Abstract
Single nucleotide polymorphisms (SNPs) are the most common genetic variation in mammalian populations. Their significance is illustrated by their potential contribution to common disease but also by their potential for use in genetic association and mapping experiments. We have examined the genetic variation between commonly used inbred rat strains by using an efficient SNP discovery and typing assay based on enzyme-based (CEL I) heteroduplex cleavage. Screening of a panel of 96 different rat (sub-)strains for 100 genomic loci in 55 genes, whose human homologs are implicated in clinically relevant diseases like neurological disorder, cancer, schizophrenia, and obesity, resulted in the identification of 103 novel polymorphisms. As all strains are simultaneously genotyped in this setup, this allowed us to make an estimate of the genetic variation between and within commonly used rat inbred strains. Interestingly, we observed substantial genetic variation between colonies of the same inbred strain, maintained at different locations. Furthermore, we identified 17 non-synonymous SNPs that may have an effect on protein function and contribute to phenotypic differences between different laboratory strains.
Rat inbred strains are widely used as laboratory models in understanding basic biology and human health and disease. Currently, over 200 different rat inbred strains are used in laboratory studies, where most strains are specially selected as a model for specific human diseases (Steen et al. 1999; Kwitek-Black and Jacob 2001; http://rgd.mcw.edu). Knowledge of genetic variation between strains will be useful to obtain insight in the relationship between different strains, but also for the design of genetic mapping panels for association studies. Furthermore, as genetic variation within inbred strains does affect experimental design and interpretation, knowledge of the extent of this variation is essential. Although a strain is assumed to be inbred after at least 20 generations of subsequent brother–sister matings (Kacew and Festing 1996) and from that point on is considered to be genetically homogeneous, a small but uncharacterized degree of genetic variation will always remain in the population.
Currently, information on genetic variation in rat laboratory strains is limited to a set of microsatellite markers (Canzian 1997; Thomas et al. 2003) that are generally located in noncoding regions. In this study, we focused on single nucleotide polymorphisms (SNPs) in gene coding regions of rat inbred strains. SNPs are the most abundant form of sequence variability in the vertebrate genome (Wang et al. 1998) and polymorphisms in gene coding are likely to contribute to the phenotypic differences between strains. Obviously, SNPs in noncoding regions may also have a functional effect as suggested by evolutionary conservation of parts of such sequences (Dubchak et al. 2000; Loots et al. 2000; Hare and Palumbi 2003) and the mapping of a human disease susceptibility to a noncoding region (Ueda et al. 2003). However, the vast majority of human disease alleles that are identified up to now have been found to be caused by single nucleotide polymorphisms resulting in nonsense or missense mutations at the gene coding level (OMIM, Online Mendelian Inheritance in Man database; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM).
Different experimental methods to detect genetic variation have been established and are reviewed by Kristensen et al. (2001). For large-scale genome-wide detection of SNPs, DNA microarray technology is one of the most efficient methods and it has been successfully applied to the human (Wang et al. 1998; Cargill et al. 1999; Halushka et al. 1999) as well as to the mouse genome (Lindblad-Toh et al. 2000). To discover novel SNPs, two different approaches can be followed. First, new polymorphic positions can be predicted by an in silico approach (reviewed by Vignal et al. 2002), where all available information in databases is mined and analyzed for the occurrence of polymorphisms. This results in candidate SNPs that require experimental verification. Second, experimental methods can be used directly, where direct sequencing is currently still the most accurate and informative approach, but other methods that are more cost-effective and/or allow higher throughput are continuously under development.
In this study, we have adapted high-throughput mutation discovery technology that is based on enzymatic cleavage of heteroduplexes using the CEL I nuclease isolated (Oleykowski et al. 1998) for SNP discovery. This technology has been successfully used for reverse genetics approaches and the identification of knockouts in Arabidopsis (Till et al. 2003), zebrafish (Wienholds et al. 2003), and the rat (Smits et al. 2004), and is now commonly referred to as TILLING. The major advantage of the approach described here, is that it is well suited for simultaneous SNP discovery and genotyping using a large panel of individuals or samples. In addition, the accuracy of the method is high, the costs are relatively low, and the technology is easily scalable. We have assayed 100 coding regions in 96 rat inbred strains and substrains, scanning almost 6 × 106 base pairs, resulting in the identification and genotyping of 103 novel polymorphisms. Interestingly, we observed a considerable amount of genetic variation at the single-base level within inbred strains.
