Abstract
The difficulty of fine localizing the polymorphisms responsible for genotype-phenotype correlations is emerging as an important constraint in the implementation and interpretation of genetic association studies, and calls for the definition of protocols for the follow-up of associated variants. One recent example is the 3435C>T polymorphism in the multidrug transporter gene ABCB1, associated with protein expression and activity, and with several clinical conditions. Available data suggest that 3435C>T may not directly cause altered transport activity, but may be associated with one or more causal variants in the poorly characterized stretch of linkage disequilibrium (LD) surrounding it. Here we describe a strategy for the follow-up of reported associations, including a Bayesian formalization of the associated interval concept previously described by Goldstein. We focus on the region of high LD around 3435C>T to compile an exhaustive list of variants by (1) using a relatively coarse set of marker typings to assess the pattern of LD, and (2) resequencing derived and ancestral chromosomes at 3435C>T through the associated interval. We identified three intronic sites that are strongly associated with the 3435C>T polymorphism. One of them is associated with multidrug resistance in patients with epilepsy (χ2 = 3.78, P = 0.052), and sits within a stretch of significant evolutionary conservation. We argue that these variants represent additional candidates for influencing multidrug resistance due to P-glycoprotein activity, with the IVS 26+80 T>C being the best candidate among the three intronic sites. Finally, we describe a set of six haplotype tagging single-nucleotide polymorphisms that represent common ABCB1 variation surrounding 3435C>T in Europeans.
Comparatively few of the large number of genetic associations now reported have resulted in the identification of the polymorphisms that influence the phenotypes. In many cases of course this is because the association is a false positive and there is no causal polymorphism to be found (Lohmueller et al. 2003). Even when the associations are real, however, the association is often observed at noncausal polymorphisms that are in linkage disequilibrium (LD) with the polymorphisms directly responsible for the phenotype. Once the genomic region associated with the trait of interest has been identified based on a set of relatively coarse markers, the final identification of the causal variant is a considerable task. Relatively little work has been done on how to optimize the genetic analyses required for this identification. Here we introduce methods for fine localization, including a formalization of the concept of an associated interval (Goldstein 2003), and apply these methods to a well studied polymorphism in the ABCB1 gene.
The ABCB1 gene, also known as Multi-Drug Resistance 1 (MDR1), is a large gene (209 Kb) encoding the membrane-bound ATP-dependent pump P-glycoprotein (PGP). PGP is active at the intestinal, placental, and blood-brain barriers, and in renal and hepatic tissue (Thiebaut et al. 1987; Cordon-Cardo et al. 1989; Schinkel 2001), and mediates the efflux of a wide range of different substrates, including over 50 commonly used drugs (immunosuppressants, Lown et al. 1997; cardiac glycosides, de Lannoy and Silverman 1992; Ito et al. 1992; HIV-1 protease inhibitors, Kim et al. 1998; Polli et al. 1999; β-blockers, Spahn-Langguth et al. 1998; and many others, Bellamy 1996; Fromm 2002; Gottesman et al. 2002; Wandel et al. 2002). PGP overexpression plays an important role in the development of multi-drug resistance in cancer cells (Gottesman et al. 2002).
A silent C to T transition in exon 26 of ABCB1 (3435C>T) has been associated with differences in PGP levels and activity in Europeans (CC>CT>TT, P = 0.056 and P = 0.053, respectively, Hoffmeyer et al. 2000). The polymorphism has also been associated with PGP function, for example as reflected in digoxin uptake (Hoffmeyer et al. 2000) or rhodamine efflux (Hitzl et al. 2001), and with clinical conditions such as drug-resistant epilepsy (Siddiqui et al. 2003), susceptibility to ulcerative colitis (Schwab et al. 2003), and immune recovery after initiation of antiretroviral treatment (Fellay et al. 2002).
