Abstract
LTR-containing retrotransposons reverse transcribe their RNA genomes, and the resulting cDNAs are integrated into the genome by the element-encoded integrase protein. The yeast LTR retrotransposon Ty1 preferentially integrates into a target window upstream of tDNAs (tRNA genes) in the yeast genome. We investigated the nature of these insertions and the target window on a genomic scale by analyzing several hundred de novo insertions upstream of tDNAs in two different multicopy gene families. The pattern of insertion upstream of tDNAs was nonrandom and periodic, with peaks separated by ∼80 bp. Insertions were not distributed equally throughout the genome, as certain tDNAs within a given family received higher frequencies of upstream Ty1 insertions than others. We showed that the presence and relative position of additional tDNAs and LTRs surrounding the target tDNA dramatically influenced the frequency of insertion events upstream of that target.
Ty1 is an LTR retrotransposon in Saccharomyces cerevisiae that integrates specifically into the host genome and generates target-site duplications. Previous work has shown that the Ty1's integrase protein specifically integrates Ty1 cDNA into ∼700 bp “windows” upstream of genes transcribed by RNA polymerase III (pol III), which includes all tDNAs and some snRNA genes, such as the U6 gene (Ji et al. 1993; Devine and Boeke 1996; Bolton and Boeke 2003). This observation was made initially by analyzing de novo insertion events on a single chromosome, chromosome III (Ji et al. 1993). By subsequently placing prospective target genes on plasmids, diverse genes transcribed by RNA pol III were shown to be efficient targets, and targeting depended on transcriptional competence of the pol III gene (Devine and Boeke 1996).
The targeting of Ty1 insertion events to regions upstream of tDNAs has been interpreted as an evolutionary mechanism to protect the yeast cell from insertion events into genes (Boeke and Devine 1998), and recent work has shown that intergenic regions upstream of tDNAs in yeast are larger than other intergenic regions of the yeast genome (Bolton and Boeke 2003). Retrotransposon targeting to regions upstream of tDNAs has evolved in parallel several times in different systems. The mechanism has apparently evolved at least twice in S. cerevisiae, once for Ty1, a member of the copia family of LTR retroelements, and once for Ty3, a distantly related gypsy family member (Voytas and Boeke 2002; Sandmeyer 2003). In Dictyostelium discoideum, anorganism that, like S. cerevisiae, has a densely packed genome, the non-LTR retrotransposon TRE5-A integrates upstream of pol III genes as well, close to the position of pol III preinitiation factor TFIIIB complex (Winckler et al. 2002). Therefore, despite distinct mechanisms for reverse transcription and integration of the retrotransposons in different hosts, the host–retrotransposon relationship has converged on a similar strategy. Even if retrotransposon insertion per se is not harmful to the cell, the position of insertion events can have significant repercussions on genomic structure and profound effects on the expression level of nearby genes (Winston et al. 1984; Roeder et al. 1985; Natsoulis et al. 1989; Kinsey and Sandmeyer 1991; Bolton and Boeke 2003), necessitating the careful regulation of integration specificity.
The completion of the yeast genome sequence allowed a study of the distribution and position of existing retroelements in the yeast genome. This analysis showed the correlation between the endogenous Ty1–Ty4 element families and pol III genes (90% correlation for Ty1); Ty5 elements, in contrast, were found near sites of silent chromatin (Kim et al. 1998). Ty1, Ty2, and Ty4 elements were found in regions upstream of tDNAs, whereas Ty3 insertions were always found within a few bases of RNA pol III transcription start sites (TSSs). That study also revealed some disparity in the distribution of genomic Ty elements, in that there was a higher density of retrotransposon DNA on the smaller chromosomes, I, III, and VI, whereas the larger ones, VII, XV, and IV, had a lower density of retrotransposon sequence, with some areas, such as a 434-kb region on the left arm of chromosome IV, completely devoid of Ty sequences. Interestingly, only 66% of genomic tDNAs were associated with retrotransposon sequence (Kim et al. 1998), leaving open the possibility that not all tDNAs were equally good targets.
Important aspects of the Ty3 and Ty5 targeting mechanisms have been elucidated, and in both cases, the target specificity is mediated by interactions between the transposon integrase protein and a host protein present at the target site. Ty3 integration always occurs within 1–2 bases from the TSS, and Ty3 integrase protein has been shown to interact with subunits of both the TFIIIC and TFIIIB protein complexes, preinitiation factors of RNA pol III (Yieh et al. 2000, 2002; Aye et al. 2001). Ty5 integrase mediates integration of the Ty5 transposon into silent chromatin through an interaction with the Sir4 protein (Zou and Voytas 1997; Zhu et al. 1999, 2003; Xie et al. 2001). In contrast, no such interactions have been observed with the Ty1 integrase.
In this work, we characterize the targeting preferences of Ty1 integration in more detail than previous studies by analyzing several hundred de novo insertion events upstream of two tDNA gene families, copies of which are spread throughout the genome. We use a robust PCR assay to amplify de novo insertion events. Sequencing these PCR products allowed high-resolution analysis of the positions of the new insertions. We show that, upstream of tDNAs in two different gene families, the patterns of integration events are highly nonrandom, both globally and locally, even though the coding regions within each tDNA family are identical. The pattern of insertion events upstream of the tDNAs appears to be periodic, with peaks separated by ∼80 bp. On a genomic level, certain tDNAs within the identical gene families are preferred targets for integration. We show that local genomic features, including additional tDNAs and LTR sequences and their relative positions have significant effects on the activity of specific tDNA targets.
RESULTS
A New Physical Assay for Targeted Ty1 Integration Into the Yeast Genome
To study Ty1 targeted integration, we developed a simple PCR-based assay. Using a primer complementary to the 3′ end of a specific tDNA family and combining it with a primer complementary to the end of a marked Ty1, PCR is used to amplify insertions from cell populations that have undergone transposition (Fig. 1). This method has been used to analyze genome-wide Ty1 insertions (Smith et al. 1995), but our method differs from published methods in several ways that make it more robust and versatile.
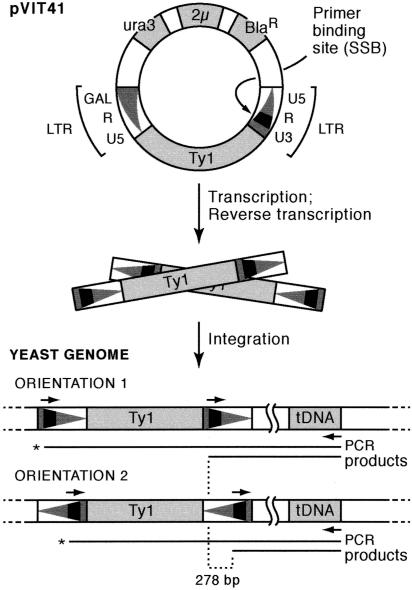
Figure 1.
The PCR assay for targeted integration upstream of tDNAs in S. cerevisiae. The pVIT41 donor plasmid (Lauermann et al. 1997) contains a unique oligonucleotide-binding site referred to as SSB. LTRs are shown as triangles, with the wide part of the triangle representing the U3 region. After induction on galactose medium to express the Ty1, the Ty1 transcript is translated, reverse transcribed, and the resulting cDNA is integrated into the yeast genome in either orientation. Insertion events are detected by using a primer complementary to the SSB in the Ty1 element in combination with a tDNA-specific primer. PCR products labeled with an asterisk (*) are >6 kb long and are not generated by the PCR conditions used in these experiments.
Previous analyses of insertions at specific targets relied on the presence of high-copy plasmid targets (Devine and Boeke 1996), the use of Southern blotting of PCR products amplified from insertions into genomic DNA (Scholes et al. 2001), or the use of an end-labeled oligonucleotide (Smith et al. 1995; Lee et al. 1998, 2000; Griffith et al. 2003). The method described here is easier, because it uses direct visualization of PCR products on an ethidium bromide-stained agarose gel and does not require blotting. To achieve this, we used a high-copy donor Ty1 construct, pVIT41 (Smith et al. 1995; Lauermann et al. 1997), containing a unique 15-bp oligonucleotide marker in the LTR called SSB after the fact that it contains SacI, SmaI, and BamHI restriction sites (Fig. 1). This sequence distinguishes the introduced donor Ty1-SSB element from endogenous Ty1s, and is absent from the yeast genome. The use of the SSB marker in combination with a target gene-specific primer for PCR allowed specific amplification of new, donor element-derived transposition events.
The general principle of the assay is shown in Figure 1. Insertions upstream of any tDNA can be amplified by using two primers, one complementary to the SSB marker, and the other to the target tDNA family of choice. The tGly family was chosen because SUF16, the Glycine tDNA on chromosome III, was previously shown to be an efficient target for Ty1 transposition (Ji et al. 1993; Devine and Boeke 1996), and its tGly(GCC) family has 16 members, all with identical coding regions, but different flanking sequences, making it a robust target for PCR. Similarly, the tThr family was chosen for comparison because it contains 11 scattered identical tDNA copies. Transposition is induced in a population of cells from which genomic DNA is prepared. The resulting genomic DNA therefore contains a population of insertions at different positions upstream of several different genomic target tDNAs. When PCR is performed and the products are run on a gel, the product lengths reflect the distance of the insertion events from the tDNA and the banding pattern reflects the target selection pattern. Therefore, analysis of populations of insertion events by this method gives a simple visual representation of target preferences and the integration window.
A combination of four primer pairs was used in the PCR analysis of integration events. In the case of both tDNA families, primers complementary to the extreme 3′ end of the tDNA worked best in the PCR. The Ty1 SSB marker is in the U3 region of the LTR. By using two complementary primers, containing either strand of the 15-mer SSB marker sequence, Ty1 insertions occurring in both orientations could be amplified. The asymmetrical placement of the SSB oligo-binding site in the LTR results in a 278-base difference in size of the PCR products generated from amplification of insertions in one orientation (orientation 1) compared with the other (orientation 2) (Fig. 1).
