Abstract
Access to the human genome facilitates extensive functional proteomics studies. Here, we present an integrated approach combining large-scale protein interaction mapping, exploration of the interaction network, and cellular functional assays performed on newly identified proteins involved in a human signaling pathway. As a proof of principle, we studied the Smad signaling system, which is regulated by members of the transforming growth factor β (TGFβ) superfamily. We used two-hybrid screening to map Smad signaling protein–protein interactions and to establish a network of 755 interactions, involving 591 proteins, 179 of which were poorly or not annotated. The exploration of such complex interaction databases is improved by the use of PIMRider, a dedicated navigation tool accessible through the Web. The biological meaning of this network is illustrated by the presence of 18 known Smad-associated proteins. Functional assays performed in mammalian cells including siRNA knock-down experiments identified eight novel proteins involved in Smad signaling, thus validating this integrated functional proteomics approach.
Functional proteomics approaches have led in the past to the description of large protein–protein interaction networks. Genome-wide studies have been achieved for viruses (Bartel et al. 1996; Flajolet et al. 2000; McCraith et al. 2000), yeast (Uetz et al. 2000; Ito et al. 2001), and bacteria (Rain et al. 2001). For higher eukaryotes, a comprehensive protein interaction map (Giot et al. 2003) as well as more focused studies on cell cycle factors (Finley Jr. and Brent 1994) or on homologs of human cancer-related proteins (http://pim.hybrigenics.com) were performed for Drosophila proteins. Studies on Caenorhabditis elegans with the establishment of protein interaction maps in DNA damage response (Boulton et al. 2002) and in DAF7/TGFβ signal transduction (Tewari et al. 2004) followed by functional validation have also been reported. However, in mammalian cells, except the recently published study on the TNFα/NF-κB signal transduction pathway (Bouwmeester et al. 2004), only specific studies focused on a limited set of proteins and interactions are available. A key limitation is the difficult handling of the large number of interactions that are identified through the yeast two-hybrid technology combined with the numerous specific functional analyses to be performed in order to demonstrate the biological function for each newly identified protein and/or interaction. An efficient strategy for functional exploration of complex proteomes would require (1) an in-depth annotation of protein–protein interaction maps with any type of available information on proteins, protein domains, and interactions; (2) an exploration tool that allows easy navigation in such a complex database; and (3) streamlined functional assays that would confirm implication of newly identified proteins in a specific biological and/or biochemical readout. In a model experiment, we applied this strategy to the human Smad signaling pathway.
Members of the TGFβ superfamily (e.g., TGFβ, activins and bone morphogenetic proteins, BMPs) are secreted signaling molecules that regulate many biological processes such as cell growth, differentiation, and morphogenesis. The disruption of components of the TGFβ superfamily pathways is associated with human diseases including fibrosis, inflammatory disorders, and cancer. The effects of these pleiotropic molecules are transduced via the ligand-induced formation of heteromeric complexes, consisting of different type I and type II serine/threonine kinase receptors (TβRI, TβRII; Massague 1998). Upon ligand binding, TβRII phosphorylates and activates TβRI, leading to the phosphorylation of receptor-activated Smads (R-Smads; Massague 1998). TGFβ and activin receptors phosphorylate Smad2 and Smad3 (Baker and Harland 1996; Macias-Silva et al. 1996; Zhang et al. 1996), whereas BMP receptors phosphorylate Smad1, Smad5, and Smad8 (Hoodless et al. 1996; Liu et al. 1996). The phosphorylation of R-Smads induces their release from the receptor complex and from SARA, a protein that recruits Smads to the cell membrane (Tsukazaki et al. 1998). Phosphorylation also stimulates R-Smads to translocate to and to accumulate in the nucleus along with their common partner Smad4 (Lagna et al. 1996). Smads then regulate target gene expression in collaboration with other transcriptional partners (ten Dijke et al. 2000).
Here we present an integrated approach for the identification of new factors implicated in this key human signaling pathway involved in several human pathologies. We screened for protein–protein interactions with 11 Smad and related proteins, we generated a complex protein–protein interaction map that is explored through a specific bioinformatics platform, and we demonstrate via cellular assays the function of eight novel proteins involved in the Smad pathway.
RESULTS
Protein Interaction Map of the Smad Pathway
To improve our understanding of Smad signaling, we combined a large-scale yeast two-hybrid-based protein–protein interaction mapping with a systematic functional validation in mammalian cells (Fig. 1). The two-hybrid screening process was performed using a highly complex placental cDNA library (10 million independent yeast clones). Eleven proteins known to be important actors in the Smad pathway (Smad1, 2, 3, 4, 5, 7, 8, SARA, Smurf2, SNIP1, and SnoN) were selected and used to generate 25 bait fragments for yeast two-hybrid screening. Twelve additional proteins, identified as prey in these primary screens, were also selected as bait to perform a total of 44 screens (rebound screens; see Table 1). The Smad interaction map was constructed using a software platform composed of a database, a Web-based graphical interface layer and various query and analysis tools (the PIMRider; see http://pim.hybrigenics.com, where all interaction data are shown). The selected interacting domains (or SID) and the functional domains of each given protein are displayed in the PIMRider. Each protein–protein interaction pair is characterized by a PBS value (Predicted Biological Score) based on a statistical model reflecting the reliability of the interaction. The PBS is divided into five categories from A (highest confidence) to E (probable false positive; Rain et al. 2001). This confidence scoring system is possible because of the availability of a large data set of >9000 interactions identified in 410 screens against the same library (see Methods).
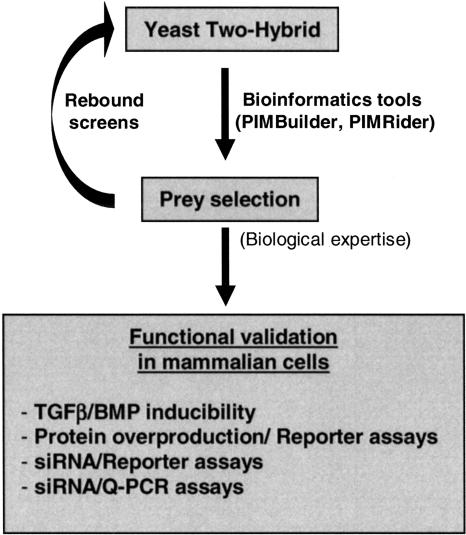
Figure 1.
