Abstract
In humans, the claudin superfamily consists of 19 homologous proteins that commonly localize to tight junctions of epithelial and endothelial cells. Besides being structural tight-junction components, claudins participate in cell–cell adhesion and the paracellular transport of solutes. Here, we identify and annotate the claudin genes in the whole-genome of the teleost fish, Fugu rubripes (Fugu), and determine their phylogenetic relationships to those in mammals. Our analysis reveals extensive gene duplications in the teleost lineage, leading to 56 claudin genes in Fugu. A total of 35 Fugu claudin genes can be assigned orthology to 17 mammalian claudin genes, with the remaining 21 genes being specific to the fish lineage. Thus, a significant number of the additional Fugu genes are not the result of the proposed whole-genome duplication in the fish lineage. Expression profiling shows that most of the 56 Fugu claudin genes are expressed in a more-or-less tissue-specific fashion, or at particular developmental stages. We postulate that the expansion of the claudin gene family in teleosts allowed the acquisition of novel functions during evolution, and that fish-specific novel members of gene families such as claudins contribute to a large extent to the distinct physiology of fishes and mammals.
The function of epithelial and endothelial cells as barriers between different compartments of tissues and organs is dependent on their ability to establish and maintain cell–cell adhesion through tight junctions (TJ; Tsukita et al. 2001). The TJ also prevents the diffusion of proteins and lipids between the apical and basolateral plasma membrane domains of epithelial and endothelial cells and is thus important for the generation and maintenance of membrane polarity in these cell types (van Meer et al. 1986). Furthermore, TJ form a continuous intercellular seal that restricts and regulates the paracellular transport of water, small solutes, and immune cells (Tsukita and Furuse 2000, 2002; Heiskala et al. 2001).
Claudins, a family of ∼20 homologous proteins that span the plasma membrane four times (see Fig. 1A; Furuse et al. 1998a; Morita et al. 1999a; Heiskala et al. 2001; Gonzalez-Mariscal et al. 2003), are structural components of TJ and major determinants of the fibrils observed by cryo-electron microscopy of freeze-fractured TJ (Furuse et al. 1998b). Homo- and heterotypic interactions between claudins on paired strands of adjacent cells provide the intercellular seal responsible for the function of TJ as paracellular diffusion barriers (Furuse et al. 1999; Inai et al. 1999). On the cytosolic face of the membrane, claudins are tethered to the actomyosin cytoskeleton via interaction with scaffolding proteins such as the zonula occludens (ZO) proteins (Itoh et al. 1999; Gonzalez-Mariscal et al. 2003). Besides epithelia and endothelia, claudins are also found at sites of intimate cell–cell contact in other cell types such as Schwann cells (Morita et al. 1999b) and keratinocytes (Brandner et al. 2002; Tsukita and Furuse 2002).
Figure 1.
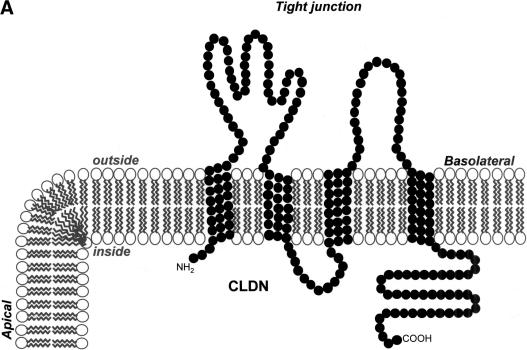
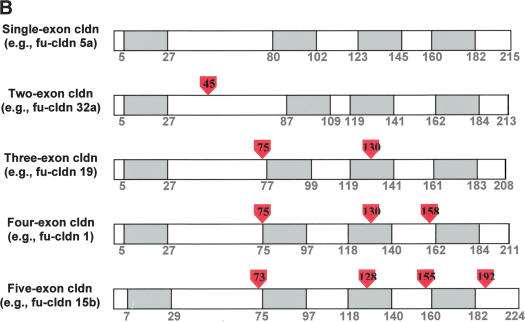
Topology and exon–intron organization of Fugu claudins. (A) Membrane topology of claudins. A diagram of the typical topology of claudins in the lipid bilayer and their location at the tight junction between the apical and lateral plasma membrane of epithelial cells is shown. (B) Exon–intron structure of typical Fugu claudin genes. The approximate location of introns in claudin genes encoded by one, two, three, or four exons is shown based on a typical example. Amino acids are numbered and show the boundaries of the four transmembrane domains (shaded in gray), which are typical for claudins. Intron position is shown by the red arrowheads (the numbers within correspond to the location in relation to the amino acid sequence).
Tissue- and cell-type-specific expression as well as the variability in homo- and hetero-oligomerization of individual claudins on neighboring cells are thought to determine the composition of the resulting intercellular complex and, in turn, the characteristic permeability properties observed for different epithelia and endothelia (Heiskala et al. 2001; Tsukita et al. 2001). While the intercellular seal provided by TJ acts as a barrier for the diffusion of large molecules, it allows the selective paracellular transport of water and, depending on the particular claudins, small solutes. Claudins 2 and 4, for example, have been linked to intercellular Na+ and K+ permeability (Van Itallie et al. 2001; Amasheh et al. 2002), whereas claudin 16 is critical for renal resorption of Ca2+ and Mg2+ (Simon et al. 1999; Muller et al. 2003), indicating that claudins can function as highly selective paracellular ion channels. Consistent with specific functions for individual claudins, mutations in an increasing number of claudins are associated with distinct diseases in humans or animal models (Gow et al. 1999; Simon et al. 1999; Weber et al. 2000; Wilcox et al. 2001; Furuse et al. 2002), supporting the idea that individual claudins mediate specific functions.