RESULTS AND DISCUSSION
SNP Discovery
We have selected a set of 55 genes that are known to play a role in neurological and endocrine processes, cholesterol metabolism, and cancer (Supplemental Table 1). Sequence variation in the human homologs of these genes may play a role in clinically relevant diseases like schizophrenia, obesity, and cancer. Primers for amplification of selected exons (total 100 loci) were designed using local genomic assemblies generated by the GENOTRACE program (Berezikov et al. 2002), as no rat genome assembly was available at this point, in combination with a Primer3-based (Rozen and Skaletsky 2000) primer design program (http://primers.niob.knaw.nl) for universal design of oligo's, allowing automated simultaneous PCR in a robotic set-up.
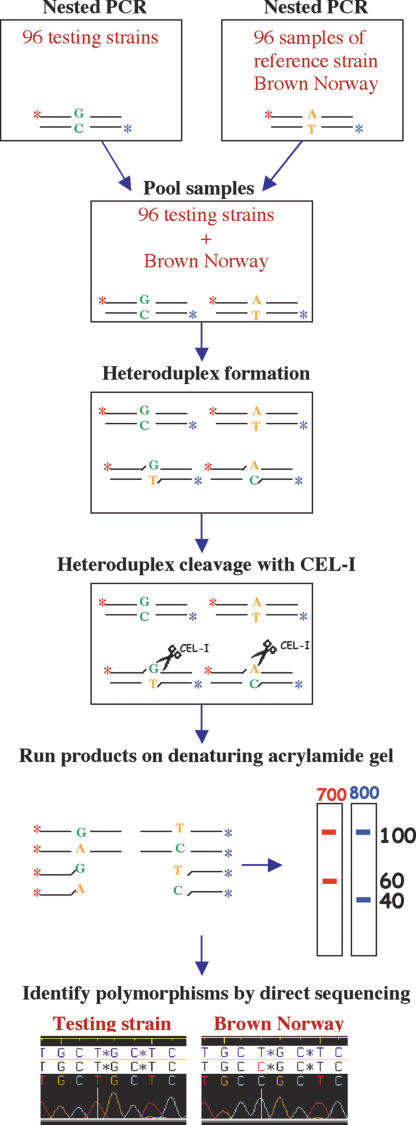
The resulting amplicons were screened for polymorphisms in a set of 96 inbred rat strains and substrains using a modified CEL I nuclease-based (Oleykowski et al., 1998) SNP detection assay (Fig. 1). Briefly, fluorescently labeled PCR products from the reference strain Brown Norway, which is being sequenced as part of the rat genome sequencing project (http://www.hgsc.bcm.tmc.edu/projects/rat/), and testing strain are mixed and allowed to form heteroduplexes that are the substrate for the CEL I nuclease. Digested products are analyzed on denaturing acrylamide gels and polymorphisms will show up as additional bands (for an example gel image see Supplemental Figure 1). Samples showing additional fragments are subsequently sequenced to reveal the molecular nature of the polymorphism. Although one has to sequence only one representative sample for every SNP, in this study we verified all polymorphisms in all amplicons by direct sequencing of the PCR product. No false positives were encountered and no novel polymorphisms were detected in the sequencing phase, indicating that the rate of false negatives is also very low.
Figure 1.
Schematic overview of the polymorphism discovery strategy. PCR on genomic DNA of 96 testing strains and on a set of 96 samples of the reference strain Brown Norway (BN/Cl) produces two sets of 96 PCR products that contain different fluorescent labels at either end (IRD700 and IRD800, indicated by red and blue stars). Pooling of tester samples with reference samples, followed by denaturing and reannealing, results in the formation of heteroduplex DNA in the case of polymorphisms between the strains used. Heteroduplex DNA is specifically cleaved by the CEL I nuclease at the site of the mismatch and the resulting fragments are separated on a denaturing polyacrylamide gel with fluorescence detection. Subsequently, representative samples for a specific polymorphism are sequenced to reveal the molecular nature of the polymorphism.