Strategy for unraveling interactions between components of the Smad pathway. This figure describes the global process from our large-scale yeast two-hybrid screening to the general functional validation assays in mammalian cells. Preys were selected by use of bioinformatics tools (PIMBuilder, PIMRider) and biological expertise.
Table 1.
Baits Used for Two-Hybrid Screening
| Protein name | Accession number | Bait fragments (amino acids) |
|---|---|---|
| Primary screens | ||
| Smad1 | NM 005900 | v1 (1-465); v2 (1-268); v3 (242-465) |
| Smad2 | NM 005901 | v1 (1-467); v2 (1-271); v4 (403-467) |
| Smad3 | NM 005902 | v1 (1-425); v2 (1-145); v3 (200-425); v4 (1-229) |
| Smad4 | NM 005359 | v1 (1-552); v2 (1-318); v3 (251-552) |
| Smad5 | U59913 | v1 (1-268) |
| Smad7 | NM 005904 | v1 (251-426) |
| Smad8 | NM 005905 | v1 (1-430); v2 (1-233); v3 (209-430) |
| SARA | AF104304 | v5 (665-1323) |
| Smurf2 | AF301463 | v1 (1-335) |
| SnoN | X15219 | v1 (1-684); v2 (1-370); v3 (290-684) |
| SNIP1 | AK022615 | v1 (1-396); v2 (1-198) |
| Rebound screens | ||
| LAPTm5 | NM 006762 | v1 (1-262); v2 (219-262) |
| PTPN12 | NM 002835 | v1 (69-457); v2 (100-336) |
| PPP1CA | NM 002708 | v1 (11-330); v2 (1-324) |
| HIPK3 | NM 005734 | v1 (667-1152); v2 (643-929) |
| p621 | NM 018179 | v1 (1-709) |
| PKD2 | XM 028271 | v1 (487-643) |
| HYPA | AF049528 | v1 (1-452) |
| LMO4 | NM 006769 | v1 (1-165) |
| RNF11 | NM 014372 | v1 (232-154); v2 (1-154) |
| KIAA1196 | XM 028968 | v1 (486-774); v2 (644-833) |
| FLJ20037 | NM 017633 | v1 (1-447); v2 (27-252) |
| ZNF8 | XM 034263 | v1 (245-436) |
Twenty-three different proteins were used as bait fragments. The GenBank accession number of each protein is given. These proteins were divided into several fragments (v1, v2, v3, v4) to generate 44 different bait clones. The amino acid positions of each fragment are given in parentheses.
Our two-hybrid screening approach identified 755 interactions, corresponding to 591 different proteins potentially connected to the Smad pathway. Of these interactions, 27 have previously been shown to be involved in Smad signaling (ten Dijke et al. 2000; Attisano and Wrana 2002). For example, Figure 2 represents the partial protein interaction map (PIM) around major members of the TGFβ pathway. Analysis of the PIM revealed an intricate network of protein–protein interactions in this pathway. Previously characterized direct interactions between R-Smads were clearly visible on the PIM. R-Smads were also found to interact with previously described partners such as Smurf1, Smurf2, SARA, SnoN, Ski, SMIF, and SIP1. Of the 99 interactions presented in this figure, 15 had been demonstrated previously. Interestingly, we identified many new interactors in the pathway such as kinases, phosphatases, transcription factors, ubiquitin ligases, and proteins of unknown function, which could be crucial for TGFβ responses (Fig. 2; PIMRider on the Web site).
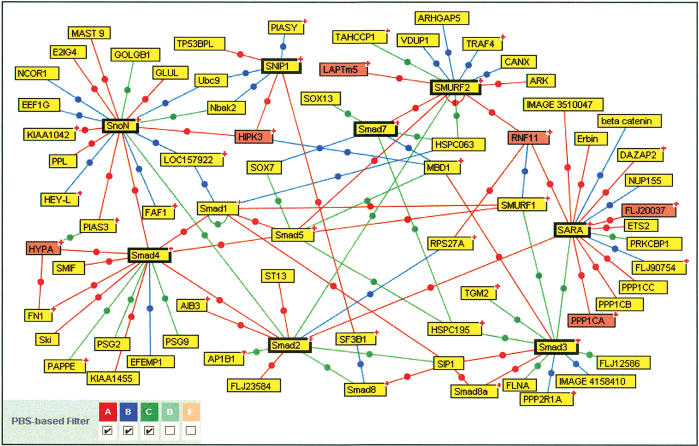
Figure 2.
Protein interaction map around some selected baits in the TGFβ pathway. PBS D and E interactions as well as protein interactions corresponding to the BMP pathway have been excluded from this map for clarity (however, all data are available on the PIMRider visualization platform at http://pim.hybrigenics.com). The following bait proteins were selected to generate a TGFβ network: Smad2, 3, 4, 7, SARA, Smurf2, SnoN, and SNIP1. These are represented in boxes with heavy outlining. Of the 14 proteins selected for further functional validation, the six proteins present in this figure are represented in red. The (+) symbol located in the upper right corner of some boxes indicates that additional partners are not represented in this figure but are visible on the Web site mentioned above.