In contrast to the ∼20 claudin genes present in mammals (Heiskala et al. 2001; Tsukita and Furuse 2002), invertebrates such as Caenorhabditis elegans or Drosophila melanogaster only possess four to five claudin-related genes (Asano et al. 2003; Behr et al. 2003). This, together with the frequently observed tissue- and cell-type-restricted expression of individual claudins in mammals (Heiskala et al. 2001), indicates that the evolution of increasingly complex tissues and organs was paralleled with the expansion of the claudin gene family, possibly to accommodate new or overlapping functions. The recent completion of the sequence of the human (Lander et al. 2001; Venter et al. 2001) and the Japanese pufferfish, Fugu rubripes (Fugu; Aparicio et al. 2002) genomes allows a detailed comparison of the complexity and organization of claudin genes between mammals and teleosts, the two most distant bony vertebrate lineages. Fugu has a compact genome of only 365 Mb (the smallest known vertebrate genome), yet it possesses a repertoire of genes similar to that of human (Brenner et al. 1993). The compactness of the Fugu genome is attributed to its short introns and compressed intergenic regions because of a paucity of repetitive sequences (Venkatesh et al. 2000), making it an ideal vertebrate genome for characterizing genes and their regulatory regions.
Here, we report on the annotation of the claudin genes in Fugu and their expression profile, and the evolution of the vertebrate claudin gene family.
RESULTS AND DISCUSSION
Claudin Gene Prediction and Annotation
A search of the complete genome sequence of Fugu (Aparicio et al. 2002), and annotation and analyses of the claudin genes identified a total of 56 claudin gene sequences, distributed on 31 scaffolds (Table 1). The annotated sequences of all the Fugu claudin genes have been submitted to GenBank (accession numbers AY554341–AY554396). The 56 claudin genes found in Fugu represents a substantial increase when compared with the total number of claudin genes in humans or mouse, consistent with the previously observed duplication of many genes in Fugu (Taylor et al. 2001, 2003; Aparicio et al. 2002; Williams et al. 2002a,b; Yu et al. 2003). The presence of additional genes in Fugu and other teleosts such as zebrafish has led to the hypothesis that a whole-genome duplication occurred in the teleost lineage (Amores et al. 1998; Taylor et al. 2003; Christoffels et al. 2004). However, in contrast to the duplication observed for most other genes in Fugu and zebrafish, the 56 claudin genes represent an almost threefold increase as compared with the 20 claudin genes in mammals, and cannot be accounted for by a single genome duplication event.
Table 1.
Nomenclature, Scaffold Location, and Exon/Intron Organization of Fugu Claudin Genes
| S/No. | Fugu scaffold number | Fugu claudin number | No. of exons in Fugu | No. of exons in human orthologa | Zebrafish orthologaexon structure |
|---|---|---|---|---|---|
| 1 | 2 | fu-cldn 5a | 1 | 1 | — |
| 2 | fu-cldn 33c | 1 | — | — | |
| 3 | 8 | fu-cldn 7a | 4 | 4 | zf-cldn 7/M |
| 4 | fu-cldn 3c | 1 | 1 | — | |
| 5 | fu-cldn 3d | 1 | 1 | zf-cldn c/S | |
| 6 | fu-cldn 27c | 1 | — | — | |
| 7 | fu-cldn 27d | 3 | — | — | |
| 8 | 102 | fu-cldn 28b | 1 | — | zf-cldn e/S |
| 9 | fu-cldn 28c | 1 | — | — | |
| 10 | fu-cldn 29a | 1 | — | zf-cldn d/S | |
| 11 | fu-cldn 29b | 1 | — | — | |
| 12 | fu-cldn 30c | 1 | — | — | |
| 13 | fu-cldn 30d | 1 | — | zf-cldn a/b/S | |
| 14 | 107 | fu-cldn 18 | 4 | 5 | — |
| 15 | 137 | fu-cldn 23b | 1 | — | — |
| 16 | 161 | fu-cldn 12 | 2 | 3 | zf-cldn 12/S |
| 17 | 196 | fu-cldn 7b | 4 | 4 | zf-cldn 7/M |
| 18 | 281 | fu-cldn 23a | 1 | — | — |
| 19 | 300 | fu-cldn 14a | 2 | 3 | — |
| 20 | fu-cldn 3a | 1 | 1 | zf-cldn h/S | |
| 21 | fu-cldn 3b | 1 | 1 | — | |
| 22 | fu-cldn 5c | 1 | 1 | — | |
| 23 | fu-cldn 6 | 1 | 1 | zf-cldn j/S | |
| 24 | 302 | fu-cldn 13 | 1 | — | — |
| 25 | fu-cldn 27a | 1 | — | — | |
| 26 | fu-cldn 27b | 1 | — | zf-cldn f/S | |
| 27 | fu-cldn 28a | 1 | — | zf-cldn e/S | |
| 28 | fu-cldn 30a | 1 | — | — | |
| 29 | fu-cldn 30b | 1 | — | zf-cldn a/b/S | |
| 30 | 415 | fu-cldn 20a | 1 | 1 | — |
| 31 | 457 | fu-cldn 31 | 1 | — | zf-cldn g/S |
| 32 | 552 | fu-cldn 1 | 4 | 4 | zf-cldn 19/M |
| 33 | 628 | fu-cldn 2 | 1 | 2 | — |
| 34 | 641 | fu-cldn 20b | 1 | 1 | — |
| 35 | 654 | fu-cldn 11b | 3 | 3 | zf-cldn 11/M |
| 36 | 696 | fu-cldn 32b | 1 | — | — |
| 37 | 812 | fu-cldn 15b | 5 | 5 | — |
| 38 | fu-cldn 8a | 1 | 1 | — | |
| 39 | 863 | fu-cldn 8b | 1 | 1 | — |
| 40 | fu-cldn 8c | 1 | 1 | — | |
| 41 | fu-cldn 8d | 1 | 1 | — | |