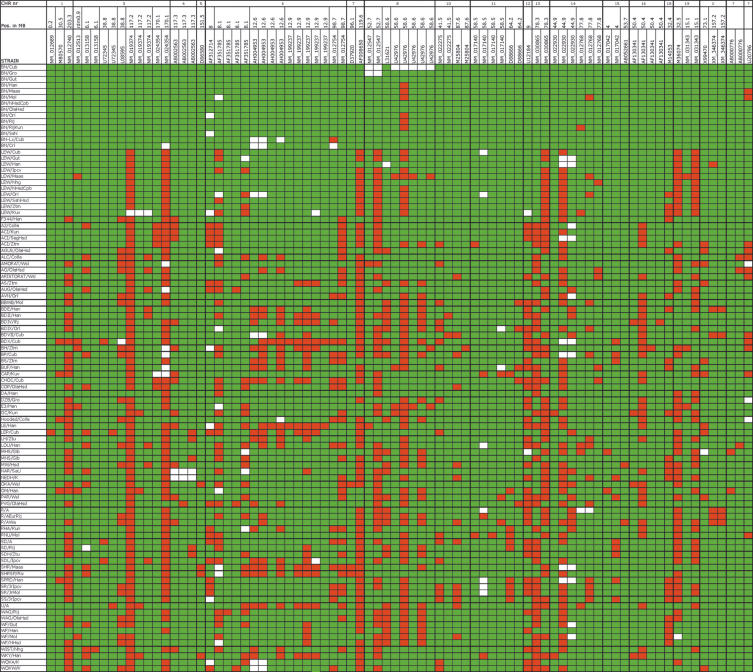
In total, we screened more than 5.8 × 106 base pairs, with 40,977 base pairs of coding and 19,198 base pairs of noncoding sequence per (sub)strain and identified 103 novel polymorphisms (Supplemental Table 1), that were simultaneously genotyped in 96 rat strains and substrains (Fig. 2). SNPs in a single gene that were found to cosegregate in our data set are represented as a single polymorphic locus in Figure 2.
Figure 2.
Genotype of the identified polymorphisms for 96 rat inbred strains and substrains. Polymorphisms are typed in reference to the Brown Norway strain (BN/Crl). The green squares represent the allele in accordance with BN/Crl. The red squares represent the presence of the alternative allele. The white squares represent genotyping failures. Different isolates of the same inbred strain are grouped together, separated by thin lines. Thick lines separate different inbred strains. SNPs that are present in the same gene and cosegregate in our data set are represented as a single locus, reducing the number of polymorphic sites to 80. The polymorphic loci are ordered on the horizontal axis in line with the map position based on the June 2003 rat genome assembly (UCSC version rn3). The map position of the last two genes is unknown.
The average failure rate per amplicon was found to be only 1.3%, and is most likely due to pipetting errors and inaccuracies introduced by the automated robotic setup of the PCR and CEL I reactions. No inconsistencies were observed between the CEL I-based and the resequencing results. Taken together, this strategy is an efficient and reliable approach for high throughput SNP discovery and simultaneous genotyping in medium to large sample sets.
SNP Characteristics and Frequency
Although the focus of this study is on polymorphisms in coding regions, we did identify 38 SNPs in noncoding regions as a result of the PCR-based strategy with primers designed in exon-flanking sequences. The calculated SNP frequency in exon, intron and untranslated (UTR) regions is one in 630, 555, and 345 base pairs, respectively (Supplemental Table 1). Generally, SNP frequencies are found to be higher in noncoding sequences compared to coding sequences. The difference in our set might be due to the method of screening, for which it has been reported that it does not efficiently detect polymorphisms close to the ends of the amplicon (Till et al. 2003). Rather crude correction of this effect by subtraction of 30 base pairs from either end of the amplicon, would result in SNP frequencies in exon-flanking intronic regions of one in 367 nucleotides (29 SNPs in 10,638 bps) and one in 269 nucleotides for UTR sequences (nine SNPs in 2419 bps), which are more in line with expectations.