Functional Validation in Mammalian Cells
To build an integrated functional proteomics platform, we combined the protein–protein interaction mapping with the systematic use of functional validation assays in mammalian cells. This “functional interaction assay” in mammalian cells allowed us to confirm the involvement of new proteins in the Smad pathway, suggested by our results of protein–protein interactions. Different interference technologies and read-out assays were thus developed to assign functions to as many proteins as possible. A set of 14 proteins was selected on the basis of interaction scores and attractive functional domain annotations or protein novelty. Rebound screens were performed with most of these 14 proteins, using the prey as bait to explore the protein network around these candidates and to confirm the interaction (Table 2). We then used the human HepG2 cell line, which has functional TGFβ and BMP signaling pathways, to test the involvement of these 14 proteins in the Smad pathway (Jonk et al. 1998). The endogenous mRNA level of each candidate was first monitored for TGFβ or BMP induction using Quantitative PCR (Q-PCR) in HepG2 cells. Only one gene was transcriptionally regulated by TGFβ (LAPTm5; see below). In addition, we used the TGFβ-RE and BMP-RE luciferase reporters, which contain elements that respond transcriptionally to TGFβ and BMP-7, respectively, to investigate the effect of the 14 selected proteins on the regulation of the transcriptional responses induced by TGFβ and BMP7 (Zawel et al. 1998; Kusanagi et al. 2000). We used these reporter constructs in transient transfection experiments to provide a read-out to allow us to monitor the effect of protein overproduction and siRNA on the Smad pathway (Table 2). The overproduction of two of the selected proteins (LAPTm5 and PPP1CA) had a significant effect on the TGFβ-responsive promoter. We also used TGFβ-RE and BMP-RE in cellular knock-down experiments using chemically synthesized specific siRNA duplexes targeting each protein. The efficiency and specificity of each siRNA duplex on the levels of the endogenous and control mRNAs were systematically tested (Supplemental Table 1). Knock-down of seven proteins (LAPTm5, PPP1CA, MAN1, HYPA, LMO4, KIAA1196, and ZNF8) had a significant effect on TGFβ and/or BMP signaling (Table 2). To assess independently the implication of these 14 selected proteins in Smad signaling, we performed similar siRNA experiments followed by Q-PCR analysis of TGFβ- or BMP-regulated endogenous markers. The chosen markers were the plasminogen-activator–inhibitor Type 1 gene (PAI-1) and the alkaline phosphatase gene (AP), which are known to be induced by TGFβ and BMP, respectively (Keeton et al. 1991; Lecanda et al. 1997). Indeed, the siRNA-mediated knock-down of several proteins (LAPTm5, MAN1, HYPA, LMO4, RNF11, KIAA1196 and ZNF8) had a significant effect on the TGFβ- or BMP-regulated Smad signaling (Table 2).
Table 2.
Functional Validation of the 14 Selected Proteins in HepG2 Cells
|
Gene reporter activity
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
mRNA induction assay
|
Protein overproduction
|
siRNA interference
|
Endogenous gene expression
|
|||||||
| Selected proteins | Bait proteins | Rebound screens | TGF β | BMP7 | TGF β | BMP7 | TGF β | BMP7 | TGF β (PAI-1) | BMP7 (AP) |
| LAPTm5 | Smurf2 | + | ▴+++ | - | ▾++ | - | ▴+ | - | ▴++ | - |
| MAPK13 | Smad4 | nd | - | - | - | - | - | - | - | - |
| PTPN12 | Smad5 | - | - | - | - | - | - | - | - | - |
| PPP1CA | SARA | + | - | - | ▴++ | - | ▾+ | - | - | - |
| HIPK3 | SNIP1 | - | - | - | nd | nd | - | - | - | - |
| SnoN | - | |||||||||
| p621 | Smad4 | - | - | - | nd | nd | - | - | - | - |
| PKD2 | Smad1 | - | - | - | - | - | - | - | - | - |
| Smad8 | - | |||||||||
| MAN1 | Smad1 | nd | - | - | - | - | - | ▾+ | - | ▾++ |
| Smad8 | nd | |||||||||
| HYPA | Smad4 | - | - | - | - | - | ▾+ | ▾+ | - | ▾+ |
| LMO4 | Smad8 | - | - | - | - | - | - | ▾+++ | - | ▾+++ |
| RNF11 | Smurf2 | + | - | - | - | - | nd | nd | ▾+++ | - |
| SARA | - | |||||||||
| KIAA1196 | Smad1 | + | - | - | nd | nd | ▾+++ | - | ▾++ | - |
| FLJ20037 | SARA | - | - | - | - | - | - | - | - | - |
| ZNF8 | Smad1 | + | - | - | - | - | - | ▾++ | - | ▾++ |
| Smad5 | - | |||||||||
| Smad8 | - | |||||||||
Proteins used as bait fragments for this selection of prey proteins in yeast two-hybrid screening are indicated (bait proteins). Rebound screening was performed for most of these proteins (nd: not done): a “+” indicates that the selected protein was used in a rebound screen and that interaction was found again. A “-” indicates that the selected protein was used in a rebound screen but that interaction was not found again. The mRNA level of each candidate was monitored for TGFβ or BMP induction using Q-PCR in HepG2 cells. TGFβ and BMP7-responsive luciferase reporters were transfected in HepG2 cells to monitor the effect of the selected proteins' overproduction or siRNA transfection. Reporter assays were normalized using the pRL-TK vector as an internal control. The endogenous expression of the PAI-1 gene or the AP gene was determined in the presence of TGFβ or BMP7 after siRNA transfection, respectively. All Q-PCR results were normalized using GUS as a standard control. Note that reporter or mRNA gene expression was detected above background in each experimental condition. The symbols (▴) and (▾) correspond to up-regulation and down-regulation by the selected protein, respectively. (nd) not done, (-) no significant effect, (+) slight but significant effect (<twofold), (++) a twofold to fourfold effect, and (+++) indicates a strong effect (>fourfold). Triplicates were performed in at least two independent experiments.
Thus, functional validation experiments confirmed that eight of the 14 proteins identified by two-hybrid screening are involved in the Smad pathway. While this work was in progress, four of these proteins (PPP1CA, ZNF8, MAN1, and RNF11) were shown by other groups to be involved in Smad signaling. PPP1CA was shown to bind to SARA and to regulate Decapentaplegic (Dpp) signaling in Drosophila melanogaster (Bennett and Alphey 2002). The mouse ZNF8 protein was shown to interact with Smad1 and to be involved in the Smad pathway (Jiao et al. 2002). The Xenopus MAN1 ortholog, SANE, was shown to interact with Smad1 and to regulate BMP signaling (Raju et al. 2003). Our results also involve these proteins in Smad signaling in a human system and emphasize that components of Smad signaling are strongly conserved in various species, although the precise role of each of them remains to be addressed. Lastly, human RNF11 was shown to restore TGFβ responsiveness, through its interaction with Smurf2 (Subramaniam et al. 2003). We also identified a new interaction between RNF11 and SARA (Fig. 2). Involvement of these four proteins in the Smad pathway was thus confirmed by other groups, thus demonstrating the specificity and the pertinence of our experimental approach.