| 42 | 1289 | fu-cldn 11a | 3 | 3 | zf-cldn 11/M |
| 43 | 1321 | fu-cldn 10d | 4 | 5 | — |
| 44 | fu-cldn 10e | 3 | 5 | — | |
| 45 | 1339 | fu-cldn 10a | 4 | 5 | — |
| 46 | 2207 | fu-cldn 32a | 2 | — | zf-cldn I/S |
| 47 | 2235 | fu-cldn 5b | 1 | 1 | — |
| 48 | 2355 | fu-cldn 19 | 3 | 4 | — |
| 49 | 2558 | fu-cldn 26 | 4 | — | — |
| 50 | 2732 | fu-cldn 25 | 4 | — | zf-cldn 10/M |
| 51 | 2743 | fu-cldn 14b | 1 | 3 | — |
| 52 | fu-cldn 33b | 1 | — | — | |
| 53 | 2835 | fu-cldn 15a | 4 | 5 | — |
| 54 | 2840 | fu-cldn 10b | 4 | 5 | — |
| 55 | fu-cldn 10c | 4 | 5 | — | |
| 56 | 3054 | fu-cldn 33a | 1 | — | — |
Each of the predicted 56 Fugu claudin genes was named according to its human ortholog (see Fig. 1), with alphabetical suffixes if there was more than one Fugu ortholog. If a human ortholog could not be identified, the identity of such genes was confirmed after phylogenetic analysis, and new numbers were assigned from 25 onward. The scaffolds on which the different Fugu claudin genes are located are listed. The exon—intron structures of Fugu genes were predicted based on their homology to known claudin genes from other organisms, and, where the boundaries were not clear, the intron acceptor and donor sites and the corresponding open reading frames were determined by visual inspection. The number of exons encoding the human orthologs is provided for reference. Where data were available, zebrafish orthologs were classified into single (S) or multiple (M) exon genes (Kollmar et al. 2001).
Where applicable/data available.
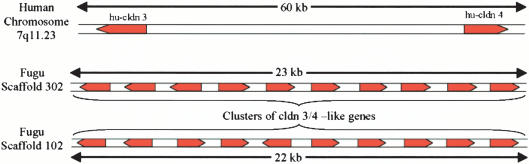
Of the 56 Fugu claudin genes, 24 are located independently on different scaffolds, and the remaining 32 are found in seven clusters containing two or more genes each (Table 1). Of particular interest are gene duplications on scaffolds 102 and 302, each containing 10 claudin genes spread over a distance of ∼23 kb. Of these 20 genes, 17 are closely related to human claudin 3 and 4 (see below), which are located 60 kb apart on human Chromosome 7q11.23.
Of the 56 claudin genes, 36 are encoded in a single exon, three by two exons, and the remaining 17 by three or more exons (Table 1). The location of introns in relation to the four transmembrane domains that are typical for claudins is shown for representative genes encoded by one or more exons (Fig. 1B). Of the 18 orthologs characterized so far in zebrafish (Kollmar et al. 2001), 10 claudin genes are encoded by a single exon and six by multiple exons, like their orthologs in Fugu (Table 1). Thus, it is likely that most of the duplications have occurred early in the teleost linage and are shared by related subgroups of this lineage. In contrast, only five of the 19 human genes are encoded by one exon, with one ortholog being encoded by two exons and the rest by three exons or more. Of the 32 orthologous genes shared between Fugu and humans, 20 Fugu genes have the same number of exons as their human counterparts. Thus, the claudin loci in Fugu and humans appear to be very dynamic in terms of “gain” and “loss” of spliceosomal introns.
Phylogenetic Analysis of Claudin Genes
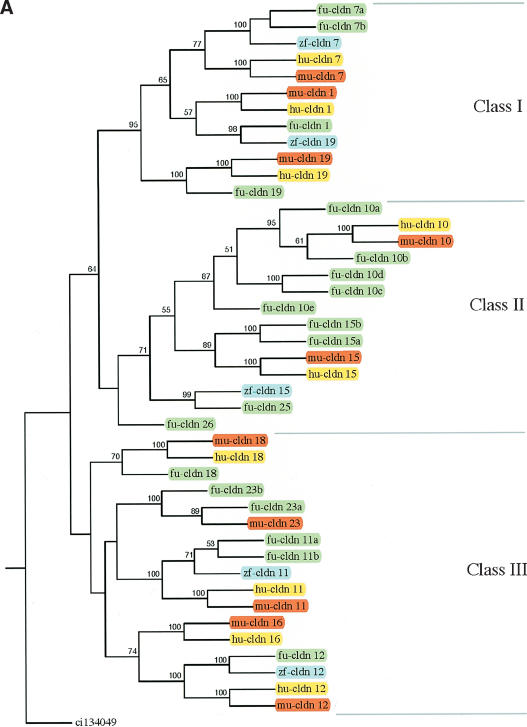
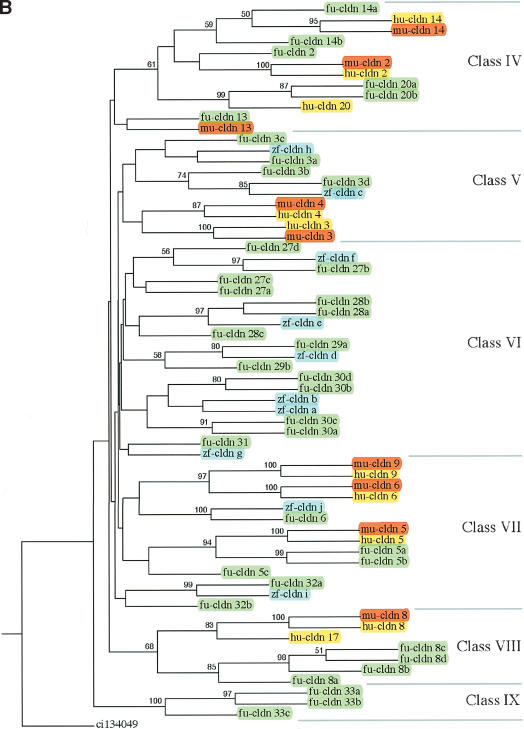
A total of 109 claudin genes identified from Fugu, human, mouse, and zebrafish were used to generate a phylogenetic tree, using a claudin-related gene from Ciona as an outgroup. Because a large number of diverse genes were involved, several preliminary rounds of alignment and tree construction with the complete set and subsets of genes were computed, allowing us to divide the genes into two major subgroups and subsequently generate their trees separately (Fig. 2A,B). For convenience, the claudin gene family was grouped into nine classes (Class I to Class IX) of related genes. The orthology of Fugu genes was assigned based on their phylogenetic relationships to the known claudin genes from humans and mouse. Duplicate copies of genes are denoted by an alphabetic suffix (a, b, etc.). Novel genes found only in Fugu were given numbers starting from 25. Overall, 37 Fugu claudin genes were found to be orthologous to 17 mammalian claudin genes, with the rest being unique to the fish lineage. Fugu does not contain the ortholog for mammalian claudin 16. With few exceptions, Fugu claudin genes encoded by a single exon clustered together (Classes I–III), whereas those with multiple exons grouped to Classes IV–IX. Single-exon claudin genes also have more paralogs, suggesting retrotransposition of a first copy followed by tandem duplications as a possible mechanism for this expansion.
