Abstract
The degradation of mRNA is an important regulatory step in the control of gene expression. However, mammalian RNA decay pathways remain poorly characterized. To provide a framework for studying mammalian RNA decay, a two-hybrid protein interaction map was generated using 54 constructs from 38 human proteins predicted to function in mRNA decay. The results provide evidence for interactions between many different proteins required for mRNA decay. Of particular interest are interactions between the poly(A) ribonuclease and the exosome and between the Lsm complex, decapping factors, and 5′→3′ exonucleases. Moreover, multiple interactions connect 5′→3′ and 3′→5′ decay proteins to each other and to nonsense-mediated decay factors, providing the opportunity for coordination between decay pathways. The interaction network also predicts the internal organization of the exosome and Lsm complexes. Additional interactions connect mRNA decay factors to many novel proteins and to proteins required for other steps in gene expression. These results provide an experimental insight into the organization of proteins required for mRNA decay and their coupling to other cellular processes, and the physiological relevance of many of these interactions are supported by their evolutionary conservation. The interactions also provide a wealth of hypotheses to guide future research on mRNA degradation and demonstrate the power of exhaustive protein interaction mapping in aiding understanding of uncharacterized protein complexes and pathways.
The level of each mRNA in a cell is a tightly regulated function of its rate of transcription, processing, and degradation. Global analysis has shown that the stabilities of different mRNAs vary widely from <10 min to many hours (Khodursky and Bernstein 2003). In addition, the half-lives of certain mRNAs are known to change dramatically throughout the cell cycle or in response to cell signaling, suggesting that mRNA degradation is a regulated process (Wilusz et al. 2001). To understand the control of gene expression, it is thus necessary to understand the regulation and mechanisms of mRNA degradation.
In Saccharomyces cerevisiae, mRNA degradation is normally initiated by removal of the poly(A) tail by a deadenylase complex (for review, see Wilusz et al. 2001). The major decay pathway involves removal of the 5′-cap by the Dcp1/Dcp2 enzyme and then degradation by the Xrn1p 5′→3′ exonuclease. Decapping also requires the Lsm1–7p complex, and all of these proteins localize to cytoplasmic P-bodies, which are believed to be the active sites of 5′→3′ decay (Sheth and Parker 2003). A minor decay pathway involves 3′→5′ degradation by the exosome, a complex of at least 10 proteins, including seven 3′→5′ exonucleases.
In mammalian cells, cytoplasmic mRNA decay is also normally initiated by deadenylation (Wilusz et al. 2001). A major human deadenylase activity was identified as PARN/DAN (Dehlin et al. 2000). However, in contrast to yeast, the predominant decay pathway appears to proceed in a 3′→5′ direction (Chen et al. 2001; Wang and Kiledjian 2001; Mukherjee et al. 2002). Several mammalian homologs of yeast decay proteins have been identified including human Dcp2 (Van Dijk et al. 2002; Wang et al. 2002), two homologs of Dcp1p (Dcp1A and Dcp1B; Lykke-Andersen 2002), and Xrn1 and Xrn2 (homologous to the yeast nuclear Rat1p 5′→3′ exonuclease). A second human decapping activity, DcpS, can only hydrolyze a free cap structure, and may therefore act on the end product of complete 3′→5′ degradation (Liu et al. 2002). In addition, human Lsm proteins (Achsel et al. 1999; Salgado-Garrido et al. 1999) and a human exosome complex have been identified (Allmang et al. 1999; Chen et al. 2001). Depletion of the human exosome from cell extracts increases the half-life of unstable mRNAs (Chen et al. 2001), and the exosome, Dcp2, and PARN are required in vivo for the degradation of at least some mRNAs (Lejeune et al. 2003). Human Dcp1, Xrn1, Lsm1–7 (Ingelfinger et al. 2002), and Dcp2 (Van Dijk et al. 2002) colocalize in cytoplasmic foci that may be equivalent to yeast P-bodies.
In most mRNAs, neither the cis-regulatory elements nor the trans-factors responsible for regulating stability are known. However, one well-studied instability element is the AU-rich element (ARE) present in the 3′-untranslated region (UTR) of a subset of unstable mammalian mRNAs. Many ARE-binding proteins (ARE-BPs) have been identified that act to destabilize (e.g., AUF1, TTP) or stabilize (e.g., HuR) an mRNA (Wilusz et al. 2001). Eukaryotic mRNAs containing a premature termination codon are rapidly destroyed in a process termed nonsense-mediated decay (NMD; for review, see Wagner and Lykke-Andersen 2002). In yeast, NMD requires the Upf1p, Upf2p, and Upf3p proteins and induces deadenylation-independent decapping and 5′→3′ degradation and, to a lesser extent, exosome-dependent 3′→5′ degradation (Mitchell and Tollervey 2003). Humans possess homologs of the Upf1p and Upf2p proteins and two Upf3p proteins: Upf3A and Upf3B (Serin et al. 2001). Tethering of any of these proteins downstream of a termination codon targets an mRNA for rapid decay (Lykke-Andersen et al. 2000), but how the Upf proteins recruit degradation enzymes is unclear.
To provide a framework for studying the mechanisms and regulation of human mRNA decay, we have used exhaustive yeast two-hybrid screening to identify interactions among predicted pathway components and to identify potential new components. Although yeast two-hybrid data are never definitive, the experimental interactions presented in this paper provide the first global view of the organization of proteins and protein complexes involved in human mRNA decay.
RESULTS AND DISCUSSION
Subunit Interactions of the Human Exosome Complex
In eukaryotic cells, degradation of an mRNA is normally initiated by removal of the poly(A) tail (Wilusz et al. 2001). In yeast, degradation then proceeds via two pathways: rapid 5′→3′ degradation following removal of the 5′-cap structure, and slower 3′→5′ degradation catalyzed by the exosome complex (for review, see Wilusz et al. 2001). In human cells, exosome-mediated 3′→5′ degradation appears to be the predominant pathway (Chen et al. 2001; Wang and Kiledjian 2001; Mukherjee et al. 2002). The human exosome is known to contain six RNase-PH domain subunits (Rrp41, Rrp42, Rrp46, PM/Scl-75, Mtr3, Oip2/Rrp43), S1 (and KH) RNA-binding domain subunits (Rrp4, Rrp40, Csl4), an RNase D-like subunit (PM/Scl-100), a putative helicase (Kiaa0052), and a protein that is specifically phosphorylated in the M phase of the cell cycle (Mpp6; Allmang et al. 1999; Chen et al. 2001).