New Factors Involved in the Smad Pathway
In addition to these four proteins, we identified four other novel proteins that are involved in the Smad pathway: LMO4, HYPA, KIAA1196, and LAPTm5.
LMO4
We identified, by yeast two-hybrid, an interaction between LMO4 and Smad8, a protein involved in the BMP pathway (see PIMRider). This result suggests that they may interact in mammalian cells, but further evidence is needed to confirm this finding. Q-PCR and reporter experiments showed that the LMO4-targeting siRNA duplex specifically repressed BMP-induced AP mRNA levels and BMP-induced reporter activity (Fig. 3A). These findings demonstrate that this protein is involved in the regulation of the BMP pathway.
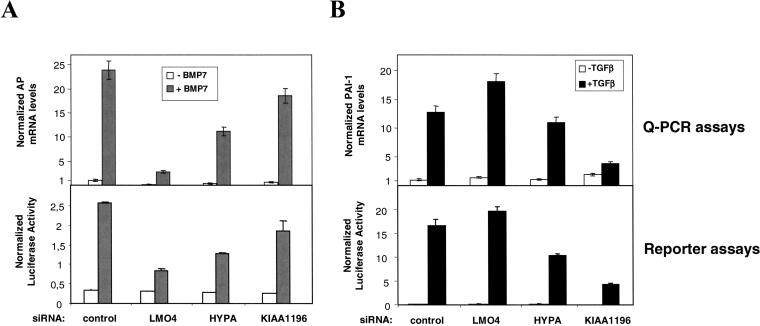
Figure 3.
Involvement of LMO4, HYPA, and KIAA1196 in the Smad pathway. (A) Following siRNA-mediated cellular knock-down targeting LMO4, HYPA, and KIAA1196, Q-PCR and reporter assays were used to analyze endogenous levels of human alkaline phosphatase (AP) mRNA and BMP-induced reporter (BMP-RE) activity in HepG2 cells, respectively. AP mRNA levels and BMP-RE activity were determined in untreated (white) and BMP7-treated (gray) HepG2 cells. (B) Following siRNA-mediated cellular knock-down targeting LMO4, HYPA, and KIAA1196, Q-PCR and reporter assays were used to analyze endogenous levels of PAI-1 mRNA and TGFβ-induced reporter (TGFβ-RE) activity in HepG2 cells, respectively. PAI-1 mRNA levels and TGFβ-RE activity were determined both in untreated (white) and TGFβ-treated (black) HepG2 cells. In all Q-PCR experiments, mRNA levels were normalized according to an internal GUS control. The specific luciferase activity was normalized using the pRL-TK vector. All results are mean values ±SE calculated from triplicates performed in at least two independent experiments.
HYPA
We identified an interaction between HYPA and Smad4, suggesting that they may interact in mammalian cells (see Fig. 2). We also demonstrated that the HYPA-targeting siRNA duplex repressed the induction of luciferase reporter activity by TGFβ and BMP (Fig. 3A,B). This is in agreement with the fact that Smad4 is a common Smad involved in both TGFβ and BMP signaling.
KIAA1196
The hypothetical zinc finger protein KIAA1196 was shown to interact with Smad1, and this finding was confirmed in rebound screen (see PIMRider). Q-PCR and reporter experiments showed that the KIAA1196–targeting siRNA duplex strongly and specifically repressed PAI-1 mRNA levels and reporter activity in a TGFβ-dependent manner (Fig. 3B). Silencing TGFβ signaling by use of KIAA1196 cellular knock-down demonstrated that this protein is an essential regulator of this pathway.
LAPTm5
Finally, we showed that LAPTm5 interacts with Smurf2 (see Fig. 2), an E3-ubiquitin ligase targeting various actors in the Smad pathway for ubiquitin-mediated proteasomal degradation (Izzi and Attisano 2004). The region of LAPTm5 that interacts with Smurf2 corresponds to the 16 C-terminal amino acids of LAPTm5. This domain contains a PPXY motif, responsible for binding with proteins containing a WW motif, like those present in Smurf2 (Fig. 4A). To determine the functional role of LAPTm5 in the Smad pathway, we evaluated the endogenous level of LAPTm5 mRNA in HepG2 cells. Following TGFβ treatment, we observed a 20-fold increase in the amount of LAPTm5 mRNA (Fig. 4B). This induction was TGFβ-specific as it was totally abolished in the presence of a TβRI–targeting siRNA duplex (Fig. 4B) shown to inhibit TGFβ signaling (Fig. 4D). Furthermore, BMP treatment had no effect on LAPTm5 mRNA levels in HepG2 cells (data not shown). We next investigated the effect of LAPTm5 overproduction on TGFβ and BMP-responsive reporters. The overproduction of LAPTm5 resulted in a dose-dependent decrease in TGFβ reporter luciferase activity in HepG2 cells. The overproduction of LAPTm5 had no effect on the BMP reporter or on a control reporter, demonstrating the specificity of LAPTm5 for TGFβ signaling (Fig. 4C). Consistent with our data in the overexpression system, Q-PCR experiments in HepG2 cells showed that LAPTm5–targeting siRNA duplexes enhanced the TGFβ-mediated up-regulation of PAI-1 mRNA (Fig. 4D). Taken together, these results demonstrate that LAPTm5 is a negative regulator of the TGFβ pathway.
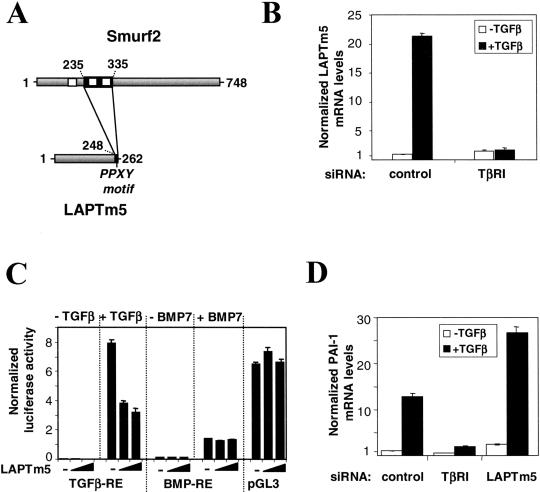
Figure 4.
