Abstract
We have performed X-inactivation and sequence analyses on 350 kb of sequence from human Xp11.2, a region shown previously to contain a cluster of genes that escape X inactivation, and we compared this region with the region of conserved synteny in mouse. We identified several new transcripts from this region in human and in mouse, which defined the full extent of the domain escaping X inactivation in both species. In human, escape from X inactivation involves an uninterrupted 235-kb domain of multiple genes. Despite highly conserved gene content and order between the two species, Smcx is the only mouse gene from the conserved segment that escapes inactivation. As repetitive sequences are believed to facilitate spreading of X inactivation along the chromosome, we compared the repetitive sequence composition of this region between the two species. We found that long terminal repeats (LTRs) were decreased in the human domain of escape, but not in the majority of the conserved mouse region adjacent to Smcx in which genes were subject to X inactivation, suggesting that these repeats might be excluded from escape domains to prevent spreading of silencing. Our findings indicate that genomic context, as well as gene-specific regulatory elements, interact to determine expression of a gene from the inactive X-chromosome.
X inactivation is a unique form of gene regulation resulting in the transcriptional silencing of most genes on one X-chromosome in each somatic cell of mammalian females. Despite the chromosome-wide nature of this phenomenon, 10% to 20% of genes on the human X-chromosome have been found to escape inactivation (Carrel et al. 1999). Many epigenetic and functional features have been identified that distinguish genes subject to inactivation from those that escape inactivation, such as differences in replication timing, CpG island methylation, and histone modifications (for review, see Disteche et al. 2002; Brown and Greally 2003), yet the mechanisms that underlie the phenomenon of escape remain poorly understood. X inactivation is initiated by the XIST gene, followed by spreading of silencing along the X-chromosome, and maintenance of the inactive state with its associated epigenetic changes (Plath et al. 2002). Escape from inactivation could be caused by a lack of initial silencing at the onset of inactivation that occurs early in development. Alternatively, genes that escape may be initially silenced, but may fail to maintain the inactive state. Our studies that have followed expression of Smcx through the course of mouse development indicate that escape appears to be caused by failure to maintain the inactive state (Lingenfelter et al. 1998).
The factors that are responsible for escape from X inactivation may reside at the level of individual genes, or they may occur at the higher-order level of chromosomal domains. There are many examples of regions of the genome that exhibit coordinate regulation of gene expression within chromosomal domains (Caron et al. 2001; Lercher et al. 2002; Spellman and Rubin 2002), including imprinted regions (Bartolomei and Tilghman 1997), homeobox gene clusters, and globin gene clusters (Dillon and Sabbattini 2000; Calhoun and Levine 2003). Elements physically near individual genes that could contribute to their escape from inactivation include unique promoter or enhancer characteristics. Alternatively, the ability to escape X inactivation may be dictated by elements that prevent the spread of the inactivation signal within a given region of the X-chromosome, or the lack of elements in certain regions of the X-chromosome that are necessary to facilitate the spreading of inactivation (Gartler and Riggs 1983; Lyon 1998; Bailey et al. 2000). In the human genome, the clustering of some genes that escape inactivation suggests that expression of these genes from the inactive X is controlled at the domain level (Disteche 1995; Carrel et al. 1996; Miller and Willard 1998). In mouse, the number of genes appreciated to escape inactivation is substantially lower than for the human X, and these genes appear to be scattered individually along the mouse X-chromosome. The difference in the size of domains of escape between different regions of the human X, and between mouse and human conserved segments, indicate that both gene-specific and regional regulatory elements dictate expression from the inactive X-chromosome (Tsuchiya and Willard 2000).
The observation that the inactivation status of many X-linked genes differs between mouse and human may shed light on the regulatory elements that are involved in the process of escape from inactivation. Cross-species functional and sequence comparisons have been used to identify key regulatory elements in other regions of the genome. For example, comparative sequence analysis led to the identification of novel regulatory elements within the α-globin gene cluster and the Bruton's tyrosine kinase locus (Oeltjen et al. 1997; Flint et al. 2001). Comparative genome analysis has been used to delineate imprinted gene clusters, as well as the X-inactivation center, which includes the XIST gene (Engemann et al. 2000; Chureau et al. 2002). This strategy has also been used to determine the basis for species-specific imprinting (Okamura et al. 2000).
A cluster of genes that escape inactivation in human Xp11.2, including SMCX, has previously been identified (Miller and Willard 1998). Initial characterization of the region of conserved synteny in mouse demonstrated highly conserved gene content and order, but differences in the inactivation status of most of the genes, with the result that SMCX/Smcx was the only gene in this region found to escape inactivation in both species (Tsuchiya and Willard 2000). Subsequent completion of genomic sequencing of this region in the human and mouse has allowed us to perform a more complete comparative analysis for the two species, including the identification of additional transcripts and their X-inactivation status. This comparative genomic characterization provides a framework for generating and testing different hypotheses to explain the phenomenon of escape from X inactivation.
RESULTS
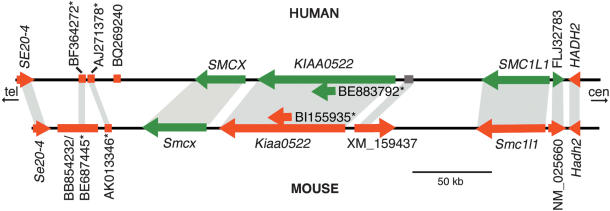
We compared 350 kb of genomic sequence from human Xp11.2 containing a cluster of genes that escaped inactivation, with the region of conserved synteny in mouse. The genomic organization of this region in terms of gene content, gene order, and transcriptional orientation was highly conserved between the two species. We identified new genes and expressed sequences in both mouse and human using gene prediction programs, homology searches against the databases, and alignment of genomic sequence between the two species to identify highly conserved segments (Supplemental Fig. S1). We studied six new transcripts in human, five of which had mouse orthologs. X-inactivation analysis of these new transcripts, in combination with previously published results, revealed an escape domain of at least five loci in human contained within 235 kb of sequence. In contrast, Smcx remained the only mouse gene in this region that escaped inactivation (Fig. 1).
Figure 1.
Schematic diagram of the human domain of escape from Xp11.2 and the corresponding region of conserved synteny in the mouse. Loci that escape are represented by green arrows or lines and those that are inactivated are in red. The X-inactivation status of the human XM_159437-like locus could not be determined (see Results). When known, the transcriptional orientation is indicated by the direction of the arrow. Orthologs are indicated by shaded regions. Sequences designated with an asterisk may not represent independent transcripts (see Results). Alternative names for transcripts/ESTs are as follows: SE20–4/Se20–4 = DXHXS1008E = TSPX/Tspx; AK055575 = SMC1L1 = SB1.8; ADS13 = FLJ32783; W08639 = NM_025660; ADS9 = HADH2.
Characterization of New Transcripts
Human and Mouse KIAA0522/Kiaa0522
We identified the DNA sequence coding for the predicted KIAA0522 protein (accession no. AB011094) by GENSCAN analysis of human genomic sequence (Burge and Karlin 1997). The gene coding for KIAA0522 demonstrated ubiquitous expression based on UniGene expression information and our own expression analyses (Table 1). The mouse Kiaa0522 gene was not annotated at the time of our analysis, but there were multiple mouse ESTs highly similar to the human cDNA sequence, and comparative sequence analysis using our own sequence data (GenBank accession no. AY451401) and a percent identity plot (PIP) demonstrated conservation of all exons between human and mouse (Supplemental Fig. S1). The KIAA0522 protein consisted of 1560 amino acids and contained a region of similarity to a domain found in the yeast Sec7 protein that has been known to function as a guanine nucleotide exchange factor and to be necessary for proper protein transport through the Golgi. KIAA0522 also contained a Pleckstrin homology domain commonly found in eukaryotic signaling proteins.