The Pattern of PCR Products Suggests Periodic Insertion
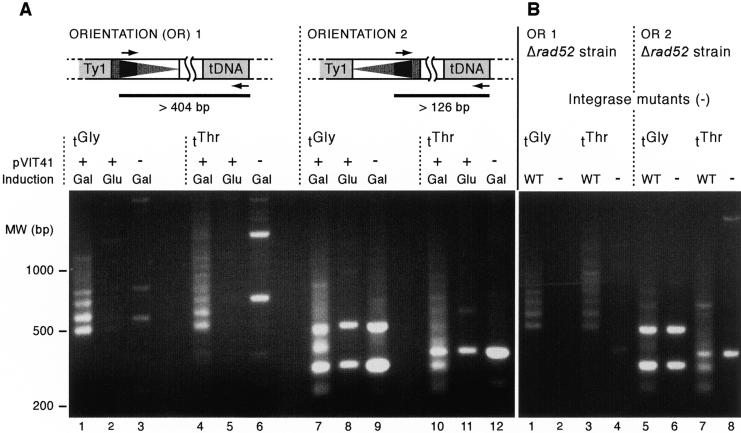
Insertions in both orientations were amplified from genomic DNA prepared from a population of galactose-induced yeast cells using combinations of the SSB-containing primers and tDNA primers (Fig. 2A). Using the tGly primers, the products show a clear ladder pattern, indicating periodicity of insertion. The bands appear separated by ∼80 bp, consistent with previous observations that insertions upstream of a plasmid-borne copy of SUF16, a Glycine tDNA, were also periodic, with peaks separated by 80 ± 10 bp (S.E. Devine and J.D. Boeke, unpubl.). The observed banding pattern only occurred when Ty1 expression was induced with galactose (Fig. 2A, lane 1) and not when grown on glucose or in the absence of the Ty1 plasmid (lanes 2 and 3). To test the generality of the pattern, we also analyzed insertions upstream of genomic tThr genes. The pattern is similar for insertions upstream of tGly and tThr in the genome (Fig. 2A, cf. lane 1 with lane 4) although the bands for tThr were less distinctly spaced.
Figure 2.
The insertion pattern upstream of the tGly and tThr gene families is periodic and depends on the expression of the marked Ty1 element and the Ty1 integrase protein. Four primer pairs were used to identify Ty1 insertions upstream of tGly and tThr genes in both orientations. (A) PCR products from each primer combination resulting from galactose induction with the pVIT41 Ty1 donor plasmid, without induction or without the plasmid. (B) The periodic insertion pattern depends on the presence of wild-type Ty1 integrase and does not require the homologous recombination pathway. Inductions were completed in a Δrad52 strain. The pNB19 plasmid containing a mutation in the catalytic site of integrase (–) or pVIT41 (WT) were used as the source of Ty1.
Analysis of insertions using the Ty1 SSB primer that generates the shorter PCR product (orientation 2) shows that the pattern of insertions is similar to that seen with the orientation 1 Ty1 primer (Fig. 2A, cf. lanes 1 and 7), except that, as expected, the bands are smaller by 278 bp, due to the asymmetrical position of the SSB marker in the LTR. However, the orientation 2 primer shows a higher background of nonspecific amplification products (Fig. 2A, lanes 8,9,11,12). These bands represent amplification from endogenous elements, and running the gel longer shows that these bands are different sizes from the bands in the experimental lane. The orientation 2 PCRs also show the same periodicity of insertion seen using the orientation 1 primer, meaning that the pattern and approximate frequency of insertions is the same in both orientations, consistent with our earlier studies (Natsoulis et al. 1989; Ji et al. 1993; Devine and Boeke 1996; Bolton and Boeke 2003). Because the PCR amplifies part of the LTR and the tDNA, the minimum expected size of the PCR product (generated if the insertion occurred into the TSS) is 404 bp for orientation 1 and 126 bp for orientation 2. The minimum observed PCR product size, however, is 80–90 bp larger than this (Fig. 2A, lanes 1,4,7,10), indicating that insertion at the right edge of the integration window is 80–90 bases upstream of the TSS, as expected from previous studies (Ji et al. 1993; Devine and Boeke 1996; Bolton and Boeke 2003). We performed the same assays in the presence of catalytic mutants in integrase (IN), which showed that the observed PCR products depend on functional IN (Fig. 2B). As expected, the background bands generated by use of the orientation 2 primer were not dependent on integrase, as they represent amplification of endogenous elements. Integrase mutant experiments were performed in a Δrad52 strain to show that the resulting pattern is due to integrase-mediated insertion events, and not by insertion via the homologous recombination pathway (Fig. 2B). Secondly, deletion of RAD52 reduces reversion of the plasmid-borne integrase mutant to wild type by preventing recombination with genomic Ty1 elements.
Sequence Analysis Reveals Unequal Genomic Distribution of Insertions
We cloned and sequenced hundreds of PCR products from insertion events in two different strains to confirm and study the fine structure of the PCR products. This analysis allowed us to determine both the genomic distribution of insertions and to plot their distribution pattern upstream of the tDNAs.
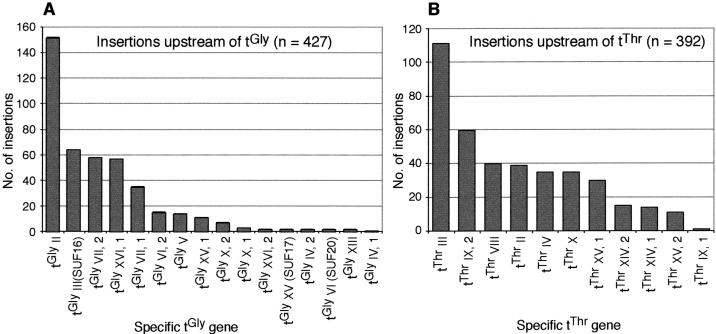
Although it has been known for some time that de novo Ty1 insertion events occur upstream of genes transcribed by RNA pol III, it is unknown whether new insertion events prefer specific pol III genes. In Saccharomyces cerevisiae, tDNAs are spread throughout the genome and are not clustered in repeat regions as they are in some other organisms. Our analysis allows for a detailed comparison of target preferences within multicopy gene families and revealed distinct genomic preferences. Within both the tGly and tThr gene families, the majority of insertion events occurred upstream of only a few tDNAs, whereas the others received strikingly fewer insertion events (Fig. 3A,B). A total of 77% of the insertions upstream of tGly genes occurred upstream of only 25% of the family's members, and 53% of all tThr upstream insertions occurred at 25% of the tThr genes.
Figure 3.
Number of insertions upstream of each member of the tGly and tThr gene families. (A) Number of insertions upstream of each of the 16 genomic tGly genes. All tGly genes assayed had GCC as their anticodon. Roman numerals represent chromosome on which the copy exists; arabic numerals after the comma represent the copy number in cases where more than one tGly exists on the same chromosome. For example, tGly IV, 1, represents the first (5′-most on the Watson strand) of the two copies of tGly on chromosome IV. The name for this gene in the Saccharomyces genome database (http://www.yeastgenome.org/) is tG(GCC)D1. (B) Number of insertions upstream of each of the 11 genomic tThr genes. All tThr assayed had AGU as their anticodon and were named by the same conventions described for tGly.
Sequence Analysis Confirms Periodicity of Insertion Upstream of the tDNA
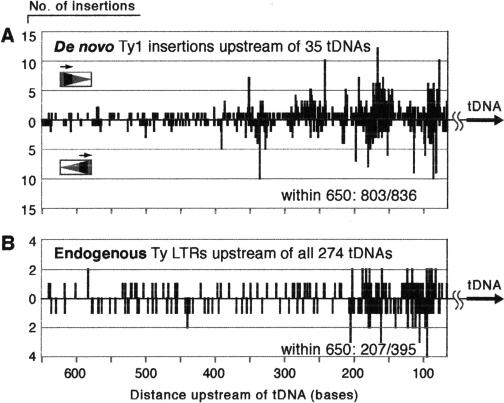
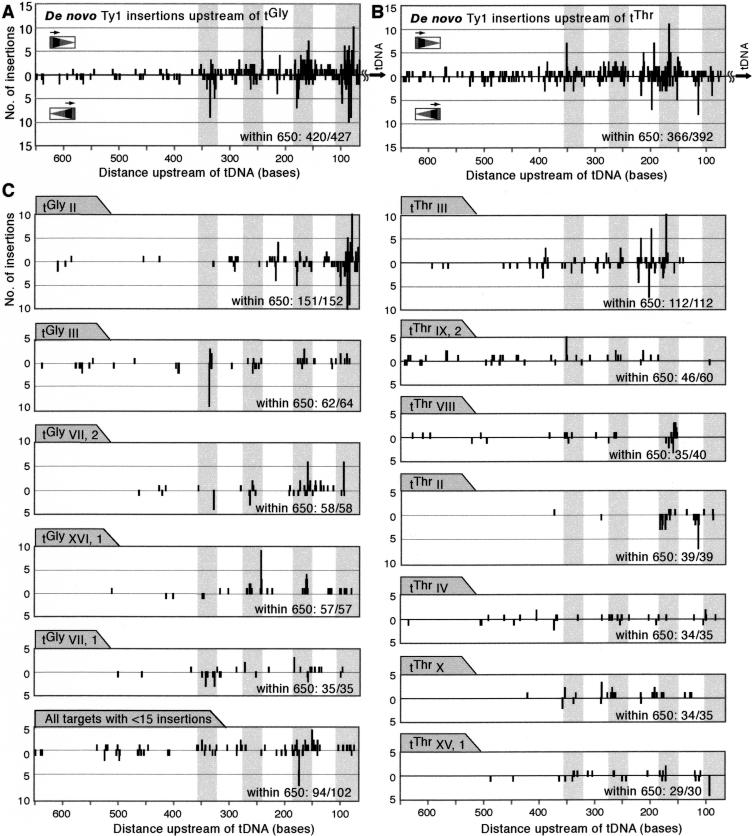
The agarose gel-banding pattern (Fig. 2) shows that the composite pattern of a population of insertions is periodic upstream of tDNAs. When each insertion event is analyzed independently and all of the insertion positions are plotted versus the distance upstream of the tDNA start site, a similar periodic pattern emerges (Fig. 4A). There are discrete peaks around 85, 170, 265, and 335 bases upstream of the tDNA (Fig. 4A). PCR products were generated from insertion events in two different strains, JB1217 (GRF167 background, n = 651 insertion events) and BY4741 (S288C background n = 185; Table 1), in which tDNA surrounding regions are largely identical (one exception will be noted). The insertion pattern in each strain was very similar (see Supplemental data). Figure 4A shows a composite of insertions upstream of 35 different tDNAs with disparate upstream sequences. In addition to amplifying insertions upstream of the 16 copies of tGly, the tGly primer also amplified eight insertions upstream of four different tAla genes and one insertion upstream of a tGly isoacceptor variant, SUF5 (Table 1). Similarly, the tThr primer amplified eight insertions upstream of three different tSer genes in addition to insertions upstream of the 11 tThr genes. All insertions within 650 bases upstream of the tDNA are included in Figure 4A. Each peak represents insertions into a single nucleotide position, and multiple insertions into the same nucleotide position are from different PCR reactions. A total of 108 independent PCR reactions were performed, and 12 or 24 products were cloned and sequenced from each reaction. To ensure against overestimating integration events into identical nucleotide positions by counting PCR siblings, insertions into the same nucleotide position were only counted if they were from independent PCR reactions (see Methods for details).
Figure 4.