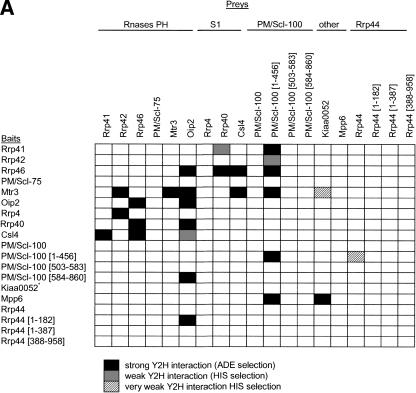
To further investigate the structure of the human exosome, a yeast two-hybrid (Y2H) system was used to test for protein–protein interactions among all subunits. Although not all Y2H interactions will represent physiological interactions, and not all physiological interactions will be detected by Y2H, we aimed to use Y2H data to generate an experimental model of subunit interactions within the human exosome complex. All of the interactions detected are listed in Figure 1A and illustrated in an exosome model in Figure 1B.
Figure 1.
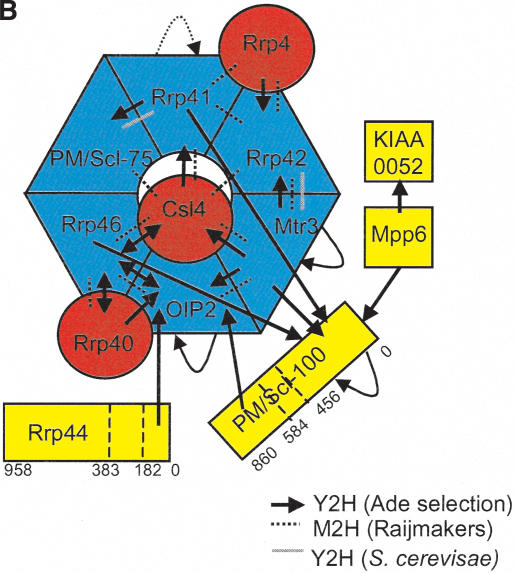
Subunit interactions of the human exosome. (A) Y2H interactions detected on stringent (–Ade) and less-stringent (–His) selection are indicated. The amino acid residues included in protein fragments are shown in brackets. (*) Self-active bait. (B) Stringent Y2H interactions are superimposed on a model of the exosome. The Rrp41-PM/Scl-75 interaction was only detected by library screening (see Table 2). Interactions previously detected by mammalian two-hybrid (M2H) and between the homologous yeast proteins are indicated. PH-domain subunits are blue, S1-domain subunits are red, and other subunits are yellow.
The exosome PH domain subunits have been proposed to assemble into a hexameric ring because of their homology to bacterial PNPase, which contains two Rnase-PH domains and crystallizes as a trimer of dimers (Symmons et al. 2002). However, the exosome contains six different Rnase-PH domains, and there are 120 ways to order these into a hexameric ring structure. The Y2H interactions identified in this study are consistent with a hexameric ring structure, and predict a subunit ordering that is unambiguous except for the relative orientation of the Rrp41-PM/Scl-75 dimer (Fig. 1B). While this work was in preparation, a study was published describing interactions between some exosome subunits detected using a mammalian two-hybrid (M2H) system (Raijmakers et al. 2002). Each method identified one unique PH–PH interaction, such that the combined results provide an unambiguous PH subunit ordering that strongly supports the ordering shown in Figure 1B, and not that proposed by the in silico modeling of Aloy et al. (2002). Moreover, the novel self-interactions of Mtr3 and Oip2 support the proposal that some endogenous exosome complexes may contain two rings of PH subunits stacked using homomeric interactions between the PH subunits (Raijmakers et al. 2002). By combining our Y2H data with the M2H data of Raijmakers et al. (2002), it is possible to predict the position of each of the S1 domain proteins relative to the PH hexamer (Fig. 1B). The more extensive interactions of Csl4 suggest it may be positioned more centrally than Rrp4 and Rrp40. The trypanosome exosome S1 domain proteins interact with each other (Estevez et al. 2003), supporting the positioning of the S1 domain proteins on the same face of the PH-subunit ring in this model. As these three subunits all contain RNA-binding motifs, it is possible that they may aid substrate recognition and delivery to the exonucleases of the PH ring.
The Y2H results also predict how peripheral subunits may associate with each other and with the exosome core. Kiaa0052 was found to interact with Mpp6, which is the first evidence for association between these two proteins other than their copurification with the exosome (Allmang et al. 1999; Chen et al. 2001). A weak interaction between Mtr3 and Kiaa0052 may partially mediate their association with the exosome core. As Kiaa0052 is a putative helicase, its association with the exosome may be important for unwinding RNA substrates before they are delivered to the catalytic core. Also, Mpp6 is known to be specifically phosphorylated in the M phase of the cell cycle (Matsumoto-Taniura et al. 1996) and so may be involved in the cell-cycle-dependent regulation of Kiaa0052 and exosome activity. As in previous M2H studies (Raijmakers et al. 2002), no strong interactions were detected between Rrp6 or Rrp44 (which is exosome-associated in yeast) and other subunits. Sometimes a protein interaction is only detectable by Y2H using a fragment of a protein, because the full-length protein may not express well in yeast, or because an interaction only occurs when a conformational change in the full-length protein exposes a subdomain (Semple et al. 2002). Therefore, for proteins where no interactions were detected using full-length constructs, protein fragments corresponding to predicted structural domains were used as Y2H baits and preys. Using this approach, interactions were detected between Oip2 and both the C terminus of PM/Scl-100 (amino acids 584–860) and the PinC domain of Rrp44 (amino acids 1–182). The exonuclease domain of PM/Scl-100 (amino acids 1–456) interacted with Mtr3 and weakly with Rrp44 as a bait construct, and with four PH subunits, itself, Mpp6, and Csl4 as a prey construct. PM/Scl-100, unlike other peripheral subunits, seems to be essential for the stability of the trypanosome exosome core (Estevez et al. 2003), suggesting the extensive interactions of the exonuclease domain may have a role in exosome assembly or maintenance. These interactions are the first indication of how peripheral subunits may associate with the exosome core, and the only evidence that Rrp44 can associate with the human exosome.