Table 1.
Summary of Genes and ESTs in and Flanking the Human Xp11.2 Escape Domain, and Comparison With Mouse Orthologs
| Human gene/EST | X-inactivation status | Expression | Mouse gene/EST | X-inactivation status | Expression | CpG island human/mouse | Nucleotide similarity | Amino acid identity |
|---|---|---|---|---|---|---|---|---|
| HADH2 | Inactivated | Ubiquitous | Hadh2 | Inactivated | Ubiquitous | Yes/no | 76%-92% | 86% |
| FLJ32783 | Escapes | Ubiquitous | NM_025660 | Inactivated | Ubiquitous | Yes/yes | 74%-90% | 70% |
| SMC1L1 | Escapes | Ubiquitous | Smc1l1 | Inactivated | Ubiquitous | Yes/yes | 92% | 99% |
| XM_159437-like | NDa | Brain/testis | XM_159437 | Inactivated | Brain | No/no | 77% | NDb |
| KIAA0522 | Escapes | Ubiquitous | Kiaa0522 | Inactivated | Ubiquitous | Yes/yes | 85%-92% | 74% |
| BE883792 | Escapes | Ubiquitous | BI155935 | Inactivated | Ubiquitous | No/no | 87%-94% | NDb |
| SMCX | Escapes | Ubiquitous | Smcx | Escapes | Ubiquitous | Yes/yes | 90% | 94% |
| BQ269240 | Inactivated | Ubiquitousc | — | — | — | No/— | — | — |
| AJ271378 | Inactivated | Ubiquitousc | AK013346 | Inactivated | Ubiquitous | No/no | 78%-90% | NDb |
| BF364272 | Inactivatedd | Ubiquitousc | BE687445 | Inactivated | Ubiquitous | No/no | 84% | NDb |
| TSPX | Inactivated | Ubiquitous | Tspx | Inactivated | Ubiquitous | Yes/yes | 81%-85% | 70% |
ND: Not determined.
X-inactivation status could not be determined because of tissue-restricted expression and lack of expression in fibroblasts.
Amino acid identity was not determined for small ESTs.
Expressed in liver and fibroblasts; other tissues not tested.
Variable, low-level escape.
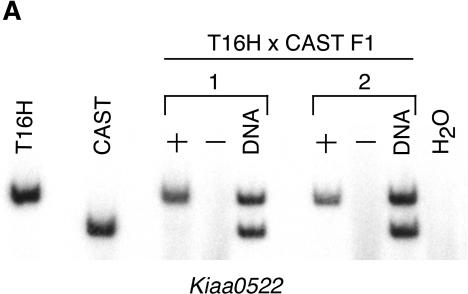
The gene coding for KIAA0522 was located <8 kb from the 5′-end of the SMCX/Smcx gene in both mouse and human (Fig. 1). Despite its close physical proximity to Smcx, which escaped inactivation, we determined that the Kiaa0522 gene was subject to inactivation in mouse using an X/autosome translocation system in which one allele (from Mus castaneus) was always inactivated. Lack of expression of the castaneus Kiaa0522 allele (CAST) in females who carried the T(X;16)16H translocation indicated that the gene was inactivated (Fig. 2A). In contrast, this gene escaped inactivation in human, as shown by positive expression in somatic cell hybrid lines that retained an inactive human X-chromosome on a Chinese hamster background (Fig. 3).
Figure 2.
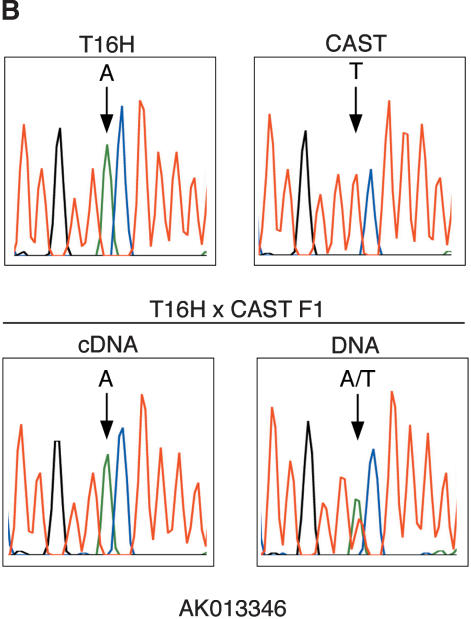
X-inactivation analyses of representative mouse transcripts. (A) Inactivation status of Kiaa0522 was determined by digesting RT-PCR products with BlpI to distinguish T16H from CAST alleles. T16H and CAST are parental controls. + and – represent RT-PCR reactions performed with and without reverse transcriptase. Only the T16H (active X) allele is expressed in T16H × CAST F1 females, demonstrating inactivation of the CAST allele of Kiaa0522. DNA from the F1 females was also amplified and digested to verify the presence and equal amplification of both alleles. Results from two different F1 females are shown. (B) Inactivation status of AK013346 was determined by sequence analysis of RT-PCR products to distinguish the T16H and CAST alleles, which differed by an A/T polymorphism. DNA sequence from a T16H × CAST F1 female confirms the presence of both alleles, whereas cDNA sequence from the same female reveals expression of only the T16H (active X) allele, consistent with inactivation of AK013346.
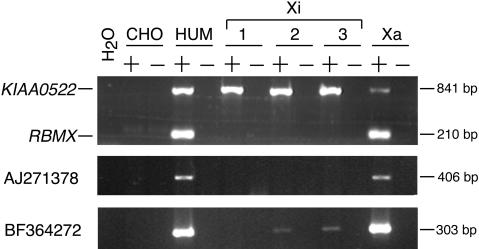
Figure 3.
X-inactivation analyses of representative human transcripts (KIAA0522, AJ271378, BF364272) in human × rodent somatic cell hybrids. Xi and Xa designate somatic cell hybrids containing an inactive or an active human X-chromosome on a Chinese hamster background, respectively. + and – indicate reactions performed with or without reverse transcriptase. The CHO (Chinese hamster ovary cell line) control shows that primers are specific for human sequence, and the human cDNA (HUM) serves as a positive control. RBMX is a control gene previously shown to be subject to X inactivation (Lingenfelter et al. 2001).
The human KIAA0522 gene contained a 36-kb intron between exons 2 and 3. Within this intron, there was a transcript (XM_098990) predicted by computational analysis of human genomic contig NT_011799 that matched multiple human and mouse ESTs from different tissues. The longest EST was 830 bp in human (BE883792, BC038213) and 806 bp in mouse (BI155935 + BY764207). The longest open reading frame from the human sequence predicted a 183-amino-acid protein with no significant similarity to known proteins. A single human cDNA clone derived from a tumor (AK095232) that partially overlapped both KIAA0522 and BE883792 indicated that this intronic transcript might represent a rare alternative splice form of KIAA0522. However, our data suggested that KIAA0522 and BE883792 were independent genes. Sequence that we generated from RT-PCR products amplified from mouse liver cDNA using primers that flanked the third intron of Kiaa0522 did not include sequence from BI155935. In addition, we could not amplify a product from mouse brain, testis, embryonic stem cell, liver, spleen, or kidney cDNA using a combination of forward primers from BI155935 and reverse downstream primers from mouse Kiaa0522, although both expressed sequences could be amplified independently from these samples (data not shown). Although it is unusual for an intronic gene to share the same transcriptional orientation as the gene within which it is embedded, this phenomenon has been described for the X-linked gene, Nap1l2 (Chureau et al. 2002). Regardless of whether or not the intronic transcript and KIAA0522 were separate genes, our X-inactivation analyses demonstrated that human BE883792 escaped inactivation, whereas mouse BI155935 was subject to inactivation (data not shown). These sequences thus have the same X-inactivation profile as human and mouse KIAA0522/Kiaa0522.