Figure 2.
Consensus phylogenetic tree of claudin proteins. The trees were generated using 109 claudin protein sequences from human (hu, yellow), mouse (mu, red), Fugu (fu, green), and zebrafish (zf, blue) and using a related gene from Ciona (ci) as an outgroup gene. To reduce the complexity of phylogenetic trees, claudin genes were first divided into two groups of related proteins based on the results of preliminary phylogenetic analysis, and phylogenetic trees were generated separately for the two groups. Based on the origin and phylogenetic relationships, vertebrate claudin genes were classified into nine classes (A, Class I–III; B, Class IV–IX). Bootstrap values >50% are given.
Multiple Duplications of the Ancestral Claudin 3 and Claudin 4 Gene Cluster in Teleosts
The phylogenetic tree for the Class V group of claudin genes (Fig. 1B) reveals that mammalian claudin 3 and 4 are duplicate genes that arose in the mammalian lineage. The close linkage of claudin 3 and claudin 4 genes on human Chromosome 17 indicates that they arose as the result of tandem duplication (Fig. 3). In Fugu, 17 claudin genes found in clusters on scaffolds 102 and 302 (Class V and VI genes) are closely related to mammalian claudin 3 and 4 (Fig. 2B). The genomic organization of the Fugu genes suggests that the Fugu ortholog of the human claudin 3 and 4 genes has undergone several successive rounds of cis and trans duplications to give rise to two clusters of 17 genes. Because orthologs for some of these Fugu genes are also present in zebrafish (Fig. 2B), a distantly related teleost to Fugu, the duplications appear to have occurred very early during the evolution of teleosts, implying that the majority of teleosts possess this complement of claudin genes.
Figure 3.
Extensive duplication of an ancestral claudin 3 and 4 gene in Fugu. The human CLDN3 and CLDN4 locus on Chromosome 7 is compared with the cldn3/cldn4-like gene loci on scaffolds 102 and 302 of Fugu. The location and orientation of the genes are indicated.
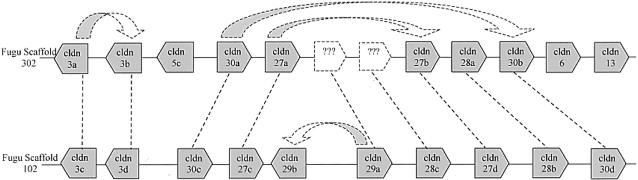
The results of the gene prediction and phylogenetic analysis also offer insight into the possible order of duplications that occurred in scaffolds 102 and 302 (Fig. 4). With the exception of cldn5c, cldn6, and cldn13, the remaining 17 genes are all closely related to human CLDN3 and CLDN4, and the pair of claudin genes that are direct paralogs in these scaffolds can be identified. Chromosomal fragments represented by scaffold 102 or 302 could have been more ancient, and intrachromosomal duplications (dotted arrows) on the ancestral chromosomal fragment had already resulted in the presence of at least seven genes. A single duplication event, presumably the postulated whole-genome duplication, then caused a duplication of this fragment (dotted lines). This event is supported by the conservation of the order and orientation of paralogous genes on the two scaffolds. The paralogs of claudin 29a and 28c (scaffold 302) were subsequently lost on scaffold 302, whereas claudin 29a duplicated once more within scaffold 102.
Figure 4.
Proposed order of duplication of the claudin 3- and 4-like genes in Fugu. The location and orientation of the different genes on the two scaffolds are indicated. The dotted arrows denote intrachromosomal gene duplications; the dotted lines indicate duplication of the chromosomal segment, presumably as a result of whole-genome duplication.
Besides the extensive duplication of genes on scaffolds 102 and 302, there are several other Fugu duplications that presumably arose as part of the proposed whole-genome duplication in the teleost lineage. These include fu-claudin 7a and 7b (Class I); claudin 15a and 15b (Class II); claudin 11a and 11b (Class III); claudin 20a and 20b (Class IV); claudin 5a and 5b (Class VII); claudin 8c and 8d (Class VIII); and claudin 33a and b (Class IX).
Independent Claudin Gene Duplication in Mammals
Independent gene duplications have occurred in the mammalian lineage as well. For example, human claudin 17 and 8 (Class VIII) are the result of gene duplication in the mammalian lineage. Similarly, mammalian claudin 3 and 4 (Class V) and claudin 9 and 6 (Class VII) are the results of duplications in the mammalian lineage.