Subunit Interactions of the Lsm Complexes
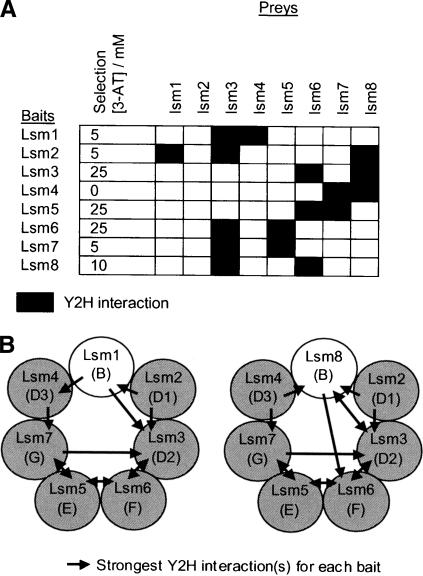
S. cerevisiae contains two heptameric complexes of Lsm proteins, the nuclear Lsm2–8 complex associated with the U6 snRNA and the cytoplasmic Lsm1–7 complex required for mRNA decapping (He and Parker 2000). Humans possess orthologs of all eight Lsm proteins, and an equivalent Lsm2–8 complex has been isolated and found to associate with U6 snRNA (Achsel et al. 1999). By EM imaging, the human Lsm2–8 complex appears as a doughnut-shaped ring (Achsel et al. 1999). In this study, the interactions between human Lsm proteins were investigated using a Y2H matrix approach. As observed with the yeast proteins (Fromont-Racine et al. 2000), each human Lsm protein was capable of interacting with multiple other Lsm proteins (Table 1). Indeed, in vitro, Lsm3 can form homo-oligomeric ring structures (Collins et al. 2003). However, these interactions conflict with evidence that in yeast each Lsm complex contains seven Lsm proteins (Salgado-Garrido et al. 1999). If, in vivo, the Lsm proteins are arranged in a single ring configuration in each Lsm complex, then they would be expected to contact two other Lsm proteins from each complex. To further investigate the assembly of subunits within the human Lsm complexes, the strength of each Lsm–Lsm Y2H interaction was tested using 3-aminotriazole (3-AT) as a competitive inhibitor of the HIS3 reporter. Figure 2A lists the interactions made by each Lsm bait at a reporter stringency at which each bait does not interact with more than two other Lsm proteins from each expected complex (Lsm1–7 or Lsm2–8). Aside from the interactions of the Lsm8 bait (which is weakly self-active in the Y2H assay) and Lsm3 (which interacts most promiscuously with other Lsm proteins), the results are only consistent with a single heptameric ring that contains either Lsm1 or Lsm8 (Fig. 2B). Remarkably, these interactions are exactly homologous to those detected between the related Sm proteins (indicated in brackets in Fig. 2B; He and Parker 2000), which strongly supports the likelihood that this is the correct ordering of subunits in the Lsm complexes. Although the strength of a Y2H interaction is not always equivalent to the affinity of an interaction, these results predict that the in vivo assembly of native Lsm complexes is controlled so that each Lsm protein contacts the two Lsm proteins to which it binds with highest affinities. However, because many other potential Lsm–Lsm interactions are possible, we can-not exclude that different cellular Lsm complexes may also exist with different specificities or functions.
Table 1.
Protein Interactions Detected by Y2H Matrix Experiments
| Description | Accession | Bait constructa | Matrix interactions (Ade/His selection)d |
|---|---|---|---|
| Exosome: PH | NM_019037 | Rrp41 | PM/Scl-100[1-456], Rrp40, Dom3, Ski2[1-708] (weak) |
| PH | NM_015004 | Rrp42/KIAA0116 | PM/Scl-1000[1-456], Dom3 (weak) |
| PH | NM_020158 | Rrp46c | Rrp40, Oip2,Csl4,PM/Scl-100[1-456] |
| PH | NM_005033 | PM/Scl-75/Rrp45 | — |
| PH | NM_058219 | Mtr3 | Rrp42,Oip2,Csl4, Mtr3, PM/Scl-100[1-456], Dcp1B (weak) Kiaa0052 (weak), TTP (weak) |
| PH | XM_085017 | Oip2 | Rrp46, Oip2, TTP |
| S1/KH | NM_014285 | Rrp4c | RRp42, Znf408 |
| S1/KH | NM_016042 | Rrp40 | Rrp46,Oip2 |
| S1 | NM_016046 | Csl4 | Rrp46, Rrp41,Oip2 |
| RNase D-like | NM_002685 | PM/Scl-100/Rrp6 exonuclease [1-456] | PM/Scl-100[1-456] (weak) |
| PM/Scl-100[1-456], Dom3, Ski2[1-708[, Rrp44 (weak), Mtr3 (weak) | |||
| HRDC [503-583] | — | ||
| [584-860] | Oip2 | ||
| Putative helicase | NM_015360 | Kiaa0052b | — |
| NM_005792 | Mpp6c | Kiaa0052, PARN, PM/Scl-100[1-456] | |
| Homolog of yeast exosome associated | NM_014953 | Rrp44 | — |
| PinC [1-182] | Oip2 | ||
| PinC+CSP [1-387] | — | ||
| RNase II + RNB [388-958] | — | ||
| Homolog of yeast exosome | NM_006929 | Ski2 helicase [1-708]b | — |
| [709-1246] | — | ||
| Lsm proteins | NM_014462 | Lsm1 | Lsm3, Lsm2, Lsm4, Mtr3(weak) Oip2 (weak) |
| NM_021177 | Lsm2 | Lsm3, Lsm8, Znf408, PM/Scl-100[1-456], Lsm1, Lsm7 | |
| NM_014463 | Lsm3 | Lsm1, Lsm2, Lsm5, Lsm6, Lsm7, Lsm8, Xrn2[1-280] | |
| NM_012321 | Lsm4 | Lsm7, Lsm8 | |
| NM_012322 | Lsm5 | Lsm3, Lsm6, Lsm7 | |
| NM_007080 | Lsm6 | Lsm3, Lsm5, Lsm7, Lsm1, Lsm8 | |
| NM_016199 | Lsm7 | Lsm3, Lsm5, Mtr3, Lsm2, Lsm4, Lsm6, Oip2 (weak), | |
| NM_016200 | Lsm8c | Lsm2, Lsm3, Lsm6, PM/Scl-100[1-456], Lsm4, Lsm7, Mtr3, Dcp1A, Upf1[702-1119] | |
| Decapping factors | NM_152624 | Dcp2 | Dcp1A,Dcp1B |
| Dcp2 (Δ exons 8 and 9) | Dcp1A, Dcp1B | ||
| NM_018403 | Dcp1Ab | — | |
| NM_152640 | Dcp1Bb | — | |
| NM_014026 | DcpS | — | |
| 5′→3′ exonucleases | XM_033181 | Xrn1/Sep1 | |
| Exonuclease [1-300] | Dep1A, Dom3, Ski2[1-708], PM/Scl-100[1-456], Lsm4 (weak) | ||
| [578-1470] | |||
| Oip2, Mtr3 (weak), Dcp1B (weak), TTP (weak), Kiaa0052 (weak), Csl4 (weak) | |||
| RBD [1440-1707] | Oip2 | ||
| Rat1p homolog | NM_012255 | Xrn2/Rat1 | Dom3, PM/Scl-100[1-456] |
| Exonuclease [1-280] | Oip2 (weak) | ||
| Rai1p homolog | NM_005510 | Dom3 | Dom3, PM/Scl-100[1-456] |
| Poly(A) ribonuclease | NM_002582 | PARN | — |
| ARE-BPs | NM_003407 | TTPb | — |
| NM_002138 | Auf1/hnRNPDc (variant 3) | — | |
| NM_001419 | HuR | HuR | |
| NMD factors | NM_002911 | Upf1 [1-415] | — |
| Helicase [416-701] | — | ||
| [702-1119] | Dom3 | ||
| NM_080599 | Upf2 [1-167] | Oip2, Mtr3, TTP (weak), Dcp1B (weak), Csl4 (weak), Kiaa005 (weak) | |
| MIF4G1+2 [166-758] | Upf2[1-167] | ||
| [757-1272]c | Oip2, Dcp2, Xrn2[1-280], PM/Scl-100[1-456], Znf408, Lsm1, Upf2[757-1272], Mtr3 (weak) | ||
| NM_023011 | Upf3A (variant2) | — | |
| NM_080632 | Upf3B | Upf2[757-1272] | |
| Novel | NM_024741 | Znf408 | Znf408, Upf2[757-1272], Rrp4 |
Previously published interactions are in bold, and interactions detected between the orthologous yeast or Drosophila proteins in genome-wide data sets are underlined.