Human-Expressed Actin Pseudogene
We searched the NCBI database by BLAST using a predicted human transcript (XM_093106) located ∼50 kb 3′ of SMCX, and identified several human ESTs with identical or highly similar sequence. The longest of these sequences, BQ269240, was a 610-bp EST that was identical to an uninterrupted stretch of human genomic sequence 3′ of SMCX (Fig. 1). This sequence was highly homologous to many actin genes, and appeared to represent a retrotransposed actin pseudogene. We designed primers specific for this X-linked, actin pseudogene by alignment with other actin sequences in the database. RT-PCR performed on human × rodent somatic cell hybrids retaining either an active or an inactive X-chromosome showed that this transcript was subject to inactivation (data not shown). This locus was particularly informative, as it delineated the telomeric boundary of the human domain of escape. The transcript was not conserved with any sequence in the mouse syntenic region, and consequently underwent retrotransposition after the divergence of rodents and primates.
Human- and Mouse-Expressed LIMEd LINE
Genomic sequence alignment between mouse and human revealed a highly conserved region adjacent to human BQ269240 (Supplemental Fig. S1). Using this conserved sequence to perform BLAST searches, we identified the human cDNA clone AJ271378. This cDNA was highly similar to several mouse EST sequences and to a 1469-bp mRNA sequence (AK013346) that perfectly matched mouse genomic sequence 3′ of Smcx (Fig. 1). Additional nearby expressed sequences were also present in human (BF364272) and mouse (BE687445, BB854232). The sequence conservation in this region could be attributed to the bulk of these sequences being composed of an L1 long interspersed nuclear element (LINE). The subclassification of the LINE as an LIMEd element indicated that this element entered the genome prior to divergence of the primate lineage (Smit et al. 1995), which was consistent with its presence in both species. We took advantage of the expression of these transcripts to test for X-inactivation status, and showed that they were predominantly inactivated. The mouse-expressed sequences showed no evidence of escape (Fig. 2B). In human, AJ271378 and BF364272 also showed no evidence of escape after 30 cycles of cDNA amplification. After 35 cycles, BF364272, but not AJ271378, demonstrated very low level escape in two of three inactive X hybrids, even though these ESTs might be part of the same expressed L1 element (Fig. 3). This difference could be due to increased amplification efficiency using BF364272 primers. Alternatively, these sequences might represent separate transcripts. To ensure that amplification of other expressed L1 elements in the genome did not occur, we designed PCR primers from unique sequence. Sequencing of RT-PCR products also confirmed specificity of the primers for amplification of the expressed L1 in this region.
XM_159437, a Human and Mouse Transcript With Tissue-Limited Expression
A highly conserved region between human and mouse located centromeric to KIAA0522 (Supplemental Fig. S1) contained a mouse transcript (XM_159437) predicted by the EST clustering, Geneid, and NCBI gene prediction programs (Fig. 1). We failed to identify any corresponding human or mouse ESTs using BLAST searches; however, RT-PCR of multiple mouse tissues revealed expression almost exclusively limited to brain (Table 1). The transcript demonstrated no expression, or only very weak expression, in liver, kidney, heart, testis, and embryonic stem cell cDNA after 35 cycles of amplification (data not shown). Analysis of brain tissue from an F1 female balanced X/autosome translocation carrier showed that this transcript was subject to X inactivation (data not shown).
We aligned mouse XM_159437 to the human genomic sequence and designed human primers within the most highly conserved sequence. RT-PCR with these primers demonstrated expression in testis and brain, but not liver (Table 1). We also did not detect any expression in the somatic cell hybrids, precluding analysis of the X-inactivation status of the human transcript.
Characterization of Previously Published Genes
Human and Mouse SMCX/Smcx
SMCX (NM_004187) and Smcx (NM_013668) are ubiquitously expressed and escape inactivation in human and mouse (Table 1; Agulnik et al. 1994b; Wu et al. 1994). The Y homolog, SMCY, is also widely expressed and is conserved on the Y-chromosome in a broad range of species (Agulnik et al. 1994a,b; Kent-First et al. 1996). Based on SIM4 alignment of cDNA to genomic sequence, we determined that SMCX/Smcx was comprised of 26 exons in both human and mouse. Mouse Smcy also contains 26 exons and shows a high degree of conservation of exon–intron boundaries with human SMCY (Agulnik et al. 1999). In human, SMCX was found to span a genomic distance of ∼33 kb, whereas the mouse gene spanned 40 kb of genomic sequence (Fig. 1). The greater genomic distance covered in mouse was primarily due to the length of introns 4, 5, 10, and 14. There were short stretches of significant sequence similarity throughout the first and second introns and the promoter region that were suggestive of the presence of regulatory elements (Supplemental Fig. S1).
Human and Mouse TSPX/Tspx (DXHXS1008E, SE20–4/Se20–4)
DXHXS1008E (NM_145936) had previously been shown to be inactivated in both human and mouse (Fig. 1; Table 1; Miller and Willard 1998; Tsuchiya and Willard 2000). The mouse BC004006 transcript was found to represent a more complete version of DXHXS1008E, including additional exons. The human BC024270 ortholog (cutaneous T-cell lymphoma-associated tumor antigen, differentially expressed nucleolar transforming growth factor-β 1 target) is a member of the TSPY/SET/NAP-1 superfamily (Ozbun et al. 2001) and has been recently identified as TSPX (Delbridge et al. 2004).
Human and Mouse SMC1L1/Smc1l1
SMC1L1, also known as SB1.8 (DXS423E), belongs to a highly conserved family of proteins including the yeast SMC1 protein that is required for chromosome cohesion and segregation (Rocques et al. 1995). SMC1L1/Smc1l1 has previously been shown to escape inactivation in human, whereas it is subject to inactivation in mouse (Table 1; Brown et al. 1995; Sultana et al. 1995). We determined that both the mouse and human genes covered a genomic distance of ∼45 kb (Fig. 1). There was 92% sequence similarity between mouse and human over the entire coding sequence of the gene, which had been noted previously for a shorter stretch of coding region (Sultana et al. 1995).
We also identified a human lung cDNA (AK055575) in GenBank identical to genomic sequence just downstream from the SMC1L1 gene. There were multiple other mRNAs and ESTs that overlapped AK055575, one of which (AK091458) extended the 5′-end of AK055575, resulting in 50 bp of overlap with the 3′-end of SMC1L1 (D80000). The latter finding indicated that AK055575 was a read-through transcript of SMC1L1. We were unable to identify a mouse homolog by BLAST searches, or by RT-PCR using primers designed from mouse genomic sequence in a region of high sequence similarity (nucleotides 2690–2897 of AK055575) between human and mouse. With the rationale that the determination of the X-inactivation state of any transcribed sequence might help to elucidate mechanisms involved in escape, we tested the inactivation status of this transcript. Like SMC1L1, the presumed read-through transcript escaped inactivation in human (data not shown).