Completely random mutations will be divided into transitions and transversions in a 1:2 ratio. However, biological data sets tend to have a strong bias toward transitions due to DNA methylation, chemical differences between bases, and differences in DNA repair efficiency for different types of nucleotide mismatches. As for the mouse (roughly 66.7% transitions; Lindblad-Toh et al. 2000) and for human (64% transitions; Halushka et al. 1999), our data set shows a similar bias towards transitions (78.4%). It is not likely that this bias is a result of a decreased sensitivity of CEL I for certain mismatches, since results obtained by others (Oleykowski et al. 1998) and by us (Wienholds et al. 2003; E. Cuppen, unpubl.) show that all possible heteroduplex mismatches are recognized efficiently by CEL I.
SNPs Between Inbred Strains
Previous analysis on microsatellites suggested that Brown Norway (BN) was genetically most similar to wild rats and furthest away from laboratory rat strains (Canzian 1997; Thomas et al. 2003). Although the current genotyping data is not sufficient to build a reliable phylogenetic tree, our results confirm that Brown Norway is clearly most distant from the other inbred strains tested. Furthermore, some clear relationships between strains can be observed. For example, DA/Han and LEW/Cub have 96% (77 of 80) polymorphic loci identical (Fig. 2), which indicates that these strains are closely related. In rat strain databases, the DA strain is suggested to be closely related to the COP strain (http://rgd.mcw.edu/strains/; http://www.mh-hannover.de/institute/tierlabor/da.html), but in our set DA and COP share only 85% of the alleles. Of course, many more polymorphic loci should be explored to reliably trace the history of rat inbred strains, as illustrated by the results of Canzian (1997) and Thomas et al. (2003), showing close relationship between DA and COP using 995 and 4800 microsatellite markers, respectively.
SNPs Within Inbred Strains
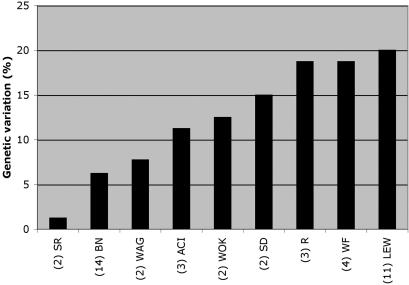
Individual animals from inbred strains are commonly considered to be genetically homogeneous and are called isogenic. However, our genotyping results using independent isolates of inbred strains, revealed significant heterogeneity (Figs. 2, 3). Statistically, at least 98.6% of the loci in each animal from an inbred population should be homozygous (Davisson 1997; Beck et al. 2000). We have genotyped 14 Brown Norway substrains and found that five of the 80 loci were variable, representing 6% of genetic variability on single-base level. In three substrains of ACI, we found variation in nine of the 80 loci, representing 11% of genetic variation. Remarkably, in the widely used Lewis (LEW) strain, 16 out of the 80 loci were found to be polymorphic, representing genetic variation of 20%. In Figure 3, variation percentages were plotted for nine inbred strains, for which multiple individuals were typed.
Figure 3.
Genetic variation between substrains of nine frequently used rat inbred strains. Percentages were calculated as percentage of variation of the total amount of loci for which the substrains were typed. The number below the name of the strain represents the number of substrains that were typed.
The observed genetic variation within inbred strains may be the result of spontaneous mutations that occurred during breeding, inaccuracies in breeding programs, or a reflection of residual heterogeneity at the time of splitting breeding populations. The first possibility would result in novel SNPs, unique to the (sub)strain, whereas the latter two possibilities would result in nonunique SNPs shared with other strains. We only identified SNPs shared with other inbred strains, excluding spontaneous mutations as a source.
It should be noted that from these results no conclusions can be drawn about the degree of isogenicity within specific breeding colonies of inbred strains, but it illustrates the potential variation between different isolates worldwide that should be taken into account when comparing experimental results from different labs using the same inbred lines.