Independent Claudin Gene Loss in Mammals and Teleosts
In addition to gene duplications, claudin genes have been lost in both mammalian and fish lineages. From the phylogenetic tree (Fig. 1A,B), it can be inferred that orthologs of Fugu claudin 32 (Class VII) and claudin 25 (Class II) have been lost in the mammalian lineage. Interestingly, whereas the ortholog of Fugu claudin 13 (Class IV) is present in mouse, there is no ortholog in humans. On the other hand, the fish ortholog of mammalian claudin 16 (Class III) has been lost in the Fugu lineage.
Tissue- and Developmental-Stage-Specific Expression of Fugu Claudin Genes
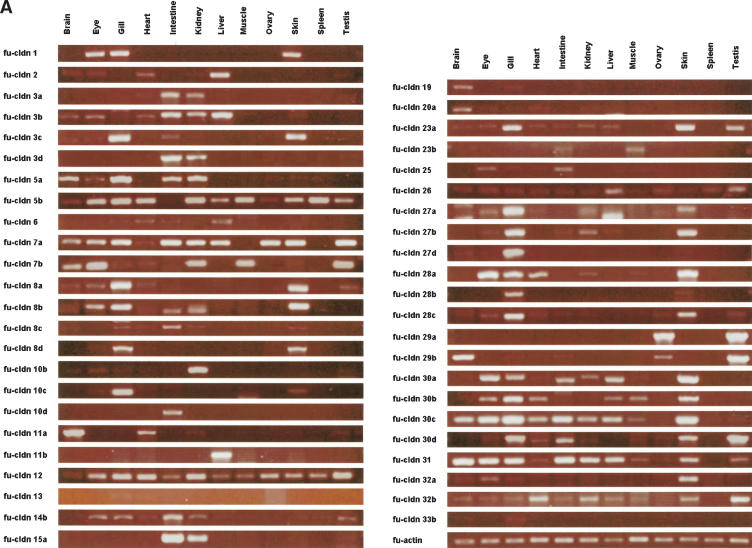
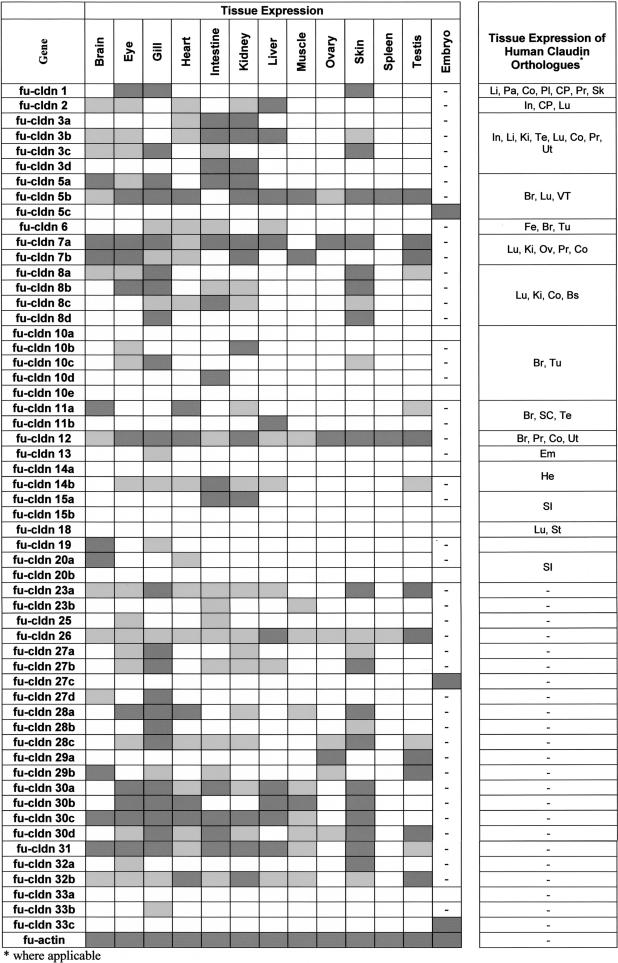
Given the unexpected expansion of the claudin gene family in Fugu, it was of interest to determine how many of these genes are actually expressed, and how many of the duplicate copies have an identical expression pattern, which could suggest redundant function. For this purpose, we did RT-PCR using primers specific to each gene (see Supplemental material) to check the expression in various adult Fugu tissues (Fig. 5A). Surprisingly, most of the Fugu claudin genes were found expressed in a more or less tissue-specific manner (Table 2), indicating that they are functional and have distinct expression patterns. cldn12 and cldn26 were expressed in all Fugu tissues analyzed. Expression of human CLDN12 has been reported for brain, prostate, colon, and uterus (Heiskala et al. 2001).
Figure 5.
Expression profiling of the Fugu claudin genes. Expression of the different claudin genes in (A) adult tissues and (B) an early somite embryonic stage of Fugu was analyzed by semiquantitative RT-PCR. The amplification of fu-actin was monitored as a positive control.
Table 2.
Tissue and Developmental Expression of Fugu Claudin Genes
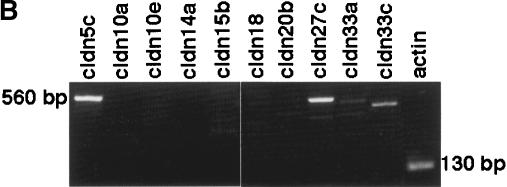
Transcripts for only 10 Fugu claudin genes (cldn5c, cldn10a, cldn10e, cldn14a, cldn15b, cldn18, cldn20b, cldn27c, cldn33a, and cldn33c) could not be detected in the tissues analyzed. With the exception of cldn27c, cldn33a, and cldn33c, all these genes have human orthologs that are expressed in brain, lung, and various other tissues (CLDN5); brain (CLDN10); heart (CLDN14); intestine (CLDN15 and CLDN20); or lung and stomach (CLDN18; Table 2; Heiskala et al. 2001). Because, except for cldn18, these claudin genes have undergone duplication in teleost and duplicated genes were expressed in the corresponding tissues (i.e., cldn5b and cldn10b in brain, cldn14b in heart, cldn15a in intestine), it is conceivable that cldn5c, cldn10a, cldn10e, cldn14a, and cldn15b are pseudogenes. Alternatively, however, the claudins we failed to detect may be expressed in tissues other than those analyzed, or during earlier stages of Fugu development. Indeed, three out of the 10 claudins (i.e., cldn5c, cldn27c, and cldn33c) not detected in the adult tissues analyzed were expressed in an early somite stage of the Fugu embryo (Fig. 5B; Table 2), indicating that at least some of the claudin genes are subject to developmental regulation.