FLJ32783 Predicted Gene
Centromeric to SMC1L1 was the coding sequence for the hypothetical protein FLJ32783 (NM_144968) that was also conserved in mouse (2610028I09rik, NM_025660; Fig. 1). In both species, this gene appeared to be associated with the same CpG island as the SMC1L1 promoter. The longest open reading frame encoded a 379-amino-acid protein whose function was unknown. The mouse cDNA contained W08639 that had previously been shown to be subject to X inactivation (Tsuchiya and Willard 2000). Human ADS13, which escaped inactivation (Miller and Willard 1998), was contained within an intron of the gene coding for human FLJ32783, and was probably identified from unspliced heteronuclear RNA. We assessed the X-inactivation status of the gene itself in the somatic cell hybrid lines using primers within coding sequence, and found that the gene proper also escaped X inactivation (data not shown).
Human and Mouse HADH2/Hadh2
The gene coding for hydroxyacyl-Coenzyme A dehydrogenase, type II (short chain L-3-hydroxyacyl-CoA dehydrogenase, ADS9), has previously been shown to be subject to X inactivation in both mouse and human (Carrel et al. 1999; Tsuchiya and Willard 2000). In human, HADH2 (NM_004493) and the gene that codes for FLJ32783 differ in their X-inactivation status, despite their close proximity (Fig. 1; Table 1). HADH2 thus represents the centromeric boundary of the escape domain in human. Although we found mouse Hadh2 to lack a CpG island at its promoter, the human ortholog had this sequence feature, despite both loci being subject to X inactivation.
Sequence Analysis
Human/Mouse Sequence Comparison
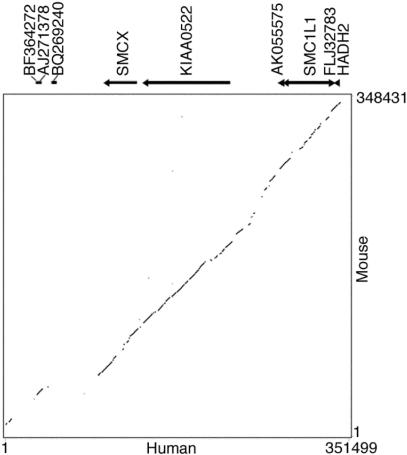
Alignment of the human and mouse genomic sequences revealed extensive conservation across the domain of human escape from HADH2 to SMCX (Fig. 4). Sequence similarity fell off telomeric to SMCX, with this region containing an active pseudogene (BQ269240) in human but not in mouse (Fig. 4). The majority of highly conserved sequences represented orthologous genes or expressed sequences between the two species (Table 1; Supplemental Fig. S1), although there was also sequence conservation within intergenic sequences in this region (Fig. 4). High sequence similarity between the two species extended into the intergenic region between HADH2/Hadh2 and the gene coding for FLJ32783 (73% identity), even though this region represented a transition characterized by a difference in X-inactivation status between human and mouse. Therefore, the difference in X inactivation between the two species at this boundary could not be explained by marked divergence in DNA sequence.
Figure 4.
Dot plot of human versus mouse genomic sequence across the human Xp11.2 escape domain, generated using Pip analysis software (http://bio.cse.psu.edu/pipmaker). Human genes, ESTs, and direction of transcription are shown above the plot.
The overall (G+C) content of the human genome (41%) is slightly lower than that of mouse (42%; Waterston et al. 2002). In contrast, the (G+C) content of the human X-chromosome is slightly higher compared with the mouse X (39.4% vs. 39.0%; Waterston et al. 2002). Our comparison of the human 350-kb region with the mouse conserved segment also revealed a higher (G+C) content in human (46.3% vs. 42.9%), although the (G+C) content in both species in this region was greater than the rest of the X-chromosome. Ke and Collins (2003) found that significantly fewer human genes escaping X inactivation contained CpG islands compared with those subject to X inactivation. We did not observe this phenomenon (Table 1). Instead, we found that the majority of genes in the human escape domain possessed a 5′ CpG island.
Repeat Distribution
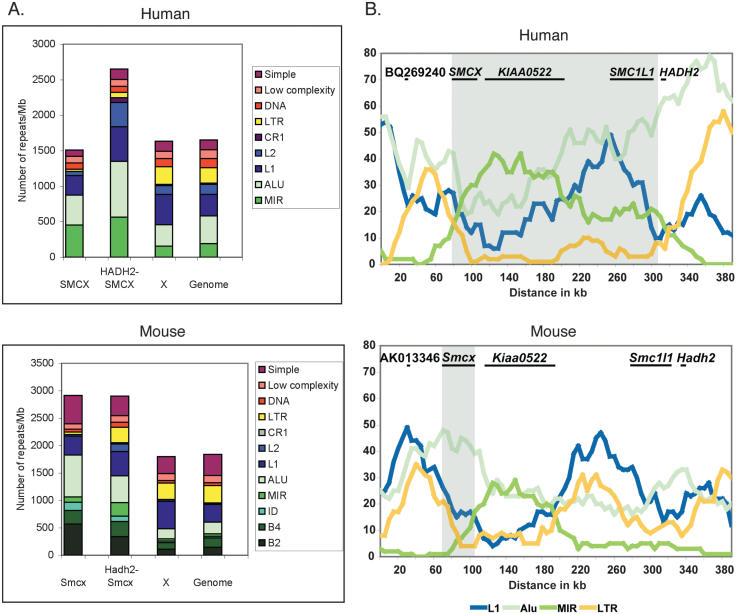
Having defined the extent of the escaping regions in human and mouse, we determined whether certain sequence features correlated with the ability of genes to escape X inactivation. This approach is based on the hypothesis that certain sequences might be required to propagate X inactivation in cis (Gartler and Riggs 1983), such as L1 LINE elements (Lyon 1998), that are unusually abundant on the mammalian X (Korenberg and Rykowski 1988; Boyle et al. 1990; Waterston et al. 2002), but may be reduced in regions harboring genes that escape inactivation (Bailey et al. 2000). We determined the numbers of individual repetitive elements (normalized per megabase) in the SMCX/Smcx gene region, the remainder of the human escape domain and the corresponding mouse region (between SMCX/Smcx and HADH2/Hadh2), the whole X-chromosome, and the entire genome (Fig. 5A; Table 2).
Figure 5.
Repetitive DNA content in the domain of escape. (A) Number of repetitive elements in the SMCX/Smcx gene region, the remainder of the human escape domain (between SMCX and HADH2, these two genes being excluded) and the equivalent mouse region, the whole X-chromosome, and the entire genome (see Table 2). Repeats are displayed as number normalized per megabase of DNA. (B) Sliding-window analysis of the distribution of repeats across the region of escape (shaded) and flanking inactivated regions.
Table 2.