Protein fragments are indicated by the residue numbers covered and a domain name (where known).
Self-active bait.
Weakly self-active bait (— His selection).
Interactions only selected on — His media are shown in italics, weak but reproducible interactions (<50% growth) are indicated as “weak.”
Figure 2.
Subunit interactions of the human Lsm complexes. (A) Each potential Lsm–Lsm interaction was assayed on selective media lacking His and containing 0 mM, 5 mM, 10 mM, 25 mM, or 50 mM 3-AT, a competitive inhibitor of the HIS3 reporter. The interactions for each Lsm bait construct are shown at the indicated concentration of 3-AT, whereby no more than two interactions are seen with proteins from either the expected Lsm1–7 or Lsm2–8 complexes. (B) These interactions are shown on models of the Lsm1–7 and Lsm2–8 complexes. The most similar Sm protein is indicated in brackets for each Lsm protein.
Interactions Between mRNA Decay Factors
The successful use of Y2H to dissect the exosome and Lsm complex subunit interactions suggested that a similar strategy could be used to test for interactions between other proteins predicted to have a role in human mRNA decay. Therefore, a total of 54 bait and 54 prey yeast strains were constructed, each expressing a full-length protein or a protein domain. These included decapping factors (Dcp2, Dcp1a, Dcp1b, DcpS; Liu et al. 2002; Lykke-Andersen 2002; Van Dijk et al. 2002; Wang et al. 2002), 5′→3′ exonucleases (Xrn1, Xrn2/Rat1), poly(A) ribonuclease (PARN; Dehlin et al. 2000), representative AU-rich element (ARE)-binding proteins known to bind and regulate the stability of certain unstable mRNAs (TTP, HuR, Auf1; Chen et al. 2001), proteins required for nonsense-mediated decay (NMD) of mRNAs containing a premature stop codon (Upf1, Upf2, Upf3A, Upf3B; Lykke-Andersen et al. 2000), and three other proteins predicted to associate with decay factors (Ski2, Dom3/Rai1, Znf408; Jacobs et al. 1998; Xue et al. 2000). We used full-length proteins as Y2H baits and preys, except for large multidomain proteins without an available full-length cDNA clone. In these cases, we used protein fragments designed to cover every predicted structural domain in the protein. Each protein/domain was individually tested in duplicate for the ability to interact with every other protein/domain using an Y2H matrix approach to analyze a total of >2900 possible interactions. The interactions are listed in Table 1 and illustrated as a network in Figure 3A.
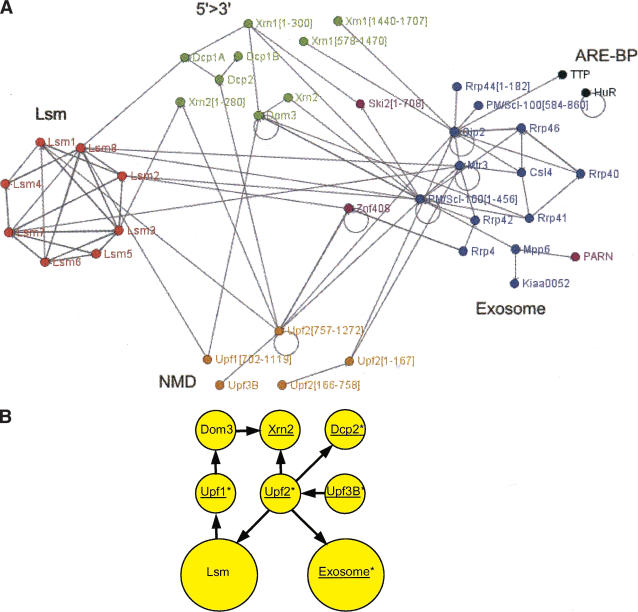
Figure 3.
Protein–protein interactions detected between proteins predicted to function in human RNA decay. (A) Each Y2H interaction (selected on media lacking Ade or His) is indicated as an arrow from bait to prey. The amino acid residues included in protein fragments are indicated in brackets. Very weak interactions are not shown. The figure was drawn using BioLayout (http://maine.ebi.ac.uk:8000/services/biolayout/; Enright and Ouzounis 2001). (B) The interactions of the Upf proteins suggest how both nuclear and cytoplasmic 5′→3′ and 3′→5′ decay factors may be recruited during NMD. Y2H interactions are shown as arrows, proteins required for NMD are indicated by *, and proteins that coprecipitate from cell extracts are underlined (Lejeune et al. 2003).
To our knowledge, direct evidence for only two of these interactions, Upf2–Upf3 (Serin et al. 2001) and Dcp1a–Dcp2, has been reported previously (Lykke-Andersen 2002). However, we did not detect one previously reported interaction between Upf1 and Upf2 (Serin et al. 2001).