Involvement of LAPTm5 in the TGFβ pathway. (A) Interaction between Smurf2 and LAPTm5. The full-length proteins are represented in gray; black boxes correspond to the interaction domains. Positions are indicated in amino acids. White boxes correspond to WW motifs in Smurf2. The PPXY motif of LAPTm5 is indicated. (B) Endogenous levels of LAPTm5 mRNA were determined in HepG2 cells by Q-PCR in the absence (white) and the presence (black) of TGFβ for 18 h with or without a TβRI–targeting siRNA duplex. (C) The effect of LAPTm5 overproduction was studied using the following luciferase reporter vectors: a TGFβ-responsive element (TGFβ-RE), a BMP-responsive element (BMP-RE), and an unrelated reporter (pGL3 control). The effect was measured in the presence and the absence of TGFβ or BMP7. HepG2 cells were transfected with 0, 2, or 10 ng of pV3-LAPTm5. The specific luciferase activity was normalized using the pRL-TK vector. (D) Following siRNA-mediated cellular knock-down targeting TβRI and LAPTm5, Q-PCR was used to analyze the endogenous levels of PAI-1 mRNA both in untreated (white) and TGFβ-treated (black) HepG2 cells. mRNA levels were normalized according to an internal GUS control. All results are mean values ±SE calculated from triplicates performed in at least two independent experiments.
DISCUSSION
Here, we describe an integrated study of a human signaling pathway starting from protein interaction map to functional assays in mammalian cells. We identified a network of 755 protein–protein interactions in the Smad pathway. The use of PIMRider, a dedicated navigation tool accessible through our Web site (http://pim.hybrigenics.com), dramatically improved the exploration of such complex interaction databases. Our functional proteomic mapping of this pathway resulted in the biological validation of eight proteins connected to this signaling. In this set of proteins, four were recently shown to be involved in Smad signaling by other groups. We have shown the involvement of four additional proteins in regulation of TGFβ and/or BMP pathways.
LMO4, an Smad8-interacting partner, is down-regulated in prostate cancer and overexpressed in human breast cancer cell lines and in primary invasive breast carcinomas (Visvader et al. 2001). LMO4 also represses BRCA1-mediated transcriptional activation in mammalian cells, suggesting a repression of BRCA1 activity in breast tissue (Sum et al. 2002). An additional role in transcriptional regulation can now be assigned to LMO4 because we demonstrated the involvement of LMO4 in the regulation of the BMP pathway.
HYPA has been shown to interact with huntingtin, a protein implicated in Huntington's disease. This protein, also called FBP11, is the human homolog of the essential pre-mRNA splicing factor PrP40 (Faber et al. 1998). HYPA was shown here to be involved in both TGFβ and BMP signaling, presumably by interacting with Smad4, the Smad protein common to both pathways.
KIAA1196 is an example of the numerous genes present in our interaction database with no known function so far. As KIAA1196 displays seven zinc fingers and a leucine zipper, it may act as a transcription factor. We demonstrated that knocking down this putative transcription factor leads to a strong inhibition of the TGFβ pathway. However, additional experiments are required to explain the specific modulation of TGFβ signaling by this Smad1-interacting protein.
The LAPTm5 protein contains five transmembrane domains and is located in the lysosomal membrane (Adra et al. 1996). LAPTm5 is up-regulated in Sjögren's syndrome, a chronic autoimmune disease (Azuma et al. 2002), and in malignant B lymphomas (Seimiya et al. 2003). LAPTm5 is also coexpressed with activated macrophage genes in rheumatoid arthritis (Walker 2002). GCD-10, the LAPTm5 ortholog in rats, was found to be activated in response to neuronal apoptosis (Origasa et al. 2001). Recently, inactivation of the LAPTm5 gene was observed in human multiple myeloma (Hayami et al. 2003). Although this protein is structurally related to a family of lysosomal transporter proteins that regulate cellular multidrug resistance (Cabrita et al. 1999), its function remained unknown. We identified LAPTm5 as a Smurf2-interacting partner. Smurf2 is an E3 ubiquitin ligase involved in the degradation of TGFβ signaling components, including receptor and Smad proteins (Izzi and Attisano 2004). In addition, rebound screening experiments using LAPTm5 as bait confirmed the interaction between Smurf2 and LAPTm5 and allowed us to identify new LAPTm5 protein partners, including ubiquitin, other E3 ubiquitin ligases, and proteins involved in endocytosis. Many of these partners were also found when the putative E3 ubiquitin ligase, RNF11, was used as bait. These results describe an intimate network of protein interactions between LAPTm5 and proteins involved in protein degradation. It has been suggested that Smurf2 targets TGFβ receptors and Smad7 to lysosomal and proteasomal degradation (Kavsak et al. 2000). Our results suggest that LAPTm5 might be a smurf2 receptor at the lysosomal membrane and might address some TGFβ signaling members to the lysosomal compartment to induce their degradation, resulting in inhibition of the TGFβ pathway. In addition, we show that LAPTm5 is transcriptionally activated by TGFβ, suggesting a potential feedback mechanism. In accordance with this induction by TGFβ, we identified two putative consensus Smad binding elements (Dennler et al. 1998) in the LAPTm5 promoter at positions –1664 and –465 with respect to the ATG start site.
These results give many insights about the involvement of several proteins in the Smad pathway, but the precise role and mechanism of action of each of them remains to be further characterized. For this study, we used functional assays based on Smad transcriptional responses. Other functional assays, addressing different biological mechanisms such as Smad or receptor phosphorylation, protein stability, or proliferation rate, could be used to validate the other proteins identified in our Smad-interaction database and available through the PIMRider. Our results demonstrate that a large-scale protein–protein interaction map combined with general functional validation in mammalian cells is of great help in identifying new components of the Smad signaling. This experimental strategy can be applied to other human transduction signals and could considerably increase our knowledge of many of the numerous recently sequenced genes.