Density of Repeat Elements in the Smcx/SMCX Region in Mouse and Human
|
Mouse
|
Human
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Smcxa | Smcx-Hadh2b | Xc | Genomed | SMCXa | SMCX-HADH2b | Xc | Genomed | |
| SINE | ||||||||
| B2 | 568e | 341 | 110 | 145 | ||||
| B4 | 247 | 275 | 121 | 173 | ||||
| ID | 148 | 96 | 25 | 24 | ||||
| MIR | 99 | 249 | 45 | 47 | 453 | 564 | 155 | 192 |
| ALU | 766 | 489 | 183 | 217 | 423 | 784 | 302 | 390 |
| LINE | ||||||||
| L1 | 346 | 441 | 501 | 320 | 272 | 490 | 425 | 303 |
| L2 | 25 | 136 | 26 | 24 | 60 | 343 | 126 | 140 |
| CR1 | 0 | 22 | 4 | 3 | 0 | 64 | 18 | 18 |
| LTR | 49 | 284 | 300 | 316 | 30 | 78 | 246 | 217 |
| DNA | 49 | 92 | 46 | 50 | 91 | 83 | 119 | 130 |
| Low complexity | 99 | 122 | 129 | 137 | 91 | 98 | 98 | 122 |
| Simple | 519 | 354 | 311 | 384 | 91 | 147 | 145 | 139 |
| GC (%) | 42.6 | 43 | 39 | 42 | 48.4 | 47.4 | 39.4 | 41 |
| Size (Mb) | 0.040 | 0.228 | 150 | 2500 | 0.033 | 0.204 | 154 | 3000 |
Smcx/SMCX genomic region.
Smcx-Hadh2 or SMCX-HADH2: genomic region between these two genes, excluding them.
Whole X-chromosome.
Whole genome.
Repeats in number per megabase of DNA.
We found that L1 density was reduced in the SMCX/Smcx gene region compared with the whole X-chromosome, but not throughout the remainder of the human escape domain (Fig. 5A; Table 2). We also observed an increase in all SINE elements in the SMCX/Smcx gene region compared with the whole X-chromosome and the entire genome in both human and mouse. This increase in SINE content extended through the remainder of the escape domain in human; however, there was also an increase in SINEs in the corresponding mouse region in which genes were subject to X inactivation.
A repetitive sequence feature that did appear to demonstrate an association with the difference in X-inactivation status between mouse and human was LTR content (Fig. 5A; Table 2). In human, we found that LTR density was reduced in the escape domain compared with the whole X-chromosome, whereas in the conserved mouse segment adjacent to Smcx in which all genes were inactivated, there was no decrease in LTR content. In mouse, a decrease in LTR density was only observed in the Smcx gene region that escaped. In agreement with the analysis based on total number of repeats, a sliding-window analysis using a 50-kb window and 5-kb slide also showed a decrease in LTRs throughout the human domain of escape compared with flanking regions that undergo X inactivation (Fig. 5B). In mouse there was also a relative decrease in LTRs around the 5′-end of Smcx.
DISCUSSION
We have compared 350 kb of sequence from human Xp11.2, a region containing a cluster of genes that escaped inactivation, with the mouse region of conserved synteny. In addition to previously known genes in this region we have identified six new transcripts in human, five of which were conserved in mouse. X-inactivation analyses revealed a cluster of five loci that escaped inactivation in human, whereas Smcx was the only orthologous gene that escapes in the mouse. Comparative sequence analyses showed a high degree of sequence similarity between the species, despite differences in X inactivation within the region. Analyses of the distribution of repeats revealed a lower density of LTRs in the region of escape in both human and mouse.
The cluster of genes that escape inactivation in human is uninterrupted by genes that are subject to inactivation, resulting in a 235-kb domain of escape. In contrast, the mouse Smcx gene that escapes occupies a genomic region of only 40 kb. The difference in X-inactivation status between human and mouse genes observed here may reflect evolutionary processes that have differentially shaped the human and mouse sex chromosomes, following attrition and divergence of the Y-chromosome (Jegalian and Page 1998; Disteche et al. 2002). Another factor that may contribute to the difference in X inactivation between mouse and human could be the location of the X centromere, which may act as a barrier to the spread of inactivation into the short arm of the human X-chromosome (Disteche 1999; Disteche et al. 2002).
The molecular mechanisms that control expression of genes from the inactive X still remain unknown. Gartler and Riggs proposed the existence of “way-stations” that would facilitate spreading of inactivation along the X-chromosome (Gartler and Riggs 1983). The enrichment of L1 elements on the human X compared with autosomes has led to the hypothesis that these repeats may serve as the “way-stations” (Lyon 1998). A prediction from this hypothesis is that a relative deficiency of L1 elements in certain regions of the X-chromosome may contribute to a propensity to escape inactivation. This prediction has been supported in one previous study (Bailey et al. 2000), but refuted in another (Ke and Collins 2003). We only found a decrease in L1 content in the SMCX/Smcx gene region, but not in the remainder of the human domain of escape. In their study, Bailey and colleagues analyzed multiple genomic segments containing genes that escaped from both Xp11 and Xp22 in aggregate, whereas our analysis consisted of only one of the escape regions, representing a much smaller segment of genomic sequence. In addition, the genomic sequence of the X-chromosome was not completed at the time of the study by Bailey et al. (2000), and the boundaries of domains of escape were not precisely defined. The latter may have resulted in the inclusion of genomic sequence in domains of escape that really belonged to regions subject to inactivation. It could also be that a decrease in L1 content is necessary for SMCX/Smcx to escape in both human and mouse, but that the adjacent genes are transcribed from the inactive X in human only, owing to a change in chromatin environment propagated from SMCX by a mechanism independent of L1 content. Other regions of escape will need to be fully characterized to understand the role of L1 elements in X inactivation and escape.
Our analysis also uncovered an unusual enrichment for SINE elements throughout the human domain of escape and the corresponding mouse region. Although this region displays a considerable increase in GC content that has been shown to correlate with increasing SINE content (Smit 1999), recent data indicate that SINE density is determined by factors in addition to GC content (Waterston et al. 2002). Alu SINEs have been found to be sparse on the X-chromosome (Bailey et al. 2000), and are also significantly less abundant at imprinted loci in the human genome (Greally 2002; Allen et al. 2003). Genomic imprinting is similar to X inactivation in that it is characterized by the silencing of one allele. These data suggest that SINE content could be involved in regional control of gene expression, and that the accumulation of SINEs in the Xp11.2 domain may contribute to escape from X inactivation. However, if SINEs play a role in escape, the relationship appears to be complex because the increase in SINE content also extends through the equivalent mouse region that contains genes that are subject to X inactivation. In contrast to our findings, Ke and Collins (2003) have reported a decrease in SINE MIR elements associated with genes that escape inactivation in human. However, SMCX was the only escape gene that they included in their analysis from this region; thus, this difference may reflect alternative mechanisms of escape between clusters of genes and individual genes.
A potentially interesting finding in our study is that LTR density in the region examined appears to reflect a difference in the inactivation status of genes between human and mouse. Indeed, we observed that LTR density was decreased in the human escape domain, but not throughout the conserved mouse segment in which genes were inactivated. X inactivation is associated with the formation of heterochromatin characterized by specific histone modifications and by DNA methylation at the CpG island (Plath et al. 2002). Genes that escape lack these epigenetic modifications (Disteche et al. 2002; Brown and Greally 2003). One possibility is that LTRs may facilitate the formation of silenced chromatin, hence their different distribution in the human and mouse regions. Compared with the human genome, the mouse genome is enriched in active retrotransposable elements including LTRs (Waterston et al. 2002), which could potentially contribute to the diminishing size of escape domains in mouse.