Effect of SNPs
For the 65 SNPs in coding regions, we find that about 26% of the polymorphisms (17) result in the replacement of an amino acid in the encoded protein product (nonsynonymous; Supplemental Table 1). A recent study showed that functional polymorphisms (nonsynonymous) are overrepresented in a set of human SNPs that have a minor allele frequency lower than 6% (Wong et al. 2003). Because a selection of inbred strains and substrains, as used in this study, does not resemble a random (outbred) population and is limited in the number of individual samples, we used another approach to investigate if there is correlation between SNPs being nonsynonymous and their minor allele frequency. We defined three bins with minor allele frequencies between 15% and 50%, between 5% and 15%, and below 5%, similar as used by Cargill et al. (1999). In line with human functional SNP data, nonsynonymous SNPs appear more frequently in the latter class (47.1%) when compared to synonymous SNPs (27.1%) and noncoding SNPs (37.1%; see Supplemental Table 2 for details).
For estimating the direct effect of SNPs, we checked the impact of all nonsynonymous SNPs on the subsequent protein product by SIFT (Ng and Henikoff 2001) and PolyPhen (Ramensky et al. 2002). These programs use phylogenetic conservation of protein (domains), chemical properties of the polymorphic amino acids, and predictions on secondary protein structure to estimate the potential effect of a polymorphism on protein function. Although results produced by these prediction programs should be used with care, SIFT correctly distinguishes between deleterious and neutral alleles for 18 out of 22 SNPs in five known human disease genes (Ng and Henikoff 2002). One of the 17 nonsynonymous SNPs in our study (C3177G of XM_237485 in NCBI database, changing His to Gln) was predicted to be damaging by both methods and four others were found to be probably/possibly damaging by PolyPhen (CT1157/1158AC of XM_219840, C1277A of NM_017140, A671G of M36074, G646A of AF130341), but were called tolerant by SIFT, of which two were diagnosed at low confidence by SIFT. The other 11 SNPs were not predicted to be dramatically deleterious for the protein. The five polymorphisms that are predicted to potentially affect protein function were found in the FSH receptor (FSHR), G protein-coupled receptor 50 (Gpr50), dopamine receptor 3 (Drd3), mineralocorticoid receptor (Mr), and melatonine receptor (Mt1), respectively. Interestingly, four of these SNPs have minor allele frequencies lower than 6%, but the SNP in Mr is abundantly present with a minor allele frequency of 43.5%. Since these SNPs may impair the function of the protein, the strains harboring them might have aberrant characteristics in processes in which the corresponding genes are involved. Although it is currently difficult to prove that these polymorphisms account for phenotypic differences between the strains, it is clear that (combinations of) genetic polymorphisms in general do influence experimental results. For example, in the human population, the interindividual variability in drug response is a major problem for effective drug treatment and it is believed that this is caused by heritability of certain uncharacterized functional polymorphisms (Nebert 1999). Pharmacogenomic approaches in model organisms are expected to contribute considerably to the understanding of the function of specific genes in relation to drug response (Watters and McLeod 2002). Characterization of the naturally occurring genetic variation at such loci, for example in the many rat inbred strains that are now known, could contribute equally to such understanding.
METHODS
Genomic DNA Isolation and Origin of Genomic DNA
Genomic DNA from reference strain BN/Crl was isolated using QIAGEN DNeasy 96 Tissue Kit (4). Genomic DNA from 96 inbred strains and substrains was made available through the Department of Laboratory Animal Science, Faculty of Veterinary Medicine, Utrecht University, The Netherlands. More detailed information on the origin of these strains can be found in Supplemental Table 3. Five microliters of genomic DNA (concentration of ∼2 ng/μl per sample), was gridded-out in 384-well plates. In total, two sets of 96 testing strains and two sets of 96 reference strain DNA (BN/Crl), were transferred into a single 384-well plate using a 96-channel pipettor (Hydra-96).
Primer Design
Genomic organization and exon-flanking intron sequences of Rattus norvegicus target genes were determined using GENOTRACE. The published cDNA sequence was used as input. Nested sets of oligonucleotides for amplification of exon sequences were automatically designed using a PRIMER3-based Web application (http://primers.niob.knaw.nl) with optimal melting temperatures of 58°C. For this study we have designed amplicons for only the large exons (>300 bp) in genes of our specific interests. As a result, coding regions for some of the genes are fully covered, whereas others are only partially screened, resulting in an average coverage percentage of about 56%. Detailed primer information can be obtained from the authors upon request.