METHODS
Yeast Two-Hybrid Screening
Baits were PCR-amplified (Pfu; Stratagene) and cloned into a plasmid derived from pBTM116 (Vojtek and Hollenberg 1995). The fragments corresponding to the initial baits are described in Table 1. Randomly primed cDNA libraries from human placenta poly(A)+ RNA were constructed in a prey plasmid derived from pGADGH (Bartel 1993). The libraries were used to transform the Y187 yeast strain, and 10 million independent yeast colonies were collected, pooled, and stored at –80°C as equivalent aliquot fractions of the same library. The mating protocol was as described previously (Rain et al. 2001). Each screen was performed to ensure that a minimum of 50 million interactions were tested. The prey fragments of the positive clones were PCR-amplified and sequenced at their 5′ and 3′ junctions on a PE3700 Sequencer. The resulting sequences were used to identify the corresponding gene in the GenBank database (NCBI) using a fully automated procedure.
Plasmid Construction
The pV3 and pV7 expression vectors, prepared from pfastbac1 vectors (Invitrogen), contain a CMV promoter that controls the production of our proteins of interest fused to the FLAG or GST epitope, respectively. All genes cloned in pV3 were obtained by PCR amplification from the placental cDNA library. The MLP minimal promoter from an adenovirus Major Late gene, containing a TATA-box and an initiator element, was cloned into the pGL3 basic vector (Promega) to generate the MLP-Luc plasmid. To construct TGFβ-RE, eight copies of the GTCT-box, a TGFβresponsive sequence (Zawel et al. 1998), were cloned into the MLP-Luc plasmid. To construct BMP-RE, 12 copies of the GC-box, a BMP-responsive sequence (Kusanagi et al. 2000), were cloned into the MLP-Luc plasmid. All these constructs were sequenced.
Luciferase Assays
HepG2 cells were transiently transfected using Fugene 6 (Roche) according to the manufacturer's recommendations. Then, 18 h after TGFβ1 or BMP7 stimulation (10 ng/mL or 50 ng/mL, respectively), luciferase activity was measured using the Dual Luciferase reporter assay kit (Promega) in a luminometer (BMG). Values were normalized with respect to the Renilla luciferase activity expressed from pRL-TK.
Quantitative PCR Assays
HepG2 cells were lysed and RNA was extracted according to the manufacturer's recommendations (QIAGEN). The reverse transcription step was performed in 96-well plates using the TaqMan reverse transcription kit (Applied Biosystems) according to the manufacturer's recommendations. The cDNA was quantified by the SyBR green method using a SyBR Green PCR master Mix kit (Applied Biosystems). Experiments were carried out using an Applied Biosytems 7000 SDS. Oligonucleotides were validated by Q-PCR experiments to obtain a quantitative measurement (quantification of serially diluted cDNA and determination of PCR efficiency). The marker used for TGFβ signaling was the plasminogen activator–inhibitor type 1 gene (PAI-1). The marker used for BMP signaling was the alkaline phosphatase gene (AP). Values were normalized according to the β-glucoronidase (GUS) gene, which was measured as internal control.
siRNA Design and Transfection
siRNAs synthesized chemically using RNA phosphoramidites were purchased from Proligo. All siRNA duplexes were designed as described by Tuschl and colleagues (Elbashir et al. 2001). As a validation experiment, the efficiency of these siRNAs for their targeted mRNAs was tested by Q-PCR. All siRNAs inhibited the amount of their targeted mRNA by between 65% and 95%, and none of them affected significantly another unrelated mRNA (GUS). The specific silencing of the targeted genes was confirmed by at least two paired triplicate experiments (Supplemental Table 1). siRNAs targeting the validated proteins (LMO4, HYPA, and LAPTm5) were also checked for their efficiency and specificity by Western blot after cotransfection of a vector encoding for the GST-protein of interest (500 ng per well) and the corresponding siRNA (40 nM per well) in the HEK293 cell line (Supplemental Fig. 1). The blots were probed with anti-GST (Ref: 27-4577-01; Amersham) and anti-actin (Ref: A2066; Sigma) antibodies. On the four validated proteins, only siRNA targeting KIAA1196 was not tested on a GST-KIAA1196 protein because it targets the N-terminal domain of KIAA1196 (nucleotides 156–174), outside of the cloned prey fragment (nucleotides 1929–2499). Reporter plasmids and siRNAs were cotransfected using Lipofectamine 2000 (Invitrogen) as described by the manufacturer for adherent cell lines using 40 nM siRNA per well. For Q-PCR experiments, 40 nM siRNA duplex per well was transfected using Oligofectamine (Invitrogen).
Bioinformatics
The protein interaction mapping was supported by a dedicated Laboratory Information Management System (LIMS), the PIMBuilder. For every yeast two-hybrid screen, 5′ and 3′ sequences of all positive clones were determined and filtered by use of PHRED (Ewing and Green 1998) and by masking ALU repeats. Sequence contigs were then built using CAP3 (Huang and Madan 1999) and compared with the latest release of GenBank using BLASTN (Altschul et al. 1997). If GenBank entries corresponding to the complete mRNA are found, the best annotated entry, preferentially from the RefSeq division of GenBank, is assigned to every overlapping prey fragment family. The SID (Selected Interacting Domain) is the common part of all fragments in a family. This region contains the domain that interacts with the bait. Each two-hybrid screening experiment generates a list of bait–SID interactions. The PIM (Protein Interaction Map) is then built by pooling all the bait–SID interactions from several screens. The method for calculating PBS, previously described for genomic libraries (Rain et al. 2001), was adapted for randomly primed cDNA libraries. The PBS value represents the probability of a protein–protein interaction being nonspecific. It ranges from 0 to 1, but was grouped in five categories (A, B, C, D, and E) for user convenience. Intercategory thresholds were chosen manually with respect to a training data set containing known true–positive and false–positive interactions (data not shown): A < 1e-1 0 < B < 1e-5 < C < 1e-2.5 < D < 1; the E category corresponds to SIDs nonspecifically selected by baits (Rain et al. 2001) and for which the corresponding PBS was set to 1. A PBS confidence score was assigned to each protein–protein interaction in the map. In addition, proteins connected in the PIM were automatically annotated in terms of structural and functional domains by use of bioinformatics algorithms: TMHMM (Krogh et al. 2001) and SignalP (Nielsen et al. 1997) for the detection of transmembrane helices and signal peptides, respectively, and IpScan for the prediction of InterPro domains (Apweiler et al. 2001). Predicting InterPro domains allows proteins to be clustered into functional categories like those defined in the Gene Ontology (GO) classification (Ashburner et al. 2000). Finally, the annotated PIM can be explored by use of the PIMRider visualization platform, developed in-house, which includes four viewers: the ProteinViewer, which displays textual annotations for a given protein, the list of interacting partners, and the sequence (Supplemental Fig. 2); the PIMViewer, which displays a graphical and dynamic view of protein interaction networks (Fig. 2; Supplemental Fig. 3); the InteractionViewer, which gives access to raw experimental data on prey, bait, and SID sequences (Supplemental Fig. 4); and the DomainViewer, which displays and makes it possible to compare, for one protein and all its partners in the map, domains and motifs extracted from both experimental (bait and SID) and calculated (transmembrane segments, signal peptides, and functional InterPro domains) analyses (Supplemental Fig. 5).