In yeast, it has recently been shown that LTRs are required for full repression of nearby meiotically induced genes through an RNA interference pathway, in which LTRs can serve as nucleation points for methylation of histone H3 on lysine 9 and the formation of silent chromatin (Schramke and Allshire 2003). Interestingly, the LTRs must be in close proximity to a gene (within 7 kb) to initiate the spread of heterochromatin. If a similar pathway were also active in mammalian cells, then it is conceivable that LTRs could serve as way-station elements for the spreading of silencing. However, because we did not observe differences between the density of LTRs on the whole X-chromosome versus the whole genome, these repeats per se would not induce silencing. Rather, the role of LTRs may depend on additional X-chromosome specific signals (e.g., XIST RNA and/or associated proteins) not present on autosomes. In that case, the paucity of LTRs we observed in regions of escape could reflect a need to maintain these domains free of repeats that might otherwise induce nucleation of heterochromatin once activated by an X-specific signal.
In addition to repetitive DNA, other elements such as insulators could contribute to the formation of domains that escape X inactivation. An insulator in this capacity might act by shielding domains from the establishment of stable X inactivation. CTCF protein is one such insulator that is highly conserved, ubiquitously expressed, and possesses versatile regulatory functions (Ohlsson et al. 2001). The finding of CTCF-binding sites in insulators at a variety of genetic loci in different species suggests that CTCF is a conserved functional component of domain boundaries (Bell and Felsenfeld 1999). We have shown here that in mouse, the gene coding for Kiaa0522 is subject to X inactivation, but is located <8 kb from the 5′-end of Smcx that escapes. Based on these findings, we have recently identified functional CTCF binding sites at the 5′-end of mouse Smcx, suggesting that this protein may also play a role in escape from X inactivation (G.N. Filippova, J.-P. Truong, J.M. Moore, M.K. Cheng, Y.J. Hu, D.K. Nguyen, K.D. Tsuchiya, and C.M. Disteche, in prep.). Species-specific differences in the distribution of CTCF binding sites could contribute to repositioning of escape domain boundaries.
It is unlikely that one single mechanism is responsible for a gene or region of the X-chromosome escaping inactivation. This process undoubtedly involves interactions between several factors including promoter characteristics of a gene, cis-acting regulatory elements, and density of specific repetitive DNA sequences. Contribution from each of these elements is likely to vary between multigene domains of escape and isolated genes that escape. The sequence characteristics responsible for escape may also differ between mouse and human. In-depth sequence comparison and functional studies of other regions of the X that contain genes that escape in both mouse and human are necessary to fully understand this highly complex and unique form of gene regulation.
METHODS
Sequence and Expression Analyses
The human genomic sequence extending from BF364272 to HADH2 was obtained from GenBank (http://www.ncbi.nlm.nih.gov/, accession no. NT_011799, version gi:29803920). The genomic sequence from the region of conserved synteny in mouse was obtained from contig NT_039719 (gi:28529857). Human and mouse genomic sequence was aligned using PipMaker (http://bio.cse.psu.edu/pipmaker/; Schwartz et al. 2000). We assigned exons by aligning cDNA sequences to genomic sequence using SIM4 (Florea et al. 1998). Open reading frames were predicted using ORF finder (http://www.ncbi.nlm.nih.gov/). Expressed nucleotide sequences were compared with protein databases with BLAST X (Altschul et al. 1997). CpG islands were determined using GrailEXP (http://grail.lsd.ornl.gov/grailexp/).
Expression of genes and ESTs was based on RT-PCR analysis of human liver, testis, and fibroblast RNA, and mouse liver, spleen, kidney, brain, heart, and testis RNA, and on information from UniGene and GeneCards (http://bioinformatics.weizmann.ac.il/cards/).
Repeat data for the human and mouse were downloaded from the UCSC Genome Bioinformatics Server (http://genome.ucsc.edu/) using the July 2003 freeze of the human genome database and the February 2003 freeze of the mouse genome database. The human coordinates used were nucleotide positions 52188244–52221395 for the SMCX gene region, and 52221386–52425000 for the rest of the escape domain (between SMCX and HADH2). The mouse coordinates were 132121944–132162468 for the Smcx gene region and 131893114–132121945 for the mouse equivalent of the remainder of the human escape domain (between Smcx and Hadh2). Repetitive elements for each region, the entire X-chromosome, and the entire genome were normalized per megabase of sequence for comparison between regions. Sliding-window analysis was also performed on the July 2003 freeze of the human genome and the February 2003 freeze of the mouse genome. A 50-kb window with a 5-kb slide was used to perform the analysis.
Mouse Kiaa0522 Sequence
The complete mouse cDNA sequence for Kiaa0522 was generated from the following mouse ESTs that aligned with the human KIAA0522 gene sequence: AW226526, AI390800, BI250642, BE860919, AI451834, BF226420, BM935728, CB236789, BU523772, CB525954, and BE647455. Remaining gaps were filled in by sequencing RT-PCR products. The complete sequence was deposited in GenBank (accession no. AY451401).
X-Inactivation Assays
The X-inactivation status of mouse genes and ESTs was assessed in F1 females resulting from matings between females carrying the Searle's translocation, T(X;16)16H (T16H), and Mus musculus castaneus (CAST) males. This strategy has been previously described in detail (Adler et al. 1991; Carrel et al. 1996). In F1 females carrying a balanced translocation, the paternal (CAST) X-chromosome is always inactivated. A gene that is subject to inactivation results in expression of only the T16H allele, whereas a gene that escapes inactivation results in expression of both the T16H and CAST alleles. Expression of alleles from the active and inactive X-chromosomes is discriminated by using polymorphisms between T16H and CAST. We identified polymorphisms by sequencing RT-PCR products from T16H and CAST parental strains (Vanderbilt Ingram Cancer Center Sequencing Core Facility). RNA was isolated using Trizol (Invitrogen) and treated with DNase I (Invitrogen, 0.5 U/μg of RNA) according to the manufacturer's protocol, followed by organic extraction and precipitation. First-strand cDNA synthesis was performed with 5 μg of RNA in a 20-μL reaction volume using oligo(dT) primers and SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Identical reactions without reverse transcriptase were performed as controls for DNA contamination. Then 1 μL of the reverse transcription reaction was amplified for 30–35 cycles in a 25-μL reaction volume.
For each gene or EST except XM_159437, tissues from at least three different F1 balanced translocation carrier females were analyzed, including newborn liver, and 3-wk-old liver, heart, or kidney. For XM_159437, brain tissue from one F1 balanced translocation carrier female was analyzed. We also analyzed chromosomally normal F1 female littermates to confirm that biallelic expression was present in females that did not carry the translocation. The X-inactivation status of AK013346 and XM_159437 was determined by sequencing RT-PCR products. PCR products generated from amplification of genomic DNA from the same mice were also sequenced to verify the presence of both alleles. The X-inactivation status of the other mouse transcripts was assessed by restriction enzyme digestion of RT-PCR products. Again, genomic DNA from the same mice was amplified and digested with the appropriate restriction enzyme to verify the presence of both alleles. The polymorphism identified in the BI155935 EST creates an endogenous restriction site. The polymorphisms in the AI390118 (KIAA0522) and BE687445 ESTs do not result in endogenous restriction sites. For these two sequences, a restriction site was created by incorporation of a mismatch near the 3′-end of one primer, and assays were performed as described previously (Carrel and Willard 1999; Tsuchiya and Willard 2000). Restriction digests were carried out in PCR reaction buffer for BE687455 and AI390118. For BI155935, PCR products were first purified using the Qiaquick PCR purification kit (QIAGEN) according to the manufacturer's protocol, and restriction digests were performed in NEBuffer 4 (New England BioLabs). All primers, polymorphisms, and restriction enzymes are listed in Supplemental Table S1.