It is important to note that many of the Y2H interactions reported in this study are strongly supported by evidence from a range of complementary studies. For example, the subnetwork of interactions between putative 5′→3′ processing factors (Lsm proteins, Xrn1, Xrn2, Dcp1, and Dcp2) is supported both by the colocalization of Xrn1, Dcp1, and Dcp2 in cytoplasmic foci (Ingelfinger et al. 2002; Van Dijk et al. 2002) and by the coprecipitation of Dcp2, Xrn1, and Xrn2 from human cells (Lejeune et al. 2003). As such, it would appear that these proteins may be involved in a coordinated 5′→3′ decay pathway that is similar to that described for their yeast orthologs (Wilusz et al. 2001).
A second intriguing subnetwork of interactions was centered on the NMD proteins, Upf1, Upf2, and Upf3B (Fig. 3B). Upf1, Upf2, and Upf3B are recruited to mRNAs targeted for NMD, but how the mRNA decay machinery is recruited to these tagged mRNAs is unclear (Wagner and Lykke-Andersen 2002). The interactions of Upf1 and Upf2 with Xrn2, Dcp1A, Dcp2, Lsm proteins, and exosome subunits suggest that degradation of mRNAs containing premature termination codons may result from direct recruitment of decay factors by Upf1 and Upf2. This model is also supported by the coprecipitation of Upf proteins, Dcp2/Dcp1a (Lykke-Andersen 2002), Xrn1, Xrn2, PARN, and exosome components from cells (Lejeune et al. 2003). Our Y2H interaction network contains many connections between 5′→3′ and 3′→5′ decay proteins. Together with the RNAse-insensitive coprecipitation of NMD and 5′→3′ and 3′→5′ decay factors (Lejeune et al. 2003), these interactions suggest that large complexes of RNA decay factors may exist to coordinate 5′→3′ and 3′→5′ mRNA decay in human cells.
Another interaction of particular interest is that observed between the deadenylase PARN and the peripheral exosome subunit Mpp6. This interaction may mediate the recruitment of the exosome to deadenylated mRNAs for subsequent 3′→5′ degradation. Interestingly, this could explain the predominance of the 3′→5′ decay pathway in mammalian cells, whereas in yeast, which does not have the Mpp6 component of the exosome, the 5′→3′ pathway predominates.
Interactions With Other Proteins
To identify new proteins with a potential role in mRNA degradation, we identified new interaction partners for each bait by performing Y2H screens against a high-complexity cDNA library. To maximize the specificity of the Y2H system, we applied stringent criteria for the selection of interactions. These screens were performed by mating to ensure saturation screening of the library, and interactions were identified using stringent criteria, to minimize the number of false-positive interactions reported. To be considered positive, each Y2H interaction had to activate two independent Y2H reporters. This procedure eliminates interactions that result from nonspecific promoter activation (James et al. 1996). Most importantly, every positive interaction was also retested in fresh yeast cells to eliminate interactions that result from nonspecific host or vector mutations. To do this, each prey strain was reconstructed by gap-repair recombination cloning in fresh yeast. The fresh prey strains were then retested both for the ability to interact with the bait that isolated them from the library, and also for bait specificity by mating with an irrelevant bait. We have previously demonstrated that this approach dramatically reduces the number of false-positive interactions detected in Y2H screens (Lehner et al. 2004).
In total, we identified 247 reconfirmed Y2H interactions, including 26 detected by the matrix experiment. This is equivalent to an average of 4.5 interactions per bait, which compares favorably to the number of interactions per bait in genome-wide Y2H studies (∼1 interaction per bait; see Semple et al. 2002), and thus demonstrates the additional information that can result from performing comprehensive pathway-focused interaction mapping studies in addition to genome-wide projects (Walhout et al. 2000). As with the results of any high-throughput interaction assay, the interactions listed here should be considered as putative or predicted interactions. To estimate the number of these interactions that may be expected to be true positives, we analyzed the results of a previously published study that used the same stringent high-throughput Y2H system to identify binding partners for a set of well-characterized human proteins with known interaction profiles (Lehner et al. 2004). In that study, for previously characterized proteins with at least one interaction reported, 14 of the 26 (∼54%) Y2H interactions that we detected had been independently verified by a method other than Y2H (Lehner et al. 2004). Hence, by extrapolation we would expect that the inherent stringency of the assay is such that >50% of the interactions reported here will prove to be true positives. However, it is important to understand that although verification of protein interactions in different assay systems does increase confidence in the validity of a protein interaction, it does not absolutely define physiological relevance. With respect to the false-negative rate of our data, we detected 9 of 15 (∼60%) of previously reported interactions. Table 2 lists both the reconfirmed interacting preys isolated with each bait and the number of times each prey was isolated in a screen. A more detailed description of each prey is available from http://www.hgmp.mrc.ac.uk/Research/RNA.
Table 2.