CEL I-Based Polymorphism Detection
All PCR, pooling, and CEL I digestion pipetting steps were done on a Genesis Workstation 200 (Tecan). Target sequences were amplified by nested PCR in 384-well plates. The first PCR was done with gene-specific primers and carried out using a touch-down cycling program (92°C for 60 sec; 30 cycles of 92°C for 20 sec, 65°C for 20 sec with a decrement of 0.5°C per cycle, 72°C for 60 sec; followed by 10 cycles of 92°C for 20 sec, 58°C for 20 sec, and 72°C for 60 sec; 72°C for 180 sec; GeneAmp9700, Applied Biosystems). PCR reaction mixes contained 5 μl genomic DNA, 0.2 μM forward primer, and 0.2 μM reverse primer, 200 μM of each dNTP, 25 mM Tricine, 7.0% Glycerol (w/v), 1.6% DMSO (w/v), 2 mM MgCl2, 85 mM Ammonium acetate (pH 8.7), and 0.2 U Taq Polymerase in a total volume of 10 μl.
Of the first PCR reaction, 1 μl was used as template for the second PCR reaction. The second PCR reaction contained gene specific primers, at their 5′ end elongated with universal M13 adaptor sequences (forward 0.08 μM and reverse 0.04 μM). Additionally, the reaction mixture contained corresponding universal M13-forward primer (5′-TGTAAAACGACGGCCAGT, 0.12 μM) and universal M13-reverse primer (5′-AGGAAACAGCTAT GACCAT, 0.16 μM). The universal M13 primers were labeled with fluorescent dyes, IR Dye 700 and IR Dye 800, respectively. Furthermore, the nested PCR mixture contained 200 μM of each dNTP, 25 mM Tricine, 7.0% Glycerol (w/v), 1.6% DMSO (w/v), 2 mM MgCl2, 85 mM Ammonium acetate (pH 8.7), and 0.2U Taq Polymerase in a total volume of 5 μl. Standard cycling conditions were used for the nested PCR reactions (30 cycles of 92°C for 20 sec, 58°C for 40 sec, and 72°C for 60 sec).
After the second PCR reaction, 2.5 μl of each sample of the 96 testing strains was mixed in a fresh 384-well plate with 2.5 μl of the reference strain PCR, followed by heteroduplex formation (99°C for 10 min and 70 cycles of 70°C for 20 sec with a decrement of 0.3°C per cycle). Specific heteroduplex cleavage was performed by adding to each sample 10 μl of CEL I mixture, containing 10 mM HEPES (pH 7.0), 10 mM MgSO4, 10 mM KCl, 0.002% Triton X-100, 0.2 μg BSA and 0.01 μl CEL I enzyme solution (isolated from celery according to Oleykowski et al. (1998) with minor modifications; protocol available at http://www.niob.knaw.nl/researchpages/cuppen) and incubation at 45°C for 15 min. The CEL I reactions were stopped by addition of 5 μl 75 mM EDTA (pH 8.0).
Fragments were purified using Sephadex G50 (medium coarse, Sigma) mini-columns in 96-well filter plates (Multiscreen HV, Millipore) and eluted into plates prefilled with 5 μl formamide loading buffer (37% [v/v] de-ionized formamide, 4 mM EDTA, pH 8.0, 90 μg/ml bromophenol blue) per well. Samples were concentrated to about 1.5 μl by heating at 85°C for 45 to 60 min without cover. Three-tenths of a microliter (0.3 μl) was applied to a 96-lane membrane comb (The Gel-company) and loaded on 25-cm denaturing 6% polyacrylamide gels on LI-COR 4200 DNA analyzers. Raw TIFF-images produced by the analyzers were modified and visualized using Adobe Photoshop and potential polymorphisms were detected and scored manually.
Verification and Identification of Polymorphisms by Sequencing
PCR reactions for polymorphism identification were done by using the same conditions as for CEL I mediated polymorphism detection. In the nested PCR, universal M13 oligo's were omitted and only gene specific primers were used, both in the concentration of 0.2 μM. Nested PCR products were diluted with 30 μl water and 1 μl was used as template for the sequencing reactions. Sequencing reactions, containing 0.5 μl DYEnamic ET Terminator (Amersham Pharmacia Biotech), 3.5 μl ET Terminator dilution buffer (Amersham Pharmacia Biotech) and 0.5 μM nested gene specific forward primer or reverse primer in a total volume of 10 μl, were performed using cycling conditions recommended by the manufacturer. Sequencing products were purified using Sephadex G50 (superfine, coarse, Sigma) mini-columns and analyzed on a 96-capillary 3700 DNA analyzer (Applied Biosystems). Sequences were analyzed for polymorphisms using polyphred (Nickerson et al. 1997).