The X-inactivation status of human genes and ESTs was determined by RT-PCR using human × Chinese hamster somatic cell hybrid cell lines containing an active or inactive human X-chromosome as described previously (Lingenfelter et al. 2001). The three somatic cell hybrids containing an inactive X-chromosome (X8.6T2H1, 8121-TGRD, and THX88) and an active X hybrid (Y162.HC) that were used in this study have been described elsewhere (Ledbetter et al. 1991; Ellison et al. 1993; Hansen et al. 1996). These hybrid cell lines have been extensively used to determine the activity of X-linked genes (Agulnik et al. 1994b; Hornstra and Yang 1994; Hansen et al. 1996; Esposito et al. 1997). We performed RT-PCR of Chinese hamster (CHO) RNA for each expressed sequence as a control for human-specific amplification. RT-PCR for RBMX, an X-linked gene that has been shown to be inactivated (Lingenfelter et al. 2001), was performed on each hybrid to verify the inactivation status of the X-chromosome (Fig. 3). Primers and reaction conditions for human sequences assayed in this study are listed in Supplemental Table S1.
Acknowledgments
This work was supported by National Institutes of Health, National Institute of Child Health and Human Development grant HD01177 (K.D.T.), and National Institute of General Medicine grant GM46883 and GM61948 (C.M.D.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2575904. Article published online before print in June 2004.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession no. AY451401. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: G. Filippova and M. Delbridge.]
References
- Adler, D.A., Bressler, S.L., Chapman, V.M., Page, D.C., and Disteche, C.M. 1991. Inactivation of the Zfx gene on the mouse X chromosome. Proc. Natl. Acad. Sci. 88: 4592–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agulnik, A.I., Mitchell, M.J., Lerner, J.L., Woods, D.R., and Bishop, C.E. 1994a. A mouse Y chromosome gene encoded by a region essential for spermatogenesis and expression of male-specific minor histocompatibility antigens. Hum. Mol. Genet. 3: 873–878. [DOI] [PubMed] [Google Scholar]
- Agulnik, A.I., Mitchell, M.J., Mattei, M.G., Borsani, G., Avner, P.A., Lerner, J.L., and Bishop, C.E. 1994b. A novel X gene with a widely transcribed Y-linked homologue escapes X-inactivation in mouse and human. Hum. Mol. Genet. 3: 879–884. [DOI] [PubMed] [Google Scholar]
- Agulnik, A.I., Longepied, G., Ty, M.T., Bishop, C.E., and Mitchell, M. 1999. Mouse H-Y encoding Smcy gene and its X chromosomal homolog Smcx. Mamm. Genome 10: 926–929. [DOI] [PubMed] [Google Scholar]
- Allen, E., Horvath, S., Tong, F., Kraft, P., Spiteri, E., Riggs, A.D., and Marahrens, Y. 2003. High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes. Proc. Natl. Acad. Sci. 100: 9940–9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Carrel, L., Chakravarti, A., and Eichler, E.E. 2000. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc. Natl. Acad. Sci. 97: 6634–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei, M.S. and Tilghman, S.M. 1997. Genomic imprinting in mammals. Annu. Rev. Genet. 31: 493–525. [DOI] [PubMed] [Google Scholar]
- Bell, A.C. and Felsenfeld, G. 1999. Stopped at the border: Boundaries and insulators. Curr. Opin. Genet. Dev. 9: 191–198. [DOI] [PubMed] [Google Scholar]
- Boyle, A., Ballard, G., and Ward, D. 1990. Differential distribution of long and short interspersed element sequences in the mouse genome: Chromosome karyotyping by fluorescence in situ hybridisation. Proc. Natl. Acad. Sci. 87: 7757–7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, C.J. and Greally, J.M. 2003. A stain upon the silence: Genes escaping X inactivation. Trends Genet. 19: 432–438. [DOI] [PubMed] [Google Scholar]
- Brown, C.J., Miller, A.P., Carrel, L., Rupert, J.L., Davies, K.E., and Willard, H.F. 1995. The DXS423E gene in Xp11.21 escapes X chromosome inactivation. Hum. Mol. Genet. 4: 251–255. [DOI] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Calhoun, V.C. and Levine, M. 2003. Coordinate regulation of an extended chromosome domain. Cell 113: 278–280. [DOI] [PubMed] [Google Scholar]
- Caron, H., van Schaik, B., van der Mee, M., Baas, F., Riggins, G., van Sluis, P., Hermus, M.C., van Asperen, R., Boon, K., Voute, P.A., et al. 2001. The human transcriptome map: Clustering of highly expressed genes in chromosomal domains. Science 291: 1289–1292. [DOI] [PubMed] [Google Scholar]
- Carrel, L. and Willard, H.F. 1999. Heterogeneous gene expression from the inactive X chromosome: An X-linked gene that escapes X inactivation in some human cell lines but is inactivated in others. Proc. Natl. Acad. Sci. 96: 7364–7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel, L., Clemson, C.M., Dunn, J.M., Miller, A.P., Hunt, P.A., Lawrence, J.B., and Willard, H.F. 1996. X inactivation analysis and DNA methylation studies of the ubiquitin activating enzyme E1 and PCTAIRE-1 genes in human and mouse. Hum. Mol. Genet. 5: 391–401. [DOI] [PubMed] [Google Scholar]
- Carrel, L., Cottle, A.A., Goglin, K.C., and Willard, H.F. 1999. A first-generation X-inactivation profile of the human X chromosome. Proc. Natl. Acad. Sci. 96: 14440–14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chureau, C., Prissette, M., Bourdet, A., Barbe, V., Cattolico, L., Jones, L., Eggen, A., Avner, P., and Duret, L. 2002. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 12: 894–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbridge, M.L., Longepied, G., Depetris, D., Mattei, M-G., Disteche, C.M., Marshall Graves, J.A., and Mitchell, M.J. 2004. TSPY, the candidate gonadoblastoma gene on the human Y chromosome, has a widely expressed homologue on the X—Implications for Y chromosome evolution. Chromosome Res. (in press). [DOI] [PubMed]
- Dillon, N. and Sabbattini, P. 2000. Functional gene expression domains: Defining the functional unit of eukaryotic gene regulation. Bioessays 22: 657–665. [DOI] [PubMed] [Google Scholar]
- Disteche, C.M. 1995. Escape from X inactivation in human and mouse. Trends Genet. 11: 17–22. [DOI] [PubMed] [Google Scholar]
- Disteche, C.M. 1999. Escapees on the X chromosome. Proc. Natl. Acad. Sci. 96: 14180–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche, C.M., Filippova, G.N., and Tsuchiya, K.D. 2002. Escape from X inactivation. Cytogenet. Genome Res. 99: 36–43. [DOI] [PubMed] [Google Scholar]
- Ellison, K.A., Roth, E.J., McCabe, E.R., Chinault, A.C., and Zoghbi, H.Y. 1993. Isolation of a yeast artificial chromosome contig spanning the X chromosomal translocation breakpoint in a patient with Rett syndrome. Am. J. Med. Genet. 47: 1124–1134. [DOI] [PubMed] [Google Scholar]
- Engemann, S., Strodicke, M., Paulsen, M., Franck, O., Reinhardt, R., Lane, N., Reik, W., and Walter, J. 2000. Sequence and functional comparison in the Beckwith-Wiedemann region: Implications for a novel imprinting centre and extended imprinting. Hum. Mol. Genet. 9: 2691–2706. [DOI] [PubMed] [Google Scholar]