Protein Interactions Detected by Y2H Library Screens
| Bait construct | Library screen interactionsd |
|---|---|
| Rrp41 | PM/ScI-75 (5), FLJ20297 (3), C1or19 (3), DKFZp566J2046 (3), FLJ33418, EEF1A1, NEK1, BC006102, BM770606, AKR1A1, GTF2IRD1, LRRC5, FLJ21047, PLINP |
| Rrp42/KIAA0116 | — |
| Rrp46c | OIP2 (94), Lsm5 (8), Rrp40 (4), EBP1 (3), NUP210 (2), CsI4, PKM2, SFPQ, FLJ30932, DCK5RAP1, POLR2L |
| PM/ScI-75/Rrp45 | — |
| Mtr3 | EIF3S2 (5), Rrp42 (5), GRIM19 (2), SF2p32, SFRS3, CPSF5, RAB3-GAP150, PSMB1, ERRalpha, AD023, MOCS3, FLJ22678, WBSCR18, KIAA0182 |
| Oip2 | FHOD1 (11), RASSF1 (9), MKRN1 (8), KIAA1604, AI685517, FOXN3 |
| Rrp4c | Rrp42 (49), Znf408 (6), RNF8 (2), FLJ40491 |
| Rrp40 | Rrp46, CDK5RAP1 |
| CsI4 | NUP160 (17), Rrp41 (3), AV753896 |
| PM/ScI-100/Rrp6 | CIB1 (3), C14orf92, SCRIB, PTGES2, BM451300, |
| exonuclease [1-456] | PM5 (2), GA17, FACT-80, USP16, EPPB9, H17, FLJ22678, FLJ23784, AL110135, AL512687 |
| HRDC [503-583] | FLJ11934 (2), TARDBP, RPE, URP2, |
| [584-860] | RUVBL2, AJ431196, AI620703 |
| Kiaa0052b | — |
| Mpp6c | EIF3S4 (9), FTL (4), SNX9 (4), THOP1, FLJ14502, DNLC2A, MT2P1, BM806939, RPS20, BRG1, DYN2, AATF, MacGAP |
| Rrp44 | — |
| PinC [1-182] | AD-003 (2), ZNF151 |
| PinC + CSP [1-387] | — |
| RNase II + RNB [388-958] | — |
| Ski2b | — |
| helicaseb [1-708] | — |
| [709-1246] | SmB (5), AL517440 (2), FLJ34136, GALNT13, BF248318, U80752, WDR1, CDC2L2 |
| Lsm1 | VPS11 (20), Lsm2 (12), NARS (9), PSMB8 (7), Lsm3 (5), PSMB5 (2), FLJ20254 (2), UXT, N4BP1, GRHPR, HSPC1, DKFZP564J0123, AF090094, AW27564, AW974116, AW975586, KIAA0179 |
| Lsm2e | Lsm8 (88), Lsm3 (75), SmE (5), Znf408 (3), DNAJL2 (2), Lsm7, SmD2 |
| Lsm3 | Lsm2 (51), Lsm8 (36), Lsm10 (21), SmD3 (14), SmE (8), Lsm7 (8), XPC, BC009518 |
| Lsm4 | Lsm8 (32), Lsm7 (27), FLJ13289 (18), SmE (11), Nmi (4), PEPP-2 |
| Lsm5 | Lsm7 (75), SmF (13), SmE, Lsm3 |
| Lsm6 | Lsm5 (40), SmD2 (37), Lsm7 (32), Lsm3 (7), MCRS1 (4), POLD2 |
| Lsm7 | Lsm5 (61), SmD3 (3), Lsm4 (3), RAB7L1 (2), NPY1R, MYH13 |
| Lsm8c | Lsm2 (17), NY-REN-45 (8), FLJ23534 (8), LSM4 (5), TALDO1 (4), SmD1 (3), PDCD2 (3), DKFZp565A176 (3), PGD (2), ASK (2), FLJ23469 (2), RPL24, SMBP2, PNUTL1, MTHFR, DKFZP434F091, MGC10966, GTF21, ITSN2, FLJ11280, DKFZp434K0621, IFNLR1 |
| Dcp2 | POLA2 (7), NAGK, PSMB1 |
| Dcp28 (Δ exons 8 and 9) | RCD-8 (7) |
| Dcp1Ab | — |
| Dcp1Bb | — |
| DcpS | — |
| Xrn1/Sep1 | BAT3 (2), LDHA (2), PABPC4, BC001200, ALDOA, MGC13198, AP1G2, STAM2A, CHD4, |
| Exonuclease [1-300] | SMARCD2, CCT5, GRIM19, FLJ10359, FLJ21347, AI821928 |
| [578-1470] | — |
| RBD [1440-1707] | hnRNPA1 (2), KIAA0261 (2), DPP2 (2) and many others |
| Xrn2/Rat1 | PSMA3 (6), EIF3S6IP, AJ420488, AW977134 |
| Exonuclease [1-280] | EIF6 (2), EIF5A, DKFZP434J214, COMT, MOCS3, TOLLIP, TARDBP |
| Dom3d | AI580336 |
| PARN | DKFZp434N0650 (12) |
| TTPb | — |
| Auf1/hnRNPDc (variant 3) | SF2p32 (30), Rrp41, Lsm5 plus many others |
| HuR | — |
| Upf1 [1-415] | — |
| Helicase [416-701] | — |
| [702-1119] | HIRA (2), BC040175, N21259, PLEKHB2, FLJ10631, ACAS2, BAT5, PEPP2, KIAA0460, GNPTAG |
| Upf2 [1-167] | — |
| MIF4G1 + 2 [166-758] | — |
| [757-1272]c | RPS15 (12), RPL5 (5), REA (3), MAP1A (3), RPS25 (2), ART4 (2), UPF3B + many others |
| Upf3A (variant2) | GSK3B (2), BM310104 (2), IFITM2 |
| Upf3B | MCRS1, FLJ20343 |
| Znf408 | RGS19IP1 (6), RIL (2), NOH61 (2), Lsm2, Znf408, NOA36, DKFZP434G156, BG403593, BQ069192, MIF, TOP3B, CAF1B |
aFor descriptions of baits, see Table 1.
Previously published interactions are in bold, and interactions detected between the orthologous yeast or Drosophila proteins in genome-wide data sets are underlined.
Self-active bait.
Weakly self-active bait (— His selection).
The number of colonies isolated for each bait is indicated in parentheses when greater than 1. Singly isolated preys are not listed for baits that isolated >20 different preys. A few interactions with extracellular proteins or those known to localize to the secretory pathway or mitochondria are not listed, because they are unlikely to represent physiological interactions. A full list of interacting preys, accessions, and descriptions are available from http://www.hgmp.mrc.ac.uk/Research/RNA and as Supplemental material.
Lsm2 and Dom3 library screen interactions were previously reported (Lehner et al. 2004).
These interactions provide a wealth of data that can be used to explain previous observations and to guide future research on RNA decay. For example, it has been demonstrated that the ubiquitin–proteasome system is required for degradation of ARE-containing mRNAs, but its precise role is unclear (Laroia et al. 2002). In the Y2H data there are multiple connections between RNA decay proteins and components of the ubiquitin–proteasome system, including E3 ubiquitin ligase enzymes (Oip2-Makorin1, Rrp4-RNF8), as well as direct interactions with specific proteasome subunits (Lsm1–PSMB5, PSMB8). Future work should address how these interactions may regulate the exosome and Lsm complexes during RNA decay.
Interactions were also observed with transcription factors (e.g., Oip2-FOXN3, Rrp41-GTF2IRD1), splicing and mRNA-processing factors (e.g., Rrp46-SFPQ, Mtr3-SF2p32, Mtr3-CPSF5, Mtr3-SFRS3, Xrn1[1–300]-PABPC4), and translation factors (e.g., Rrp41-EEF1A1, Xrn2-EIF3S6IP, Xrn2[1–280]-EIF6). These interactions suggest that mRNA degradation may be coupled to other steps in gene expression. Indeed, in yeast, the exosome is needed for quality surveillance of nascent transcripts at transcription sites (Hilleren et al. 2001), and many functional connections between translation and mRNA stability have been reported (Wilusz et al. 2001). It is likely that some of the interactions reported here mediate these couplings between mRNA decay and other steps in gene expression. In archael genomes, the majority of predicted exosome subunits are encoded in a superoperon that also encodes proteasome subunits, ribosomal proteins, and an RNA polymerase subunit (Koonin et al. 2001). Our interaction map suggests that a tight functional coupling between translation, RNA processing and degradation, and protein degradation may have been conserved since the divergence of archael and eukaryotic genomes.