Distribution of de novo Ty1 insertions and endogenous Ty retrotransposon sequences upstream of tDNA targets in the yeast genome. (A) Composite diagram of all sequenced de novo insertion events within 650 bases upstream of the TSS of 35 different genomic tDNAs (n = 803). Each line represents insertion events into a single base position. The frequency of insertion events into a position is shown vs. the distance upstream of the tDNA target. Insertions begin 65 bases upstream of the tDNA, shown at the far right, and distance increases leftward across the x-axis. Ty1 insertions amplified using the orientation 1 primer are shown above the x-axis; those amplified with the orientation 2 primer are shown below the line. (B) Position of endogenous Ty1, Ty2, and Ty4 LTRs in the yeast genome upstream of the mature 5′ processed end of the tDNA target.
Table 1.
Summary of PCR Product Sequences
| Strain BY4741 | Strain JB1217 | Insertions within 650 bases | Total insertions | |
|---|---|---|---|---|
| tGly (16 genes) | 94 | 333 | 420 | 427 |
| tThr (11 genes) | 82 | 310 | 366 | 392 |
| Subtotal (27 genes) | 176 | 643 | 786 | 819 |
| Othera (8 genes) | 9 | 8 | 17 | 17 |
| Total (35 genes) | 185 | 651 | 803 | 836 |
Eight insertions upstream of three different tSer, eight insertions upstream of four tAla and one insertion upstream of SUF5 (tGly isoacceptor).
Using the yeast genome sequence (http://www.yeastgenome.org/), we can compare the endogenous positions of tDNA-proximal LTRs to the de novo insertion pattern. Using the tDNA database created by Bolton (http://www.bs.jhmi.edu/MBG/boekelab/Supplements/Supplements.html), we plotted the positions of LTRs (Ty1, Ty2, and Ty4) relative to the mature processed 5′ end of the tDNA target (Fig. 4B). There are 274 tDNAs and 395 LTRs in the yeast genome (Bolton and Boeke 2003). A total of 207/395 (52%) of LTRs are within 650 bases from a tDNA. Interestingly, when the composite of all existing LTR positions is examined, it looks surprisingly similar to the composite insertion pattern of de novo insertions. The peaks around –85 and –170 appear to be generally favorable positions for insertions upstream of tDNAs.
Integration Pattern Upstream of Two Different tDNA Families and Specific tDNA Copies Is Also Periodic
The data can be examined individually to show insertion patterns upstream of each tDNA family, and further to show insertions upstream of individual tDNAs. When insertions upstream of tGly and tThr genes are plotted separately and their patterns are compared, both exhibit the periodic integration pattern observed in the composite, but some differences emerge (Fig. 5A,B). Both data sets share the peaks around –170, –265, and –335, but the peak around 85 bases upstream of the tDNA is prominent when insertions occur upstream of the tGly genes, whereas it is almost nonexistent in the tThr data. Comparison of the number of insertions between –65 and –127 upstream of each tDNA family using χ2 analysis shows a statistically significant difference between number of insertion events in this area upstream of tGly compared with tThr (Table 2) (P = 1.8 × 10–14). Analysis of the number of insertion events surrounding each of the other prominent peaks shows no statistically significant differences between the number of insertions into each of these regions (Table 2). Thus, whereas the positions of the peaks of integration events are strikingly similar upstream of tGly and tThr genes, the distribution of insertion events among the peaks may vary. In addition, the insertions upstream of tGly genes are closer to the tDNA itself (Fig. 5C). Seven percent of insertions upstream of tThr genes (26/392) (Fig. 5C) occur >650 bp upstream of the tDNA TSS, compared with only 2% of the insertions upstream of tGly TSS (7/427) (Tables 1, 2). Eighty-eight percent (377/427) of insertions upstream of tGly genes are within 350 bases from the tDNA, whereas only 70% (275/392) of insertions upstream of tThr fall within this region. Comparison of the number of insertions within 650 bases and 350 bases from the tDNA between the two tDNA families using a χ2 test reveals a statistically significant difference between the populations (P = 0.0004 and P = 1.1 × 10–10 for insertions within 650 and 350 bases, respectively). Although it could be argued that this difference is caused by differential amplification of long PCR products in the tThr and tGly PCRs, it may also represent true target preferences. Analysis of endogenous LTRs upstream of these two gene families shows that 67% (14/21) genomic LTRs upstream of tGly genes are within 650 bases from the tDNA, whereas only 53% (10/19) of endogenous LTRs upstream of tThr genes are within 650 bases, although these differences are not statistically significant (χ2 test, P = 0.37).
Figure 5.
Distribution of de novo insertions within 650 bases upstream of the TSSs of tGly and tThr gene families and individual preferred tDNAs. (A) Insertions upstream of the tGly family. (B) Insertions upstream of the tThr family. (C) Insertions upstream of individual tDNAs that received more than 15 insertion events. (Bottom, left) Composite of insertions upstream of all tDNA targets that received 15 or less insertion events. Insertions are plotted as described in the legend to Figure 4. Gray bars highlight the 36 bases (18 bases on each side) around each integration peak (–85, –170, –265, and –335).
Table 2.
Quantitative Analysis of Insertion Events Occurring Into Specific Areas Upstream of tGly vs. tThr Genes
|
Bases upstream of tDNA
|
||||||
|---|---|---|---|---|---|---|
| -65 to -127 | -128 to 217 | -218 to -300 | -301 to -370 | Within 350 bases | Within 650 bases | |
| tGly (427 total) | 138 | 128 | 67 | 48 | 377 | 420 |
| tThr (392 total) | 40 | 140 | 59 | 48 | 275 | 366 |
| χ2 test p-valuea | 1.7 × 10-14 | 0.08 | 0.79 | 0.21 | 1.2 × 10-12 | 0.0004 |
| Significant difference between tGly and tThr | yes | no | no | no | yes | yes |
Frequency of integration into each region upstream of tGly was compared to that of integration upstream of tThr. See Methods for details.
We plotted the insertions upstream of specific tDNA targets to determine whether the periodic insertion pattern was observable upstream of individual tDNAs and to rule out the possibility that the periodic pattern was generated by insertions upstream of only the few preferred target tDNAs. Analysis of integration events upstream of each of the preferred targets, as well as a composite of insertions upstream of the cold targets (15 or less insertions) shows that a similar periodic pattern is observable upstream of single tDNAs (Fig. 5C). The positions of insertion peaks upstream of each tDNA is similar, as observed when insertions upstream of tGly and tThr were compared. Most insertions upstream of tGly occur around –85, –170, –265, and –335, and most insertions upstream of tThr show the same pattern, with fewer insertions around –85.
The single tDNA analysis confirms that within the limits of the periodic pattern, distribution of insertions to each of the peaks may vary. For example, insertions upstream of tGlyII occur primarily around the –85 position, whereas insertions upstream of tGlyIII, SUF16 are more equally distributed throughout the peaks. Similarly, upstream of certain tDNAs, the peaks may be shifted in one direction. For example, the peaks upstream of tGlyII are shifted slightly to the right, closer to the tDNA, compared with insertions upstream of most other tDNAs. Such shifts result in the broadness of the peaks in the composite (Figs. 4A, 5A,B). The presence of surrounding elements also affects the distribution of insertion events. For example, the peaks upstream of tThrIX,2 are further upstream than most of the peaks upstream of the other tThr genes, possibly because insertions are actually being targeted to the aspartic acid tDNA that lies 1.4 kb upstream of this gene. This analysis also shows specific insertion hotspots upstream of each tDNA. For example, there are 10 insertions at position –79 upstream of the tGly on chromosome II (tGly II), each from an independent PCR reaction. Similarly, nine insertions at position –244 occur upstream of tGly XVI, 1. These preferences may be based on local sequence differences.
The fact that the same areas of insertion are used for Ty1 insertion upstream of each tDNA suggests that Ty1-targeted integration could be guided by a specific physical structure generated by RNA pol III transcription. Within these constraints, the distribution of insertions to each of the peaks may vary depending on local effectors, such as neighboring genes and specific sequences.
Features That Correlate With Target Preference
The unequal genomic distribution of integration events within a tDNA family with identical coding regions was striking, and to understand the basis for these preferences, we examined possible features that may correlate with target strength. We found no correlation between targeting frequency and chromosome size or distance from telomere or centromere. Certain tDNAs have been associated with the chromatin remodeling RSC complex (Ng et al. 2002), although we found no correlation between tDNA target strength and physical association with RSC subunits (http://web.wi.mit.edu/young/RSC). Similarly, we did not find a correlation between target strength and distance from pro-ARS sequences (MCM and ORC-binding sites) (http://www.sciencemag.org/cgi/content/full/294/5550/2357/DC1), associated with LTR-containing intergenic regions that neighbor tDNAs (Wyrick et al. 2001). To address the possibility that tDNA expression level correlates with targeting ability, we note that the family of tGly (GCC) genes contains several suppressor alleles known to be expressed by virtue of their ability to suppress nonsense mutations. SUF16, SUF17, and SUF20 are all tGly (GCC) members that exhibit equal levels of nonsense suppression (Mendenhall et al. 1987), suggesting that they have similar expression levels. The tGly (GCC) genes defined by these suppressor alleles do not correlate with levels of Ty1 integration. SUF16 is a good target, whereas SUF17 and SUF20 are not (Fig. 3A). This suggests that expression level may not be a major determinant of target integration frequency.