All the remaining Fugu claudin genes showed a more or less restricted tissue-specific expression pattern. Interestingly, many Fugu claudins were only detected in organs that are in direct contact with the aquatic environment of fish. Expression of cldn1 and cldn10c was restricted to eyes, gills, and skin; cldn8d and cldn28b to gills and skin; and cldn13 was only found in gills. Several other claudins with a restricted expression pattern were detected in gills (cldn19, cldn33b, and cldn27d). Given the importance of claudins for the permeability characteristics of epithelia, it is likely that these claudins are involved in regulating the exchange of specific solutes with the aqueous environment. cldn1, expressed in the skin of Fugu, is found in the epidermis of mammals, where it is crucial for preventing water loss across the skin and dehydration (Furuse et al. 2002). Other claudin genes showing a restricted expression profile are cldn10d (intestine), cldn11a (brain, heart, kidney, testis), cldn11b (liver), cldn23b (intestine, muscle), cldn15a (intestine, kidney), and cldn29a (ovary and testis). In addition to gills and skin, heart, kidney, and intestine express the largest number of different claudin genes. Similar to gills and skin, kidney and intestine are also involved in maintaining osmotic homeostasis in fishes.
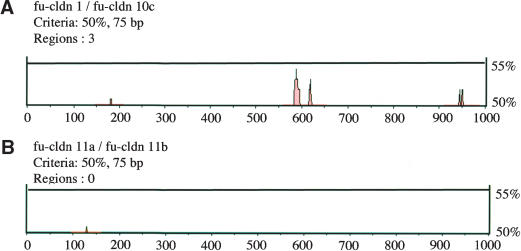
Because Fugu claudin genes with a similar tissue distribution may encode conserved regulatory elements in their 5′ noncoding region, we selected, as an example, two claudin genes with a similar expression profile, cldn1 and cldn10c (Table 2), for promoter analysis (Loots et al. 2002). The two claudin genes shared three highly conserved regions that may encode potential regulatory elements for tissue- and/or stage-specific expression in their 5′ part (Fig. 6A). In contrast, two closely related claudin genes with a different tissue distribution, cldn11a and cldn11b (Table 2), did not share conserved promoter regions (Fig. 6B), suggesting that their promoters have diverged considerably since the duplication such that they either share the expression domains of the parent gene (“subfunctionalization”) or have acquired novel expression domains (“neofunctionalization”).
Figure 6.
Promoter region analysis for two pairs of Fugu claudin genes with either similar (A) or distinct (B) tissue distributions. The 5′-noncoding regions of two claudin genes with a similar (cldn1 and cldn10c; Table 2) or different (cldn11a and cldn11b; Table 2) expression profile were analyzed. Red areas show regions along the 5′-noncoding sequence that share a homology of 50% or more.
The variability of the expression profiles observed even for closely related Fugu genes suggests that the duplicated claudin genes may have acquired new and unique functions. Comparison of the tissue expression of Fugu and mammalian claudin orthologs did not reveal an extensive correlation, possibly reflecting the acquisition of similar functions by some of the different teleost claudins. Thus, although the specificity and functionality of Fugu claudins diverged from their human orthologs, it is possible that the overall functionalities were both compensated and/or expanded by the duplicated claudins. Duplication of claudin genes with the retention of their function may also have enabled teleosts, by introducing evolutionary modifications in the promoter regions, to evolve new temporal and/or spatial expression patterns of a particular claudin gene.
In conclusion, a total of 56 unique claudin genes were identified and analyzed in Fugu. The phylogenetic analysis revealed that the claudin genes have undergone unprecedented expansion in the teleost lineage, leading to an almost threefold higher number of claudins in Fugu as compared with mammals. Although some of the additional genes arose as a result of the proposed whole-genome duplication in the fish lineage, a substantial number of new genes are the result of tandem duplications in the fish lineage. In addition to extensive tandem duplications, some of the claudin genes that were present in the ancestral vertebrate have been lost in the fish lineage. Independent duplications and deletions have also occurred in the mammalian lineage, albeit on a smaller scale as compared with the fish lineage. The bulk of the 56 Fugu claudin genes are expressed in a more or less tissue-specific fashion, suggesting that individual members of the expanded families may have acquired new or overlapping functions in teleost fish. These results show that claudins are a dynamic family of genes in vertebrates and have continued to evolve after the divergence of the common ancestors of the mammalian and fish lineages. The new genes in each lineage may have acquired novel functions, reflecting the unique tissues and cell types in the two distant vertebrates. It is likely that the large number of novel claudin genes found in fishes are involved in the exchange of specific ions with the aquatic environment and maintenance of osmotic balance. The expression of a large number of claudin genes in the gills and kidney, which play a major role in these functions in fishes, supports this view.