SIFT and PolyPhen
To predict the impact of the nonsynonymous SNPs we found, we used two different applications, SIFT (v2.0; Ng and Henikoff 2001) and PolyPhen (command-line version; Ramensky et al. 2002), both stand-alone versions. To be able to compare the data, the same databases (SWISS-PROT+TrEMBL+TrEMBL_NEW, downloaded from ftp://ftp.expasy.org) and BLAST parameters (expectation cut-off = 1E-04) were applied for the analysis.
Acknowledgments
This work was financially supported by the Dutch Ministry of Economic Affairs through the Innovation Oriented Research Program on Genomics (IOP Genomics). We thank Steve Henikoff for help setting up the TILLING technology, R. Malik and V. Guryev for bioinformatic support, and J. Mudde for technical support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2155004.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to dbSNP under accession nos. ss12588106–ss12588203.]
References
- Beck, J.A., Lloyd, S., Hafezparast, M., Lennon-Pierce, M., Eppig, J.T., Festing, M.F., and Fisher, E.M. 2000. Genealogies of mouse inbred strains. Nat. Genet. 24: 23–25. [DOI] [PubMed] [Google Scholar]
- Berezikov, E., Plasterk, R.H., and Cuppen, E. 2002. GENOTRACE: cDNA-based local GENOme assembly from TRACE archives. Bioinformatics 18: 1396–1397. [DOI] [PubMed] [Google Scholar]
- Canzian, F. 1997. Phylogenetics of the laboratory rat Rattus norvegicus. Genome Res. 7: 262–267. [DOI] [PubMed] [Google Scholar]
- Cargill, M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., Shaw, N., Lane, C.R., Lim, E.P., Kalyanaraman, N., et al. 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22: 231–238. [DOI] [PubMed] [Google Scholar]
- Davisson, M.T. 1997. Rules and guidelines for genetic nomenclature in mice: excerpted version. Committee on Standardized Genetic Nomenclature for Mice. Transgenic Res. 6: 309–319. [DOI] [PubMed] [Google Scholar]
- Dubchak, I., Brudno, M., Loots, G.G., Pachter, L., Mayor, C., Rubin, E.M., and Frazer, K.A. 2000. Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 10: 1304–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halushka, M.K., Fan, J.B., Bentley, K., Hsie, L., Shen, N., Weder, A., Cooper, R., Lipshutz, R., and Chakravarti, A. 1999. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet. 22: 239–247. [DOI] [PubMed] [Google Scholar]
- Hare, M.P. and Palumbi, S.R. 2003. High intron sequence conservation across three Mammalian orders suggests functional constraints. Mol. Biol. Evol. 20: 969–978. [DOI] [PubMed] [Google Scholar]
- Kacew, S. and Festing, M.F. 1996. Role of rat strain in the differential sensitivity to pharmaceutical agents and naturally occurring substances. J. Toxicol. Environ. Health 47: 1–30. [DOI] [PubMed] [Google Scholar]
- Kristensen, V.N., Kelefiotis, D., Kristensen, T., and Borresen-Dale, A.L. 2001. High-throughput methods for detection of genetic variation. Biotechniques 30: 318–322, 324, 326 passim. [DOI] [PubMed] [Google Scholar]
- Kwitek-Black, A.E. and Jacob, H.J. 2001. The use of designer rats in the genetic dissection of hypertension. Curr. Hypertens. Rep. 3: 12–18. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh, K., Winchester, E., Daly, M.J., Wang, D.G., Hirschhorn, J.N., Laviolette, J.P., Ardlie, K., Reich, D.E., Robinson, E., Sklar, P., et al. 2000. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat. Genet. 24: 381–386. [DOI] [PubMed] [Google Scholar]
- Loots, G.G., Locksley, R.M., Blankespoor, C.M., Wang, Z.E., Miller, W., Rubin, E.M., and Frazer, K.A. 2000. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288: 136–140. [DOI] [PubMed] [Google Scholar]
- Nebert, D.W. 1999. Pharmacogenetics and pharmacogenomics: Why is this relevant to the clinical geneticist? Clin. Genet. 56: 247–258. [DOI] [PubMed] [Google Scholar]