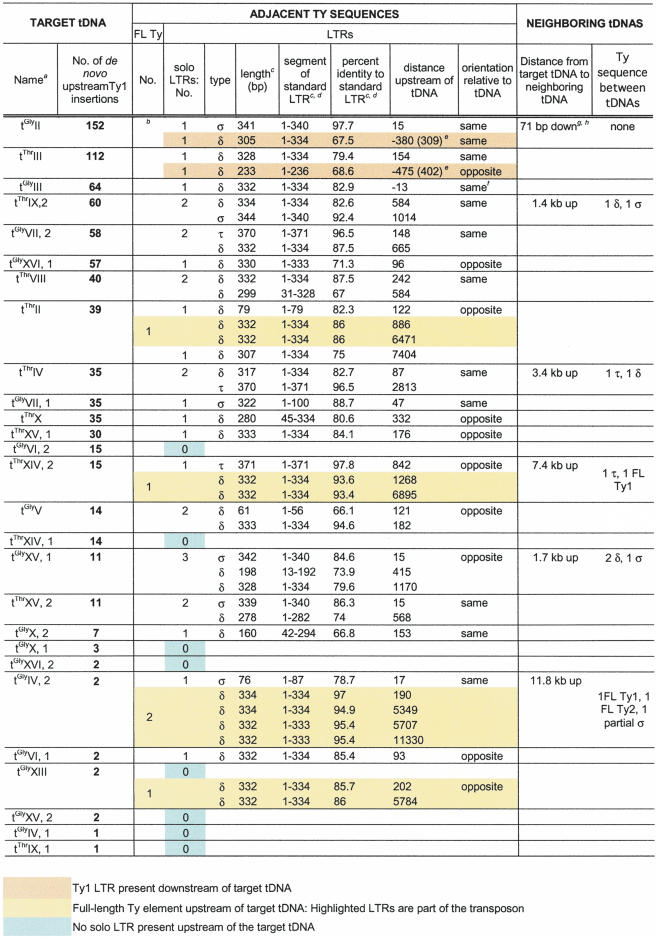
We then examined the genetic structure of each tDNA target, searching for neighboring elements that the most effective targets have in common. We organized the target in order of frequency for transposition integration and included information about neighboring genetic structures using the data set compiled by Bolton and Boeke (2003) and the Voytas lab (http://www.public.iastate.edu/∼voytas/; Table 3). This analysis allows a visual comparison of the elements surrounding a target and shows that the tDNAs in the hottest target group shared several features. The two best targets both had an LTR within 1 kb downstream of the target tDNA (denoted by orange boxes; in these two cases, distance from both the 5′ and 3′ end of the tDNA to the 5′ end of the LTR are shown). No other tDNA targets shared this characteristic. In addition, two of the high-frequency targets have a tDNA as a relatively close neighboring gene with little intervening Ty sequence. Complete absence of an upstream LTR (green boxes) correlates with low target strength. Of the 12 tDNAs with 30 or more insertions, all had at least one upstream LTR, whereas of the 14 tDNAs with 15 or less integration events, eight had no upstream LTR. This represents a statistically significant difference by χ2 comparison with a P-value of <0.002.
Table 3.
tDNA Targets and Surrounding Full-Length Ty Elements, LTR, and tDNAs, Grouped and Listed in Order of Ty1 Target Preference
Although the presence of an upstream LTR is shared by most of the strong targets, neither its distance from the tDNA nor its orientation relative to the tDNA is a good indicator of target strength (Table 3). The percent identity of the LTR to a query LTR from an active Ty (Ty1-H3 5′ LTR, Ty3-1 5′ LTR, and Ty4-2 LTR) also does not correlate with target strength (Table 3). That is, even though an LTR is likely to be present upstream of a strong tDNA target, it does not have to be a young or intact LTR. This is true for σ LTRs (derived from Ty3), δ LTRs (derived from Ty1 and Ty2), and τ LTRs (from Ty4). In addition, the presence of a σ element upstream of the tDNA does not correlate with target preference. Moreover, full-length Ty elements (yellow boxes) are not present upstream of the most preferred targets with 57 or more insertions, although their presence is not significantly correlated with low-targeting efficiency. Finally, examination of the presence, position, and expression level of pol II genes neighboring the tDNA targets also showed that their position, status for viability (whether they are essential genes or not), and expression level do not correlate with strength of target (data not shown, based on data compiled by Bolton and Boeke 2003).
Genomic Features Surrounding a Preferred Target
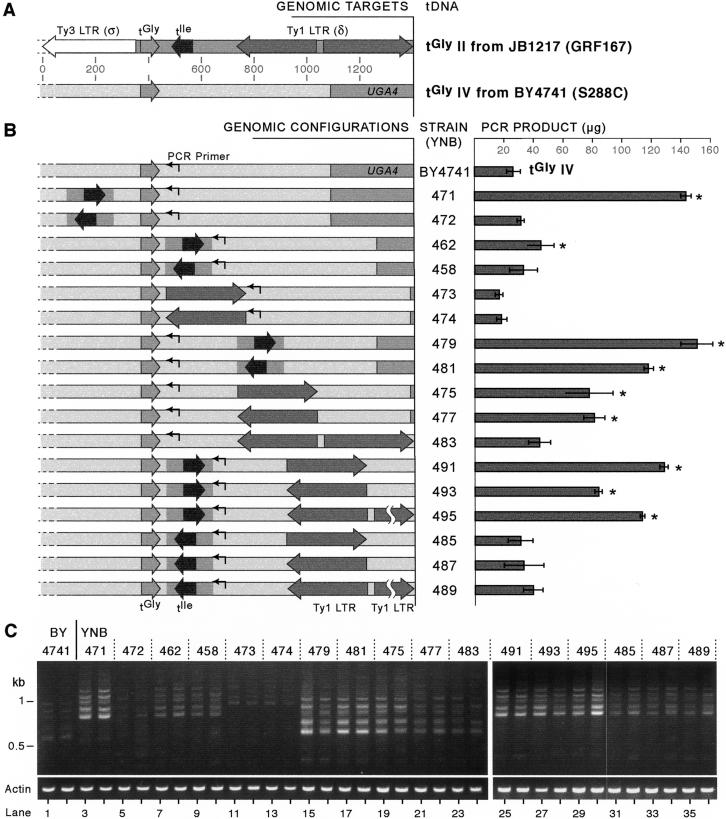
Our analysis showed that the tDNA target preferred above all others was tGly on chromosome II, tGlyII (tG(GCC)B), from JB1217, which received 152 upstream insertions. We sought to determine what special characteristic, or combination of characteristics, this gene had that made it such an attractive target for Ty1 integration. The area surrounding tGlyII is crowded with known genetic elements (Fig. 6A). Most striking is the proximity of the downstream tIle gene. The tGly and tIle are transcribed convergently, with only 52 bases of sequence between the mature 3′ ends that largely consists of As and Ts, which form the termination sequences for pol III transcription. In addition, there are two Ty1 LTRs (delta, δ) downstream of this tDNA and an upstream Ty3 LTR (sigma, σ). The sequence of this region from strain JB1217 differs slightly from the reference S. cerevisiae sequence (see below) and was deposited to GenBank (accession no. AY426826). In contrast, the coldest tGly target, one of the two tGly genes on chromosome IV, tGly IV, one [called tG(GCC)D1 at http://www.yeastgenome.org/ and hereafter referred to as tGly IV for simplicity] has no known elements nearby (Fig. 6A) and is within a region devoid of endogenous retrotransposon copies (Kim et al. 1998). The closest feature is a pol II gene, UGA4, 649 bp downstream.
Figure 6.
Comparison of features around a hot and cold tGly target and effects on integration frequency. (A) Schematic of features surrounding the most preferred tGly target, tGlyII (tG(GCC)B) from JB1217 and the least preferred target, tGlyIV (tG(GCC)D1) from BY4741. (B) Various features were added into several positions around tGlyIV. The tIle with 60 bases of upstream sequence and 46 bases of downstream sequence was amplified from JB1217 tGlyII and inserted 60 bases upstream, 25 bases downstream, or 299 bases downstream of BY4741 tGlyIV. The LTR downstream of tGlyII (68% identity to a query Ty1 LTR [http://www.public.iastate.edu/∼voytas/], Ty1 H3 5′ LTR) was inserted in both orientations either 299 or 25 bases downstream of tGly. Back-to-back LTRs (the first LTR is 68% identical and the second 87% identical to the reference LTR) were added at position 299. The LTR insertions were combined with insertion of tIle. When the 180 bases containing the tIle were added 25 bases downstream of tGly, the LTR shifts downstream by 180 bases, to 479 bases downstream of the tGlyIV. The same PCR primer was used to amplify insertions upstream of tGly in every strain, and its distance from tGlyIV shifts depending on the length of sequences inserted at the position 25 bases downstream of tGlyIV. Histogram showing amounts of PCR product generated from PCR of insertions upstream of tGlyIV in each strain after normalization to actin is shown at right. Histogram represents the mean of either eight or nine independent trials per strain (quantitation described in Methods section). Error bars, SEM. An asterisk (*) to the right of the bar indicates a statistically significant difference (P ≤ 0.01 by the Mann-Whitney U-test) in PCR product compared with the amount generated in the BY4741 strain. (C) Gels showing the products generated using the PCR assay of insertions upstream of tGlyIV. Two different transformants from every strain are shown. PCR of the actin gene was performed to control for amount of input genomic DNA.
tGlyII in BY4741 contains the upstream σ and downstream tIle and δ, but lacks the second downstream δ element. The spacing between these elements was a few bases different in the two strains. For example, there were 71 bases between the two tDNAs in BY4741, whereas there were 52 in JB1217 (JB1217 tGlyII sequence is available from GenBank and BY4741 sequence available from SGD [http://www.yeastgenome.org]). The number of insertion events upstream of tGlyII locus in BY4741 was fewer than those in JB1217, although the total number of sequences analyzed from BY4741 was far fewer. The possibility therefore exists that the target preference-conferring element is related to the JB1217 strain's sequence or configuration of features.
To determine the stimulatory features of the hot target, we placed various elements from the tGlyII region from JB1217 at several positions around tGlyIV in BY4741, and assayed transposition frequency upstream of the cold locus (Fig. 6B) in an experimental attempt to convert the cold target to a hotter one.
Features That Define Hot Targets
We added features that surround the tGlyII in JB1217 to tGlyIV in BY4741 at similar relative positions to see whether any of these additions would confer an increased targeting frequency to the cold tGlyIV target (Fig. 6B). Because downstream LTRs and neighboring tDNAs were features correlated with target strength on the basis of the analysis of genomic preferences, we inserted the tIle, a single LTR, or back-to-back LTRs at several positions relative to the tGly. The σ element was not added because both preferred and nonpreferred genomic targets harbored σ elements upstream (Table 3). We inserted the tIle with 60 bases of upstream and 46 bases of downstream flanking sequence at three discrete positions as follows: 60 bases upstream of tGlyIV, 25 bases downstream of tGlyIV (resembling its position downstream of tGlyII), and 299 bases downstream, the position of the LTR downstream of tGlyII. The first Ty1 LTR (δ) downstream of tGlyII (which has an only 68% identity to a query Ty1 LTR [Ty1-H3 5′ LTR; http://www.public.iastate.edu/∼voytas/]) was also inserted into both of the downstream positions. Insertions of each feature were made in both orientations (Fig. 6B). Because tGlyII in JB1217 has two δ LTRs downstream, we also created a strain with these two back-to-back δ LTRs (the second δ element has 87% identity to Ty1-H3 5′ LTR) inserted at the 299 bases downstream position to test whether LTR effects might be additive. Lastly, we created strains with both the downstream tIle and LTRs (Fig. 6B).