Acknowledgments
The authors thank all the staff of Hybrigenics for their contribution and especially G. Lenzen, E. Vanmerris, G. Conan, and P. Desmoucelle for yeast two-hybrid screens; J. Bianchi, C. Planquette, and L. Esteve for their technical assistance in validation assays; and G. Boissy and V. Collura for their help in bioinformatics developments. We are very grateful to A.D. Strosberg, L. Daviet, J.C. Rain, and A. Atfi for many stimulating discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2334104.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Adra, C.N., Zhu, S., Ko, J.L., Guillemot, J.C., Cuervo, A.M., Kobayashi, H., Horiuchi, T., Lelias, J.M., Rowley, J.D., and Lim, B. 1996. LAPTM5: A novel lysosomal-associated multispanning membrane protein preferentially expressed in hematopoietic cells. Genomics 35: 328–337. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D., et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29: 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano, L. and Wrana, J.L. 2002. Signal transduction by the TGF-β superfamily. Science 296: 1646–1647. [DOI] [PubMed] [Google Scholar]
- Azuma, T., Takei, M., Yoshikawa, T., Nagasugi, Y., Kato, M., Otsuka, M., Shiraiwa, H., Sugano, S., Mitamura, K., Sawada, S., et al. 2002. Identification of candidate genes for Sjogren's syndrome using MRL/lpr mouse model of Sjogren's syndrome and cDNA microarray analysis. Immunol. Lett. 81: 171–176. [DOI] [PubMed] [Google Scholar]
- Baker, J.C. and Harland, R.M. 1996. A novel mesoderm inducer, Madr2, functions in the activin signal transduction pathway. Genes & Dev. 10: 1880–1889. [DOI] [PubMed] [Google Scholar]
- Bartel, P.L. 1993. Using the two-hybrid system to detect protein–protein interactions. In Cellular interactions in development: A practical approach (ed. D.A. Hartley), pp. 153–179. Oxford University Press, Oxford, UK.
- Bartel, P.L., Roecklein, J.A., SenGupta, D., and Fields, S. 1996. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12: 72–77. [DOI] [PubMed] [Google Scholar]
- Bennett, D. and Alphey, L. 2002. PP1 binds Sara and negatively regulates Dpp signaling in Drosophila melanogaster. Nat. Genet. 31: 419–423. [DOI] [PubMed] [Google Scholar]
- Boulton, S.J., Gartner, A., Reboul, J., Vaglio, P., Dyson, N., Hill, D.E., and Vidal, M. 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295: 127–131. [DOI] [PubMed] [Google Scholar]
- Bouwmeester, T., Bauch, A., Ruffner, H., Angrand, P.O., Bergamini, G., Croughton, K., Cruciat, C., Eberhard, D., Gagneur, J., Ghidelli, S., et al. 2004. A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat. Cell Biol. 6: 97–105. [DOI] [PubMed] [Google Scholar]
- Cabrita, M.A., Hobman, T.C., Hogue, D.L., King, K.M., and Cass, C.E. 1999. Mouse transporter protein, a membrane protein that regulates cellular multidrug resistance, is localized to lysosomes. Cancer Res. 59: 4890–4897. [PubMed] [Google Scholar]
- Dennler, S., Itoh, S., Vivien, D., ten Dijke, P., Huet, S., and Gauthier, J.M. 1998. Direct binding of Smad3 and Smad4 to critical TGF β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17: 3091–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Faber, P.W., Barnes, G.T., Srinidhi, J., Chen, J., Gusella, J.F., and MacDonald, M.E. 1998. Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet. 7: 1463–1474. [DOI] [PubMed] [Google Scholar]
- Finley Jr., R.L. and Brent, R. 1994. Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc. Natl. Acad. Sci. 91: 12980–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajolet, M., Rotondo, G., Daviet, L., Bergametti, F., Inchauspe, G., Tiollais, P., Transy, C., and Legrain, P. 2000. A genomic approach of the hepatitis C virus generates a protein interaction map. Gene 242: 369–379. [DOI] [PubMed] [Google Scholar]
- Giot, L., Bader, J.S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y.L., Ooi, C.E., Godwin, B., Vitols, E., et al. 2003. A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736. [DOI] [PubMed] [Google Scholar]
- Hayami, Y., Iida, S., Nakazawa, N., Hanamura, I., Kato, M., Komatsu, H., Miura, I., Dave, B.J., Sanger, W.G., Lim, B., et al. 2003. Inactivation of the E3/LAPTm5 gene by chromosomal rearrangement and DNA methylation in human multiple myeloma. Leukemia 17: 1650–1657. [DOI] [PubMed] [Google Scholar]
- Hoodless, P.A., Haerry, T., Abdollah, S., Stapleton, M., O'Connor, M.B., Attisano, L., and Wrana, J.L. 1996. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell 85: 489–500. [DOI] [PubMed] [Google Scholar]
- Huang, X. and Madan, A. 1999. CAP3: A DNA sequence assembly program. Genome Res. 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 98: 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzi, L. and Attisano, L. 2004. Regulation of the TGFβ signalling pathway by ubiquitin-mediated degradation. Oncogene 23: 2071–2078. [DOI] [PubMed] [Google Scholar]
- Jiao, K., Zhou, Y., and Hogan, B.L. 2002. Identification of mZnf8, a mouse kruppel-like transcriptional repressor, as a novel nuclear interaction partner of Smad1. Mol. Cell. Biol. 22: 7633–7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonk, L.J., Itoh, S., Heldin, C.H., ten Dijke, P., and Kruijer, W. 1998. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 273: 21145–21152. [DOI] [PubMed] [Google Scholar]
