.V. 2000. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 17: 915–928. [DOI] [PubMed] [Google Scholar]
- Boissinot, S., Entezam, A., and Furano, A.V. 2001. Selection against deleterious LINE-1-containing loci in the human lineage. Mol. Biol. Evol. 18: 926–935. [DOI] [PubMed] [Google Scholar]
- Brouha, B., Schustak, J., Badge, R.M., Lutz-Prigge, S., Farley, A.H., Moran, J.V., and Kazazian Jr., H.H. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. 100: 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buluwela, L., Forster, A., Boehm, T., and Rabbitts, T.H. 1989. A rapid procedure for colony screening using nylon filters. Nucleic Acids Res. 17: 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton, F.H., Loeb, D.D., Voliva, C.F., Martin, S.L., Edgell, M.H., and Hutchison III, C.A. 1986. Conservation throughout mammalia and extensive protein-encoding capacity of the highly repeated DNA long interspersed sequence one. J. Mol. Biol. 187: 291–304. [DOI] [PubMed] [Google Scholar]
- Buzdin, A., Ustyugova, S., Gogvadze, E., Vinogradova, T., Lebedev, Y., and Sverdlov, E. 2002. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of L1. Genomics 80: 402–406. [DOI] [PubMed] [Google Scholar]
- Buzdin, A., Gogvadze, E., Kovalskaya, E., Volchkov, P., Ustyugova, S., Illarionova, A., Fushan, A., Vinogradova, T., and Sverdlov, E. 2003a. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 31: 4385–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzdin, A., Ustyugova, S., Gogvadze, E., Lebedev, Y., Hunsmann, G., and Sverdlov, E. 2003b. Genome-wide targeted search for human specific and polymorphic L1 integrations. Hum. Genet. 112: 527–533. [DOI] [PubMed] [Google Scholar]
- Cost, G.J. and Boeke, J.D. 1998. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37: 18081–18093. [DOI] [PubMed] [Google Scholar]
- DeBerardinis, R.J. and Kazazian Jr., H.H. 1998. Full-length L1 elements have arisen recently in the same 1-kb region of the gorilla and human genomes. J. Mol. Evol. 47: 292–301. [DOI] [PubMed] [Google Scholar]
- Dewannieux, M., Esnault, C., and Heidmann, T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35: 41–48. [DOI] [PubMed] [Google Scholar]
- Esnault, C., Maestre, J., and Heidmann, T. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24: 363–367. [DOI] [PubMed] [Google Scholar]
- Floyd, J.A., Gold, D.A., Concepcion, D., Poon, T.H., Wang, X., Keithley, E., Chen, D., Ward, E.J., Chinn, S.B., Friedman, R.A., et al. 2003. A natural allele of Nxf1 suppresses retrovirus insertional mutations. Nat. Genet. 35: 221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano, A.V. 2000. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog. Nucleic Acids Res. Mol. Biol. 64: 255–294. [DOI] [PubMed] [Google Scholar]
- Harendza, C.J. and Johnson, L.F. 1990. Polyadenylylation signal of the mouse thymidylate synthase gene was created by insertion of an L1 repetitive element downstream of the open reading frame. Proc. Natl. Acad. Sci. 87: 2531–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison III, C.A., Hardies, S.C., Loeb, D.D., Shehee, W.R., and Edgell, M.H. 1989. LINEs and related retroposons: Long interspersed repeated sequences in the eucaryotic genome. In Mobile DNA (eds. D.E. Berg and M.M. Howe), pp. 593–617. American Society for Microbiology, Washington, DC.
- International Human Genome Sequencing Consortium (IHGSC). 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Jurka, J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. 94: 1872–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberland, M.L., Divoky, V., Prchal, J., Schwahn, U., Berger, W., and Kazazian Jr., H.H. 1999. Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum. Mol. Genet. 8: 1557–1560. [DOI] [PubMed] [Google Scholar]
- Kong, A., Gudbjartsson, D.F., Sainz, J., Jonsdottir, G.M., Gudjonsson, S.A., Richardsson, B., Sigurdardottir, S., Barnard, J., Hallbeck, B., Masson, G., et al. 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241–247. [DOI] [PubMed] [Google Scholar]
- Lindtner, S., Felber, B.K., and Kjems, J. 2002. An element in the 3′ untranslated region of human LINE-1 retrotransposon mRNA binds NXF1(TAP) and can function as a nuclear export element. RNA 8: 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium (MGSC). 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Moran, J.V. and Gilbert, D. 2002. Mammalian LINE-1 retrotransposons and related elements. In Mobile DNA II (eds. N.L. Craig et al.), pp. 836–869. ASM Press, Washington, DC.
- Myers, J.S., Vincent, B.J., Udall, H., Watkins, W.S., Morrish, T.A., Kilroy, G.E., Swergold, G.D., Henke, J., Henke, L., Moran, J.V., et al. 2002. A comprehensive analysis of recently integrated human Ta L1 elements. Am. J. Hum. Genet. 71: 312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag, E.M. and Kazazian Jr., H.H. 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35: 501–538. [DOI] [PubMed] [Google Scholar]
- Ostertag, E.M., DeBerardinis, R.J., Goodier, J.L., Zhang, Y., Yang, N., Gerton, G.L., and Kazazian Jr., H.H. 2002. A mouse model of human L1 retrotransposition. Nat. Genet. 32: 655–660. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov, I., Troxel, A.B., and Swergold, G.D. 2001. Genomic characterization of recent human LINE-1 insertions: Evidence supporting random insertion. Genome Res. 11: 2050–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepelitsa-Belancio, V. and Deininger, P. 2003. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 35: 363–366. [DOI] [PubMed] [Google Scholar]
- Sheen, F.M., Sherry, S.T., Risch, G.M., Robichaux, M., Nasidze, I., Stoneking, M., Batzer, M.A., and Swergold, G.D. 2000. Reading between the LINEs: Human genomic variation induced by LINE-1 retrotransposition. Genome Res. 10: 1496–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski, J., Fanning, T.G., and Singer, M.F. 1988. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol. Cell. Biol. 8: 1385–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A.F.A., Tóth, G., Riggs, A.D., and Jurka, J. 1995. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 246: 401–417. [DOI] [PubMed] [Google Scholar]
- Wei, W., Gilbert, N., Ooi, S.L., Lawler, J.F., Ostertag, E.M., Kazazian Jr., H.H., Boeke, J.D., and Moran, J.V. 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21: 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]