To assay integration upstream of the modified tGlyIV loci, we modified the PCR assay to amplify insertions upstream of this specific tDNA target by using a primer complementary to unique sequence downstream of tGlyIV in place of the primer common to all tDNAGlys. We combined this primer with the orientation 1 Ty1 primer, to avoid the background bands generated by the orientation 2 primer. This increased the size of the smallest expected PCR product by 71 bases, to ∼560, taking into account the 80–90 base region upstream of the tDNA TSS that does not receive insertions. The PCR-banding pattern of insertions upstream of unmodified tGlyIV using the starting strain (BY4741) was visible, athough faint, and showed the same periodic pattern seen when all 16 tGly genes were assayed simultaneously (Fig, 6C, lanes 1,2). Because this PCR was in the linear range of the reaction for all samples, the bands were not brightly visible in the BY4741 samples, but the brightness of these PCR bands increased with either increased cycle number or increased input genomic DNA (data not shown). Three independent transformants from each strain were induced for transposition on galactose in the presence of the GalTy plasmid (pVIT41), and the genomic DNAs were assayed for insertions (two transformants are shown in Fig. 6C). The brightness of the PCR bands represents the level of insertion occurring upstream of tGlyIV in each strain with added features. Because the same PCR primer pair was used to amplify insertions upstream of tGlyIV in every strain, the length of the smallest PCR product changed depending on the length of sequence added at the position 25 bases downstream of tGlyIV. To ensure equal loading of genomic DNA into each PCR, and to aid in quantitation of the PCR products, PCR of the actin gene was also performed (Fig. 6C).
The PCR results show that the addition of various elements around the tGlyIV affects the frequency of insertion at this locus (Fig. 6B,C). Using a U-test (Mann-Whitney test) comparing the amount of PCR product amplified from insertions upstream of the element-free wild-type tGlyIV (BY4741) with each of the other strains, we found significant differences between frequency of integration events upstream of tGlyIV and several of our constructed configurations. Most interestingly, addition of the tIle in the same orientation as the tGlyIV confers a significant increase in integration frequency upstream of tGlyIV (YNB471, YNB462, YNB479, YNB491, YNB493, and YNB495). This phenomenon is observed when the tIle is placed either upstream or downstream of tGlyIV. Placing the tIle in the opposite orientation is significantly stimulatory only when it is inserted 299 bases downstream of tGlyIV (YNB481).
Addition of an LTR to the tGlyIV target also significantly stimulated targeted integration. The LTR was stimulatory when inserted 299 bases downstream of tGlyIV in either orientation, but not when placed closer to the tDNA. Although significant, the LTR's effect was not as stimulatory as that of the tIle. Addition of the two back-to-back LTRs downstream of tGlyIV (YNB483, YNB489, YNB495) did not have a consistent effect on target frequency compared with the presence of the single downstream LTR. The two back-to-back LTRs are different from one another in sequence. The first LTR has only 68% identity to the query Ty1-H3 LTR (see http://www.public.iastate.edu/∼voytas/), and lacks most of the characteristic protein-binding sites in LTR sequences. It does not contain binding sites for the known LTR transcription regulators, Gcr1 in the UAS (Turkel et al. 1997; Dudley et al. 1999), Gcn4 (Morillon et al. 2002), or Mot3 (Grishin et al. 1998; Madison et al. 1998). Neither of the two TATA boxes is present in the sequence (Liao et al. 1987; Coney and Roeder 1988; Dudley et al. 1999). The second of the two LTRs, however, is 87% identical to the query δ element and contains the Gcr1 site, both TATA boxes, and a consensus Gcn4-binding site, Gcn4D (Morillon et al. 2002). The second LTR was present either 630 (YNB483) or 811 (YNB489 and YNB495) bases downstream of tGlyIV in our studies. Because the effect of adding the second, high-quality LTR downstream of the first LTR was not consistent, we are not able to draw a strong conclusion about its effect, although it can be confidently stated that the presence of multiple LTRs does not have an additive effect on integration frequency.
Addition of the LTR to strains in which the downstream tIle was present resulted in a further stimulation of integration only when the two tDNAs were in tandem orientation (YNB491, YNB493, YNB495). A U-test comparing the frequency of insertion in the strain with only a tandem tIle added (YNB462) to those with both the tandem tIle and the δ element(s) (YNB491, YNB493, YNB495) showed that in each case the addition of the LTR conferred a statistically significant increase on integration frequency (P < 0.01). Contrastingly, in no case did the addition of the LTR to the convergent tDNAs (YNB485, YNB487, YNB89) result in a significant increase in integration frequency compared with the convergent tDNAs alone (YNB458). Finally, we also tested the effect of placing a pol II gene, URA3, either 15 or 60 bases upstream of the tGly and found that doing so had no significant effect on target integration frequency (data not shown).
Examination of the sizes of the PCR products shows that, surprisingly, in cases in which there were two tDNAs near one another, Ty1 elements did not insert between them (Fig. 6C). For example, the smallest PCR products from Ty1 insertion events in strains YNB471, YNB462, YNB491, YNB493, and YNB495 were all the same size, ∼660 bases, corresponding to insertion events upstream of only the 5′ tDNA gene, regardless of whether it was tGly or tIle. Moreover, tIle may be a more attractive target than tGly for Ty1 integration, suggested by the comparison of insertion frequencies in YNB471 and YNB462. Both YNB471 and YNB462 had tGly and tIle in tandem at similar distances apart (107 bases in YNB471 and 85 bases in YNB462), but more insertions occurred upstream of the tIle in YNB471 than upstream of the tGly in YNB462. Although the tDNA configurations in these strains are not identical, this observation raises the possibility that certain tDNA families could be more attractive targets for integration than others.
Genomic Examples of Adjacent tDNA Target Pairs
We examined the genomic sequence to determine whether tDNAs separated by the distances assayed in this work showed evidence of previously having suffered Ty insertion events. Examination of tDNA genomic data showed that 50 of 274 tDNAs have a tDNA as their immediate non-Ty gene neighbor, and most of these have Ty sequence(s) interposed (Table 4). On the basis of our experiments, of the 25 pairs, four are spaced too closely to receive Ty1 integration events between them (11 bases). Examination of the remaining 21 tDNA pairs showed that 17 (81%) are associated with Ty sequence with each pair receiving >1 integration event, compared with the genomic average of 66% of tDNAs with associated Ty sequence (Kim et al. 1998). When the Ty sequence was subtracted from the distance between tDNA pairs, at least 50% of the cases showed a distance between 200 and 700 bases, providing bioinformatic support that this could be the preferred separation for inviting a Ty insertion event. On the basis of analyses of genomic correlations presented in Table 3 and the comparisons of integration frequencies around a tDNA with different surrounding features (Fig. 6), we predict that tandem tDNAs separated by >100 bases or any tDNA pair separated by >300 bases would be likely to receive a high number of integration events. A total of four of 21 tDNA pairs competent for receiving integration events lacks intervening Ty sequences and our findings could be extended to these examples by comparing the upstream integration frequency of those copies to other family members. For example, two tDNAs on chromosome VII, tLys VI, 2 and tLeu VII are transcribed convergently and separated by 292 bases with no intervening Ty sequence. This pair is similar to the configuration of YNB481, and would be expected to be a preferred target compared with other tLys and tLeu genes in the genome.
Table 4.
The Genomic Configuration of tDNAs With tDNA Neighbors
| tDNA paira | Transcriptional orientation | Ty seq between tDNAs | Total distance between tDNAs | Distance w/o Ty seq | de novo insertionsb | |
|---|---|---|---|---|---|---|
| tArg X, 2 | tAsp X, 3 | Same | None | 11 | 11 | NDc |
| tAsp II | tArg II | Same | None | 11 | 11 | ND |
| tAsp IV | tArg IV | Same | None | 11 | 11 | ND |
| tAsp X, 2 | tArg X, 1 | Same | None | 11 | 11 | ND |
| tArg X, 1 | tTyr X, 1 | Same | 2 δ | 1042 | 376 | ND |
| tVal XIII, 1 | tTrp XIII | Same | 2 δ, 1 FL Ty4 | 6785 | 400 | ND |
| tLys V, 2 | tHis V, 2 | Same | 1 δ, 1 σ | 1140 | 481 | ND |
| tGln IV, 1 | tLeu IV | Same | 3 δ | 1146 | 489 | ND |
| tGly II | tIle II | Opposite, convergent | None | 71 | 71 | 152 |
| tLys VII, 2 | tLeu VII | Opposite, convergent | None | 292 | 292 | ND |
| tLys III | tMet III | Opposite, convergent | 1 δ, 1 τ | 1293 | 602 | ND |
| tTrp X | tLys X | Opposite, convergent | None | 893 | 893 | ND |
| tSer I | tLeu I | Opposite, convergent | None | 1268 | 1268 | ND |
| tAsn XIV, 2 | tPro XIV | Opposite, divergent | 1 δ, 1 τ | 682 | 196 | ND |
| tGly IV, 2 | tSer IV, 2 | Opposite, divergent | 1 σ, 1 FL Ty1, 1 FL Ty2 | 11777 | 256 | 2 |
| tAsn XIV, 1 | tGly XIV | Opposite, divergent | 1 σ, 1 τ, 1 FL Ty1 | 6477 | 264 | ND |
| tGln VII | tSer VII | Opposite, divergent | 3 δ | 1214 | 529 | ND |
| tAla X | tAsp X, 1 | Opposite, divergent | 1 δ, 1 σ, 1 FL Ty4 | 7348 | 648 | ND |
| tThr IX, 2 | tAsp IX, 1 | Opposite, divergent | 1 δ, 1 σ | 1374 | 698 | 60 |
| tAsn XV, 1 | tGly XV, 1 | Opposite, divergent | 2 δ, 1 σ | 1650 | 785 | 11 |
| tThr XIV, 2 | tPro XIV, 2 | Opposite, divergent | 1 τ, 1 FL Ty2 | 7350 | 1021 | 15 |
| tGlu II | tCys II | Opposite, divergent | 2 δ, 1 τ | 2089 | 1334 | ND |
| tLeu III | tGlu III | Opposite, divergent | 4 δ, 1 FL Ty2 | 8326 | 1584 | ND |
| tArg V | tLys V, 1 | Opposite, divergent | 3 δ | 3169 | 2195 | ND |
| tSer IV, 1 | tThr IV | Opposite, divergent | 1 δ, 1 τ | 3436 | 3282 | 36 |
tDNAs named as described in the legend to Figure 3.
Several members of these tDNA pairs are from the tGly and tThr gene families. The number of de novo insertions upstream of each of these as determined in this work is listed.
(ND) Not determined.