METHODS
Gene Prediction
The human claudin protein sequences, 19 in all, were retrieved from the National Center for Biotechnology Information (NCBI). They were searched against the “draft” Fugu genome database (3rd assembly) at http://www.fugu-sg.org using the TBLASTN program to identify homologous claudin loci in the Fugu genome. The “draft” Fugu genome sequence comprises 8597 scaffolds each >2 kb long, and represents 95% of the nonrepetitive portion of the 365-Mb Fugu genome (Aparicio et al. 2002). The identified Fugu gene loci on the scaffolds were then searched against the NCBI human genome protein database using BLASTX to confirm that they, in fact, code for claudins, and to identify their putative human orthologs and obtain an initial closest human claudin match. The exon–intron structure of Fugu genes was predicted by their homology to known claudin genes from other organisms, and where the boundaries were not clear, the intron acceptor and donor sites and the corresponding open reading frames were identified by manual inspection of the sequence.
Phylogenetic Analysis and Promoter Region Characterization
The predicted Fugu claudin protein sequences, together with those of human, and those available for mouse and zebrafish, were aligned using CLUSTALW (version 1.8.2) with default parameters (Thompson et al. 1994). The columns of alignment that contained gaps were removed and the tree topologies were generated using the PHYLIP (version 3.6) programs PROTDIST, NEIGHBOR, and CONSENSE (Felsenstein 1989). In all, 1000 bootstrap samples were used to test the reliability of the clustering within phylogenetic trees by repeated pseudo sampling with replacements of sites from the aligned data sets. Paralogs obtained aligned well in CLUSTALW. Promoter region analysis was carried out using the rVISTA software (Loots et al. 2002), with a threshold of ≥50% identity across 75-bp windows.
Nomenclature
Each of the predicted Fugu claudin genes was numbered (Claudin 1, Claudin 2, etc.) according to its human ortholog, with alphabetical suffixes (Claudin 1a, Claudin 1b, etc.) if there was more than one Fugu ortholog. If a human ortholog could not be identified unambiguously, the identity of such genes was confirmed after phylogenetic analysis and a new number commencing from 25 onward was assigned. Zebrafish claudin genes were named in accordance with a previous study (Kollmar et al. 2001).
Gene Expression Profiling
cDNA from early-somite-stage Fugu embryos was provided by Tohru Suzuki (National Research Institute of Aquaculture, Fisheries Research Agency, Mei-ken 516-0193, Japan). Total RNA from various adult Fugu tissues (brain, eye, gill, heart, intestine, kidney, liver, muscle, ovary, skin, spleen, testis) was extracted using Tizol reagent (GIBCO BRL), and the first-strand cDNA was synthesized using the SMART RACE cDNA amplification kit (Clontech). First-strand cDNA was used as template for RT-PCR together with gene-specific primers (see Supplemental material). A 130-bp fragment of actin cDNA was amplified as an internal positive control using the primers ACTF (5′-AACTGGGAYGA CATGGAGAA-3′) and ACTR (5′-TTGAAGGTCTCAAACATGAT-3′). The PCR comprised an initial denaturation step for 2 min at 95°C followed by 35 cycles of denaturation (30 sec at 95°C), annealing (60 sec at 55°C), and extension (60 sec at 72°C), and a final elongation step of 5 min at 72°C. The PCR products were separated by gel electrophoresis (1.5% agarose gel at 160 V) in the presence of ethidium bromide and visualized under ultraviolet light. The identity of PCR products was confirmed by sequencing representative PCR fragments.
Acknowledgments
We thank Tay Boon Hui for expert technical help and Tohru Suzuki for providing Fugu embryo cDNA. This work was supported by the Agency for Science, Technology and Research (A*STAR), Singapore.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2400004. Article published online before print in June 2004.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. AY554341–AY554396. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: T. Suzuki.]
References
- Amasheh, S., Meiri, N., Gitter, A.H., Schoneberg, T., Mankertz, J., Schulzke, J.D., and Fromm, M. 2002. Claudin-2 expression induces cation-selective channels in tight junctions of epithelial cells. J. Cell Sci. 115: 4969–4976. [DOI] [PubMed] [Google Scholar]
- Amores, A., Force, A., Yan, Y.L., Joly, L., Amemiya, C., Fritz, A., Ho, R.K., Langeland, J., Prince, V., Wang, Y.L., et al. 1998. Zebrafish hox clusters and vertebrate genome revolution. Science 282: 1711–1714. [DOI] [PubMed] [Google Scholar]
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310. [DOI] [PubMed] [Google Scholar]
- Asano, A., Asano, K., Sasaki, H., Furuse, M., and Tsukita, S. 2003. Claudins in Caenorhabditis elegans: Their distribution and barrier function in the epithelium. Curr. Biol. 13: 1042–1046. [DOI] [PubMed] [Google Scholar]
- Behr, M., Riedel, D., and Schuh, R. 2003. The Claudin-like megatrachea is essential in septate junctions for the epithelial barrier function in Drosophila. Dev. Cell 5: 611–620. [DOI] [PubMed] [Google Scholar]
- Brandner, J.M., Kief, S., Grund, C., Rendl, M., Houdek, P., Kuhn, C., Tschachler, E., Franke, W.W., and Moll, I. 2002. Organization and formation of the tight junction system in human epidermis and cultured keratinocytes. Eur. J. Cell Biol. 81: 253–263. [DOI] [PubMed] [Google Scholar]
- Brenner, S., Elgar, G., Sandford, R., Macrae, A., Venkatesh, B., and Aparicio, S. 1993. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature 366: 265–268. [DOI] [PubMed] [Google Scholar]
- Christoffels, A., Koh, E.G.L., Chia, J., Brenner, S., Aparicio, S., and Venkatesh, B. 2004. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol. 21: 1146–1151. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. 1989. PHYLIP—Phylogeny inference package (version 3.2). Cladistics 5: 164–166. [Google Scholar]