Evolutionary Conservation of the Interaction Network
Another method of assessing the potential physiological relevance of protein interaction data is to identify those interactions that are conserved between homologous proteins in different species. These evolutionarily conserved interactions (“interlogs”) are likely to be of fundamental importance to a eukaryotic cell (Walhout et al. 2000). Therefore, to identify evolutionarily conserved interactions in our data set, we first used the “Inparanoid” database (Remm et al. 2001; http://inparanoid.cgb.ki.se/) to identify orthologs of our bait and prey proteins. We then extracted interaction data for each bait ortholog from three independent data sets. The three data sets used were a genome-wide Y2H interaction map for Drosophila melanogaster (Giot et al. 2003), an Y2H interaction map for metazoan-specific proteins from Caenorhabditis elegans (Li et al. 2004), and a compilation of Y2H, complex purification, genetic, or in silico predicted interactions for Saccharomyces cerevisiae (von Mering et al. 2002).
The Drosophila Y2H interaction map contains 31 high-confidence interactions for the orthologs of our bait proteins. Of these, 10 (one-third) support interactions detected in this study. In particular, the interactions of the Drosophila Lsm proteins strongly support our model for the structure of the Lsm complexes. Significantly, 6 out of the 9 interactions required by our model were also represented in the Drosophila Y2H data set, with no inconsistent interactions (Supplemental Fig. 1). In addition, the Drosophila interaction map also supports the connection we report between the Lsm complex and the proteasome (Lsm1–Psmb5). In contrast, the C. elegans Y2H interaction map contains only three interactions (all novel) for our bait proteins. However, it is important to point out that the C. elegans data set was constructed using metazoan-specific proteins as baits. Most of our bait proteins are found in all eukaryotes, and thus were not included as baits in that study. Analysis of the yeast interaction data set identified interolog interactions that support 48 of our human interactions. Importantly, 22 of these interactions are supported by evidence from more than one yeast interaction data set. Because only 103 proteins in our interaction network have clear orthologs in yeast, these data strongly support the reliability of our interaction network.
In total, the physiological relevance of 50 of our interactions is supported by their evolutionary conservation. These interactions are indicated in Tables 1 and 2, and supporting evidence for each of these interactions is listed in Supplemental Table 1. As these interactions are highly conserved through evolution, it is likely that they represent fundamental points of communication between components of different RNA decay processes. However, because of the relatively low coverage of model organism interaction data sets (von Mering et al. 2002), we suspect that many other interactions may also be evolutionarily conserved. It is also important to remember that many of the proteins identified here do not have clear orthologs in invertebrates, and combined with the possibility of mammalian specific aspects of RNA decay, those protein interactions that do not have interlogs should not be dismissed.
Conclusions
In this report, we have shown how exhaustive Y2H protein–protein interaction mapping can provide internally consistent data on the arrangement of proteins within multisubunit protein complexes. When combined with structural data from homologous protein complexes, these interactions can be used to predict the structures of novel protein complexes. We have also identified a diverse range of both evolutionarily conserved and novel interactions between mRNA decay proteins. Although not all of these interactions will prove to occur physiologically, these results provide the first global insights into the possible organization of human mRNA decay pathways and their coupling with other steps in gene expression. These experimental predictions provide a wealth of data that can be used to guide future research on human RNA degradation.
METHODS
Construction of Y2H Baits and Preys
Bait and preys were constructed by gap repair recombination cloning as described (Estevez et al. 2003). Templates for PCR were either clones from the Mammalian Gene Collection (Strausberg et al. 2002); an Slb-1 cDNA library (Clonetech); or in the case of Mtr3, a clone generously provided by Ger Pruijn (University of Nijmegen, The Netherlands).
Matrix Experiments
Each bait was mated in duplicate to an array of preys overnight on YPAD media; diploids were selected and activation of the ADE2 (stringent) and HIS3 (less stringent) reporters was assayed by replicating to media lacking adenine or histidine and scoring growth at 3, 7, and 11 d. Each Lsm–Lsm diploid was also assayed for growth on media lacking histidine and containing 0 mM, 5 mM, 10 mM, 25 mM, or 50 mM 3-aminotriazole (3-AT).
Library Screens
Library screens were performed as described previously (Lehner et al. 2004), and 109 bait cells were mated with 109 cells of a Y2H K562 cell-line cDNA library, followed by selecting on media lacking Ade after 1 wk. All Ade-positive colonies were tested for activation of the lacZ reporter, and all prey inserts were identified by PCR and sequencing. Importantly, every interaction was retested in fresh yeast and tested for specificity. This was achieved by gap-repair cloning each prey PCR product into the pGAD-T7 prey vector in pJ69-4α yeast, and then remating these colonies to both the original bait and an irrelevant bait (Cobra1). Every colony isolated in a screen was analyzed, except when using Lsm baits that all isolated thousands of positive colonies. At least 95 colonies were analyzed for these screens. Prey insert sequences were identified by using the BLAST algorithm to search the UniGene database, which represents all known mRNA and EST sequences.
Acknowledgments
We acknowledge Amanda Thompson and Simon Duley for technical assistance; Duncan Campbell for support and critical reading of the manuscript; David Markie for the vectors pGBAD-B and pACTBD-B; Ger Pruijn for a cDNA clone encoding Mtr3; and Fabrice Lejeune and Lynne Maquat for sharing unpublished results and critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2122004.
Footnotes
[Supplemental material is available online at www.genome.org. All of the interaction data described in this manuscript has been submitted to the BIND database (IDs 133608–133972; http://www.blueprint.org/bind/bind.php) and is available as Supplemental material. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: G. Pruijn, D. Markie, F. Lejeune, and L. Maquat.]