DISCUSSION
A general model for transposition integration features a physical interaction between the transposon's integrase protein and a host protein that guides the integration event into its target site. As a result of these specific interactions, most transposition integration events are limited to a few discrete positions in their host's genome, where the host protein generally resides. The strict positioning of Ty3's integration at the start site of pol III transcription by virtue of its interaction with TFIIIB and TFIIIC exemplifies this model (Yieh et al. 2000, 2002; Aye et al. 2001). Similarly, Dictyostelium discoideum's non-LTR retrotransposon TRE5-A integrates specifically at position 48 (± 3) bases upstream of the TSS of tDNAs, very close to the boundary of the TFIIIB complex, and is likely guided by an interaction with TFIIIB (Beck et al. 2002). In contrast, Ty1 has a large (700–800 bp) target region. This lack of specificity suggests that Ty1's integration target site selection mechanism may be either more complex or significantly different from the canonical model. This work presents a high-resolution study of integrase-mediated genomic Ty1 integration events and reveals several novel features of Ty1 integration specificity. We have examined the positions of 836 de novo-targeted Ty1 integration events upstream of two yeast tDNA families. The results show a nonrandom distribution of insertion events, both throughout the genome and upstream of specific tDNAs. On a genomic level, within a tDNA gene family, most of the Ty1 insertion events occur upstream of only a few tDNA copies. Upstream of the tDNAs, the insertions are also non-randomly distributed, and occur in a periodic pattern, with peaks separated by ∼80 bases.
Ty1 Insertion Upstream of tDNAs in the Yeast Genome Is Periodic
The pattern of insertions within the target window upstream of two families of tDNAs in the yeast genome by PCR shows ∼80 bp periodicity with all insertions >64 bases upstream of the tDNA TSS. The dominant peaks of de novo insertions at –85 and –170, and the weaker peaks at –265 and –335 relative to the tDNA TSS are also seen in the profile of endogenous element positions as well as upstream of individual tDNA targets and, therefore, likely represent generally favorable positions for insertion upstream of the tDNA. The periodic insertion pattern is also observed when the PCR assay is performed on a single target (Fig. 6C), confirming that the banding pattern is not an artefact of assaying insertions upstream of an entire family of insertions. The distribution of insertions between the peaks varies upstream of different tDNAs or tDNA families, possibly based on local sequence or neighboring genes.
The relatively clean periodicity supports the idea that different tDNA loci may have similar general structural characteristics or constraints. Integration peaks observed at –85 and –170, and to a lesser degree, at –265 and –335 could be created by a configuration directly or indirectly specified by the pol III transcription complex. No insertion events were observed closer to the start of tDNA transcription than 64 bases upstream, although the boundary of the TFIIIB complex footprint has been mapped to 42 bases upstream of the tDNA TSS (White 1998). Structural constraints may also be responsible for the absence of insertions between –42 and –64. A pol III configuration may prohibit integration in this region, or another protein factor, perhaps integrase itself, could bind in this region and block integration.
The periodic insertion pattern of Ty1 is intriguing. tDNAs exhibit several potent genomic activities, for example, nucleosome repositioning (Morse et al. 1992), boundary element function (Donze et al. 1999), pol II transcriptional repression (Hull et al. 1994), and DNA replication pausing (Deshpande and Newlon 1996). It is therefore possible that the tDNA upstream region has specific structural features, such as a specific configuration of histones and other proteins, that mediate these activities. The RSC chromatin remodeling complex, whose subunits are all essential, binds to the regions upstream of tDNAs and it is possible that this complex could be regulating the chromatin structure of this region (Ng et al. 2002). A structural model for retroelement integration is not unprecedented. HIV integration is constrained by target structure—integration is stimulated by DNA incorporation into a nucleosome and insertions occur with a 10-base periodicity on the basis of the position of the DNA relative to the histone octamer (Pryciak and Varmus 1992; Pryciak et al. 1992; Pruss et al. 1994a,b). In a structural model for Ty1 integration, the configuration of bound factors provides the constraints on the target region, allowing insertions only into permissive positions, resulting in the periodic insertion pattern observed.
Effects on tDNA Targeting by Presence and Position of Additional tDNAs and LTRs
Analysis of the genomic distribution of Ty1 integration events showed that within the two gene families assayed, specific tDNA copies were preferred as targets, despite identical coding sequences. Several general trends emerged from analyzing features surrounding the targets. Target preferences were defined by local rather than global genomic features. For example, the target's immediate genomic neighbors had a larger effect on target frequency than their proximity to telomeres or centromeres. Highest frequency targets shared the presence of a downstream Ty1 LTR and a relatively close tDNA neighbor, and poor targets had no LTRs nearby. Direct experimentation confirmed that a target's surrounding features can affect the likelihood with which it is used for Ty1 integration. We implanted the features surrounding the strongest tDNA target, tGly II around one of the weakest targets, tGlyIV, to determine which surrounding features could stimulate Ty1 integration frequency.
The greatest effects on frequency were seen when a second tDNA was added in the same orientation as the original tGly. At all three positions tested, addition of the tandem tIle resulted in a significantly higher frequency of insertion than a single tGly, and stimulation was much greater when the tandem tDNAs were >100 bases apart. Similarly, although adding the tIle opposite to tGly had no significant effect in most positions, placing it ∼300 bases downstream of tGly did elevate integration frequency. The elevated integration is thus not a simple consequence of a higher local concentration of pol III factors. Rather, the configuration of the pol III complexes relative to one another rather critically affects targeting frequency, with intergenic distances of ≥300 bases optimal. Surprisingly, the bases downstream of the tDNA can have a great impact on integration events upstream of the tDNA.
Similarly, placement of a single degenerate LTR in either orientation 299 bases downstream of the tDNA significantly increased the frequency with which the tGly wasusedasatargetfor Ty1 integration. When present 25 bases downstream of the tDNA, it had no effect on frequency, showing that LTR presence alone is insufficient to increase target frequency, and that context and position of the LTR relative to the tDNA is important. The optimal distance between the tDNA and downstream LTR was again ∼300 bases. Solo LTRs have long been known to affect transcription from adjacent promoters (Roeder et al. 1980; Winston et al. 1984), and our results show that LTRs can affect a region >400 bases away, upstream of the tDNA.
LTRs contain promoter sequences for transcription of Ty1 elements such as Gcr1 (Turkel et al. 1997; Dudley et al. 1999) and Gcn4 (Morillon et al. 2002). LTRs are repressed by a chromatin structure at least partially maintained by Isw1 and Isw2 (Kent et al. 2001) and which requires activity of chromatin remodeling complexes such as SAGA for relief (for review, see Winston and Carlson 1992). Interestingly, the LTRs downstream of the best tDNA target was of low homology (68%) to a reference Ty1-H3 LTR, and lacked any binding sites known to regulate Ty1 transcription. The addition of a second LTR with 87% identity to the standard Ty1 δ sequence, which contained binding sites for Gcr1, Gcn4D, and both TATA elements did not have a consistently positive effect on transposition integration frequency. These results suggest that the LTR's binding sites may not control target frequency, rather in this instance, the lack of the known factors may positively affect the target structure, making it more permissive to Ty1 integration. Interestingly, when we examined the LTR downstream of the second best Ty1 target, tThrIII, we found that its sequence was also quite divergent from the standard Ty1-H3 LTR (Table 3), and similarly lacked the binding sites listed above.
The result of simultaneous insertion of the second downstream tDNA and the LTR is revealing. When combined with two tandem tDNAs, presence of the LTR 299 bases downstream of the second tDNA stimulated integration, but stimulation was not observed when the tDNAs were in opposite orientation relative to one another. The oppositely oriented second tDNA therefore abolishes the LTR's effect on the upstream tDNA. Thus, two tandem tDNAs may have a harmonious configuration receptive to downstream influences, whereas the oppositely oriented tDNAs do not.
The integration pattern upstream of the single tDNA target in each of the strains with manipulated target structure is the same as that upstream of a multicopy gene family of tDNAs (Figs. 2, 6C). The structural constraints that limit Ty1 integration events to specific positions upstream of the tDNA still operate in the configurations generated, demonstrating that features affecting insertion frequency are distinct from those that affect integration position.
Taken together, our data suggest a model in which the tDNA or the act of pol III transcription generates a specific physical configuration of proteins several hundred bases upstream. The Ty1 preintegration complex is recruited to the upstream region and integrates within the constraints of the physical structure present, resulting in a periodic integration pattern. The physical structure is subject to effects from the surrounding genomic elements, some stimulatory, some inhibitory. In this study, an intergenic distance of 300 bases was the optimal positioning of the structural elements for stabilization of the tDNA upstream region. If present 300 bases downstream, both single LTRs and an additional tDNA in either orientation provided a positive effect on integration frequency, helping generate or stabilize the configuration upstream of the tDNA. The presence of tDNAs in the same orientation had a positive effect on transposition integration frequency even when separated by only 85 bases, possibly because each stimulated directional formation of the upstream structure, and these effects were additive. When the tDNAs were arranged in this configuration, insertions did not occur between them, suggesting that their combined effects created only one upstream region permissive for integration. Within 300 bp, a second tDNA did not have a positive effect on the tDNA upstream region if the two tDNAs were oriented opposite one another, possibly because the formation of the upstream regions were competing (as in YNB472) or occurring on opposite ends of the tDNA block (as in YNB458). The need for the increased intergenic distance could suggest a requirement for physical flexibility, alleviation of potential steric hindrance, or simply more room for pol III transcription termination for the positive upstream effects to occur.
Finally, the second best tDNA target was tThr on chromosome III, which lacks a close tDNA neighbor, although it does have a downstream LTR. This tDNA is unusual in that it immediately abuts and defines the boundary of the tightly silenced HMR locus (Donze et al. 1999). HMR lies upstream of the tThr, and thus the inserting Ty1 elements actually jump into the adjacent silent chromatin. It is possible that the presence of the specialized silenced chromatin structure adjacent to the tThr, or the boundary between the tDNA structure and the silent chromatin represents a preferred substrate for Ty1 integration. Notably, earlier studies (Bryk et al. 1997; Smith and Boeke 1997) indicate that Ty1 insertion into another silent locus, the rDNA, occurs quite frequently.
METHODS
Yeast Strains, Plasmids, and Medium
A Δrad52 strain (YEB102: MATα his3Δ200 leu2Δ1 ura3-167 rad52Δ::natMX4 GAL+) was made by replacing RAD52 with natMX (Goldstein and McCusker 1999) by homologous recombination. JB1217 (BY8217) MATa his3Δ200 leu2Δ1 met15Δ0 trp1Δ63 ura3-167 RDN1::MET15 is a derivative of GRF167 and was made by selecting for spontaneous loss of URA3 in the rDNA repeat in JS306 (Smith et al. 1999). BY4741 MATa leu2Δ0 met15Δ0 ura3Δ0 his3Δ1 was made as described previously (Brachmann et al. 1998), and is derived from S288C.
Strains with targeting elements around the cold tGly target, tGly IV (tG(GCC)D1), were made using a two-step homologous recombination strategy. The first step consisted of inserting URA3 either 60 bases upstream, 25 bases downstream, or 299 bases downstream of the target tDNA in BY4741. To do this, URA3 was PCR amplified from pRS416 using primers with 60 bp homology to the target site (for insertion via homologous recombination), and the resulting PCR product was transformed into BY4741. Integrants were selected for on SC-ura plates and checked by colony PCR.