- Kavsak, P., Rasmussen, R.K., Causing, C.G., Bonni, S., Zhu, H., Thomsen, G.H., and Wrana, J.L. 2000. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF β receptor for degradation. Mol. Cell 6: 1365–1375. [DOI] [PubMed] [Google Scholar]
- Keeton, M.R., Curriden, S.A., van Zonneveld, A.J., and Loskutoff, D.J. 1991. Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor β. J. Biol. Chem. 266: 23048–23052. [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Kusanagi, K., Inoue, H., Ishidou, Y., Mishima, H.K., Kawabata, M., and Miyazono, K. 2000. Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol. Biol. Cell 11: 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagna, G., Hata, A., Hemmati-Brivanlou, A., and Massague, J. 1996. Partnership between DPC4 and SMAD proteins in TGF-β signalling pathways. Nature 383: 832–836. [DOI] [PubMed] [Google Scholar]
- Lecanda, F., Avioli, L.V., and Cheng, S.L. 1997. Regulation of bone matrix protein expression and induction of differentiation of human osteoblasts and human bone marrow stromal cells by bone morphogenetic protein-2. J. Cell Biochem. 67: 386–396. [PubMed] [Google Scholar]
- Liu, F., Hata, A., Baker, J.C., Doody, J., Carcamo, J., Harland, R.M., and Massague, J. 1996. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature 381: 620–623. [DOI] [PubMed] [Google Scholar]
- Macias-Silva, M., Abdollah, S., Hoodless, P.A., Pirone, R., Attisano, L., and Wrana, J.L. 1996. MADR2 is a substrate of the TGFβ receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell 87: 1215–1224. [DOI] [PubMed] [Google Scholar]
- Massague, J. 1998. TGF-β signal transduction. Annu. Rev. Biochem. 67: 753–791. [DOI] [PubMed] [Google Scholar]
- McCraith, S., Holtzman, T., Moss, B., and Fields, S. 2000. Genome-wide analysis of vaccinia virus protein–protein interactions. Proc. Natl. Acad. Sci. 97: 4879–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural. Syst. 8: 581–599. [DOI] [PubMed] [Google Scholar]
- Origasa, M., Tanaka, S., Suzuki, K., Tone, S., Lim, B., and Koike, T. 2001. Activation of a novel microglial gene encoding a lysosomal membrane protein in response to neuronal apoptosis. Brain Res. Mol. Brain Res. 88: 1–13. [DOI] [PubMed] [Google Scholar]
- Rain, J.C., Selig, L., De Reuse, H., Battaglia, V., Reverdy, C., Simon, S., Lenzen, G., Petel, F., Wojcik, J., Schachter, V., et al. 2001. The protein–protein interaction map of Helicobacter pylori. Nature 409: 211–215. [DOI] [PubMed] [Google Scholar]
- Raju, G.P., Dimova, N., Klein, P.S., and Huang, H.C. 2003. SANE, a novel LEM domain protein, regulates bone morphogenetic protein signaling through interaction with Smad1. J. Biol. Chem. 278: 428–437. [DOI] [PubMed] [Google Scholar]
- Seimiya, M., O-Wang, J., Bahar, R., Kawamura, K., Wang, Y., Saisho, H., and Tagawa, M. 2003. Stage-specific expression of Clast6/E3/LAPTM5 during B cell differentiation: elevated expression in human B lymphomas. Int. J. Oncol. 22: 301–304. [PubMed] [Google Scholar]
- Subramaniam, V., Li, H., Wong, M., Kitching, R., Attisano, L., Wrana, J., Zubovits, J., Burger, A.M., and Seth, A. 2003. The RING-H2 protein RNF11 is overexpressed in breast cancer and is a target of Smurf2 E3 ligase. Br. J. Cancer 89: 1538–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sum, E.Y., Peng, B., Yu, X., Chen, J., Byrne, J., Lindeman, G.J., and Visvader, J.E. 2002. The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J. Biol. Chem. 277: 7849–7856. [DOI] [PubMed] [Google Scholar]
- ten Dijke, P., Miyazono, K., and Heldin, C.H. 2000. Signaling inputs converge on nuclear effectors in TGF-β signaling. Trends Biochem. Sci. 25: 64–70. [DOI] [PubMed] [Google Scholar]
- Tewari, M., Hu, P.J., Ahn, J.S., Ayivi-Guedehoussou, N., Vidalain, P.O., Li, S., Milstein, S., Armstrong, C.M., Boxem, M., Butler, M.D., et al. 2004. Systematic interactome mapping and genetic perturbation analysis of a C. elegans TGF-β signaling network. Mol. Cell 13: 469–482. [DOI] [PubMed] [Google Scholar]
- Tsukazaki, T., Chiang, T.A., Davison, A.F., Attisano, L., and Wrana, J.L. 1998. SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell 95: 779–791. [DOI] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. 2000. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Visvader, J.E., Venter, D., Hahm, K., Santamaria, M., Sum, E.Y., O'Reilly, L., White, D., Williams, R., Armes, J., and Lindeman, G.J. 2001. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc. Natl. Acad. Sci. 98: 14452–14457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek, A.B. and Hollenberg, S.M. 1995. Ras–Raf interaction: Two-hybrid analysis. Methods Enzymol. 255: 331–342. [DOI] [PubMed] [Google Scholar]
- Walker, M.G. 2002. Z39Ig is co-expressed with activated macrophage genes. Biochim. Biophys. Acta 1574: 387–390. [DOI] [PubMed] [Google Scholar]
- Zawel, L., Dai, J.L., Buckhaults, P., Zhou, S., Kinzler, K.W., Vogelstein, B., and Kern, S.E. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell 1: 611–617. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Feng, X., We, R., and Derynck, R. 1996. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature 383: 168–172. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://pim.hybrigenics.com; PIMRider.