References
- Achsel, T., Brahms, H., Kastner, B., Bachi, A., Wilm, M., and Luhrmann, R. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18: 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang, C., Petfalski, E., Podtelejnikov, A., Mann, M., Tollervey, D., and Mitchell, P. 1999. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes & Dev. 13: 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloy, P., Ciccarelli, F.D., Leutwein, C., Gavin, A.C., Superti-Furga, G., Bork, P., Bottcher, B., and Russell, R.B. 2002. A complex prediction: Three-dimensional model of the yeast exosome. EMBO Rep. 3: 628–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.Y., Gherzi, R., Ong, S.E., Chan, E.L., Raijmakers, R., Pruijn, G.J., Stoecklin, G., Moroni, C., Mann, M., and Karin, M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464. [DOI] [PubMed] [Google Scholar]
- Collins, B.M., Cubeddu, L., Naidoo, N., Harrop, S.J., Kornfeld, G.D., Dawes, I.W., Curmi, P.M., and Mabbutt, B.C. 2003. Homomeric ring assemblies of eukaryotic Sm proteins have affinity for both RNA and DNA. Crystal structure of an oligomeric complex of yeast SmF. J. Biol. Chem. 278: 17291–17298. [DOI] [PubMed] [Google Scholar]
- Dehlin, E., Wormington, M., Korner, C.G., and Wahle, E. 2000. Cap-dependent deadenylation of mRNA. EMBO J. 19: 1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright, A.J. and Ouzounis, C.A. 2001. BioLayout—An automatic graph layout algorithm for similarity visualization. Bioinformatics 17: 853–854. [DOI] [PubMed] [Google Scholar]
- Estevez, A.M., Lehner, B., Sanderson, C.M., Ruppert, T., and Clayton, C. 2003. The roles of inter-subunit interactions in exosome stability. J. Biol. Chem. 23: 23. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., Mayes, A.E., Brunet-Simon, A., Rain, J.C., Colley, A., Dix, I., Decourty, L., Joly, N., Ricard, F., Beggs, J.D., et al. 2000. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast 17: 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot, L., Bader, J.S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y.L., Ooi, C.E., Godwin, B., Vitols, E., et al. 2003. A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736. [DOI] [PubMed] [Google Scholar]
- He, W. and Parker, R. 2000. Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol. 12: 346–350. [DOI] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Rosbash, M., Parker, R., and Jensen, T.H. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542. [DOI] [PubMed] [Google Scholar]
- Ingelfinger, D., Arndt-Jovin, D.J., Luhrmann, R., and Achsel, T. 2002. The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Jacobs, J.S., Anderson, A.R., and Parker, R.P. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17: 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodursky, A.B. and Bernstein, J.A. 2003. Life after transcription—Revisiting the fate of messenger RNA. Trends Genet. 19: 113–115. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V., Wolf, Y.I., and Aravind, L. 2001. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 11: 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia, G., Sarkar, B., and Schneider, R.J. 2002. Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc. Natl. Acad. Sci. 99: 1842–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner, B., Semple, J.I., Brown, S.E., Counsell, D., Campbell, R.D., and Sanderson, C.M. 2004. Analysis of a high throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics 83: 153–167. [DOI] [PubMed] [Google Scholar]
- Lejeune, F., Li, X., and Maquat, L.E. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12: 675–687. [DOI] [PubMed] [Google Scholar]
- Li, S., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D., Chesneau, A., Hao, T., et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Rodgers, N.D., Jiao, X., and Kiledjian, M. 2002. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 21: 4699–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22: 8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.D., and Steitz, J.A. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121–1131. [DOI] [PubMed] [Google Scholar]
- Matsumoto-Taniura, N., Pirollet, F., Monroe, R., Gerace, L., and Westendorf, J.M. 1996. Identification of novel M phase phosphoproteins by expression cloning. Mol. Biol. Cell 7: 1455–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P. and Tollervey, D. 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol. Cell 11: 1405–1413. [DOI] [PubMed] [Google Scholar]
- Mukherjee, D., Gao, M., O'Connor, J.P., Raijmakers, R., Pruijn, G., Lutz, C.S., and Wilusz, J. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21: 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raijmakers, R., Egberts, W.V., van Venrooij, W.J., and Pruijn, G.J. 2002. Protein–protein interactions between human exosome components support the assembly of RNase PH-type subunits into a six-membered PNPase-like ring. J. Mol. Biol. 323: 653–663. [DOI] [PubMed] [Google Scholar]
- Remm, M., Storm, C.E., and Sonnhammer, E.L. 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Salgado-Garrido, J., Bragado-Nilsson, E., Kandels-Lewis, S., and Seraphin, B. 1999. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 18: 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple, J.I., Sanderson, C.M., and Campbell, R.D. 2002. The jury is out on `guilt by association' trials. Briefings Func. Genom. Proteom. 1: 40–52. [DOI] [PubMed] [Google Scholar]
- Serin, G., Gersappe, A., Black, J.D., Aronoff, R., and Maquat, L.E. 2001. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol. 21: 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U. and Parker, R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg, R.L., Feingold, E.A., Grouse, L.H., Derge, J.G., Klausner, R.D., Collins, F.S., Wagner, L., Shenmen, C.M., Schuler, G.D., Altschul, S.F., et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. 99: 16899–16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons, M.F., Williams, M.G., Luisi, B.F., Jones, G.H., and Carpousis, A.J. 2002. Running rings around RNA: A superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 27: 11–18. [DOI] [PubMed] [Google Scholar]
- Van Dijk, E., Cougot, N., Meyer, S., Babajko, S., Wahle, E., and Seraphin, J. 2002. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21: 6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S.G., Fields, S., and Bork, P. 2002. Comparative assessment of large-scale data sets protein–protein interactions. Nature 417: 399–403. [DOI] [PubMed] [Google Scholar]
- Wagner, E. and Lykke-Andersen, J. 2002. mRNA surveillance: The perfect persist. J. Cell Sci. 115: 3033–3038. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. 2000. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116–122. [DOI] [PubMed] [Google Scholar]
- Wang, Z. and Kiledjian, M. 2001. Functional link between the mammalian exosome and mRNA decapping. Cell 107: 751–762. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Jiao, X., Carr-Schmid, A., and Kiledjian, M. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. 99: 12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz, C.J., Wormington, M., and Peltz, S.W. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2: 237–246. [DOI] [PubMed] [Google Scholar]
- Xue, Y., Bai, X., Lee, I., Kallstrom, G., Ho, J., Brown, J., Stevens, A., and Johnson, A.W. 2000. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol. 20: 4006–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://maine.ebi.ac.uk:8000/services/biolayout; Biolayout software.
- http://www.hgmp.mrc.ac.uk/Research/RNA; descriptions of all interacting proteins isolated in this study.
- http://inparanoid.cgb.ki.se/; The Inparanoid database of pairwise orthologs.
- http://www.blueprint.org/bind/bind.php; BIND protein interaction database.