The final strains were made by replacing URA3 with three possible elements from the region surrounding the tGlyII (tG(GCC)B) hot tDNA. First, a 1.3-kb fragment containing the tGly hotspot on chromosome II was PCR amplified from genomic DNA from JB1217 and cloned into the PCR 2.1 TA cloning vector (Invitrogen) per the manufacturer's instructions. The JB1217 GRF167 strain contained two δ LTRs downstream of the tIle gene, whereas S288C only contained one; both of these were included in the hotspot fragment. The resulting TA plasmid was used as the template for PCR of sections of the hotspot. Three separate sections were amplified—the tIle gene, the first downstream δ LTR alone and the second downstream δ LTRs together. The tIle fragment amplified and included 60 bases of upstream sequence and 46 bases of downstream sequence and was amplified with primers containing 60 bases of sequence homologous to the position of insertion (flanking URA3). No flanking sequence from the hotspot was amplified around the single or double δ LTRs, and these were also amplified using primers with 60 bases of sequence homology to the target position (flanking URA3). PCR products were transformed into the Ura+ strains and URA3 gene replacements were selected on 5-FOA plates. Correct integrants were confirmed by colony PCR.
Oligo sequences used to generate the yeast strains are presented in Supplemental data (Table 1).
pVIT41 was constructed as described (Lauermann et al. 1997). The catalytic integrase mutant plasmid pNB19 was constructed by removing the XhoI–HindIII fragment from pVIT41 (Lauermann et al. 1997) and replacing it with the XhoI–HindIII fragment from pGM119 (Eichinger and Boeke 1988; Monokian et al. 1994).
Medium was prepared by standard methods as described (Burke et al. 2000).
Transposition Induction
Haploid yeast were transformed with pVIT41 and plated on SC-Ura medium. Colonies were picked and patched on SC-Ura, four patches to a plate. These were grown at 30°C until the patch was thick (overnight to 2 d). The quarter-plate sized patches were replica plated to SC-Ura galactose plates and SC-Ura glucose plates in parallel and incubated for 2 d at 22°C to induce transposition. The patch was scraped and grown in 10 mL YPD overnight at 30°C. The next day, genomic DNA was prepared.
Genomic DNA Prep
Genomic DNA was prepared from 10 mL of overnight YPD culture using the teeny prep method as described (Boeke et al. 1985). Genomic DNA preps were treated with 20 μg of RNase A and incubated at 37°C for 30 min. Residual protein was removed by extraction with phenol/chloroform and once with chloroform. DNA was precipitated and resuspended in 100 μL of water or 0.01 M Tris-HCl (pH 8.0).
PCRs for Ty1 Targeted Integration
The PCR conditions were optimized for detection of transposon insertions upstream of two general classes of tDNA, tGly and tThr, as well as upstream of a specific tGly gene copy, tG(GCC)D1. Primer names and sequences are listed in Table 5 and optimal PCR conditions are listed in Table 6.
Table 5.
Oligo Names and Sequences for PCR Assays
| Primer target | Orientation (seeFig. 2) | Name | Sequence 5′-3′ |
|---|---|---|---|
| Ty1 SSB | 1 | JB2930 | AGAGCTCCCGGGATCCTCTACTAAC |
| 2 | JB3361 | GAGGATCCCGGGAGCTCTGATAGTTG | |
| tGly | JB2821 | TGCGCAAGCCCGGAATCG | |
| tThr | JB3084 | TGCTTCCAATCGGATTTGAACC | |
| tGly IV | JB6071 | GTCATTCACAATAAACACTGG | |
| Actin | Forward | JB4307 | TCGGTATGGGTCAAAAAGACTCC |
| Actin | Reverse | JB4312 | TCTGGGGCTCTGAATCTTTCG |
Table 6.
Optimized PCR Conditions
| Primer 1 | Primer 2 | Annealing temp | Cycle number |
|---|---|---|---|
| Ty1: JB2930 | tGly: JB2821 | 60 | 30 |
| Ty1: JB2930 | tThr: JB3084 | 54 | 30 |
| Ty1: JB2930 | tG(GCC)D1: JB6071 | 61 | 30 |
| Ty1: JB3361 | tGly: JB2821 | 60 | 30 |
| Ty1: JB3361 | tThr: JB3084 | 60 | 32 |
| Actin: JB4307 | Actin: JB4312 | 54 | 20 |
The concentration of genomic DNA purified by the method above was determined by analysis of A260. A total of 150 ng of DNA was used per PCR reaction. Primers were used at 0.5 μM final concentration. The 25-μL PCR reactions were performed in a Perkin-Elmer 9600 PCR machine with MicroAmp tubes (Perkin Elmer Applied Biosystems). In all PCR reactions, Amplitaq DNA polymerase (1.25 U/reaction; Perkin Elmer), MgCl2 [2 mM, except for tG(GCC)D1 PCRs, in which 1.8 mM was used] dNTP (2 μM each), and PCR reaction buffer (1×, Perkin Elmer) were used. The conditions of the PCR reactions were as follows: initial denaturation: 5 min at 95°C; cycles: 1 min at 95°C, 1 min at the empirically determined optimal annealing temperature (see Table 6), 1 min at 72°C; final extension: 5 min at 72°C. PCR conditions were optimized for annealing temperature, cycle number, Mg2+ concentration, and tDNA primer position. PCR reactions were also optimized for genomic DNA concentration to ensure that the PCR reactions would be in the linear range of the reaction. PCR products were run on 1× TTE 1.5% agarose gels with 0.5 μg/mL ethidium bromide.
TA Cloning and Sequencing
For the extensive sequencing analysis of the PCR products, two different strains were used. JB1217, derived from GRF167, and BY4741, derived from S288C, were transformed with pVIT41 in parallel. Transposition was induced, DNA isolated, and PCR performed as described above. A total of 19 independent PCR reactions were completed, with each primer pair on DNA prepared from galactose-induced JB1217, and eight independent reactions with each pair completed using strain BY4741, resulting in a total of 108 independent PCR reactions. PCR products from each reaction were ligated independently into the PCR Topo 2.1 vector (Invitrogen) per the manufacturer's instructions (108 independent ligations), and 1μL of resulting ligation reaction was transformed into DH5α bacteria (108 independent transformations). Either 12 or 24 white colonies were picked from each transformation (from each independent PCR reaction), arrayed into 96 well plates of LB, 10% glycerol, and 25 μg/mL Ampicillin, and allowed to grow statically at 37°C for 12 h. Plates were sent to Agencourt Bioscience Corporation for sequencing, using the T7 primer.
Data Analysis
Integration Position Determination
Sequences were analyzed using Sequencher 4.1 (Gene Codes Corp.). Vector sequences were removed, and the Ty1 LTR was identified by multiple sequence alignment. Sequences adjacent to the LTR were compared with the Saccharomyces cerevisiae genome database (http://www.yeastgenome.org/) using a BLAST search to determine the identity of the tDNA target. Sequences were included in the data set only if the LTR sequence was at a new position relative to genomic LTR positions, to ensure the presence of a de novo LTR.
Each independent PCR reaction generated between 12 and 24 sequences, and multiple insertions into the same nucleotide position upstream of the same tDNA from the same PCR reaction were removed from the data set because of the possibility that they represented PCR siblings rather than independent insertion events. Therefore, peaks with a height greater than 1 in each of the single tDNA plots in Figure 5C actually represent individual insertion events from independent PCR reactions. That is, the height of each line in these plots represents the number of PCR pools in which that event occurred at least once. If the same insertion event was observed more than once in the same pool, it was counted only once.
Distance upstream of the tDNA was determined by subtracting the length of the tDNA from this fragment. The most likely position of the tDNA transcription start site (TSS) was determined by G. Kassavetis (see Supplemental material) following the general guidelines defined by Fruscoloni et al. (1995). The distance between the mature processed 5′ end and the TSS was always 9–12 bases. The distance of the insertion from the 5′ end was adjusted to reflect this difference. The data were also adjusted to take into account the 5-bp duplication generated by Ty1 integration.
Quantitation and Statistical Analysis
The χ2 test was used to compare the frequency of insertions into different positions between insertions upstream of tGly and insertions upstream of tThr (Table 2). The test is used to compare counted frequencies where each sample is independent of the others and there are >20 samples (Freund and Perles 2003). The null hypothesis was that the relative frequency was the same between the two samples. The χ2 value was calculated using a contingency table (on the basis of expected values if expected frequencies in both samples were the same) and it corresponded to a probability (P-value) that the two frequencies are different with one degree of freedom (Freund and Perles 2003). Statistically significant differences were reported if the P-value was <0.05 (<5% chance of the two samples being the same)
For PCR upstream of tG(GCC)D1 (Fig. 6), three independent transformants from each of the YNB strains were induced for transposition, and DNA was isolated from each transformant. PCR reactions were completed three times on the samples, resulting in nine data points per strain. In the case of PCR failures, eight data points were used. The actin PCR was also conducted three times on each of the three transformants. Gels were analyzed using Quantity One software (Bio-Rad). The intensity of the bands between 400 and 1200 bp was summed, and the amount of PCR product was determined by comparison to molecular weight standards. A PCR fragment was also amplified from the actin gene to ensure that equal amounts of genomic DNA were loaded into each reaction. Relative amounts of genomic DNA was determined by the ratio of actin products, and the amount of PCR product from each tG(GCC)D1 PCR was normalized to the relative amounts of genomic DNA.
The U-test (Mann-Whitney test) was used to compare the difference between the amount of normalized PCR product amplified from insertions upstream of the control locus, tGlyIV in BY4741, with the experimental one. The U-test is a rank-sum test used to compare two independent sets of nonparametric data (data that are not normally distributed) when the distribution of the two samples being compared is the same (Freund and Perles 2003). The amount of PCR products in each sample (n = either eight or nine values per sample) was ranked, their ranks compared, generating a Z-score that corresponds to a probability (P-value) for the number of samples being compared (Freund and Perles 2003). A statistically significant difference was reported if probability of the ranks being the same between the two samples was ≤1% (P ≤ 0.01).
Acknowledgments
We thank members of the Boeke lab for helpful discussions and Bang Wong for assistance with figures, Eric Bolton for the Δrad52 strain, Ann Marie Egloff for helpful discussions about statistical analyses, and George Kassavetis for providing predicted TSS sites for tThr and tGly genes. This work was supported in part by Public Health Service grant, NIH-GM36481 to J.D.B. N.B. was supported in part by NIH training grant 5T32CA09139.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2052904. Article published online before print in June 2004.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession no. AY426826.]