- Furuse, M., Fujita, K., Hiiragi, T., Fujimoto, K., and Tsukita, S. 1998a. Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141: 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse, M., Sasaki, H., Fujimoto, K., and Tsukita, S. 1998b. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 143: 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse, M., Sasaki, H., and Tsukita, S. 1999. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J. Cell Biol. 147: 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse, M., Hata, M., Furuse, K., Yoshida, Y., Haratake, A., Sugitani, Y., Noda, T., Kubo, A., and Tsukita, S. 2002. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: A lesson from claudin-1-deficient mice. J. Cell Biol. 156: 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Mariscal, L., Betanzos, A., Nava, P., and Jaramillo, B.E. 2003. Tight junction proteins. Prog. Biophys. Mol. Biol. 81: 1–44. [DOI] [PubMed] [Google Scholar]
- Gow, A., Southwood, C.M., Li, J.S., Pariali, M., Riordan, G.P., Brodie, S.E., Danias, J., Bronstein, J.M., Kachar, B., and Lazzarini, R.A. 1999. CNS myelin and Sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell 99: 649–659. [DOI] [PubMed] [Google Scholar]
- Heiskala, M., Peterson, P.A., and Yang, Y. 2001. The roles of claudin superfamily proteins in paracellular transport. Traffic 2: 93–98. [DOI] [PubMed] [Google Scholar]
- Inai, T., Kobayashi, J., and Shibata, Y. 1999. Claudin-1 contributes to the epithelial barrier function in MDCK cells. Eur. J. Cell Biol. 78: 849–855. [DOI] [PubMed] [Google Scholar]
- Itoh, M., Furuse, M., Morita, K., Kubota, K., Saitou, M., and Tsukita, S. 1999. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell Biol. 147: 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar, R., Nakamura, S.K., Kappler, J.A., and Hudspeth, A.J. 2001. Expression and phylogeny of claudins in vertebrate primordia. Proc. Natl. Acad. Sci. 98: 10196–10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Loots, G.G., Ovcharenko, I., Pachter, L., Dubchak, I., and Rubin, E.M. 2002. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 12: 832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, K., Furuse, M., Fujimoto, K., and Tsukita, S. 1999a. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc. Natl. Acad. Sci. 96: 511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, K., Sasaki, H., Fujimoto, K., Furuse, M., and Tsukita, S. 1999b. Claudin-11/OSP-based tight junctions of myelin sheaths in brain and Sertoli cells in testis. J. Cell Biol. 145: 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, D., Kausalya, P.J., Claverie-Martin, F., Meij, I.C., Eggert, P., Garcia-Nieto, V., and Hunziker, W. 2003. A novel claudin 16 mutation associated with childhood hypercalciuria abolishes binding to ZO-1 and results in lysosomal mistargeting. Am. J. Hum. Genet. 73: 1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, D.B., Lu, Y., Choate, K.A., Velazquez, H., Al-Sabban, E., Praga, M., Casari, G., Bettinelli, A., Colussi, G., Rodriguez-Soriano, J., et al. 1999. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 285: 103–106. [DOI] [PubMed] [Google Scholar]
- Taylor, J.S., Van de Peer, Y., Braasch, I., and Meyer, A. 2001. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 356: 1661–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J.S., Braasch, I., Frickey, T., Meyer, A., and Van de Peer, Y. 2003. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res 13: 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita, S. and Furuse, M. 2000. Pores in the wall: Claudins constitute tight junction strands containing aqueous pores. J. Cell Biol. 149: 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita, S. and Furuse, M. 2002. Claudin-based barrier in simple and stratified cellular sheets. Curr. Opin. Cell Biol. 14: 531–536. [DOI] [PubMed] [Google Scholar]
- Tsukita, S., Furuse, M., and Itoh, M. 2001. Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell. Biol. 2: 285–293. [DOI] [PubMed] [Google Scholar]
- Van Itallie, C., Rahner, C., and Anderson, J.M. 2001. Regulated expression of claudin-4 decreases paracellular conductance through a selective decrease in sodium permeability. J. Clin. Invest. 107: 1319–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer, G., Gumbiner, B., and Simons, K. 1986. The tight junction does not allow lipid molecules to diffuse from one epithelial cell to the next. Nature 322: 639–641. [DOI] [PubMed] [Google Scholar]
- Venkatesh, B., Gilligan, P., and Brenner, S. 2000. Fugu: A compact vertebrate reference genome. FEBS Lett. 476: 3–7. [DOI] [PubMed] [Google Scholar]
- Venter, J., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304–1351. [DOI] [PubMed] [Google Scholar]
- Weber, S., Hoffmann, K., Jeck, N., Saar, K., Boeswald, M., Kuwertz-Broeking, E., Meij, I.I.C., Knoers, N.V., Cochat, P., Sulakova, T., et al. 2000. Familial hypomagnesaemia with hypercalciuria and nephrocalcinosis maps to chromosome 3q27 and is associated with mutations in the PCLN-1 gene. Eur. J. Hum. Genet. 8: 414–422. [DOI] [PubMed] [Google Scholar]
- Wilcox, E.R., Burton, Q.L., Naz, S., Riazuddin, S., Smith, T.N., Ploplis, B., Belyantseva, I., Ben-Yosef, T., Liburd, N.A., Morell, R.J., et al. 2001. Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell 104: 165–172. [DOI] [PubMed] [Google Scholar]
- Williams, H., Brenner, S., and Venkatesh, B. 2002a. Characterization of the platelet-derived growth factor receptor α and c-kit genes in the pufferfish Fugu rubripes. DNA Seq. 13: 263–270. [DOI] [PubMed] [Google Scholar]
- Williams, H., Brenner, S., and Venkatesh, B. 2002b. Identification and analysis of additional copies of the platelet-derived growth factor receptor and colony stimulating factor1 receptor in the pufferfish Fugu rubripes. Gene 295: 255–264. [DOI] [PubMed] [Google Scholar]
- Yu, W.P., Brenner, S., and Venkatesh, B. 2003. Duplication, degeneration and subfunctionalization of the nested synapsin-Timp genes in Fugu. Trends Genet. 19: 180–183. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.fugu-sg.org; “draft” Fugu genome database (3rd assembly).