- Ng, P.C. and Henikoff, S. 2001. Predicting deleterious amino acid substitutions. Genome Res. 11: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, P.C. and Henikoff, S. 2002. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 12: 436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, D.A., Tobe, V.O., and Taylor, S.L. 1997. PolyPhred: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 25: 2745–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleykowski, C.A., Bronson Mullins, C.R., Godwin, A.K., and Yeung, A.T. 1998. Mutation detection using a novel plant endonuclease. Nucleic Acids Res. 26: 4597–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky, V., Bork, P., and Sunyaev, S. 2002. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30: 3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Smits, B.M., Mudde, J., Plasterk, R.H., and Cuppen, E. 2004. Target-selected mutagenesis of the rat. Genomics 83: 332–334. [DOI] [PubMed] [Google Scholar]
- Steen, R.G., Kwitek-Black, A.E., Glenn, C., Gullings-Handley, J., Van Etten, W., Atkinson, O.S., Appel, D., Twigger, S., Muir, M., Mull, T., et al. 1999. A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res. 9: AP1–8, insert. [PubMed] [Google Scholar]
- Thomas, M.A., Chen, C.F., Jensen-Seaman, M.I., Tonellato, P.J., and Twigger, S.N. 2003. Phylogenetics of rat inbred strains. Mamm. Genome 14: 61–64. [DOI] [PubMed] [Google Scholar]
- Till, B.J., Reynolds, S.H., Greene, E.A., Codomo, C.A., Enns, L.C., Johnson, J.E., Burtner, C., Odden, A.R., Young, K., Taylor, N.E., et al. 2003. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 13: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, H., Howson, J.M., Esposito, L., Heward, J., Snook, H., Chamberlain, G., Rainbow, D.B., Hunter, K.M., Smith, A.N., Di Genova, G., et al. 2003. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 423: 506–511. [DOI] [PubMed] [Google Scholar]
- Vignal, A., Milan, D., SanCristobal, M., and Eggen, A. 2002. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 34: 275–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D.G., Fan, J.B., Siao, C.J., Berno, A., Young, P., Sapolsky, R., Ghandour, G., Perkins, N., Winchester, E., Spencer, J., et al. 1998. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280: 1077–1082. [DOI] [PubMed] [Google Scholar]
- Watters, J.W. and McLeod, H.L. 2002. Murine pharmacogenomics: Using the mouse to understand the genetics of drug therapy. Pharmacogenomics 3: 781–790. [DOI] [PubMed] [Google Scholar]
- Wienholds, E., van Eeden, F., Kosters, M., Mudde, J., Plasterk, R.H., and Cuppen, E. 2003. Efficient target-selected mutagenesis in zebrafish. Genome Res. 13: 2700–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, G.K., Yang, Z., Passey, D.A., Kibukawa, M., Paddock, M., Liu, C.R., Bolund, L., and Yu, J. 2003. A population threshold for functional polymorphisms. Genome Res. 13: 1873–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.hgsc.bcm.tmc.edu/projects/rat/; Rat whole-genome sequencing project, Baylor College of Medicine.
- http://rgd.mcw.edu/strains/; Rat Genome Database, information on rat inbred strains.
- http://www.mh-hannover.de/institute/tierlabor/; Institut fur Versuchstierkunde und Zentrales Tierlabor Hannover, additional information on rat inbred strains.
- http://www.niob.knaw.nl/researchpages/cuppen/; Detailed protocols for CEL I-based mutation/SNP detection.
- http://primers.niob.knaw.nl; Web-based primer-design application for the design of sets of nested (tailed) oligo's.
- http://www.ncbi.nlm.nih.gov/SNP/; dbSNP of NCBI.
- http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?dg=OMIM; Online Mendelian Inheritance in Man database.
- http://rgd.mcw.edu; Rat genome database; rat genetic and genomic information.