References
- Aye, M., Dildine, S.L., Claypool, J.A., Jourdain, S., and Sandmeyer, S.B. 2001. A truncation mutant of the 95-kilodalton subunit of transcription factor IIIC reveals asymmetry in Ty3 integration. Mol. Cell. Biol. 21: 7839–7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, P., Dingermann, T., and Winckler, T. 2002. Transfer RNA gene-targeted retrotransposition of Dictyostelium TRE5-A into a chromosomal UMP synthase gene trap. J. Mol. Biol. 318: 273–285. [DOI] [PubMed] [Google Scholar]
- Boeke, J.D. and Devine. S.E. 1998. Yeast retrotransposons: Finding a nice quiet neighborhood. Cell 93: 1087–1089. [DOI] [PubMed] [Google Scholar]
- Boeke, J.D., Garfinkel, D.J., Styles, C.A., and Fink, G.R. 1985. Ty elements transpose through an RNA intermediate. Cell 40: 491–500. [DOI] [PubMed] [Google Scholar]
- Bolton, E.C. and Boeke, J.D. 2003. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: A genomic point of view. Genome Res. 13: 254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P., and Boeke, J.D. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Bryk, M., Banerjee, M., Murphy, M., Knudsen, K.E., Garfinkel, D.J., and Curcio, M.J. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes & Dev. 11: 255–269. [DOI] [PubMed] [Google Scholar]
- Burke, D., Dawson, D., Stearns, T., and Cold Spring Harbor Laboratory. 2000. Methods in yeast genetics: A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Coney, L.R. and Roeder, G.S. 1988. Control of yeast gene expression by transposable elements: Maximum expression requires a functional Ty activator sequence and a defective Ty promoter. Mol. Cell. Biol. 8: 4009–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, A.M. and Newlon, C.S. 1996. DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Devine, S.E. and Boeke, J.D. 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes & Dev. 10: 620–633. [DOI] [PubMed] [Google Scholar]
- Donze, D., Adams, C.R., Rine, J., and Kamakaka, R.T. 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes & Dev. 13: 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley, A.M., Gansheroff, L.J., and Winston, F. 1999. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4-912δ promoter in Saccharomyces cerevisiae. Genetics 151: 1365–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger, D.J. and Boeke, J.D. 1988. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: Cell-free Ty1 transposition. Cell 54: 955–966. [DOI] [PubMed] [Google Scholar]
- Freund, J.E. and Perles, B.M. 2003. Statistics: A first course, 8th Ed. Prentice-Hall, Englewood Cliffs, NJ.
- Fruscoloni, P., Zamboni, M., Panetta, G., De Paolis, A., and Tocchini-Valentini, G.P. 1995. Mutational analysis of the transcription start site of the yeast tRNA(Leu3) gene. Nucleic Acids Res. 23: 2914–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A.L. and McCusker, J.H. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Griffith, J.L., Coleman, L.E., Raymond, A.S., Goodson, S.G., Pittard, W.S., Tsui, C., and Devine, S.E. 2003. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin, A.V., Rothenberg, M., Downs, M.A., and Blumer, K.J. 1998. Mot3, a Zn finger transcription factor that modulates gene expression and attenuates mating pheromone signaling in Saccharomyces cerevisiae. Genetics 149: 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, M.W., Erickson, J., Johnston, M., and Engelke, D.R. 1994. tRNA genes as transcriptional repressor elements. Mol. Cell. Biol. 14: 1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, H., Moore, D.P., Blomberg, M.A., Braiterman, L.T., Voytas, D.F., Natsoulis, G., and Boeke, J.D. 1993. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73: 1007–1018. [DOI] [PubMed] [Google Scholar]
- Kent, N.A., Karabetsou, N., Politis, P.K., and Mellor, J. 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes & Dev. 15: 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.M., Vanguri, S., Boeke, J.D., Gabriel, A., and Voytas, D.F. 1998. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8: 464–478. [DOI] [PubMed] [Google Scholar]
- Kinsey, P.T. and Sandmeyer, S.B. 1991. Adjacent pol II and pol III promoters: Transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res. 19: 1317–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauermann, V., Hermankova, M., and Boeke, J.D. 1997. Increased length of long terminal repeats inhibits Ty1 transposition and leads to the formation of tandem multimers. Genetics 145: 911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.S., Lichtenstein, C.P., Faiola, B., Rinckel, L.A., Wysock, W., Curcio, M.J., and Garfinkel, D.J. 1998. Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics 148: 1743–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.S., Bi, L., Garfinkel, D.J., and Bailis, A.M. 2000. Nucleotide excision repair/TFIIH helicases RAD3 and SSL2 inhibit short-sequence recombination and Ty1 retrotransposition by similar mechanisms. Mol. Cell. Biol. 20: 2436–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, X.B., Clare, J.J., and Farabaugh, P.J. 1987. The upstream activation site of a Ty2 element of yeast is necessary but not sufficient to promote maximal transcription of the element. Proc. Natl. Acad. Sci. 84: 8520–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison, J.M., Dudley, A.M., and Winston, F. 1998. Identification and analysis of Mot3, a zinc finger protein that binds to the retrotransposon Ty long terminal repeat (δ) in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 1879–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall, M.D., Leeds, P., Fen, H., Mathison, L., Zwick, M., Sleiziz, C., and Culbertson, M.R. 1987. Frameshift suppressor mutations affecting the major glycine transfer RNAs of Saccharomyces cerevisiae. J. Mol. Biol. 194: 41–58. [DOI] [PubMed] [Google Scholar]
- Monokian, G.M., Braiterman, L.T., and Boeke, J.D. 1994. In-frame linker insertion mutagenesis of yeast transposon Ty1: Mutations, transposition and dominance. Gene 139: 9–18. [DOI] [PubMed] [Google Scholar]
- Morillon, A., Benard, L., Springer, M., and Lesage, P. 2002. Differential effects of chromatin and Gcn4 on the 50-fold range of expression among individual yeast Ty1 retrotransposons. Mol. Cell. Biol. 22: 2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse, R.H., Roth, S.Y., and Simpson, R.T. 1992. A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol. Cell. Biol. 12: 4015–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsoulis, G., Thomas, W., Roghmann, M.C., Winston, F., and Boeke, J.D. 1989. Ty1 transposition in Saccharomyces cerevisiae is nonrandom. Genetics 123: 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H.H., Robert, F., Young, R.A., and Struhl, K. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes & Dev. 16: 806–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss, D., Bushman, F.D., and Wolffe, A.P. 1994a. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. 91: 5913–5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss, D., Reeves, R., Bushman, F.D., and Wolffe, A.P. 1994b. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biol. Chem. 269: 25031–25041. [PubMed] [Google Scholar]
- Pryciak, P.M. and Varmus, H.E. 1992. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell 69: 769–780. [DOI] [PubMed] [Google Scholar]
- Pryciak, P.M., Sil, A., and Varmus, H.E. 1992. Retroviral integration into minichromosomes in vitro. EMBO J. 11: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, G.S., Farabaugh, P.J., Chaleff, D.T., and Fink, G.R. 1980. The origins of gene instability in yeast. Science 209: 1375–1380. [DOI] [PubMed] [Google Scholar]
- Roeder, G.S., Rose, A.B., and Pearlman, R.E. 1985. Transposable element sequences involved in the enhancement of yeast gene expression. Proc. Natl. Acad. Sci. 82: 5428–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer, S. 2003. Integration by design. Proc. Natl. Acad. Sci. 100: 5586–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes, D.T., Banerjee, M., Bowen, B., and Curcio, M.J. 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159: 1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.S. and Boeke, J.D. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes & Dev. 11: 241–254. [DOI] [PubMed] [Google Scholar]
- Smith, V., Botstein, D., and Brown, P.O. 1995. Genetic footprinting: A genomic strategy for determining a gene's function given its sequence. Proc. Natl. Acad. Sci. 92: 6479–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.S., Caputo, E., and Boeke, J.D. 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19: 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkel, S., Liao, X.B., and Farabaugh, P.J. 1997. GCR1-dependent transcriptional activation of yeast retrotransposon Ty2-917. Yeast 13: 917–930. [DOI] [PubMed] [Google Scholar]
- Voytas, D.F. and Boeke, J.D. 2002. Ty1 and Ty5 of Saccharomyces cerevisiae. In Mobile DNA II (eds. N.L. Craig et al.), pp. 631–662. ASM Press, Washington, D.C.
- White, R.J. 1998. RNA polymerase III transcription. Landes Bioscience, Austin, TX.
- Winckler, T., Dingermann, T., and Glockner, G. 2002. Dictyostelium mobile elements: Strategies to amplify in a compact genome. Cell. Mol. Life Sci. 59: 2097–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, F. and Carlson, M. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8: 387–391. [DOI] [PubMed] [Google Scholar]
- Winston, F., Chaleff, D.T., Valent, B., and Fink, G.R. 1984. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107: 179–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick, J.J., Aparicio, J.G., Chen, T., Barnett, J.D., Jennings, E.G., Young, R.A., Bell, S.P., and Aparicio, O.M. 2001. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: High-resolution mapping of replication origins. Science 294: 2357–2360. [DOI] [PubMed] [Google Scholar]
- Xie, W., Gai, X., Zhu, Y., Zappulla, D.C., Sternglanz, R., and Voytas, D.F. 2001. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell. Biol. 21: 6606–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yieh, L., Kassavetis, G., Geiduschek, E.P., and Sandmeyer, S.B. 2000. The Brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J. Biol. Chem. 275: 29800–29807. [DOI] [PubMed] [Google Scholar]
- Yieh, L., Hatzis, H., Kassavetis, G., and Sandmeyer, S.B. 2002. Mutational analysis of the transcription factor IIIB-DNA target of Ty3 retroelement integration. J. Biol. Chem. 277: 25920–25928. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Zou, S., Wright, D.A., and Voytas, D.F. 1999. Tagging chromatin with retrotransposons: Target specificity of the Saccharomyces Ty5 retrotransposon changes with the chromosomal localization of Sir3p and Sir4p. Genes & Dev. 13: 2738–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Dai, J., Fuerst, P.G., and Voytas, D.F. 2003. Controlling integration specificity of a yeast retrotransposon. Proc. Natl. Acad. Sci. 100: 5891–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, S. and Voytas, D.F. 1997. Silent chromatin determines target preference of the Saccharomyces retrotransposon Ty5. Proc. Natl. Acad. Sci. 94: 7412–7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.yeastgenome.org/; Saccharomyces Genome Database.
- http://web.wi.mit.edu/young/RSC/; Supplemental data for Ng et al. (2002).
- http://www.sciencemag.org/cgi/content/full/294/5550/2357/DC1; Supplemental data for Wyrick et al. (2001).
- http://www.bs.jhmi.edu/MBG/boekelab/Supplements/Supplements.html; Supplemental data for Bolton and Boeke (2003).
- http://www.public.iastate.edu/∼voytas/; Voytas lab homepage.