- Esposito, T., Gianfrancesco, F., Ciccodicola, A., D'Esposito, M., Nagaraja, R., Mazzarella, R., D'Urso, M., and Forabosco, A. 1997. Escape from X inactivation of two new genes associated with DXS6974E and DXS7020E. Genomics 43: 183–190. [DOI] [PubMed] [Google Scholar]
- Flint, J., Tufarelli, C., Peden, J., Clark, K., Daniels, R.J., Hardison, R., Miller, W., Philipsen, S., Tan-Un, K.C., McMorrow, T., et al. 2001. Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the α globin cluster. Hum. Mol. Genet. 10: 371–382. [DOI] [PubMed] [Google Scholar]
- Florea, L., Hartzell, G., Zhang, Z., Rubin, G.M., and Miller, W. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8: 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartler, S.M. and Riggs, A.D. 1983. Mammalian X-chromosome inactivation. Annu. Rev. Genet. 17: 155–190. [DOI] [PubMed] [Google Scholar]
- Greally, J.M. 2002. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc. Natl. Acad. Sci. 99: 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, R.S., Canfield, T.K., Fjeld, A.D., and Gartler, S.M. 1996. Role of late replication timing in the silencing of X-linked genes. Hum. Mol. Genet. 5: 1345–1353. [DOI] [PubMed] [Google Scholar]
- Hornstra, I.K. and Yang, T.P. 1994. High-resolution methylation analysis of the human hypoxanthine phosphoribosyltransferase gene 5′ region on the active and inactive X chromosomes: Correlation with binding sites for transcription factors. Mol. Cell. Biol. 14: 1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegalian, K. and Page, D.C. 1998. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature 394: 776–780. [DOI] [PubMed] [Google Scholar]
- Ke, X. and Collins, A. 2003. CpG islands in human X-inactivation. Ann. Hum. Genet. 67: 242–249. [DOI] [PubMed] [Google Scholar]
- Kent-First, M.G., Maffitt, M., Muallem, A., Brisco, P., Shultz, J., Ekenberg, S., Agulnik, A.I., Agulnik, I., Shramm, D., Bavister, B., et al. 1996. Gene sequence and evolutionary conservation of human SMCY. Nat. Genet. 14: 128–129. [DOI] [PubMed] [Google Scholar]
- Korenberg, J.R. and Rykowski, M.C. 1988. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell 53: 391–400. [DOI] [PubMed] [Google Scholar]
- Ledbetter, S.A., Schwartz, C.E., Davies, K.E., and Ledbetter, D.H. 1991. New somatic cell hybrids for physical mapping in distal Xq and the fragile X region. Am. J. Med. Genet. 38: 418–420. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Urrutia, A.O., and Hurst, L.D. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31: 180–183. [DOI] [PubMed] [Google Scholar]
- Lingenfelter, P.A., Adler, D.A., Poslinski, D., Thomas, S., Elliott, R.W., Chapman, V.M., and Disteche, C.M. 1998. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat. Genet. 18: 212–213. [DOI] [PubMed] [Google Scholar]
- Lingenfelter, P.A., Delbridge, M.L., Thomas, S., Hoekstra, H.E., Mitchell, M.J., Graves, J.A., and Disteche, C.M. 2001. Expression and conservation of processed copies of the RBMX gene. Mamm. Genome 12: 538–545. [DOI] [PubMed] [Google Scholar]
- Lyon, M.F. 1998. X-chromosome inactivation: A repeat hypothesis. Cytogenet. Cell Genet. 80: 133–137. [DOI] [PubMed] [Google Scholar]
- Miller, A.P. and Willard, H.F. 1998. Chromosomal basis of X chromosome inactivation: Identification of a multigene domain in Xp11.21-p11.22 that escapes X inactivation. Proc. Natl. Acad. Sci. 95: 8709–8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeltjen, J.C., Malley, T.M., Muzny, D.M., Miller, W., Gibbs, R.A., and Belmont, J.W. 1997. Large-scale comparative sequence analysis of the human and murine Bruton's tyrosine kinase loci reveals conserved regulatory domains. Genome Res. 7: 315–329. [DOI] [PubMed] [Google Scholar]
- Ohlsson, R., Renkawitz, R., and Lobanenkov, V. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17: 520–527. [DOI] [PubMed] [Google Scholar]
- Okamura, K., Hagiwara-Takeuchi, Y., Li, T., Vu, T.H., Hirai, M., Hattori, M., Sakaki, Y., Hoffman, A.R., and Ito, T. 2000. Comparative genome analysis of the mouse imprinted gene impact and its nonimprinted human homolog IMPACT: Toward the structural basis for species-specific imprinting. Genome Res. 10: 1878–1889. [DOI] [PubMed] [Google Scholar]
- Ozbun, L.L., You, L., Kiang, S., Angdisen, J., Martinez, A., and Jakowlew, S.B. 2001. Identification of differentially expressed nucleolar TGF-β1 target (DENTT) in human lung cancer cells that is a new member of the TSPY/SET/NAP-1 superfamily. Genomics 73: 179–193. [DOI] [PubMed] [Google Scholar]
- Plath, K., Mlynarczyk-Evans, S., Nusinow, D.A., and Panning, B. 2002. Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 36: 233–278. [DOI] [PubMed] [Google Scholar]
- Rocques, P.J., Clark, J., Ball, S., Crew, J., Gill, S., Christodoulou, Z., Borts, R.H., Louis, E.J., Davies, K.E., and Cooper, C.S. 1995. The human SB1.8 gene (DXS423E) encodes a putative chromosome segregation protein conserved in lower eukaryotes and prokaryotes. Hum. Mol. Genet. 4: 243–249. [DOI] [PubMed] [Google Scholar]
- Schramke, V. and Allshire, R. 2003. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301: 1069–1074. [DOI] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. 2000. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 10: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A.F. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9: 657–663. [DOI] [PubMed] [Google Scholar]
- Smit, A.F., Toth, G., Riggs, A.D., and Jurka, J. 1995. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 246: 401–417. [DOI] [PubMed] [Google Scholar]
- Spellman, P.T. and Rubin, G.M. 2002. Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana, R., Adler, D.A., Edelhoff, S., Carrel, L., Lee, K.H., Chapman, V.C., Willard, H.F., and Disteche, C.M. 1995. The mouse Sb1.8 gene located at the distal end of the X chromosome is subject to X inactivation. Hum. Mol. Genet. 4: 257–263. [DOI] [PubMed] [Google Scholar]
- Tsuchiya, K.D. and Willard, H.F. 2000. Chromosomal domains and escape from X inactivation: Comparative X inactivation analysis in mouse and human. Mamm. Genome 11: 849–854. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Wu, J., Salido, E.C., Yen, P.H., Mohandas, T.K., Heng, H.H., Tsui, L.C., Park, J., Chapman, V.M., and Shapiro, L.J. 1994. The murine Xe169 gene escapes X-inactivation like its human homologue. Nat. Genet. 7: 491–496. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bio.cse.psu.edu/pipmaker/; PipMaker and MultiPipMaker.
- http://bioinformatics.weizmann.ac.il/cards/; GeneCards.
- http://ftp.genome.washington.edu/; RepeatMasker.
- http://genome.ucsc.edu/; USCS Genome Biotechnology.
- http://grail.lsd.ornl.gov/grailexp/; Computational Biology at ORNL.
- http://www.ncbi.nlm.nih.gov/; National Center for Biotechnology.