Although the silent 3435C>T polymorphism could affect PGP levels and activity through, for example, effects on mRNA stability or codon preference, four main lines of evidence suggest the possibility that the 3545C>T may not be causal, but rather a marker for one or more yet unidentified causal variants. (1) Site-specific mutagenesis experiments have demonstrated that the 3435C>T polymorphism (and the 2677G>A,T nonsynonymous substitution in exon 21) has no effect on PGP transport activity in vitro (Morita et al. 2003). (2) Genotypes at 3435C>T were not associated with an alternatively spliced form of PGP (Oselin et al. 2003a), suggesting that 3435C>T does not act as a cryptic splice site. (3) Despite a wealth of clinical studies investigating the effect of 3435C>T on ABCB1 expression and function in different populations, the correlation between 3435C>T and PGP levels and activity does not appear completely consistent, both across and within ethnic groups (Fromm 2002; Nakamura et al. 2002; Oselin et al 2003b; Sakaeda et al. 2003). (4) In some studies, two-locus haplotypes have shown a stronger correlation with activity (inferred from digoxin uptake) than the 3435C>T polymorphism alone (Johne et al. 2002). Moreover, the 3435C>T polymorphism sits within an extended track of LD, suggesting that the causal variant could be anywhere within this stretch of sequence (Tang et al. 2002; Siddiqui et al. 2003), most of which is poorly characterized for variation.
To characterize a set of candidate causal variants, we use a Bayesian approach to formalize the idea of an “associated interval,” which is the stretch of sequence surrounding the associated polymorphism having sufficiently high LD that the causal variant should reside within it (Goldstein 2003). The intention is that outside of the associated interval, variants will not have sufficiently high correlation with the 3435C>T variant to drive an association between this site and the phenotype. The approach applies to the common situation in LD mapping in which LD data are available for a coarse set of markers in control individuals, including the variant that has been associated with the phenotype. The approach provides statistical guidance about the boundaries of the associated interval on the basis of a comparison of the association between the associated variant and the phenotype on one hand, and between the markers and the associated variant on the other hand. In order not to have an unreasonably conservative definition of the interval, it is also necessary to place a priori belief on the relative risk of the causal variant.
Once boundaries of the associated interval have been assessed in this or another way, with sufficient phenotypic data to constrain them, all polymorphic loci within the associated interval should be identified, and typed in the phenotyped material. The problem then becomes one of distinguishing the polymorphisms based on the extent of association with the phenotype. Here we used the association of 3435C>T with both clinical phenotype (e.g., Siddiqui et al. 2003) and with intermediate phenotypes (Hoffmeyer et al. 2000) to assess the associated interval.
Using the formal definition of an associated interval, we find that the interval is not well defined within the region, and that by using the Bayesian approach virtually all of the single-nucleotide polymorphisms (SNPs) in the gene could be considered candidates for being causal. We expect that the poorly defined interval results from the very modest association with the clinical phenotype, and that in the case of ABCB1 it will be appropriate to refine the interval further by more accurate assessment of the association between 3435C>T and intermediate phenotypes (such as uptake of a substrate such as digoxin, or mRNA or protein levels in appropriate tissues). We have therefore concentrated our discovery efforts on a portion of the associated interval having particularly high levels of LD with 3435C>T. We then sought to identify all variants in this region that are tightly associated with 3435C>T by noting that the highest levels of association would be for SNPs that have arisen nearly simultaneously in the genealogy of the surrounding portion of the ABCB1 gene, which is required for very high levels of association (Slatkin 1994). Variants due to mutations occurring in the same part of the genealogy as the 3435C>T polymorphism can be identified by resequencing representative chromosomes that are, and are not, derived at the 3435C>T polymorphism (defined as the mutant state compared to the homologous primate sequence). Using this approach we identified and characterized three intronic polymorphisms that could be involved in regulating the expression of ABCB1. One of them was significantly associated with resistance to antiepileptic drugs and sits within an evolutionarily conserved genomic region. On current evidence, these loci are as good candidates as 3435C>T in explaining multidrug resistance in epilepsy due to ABCB1.
RESULTS AND DISCUSSION
Assessing the High-LD Interval
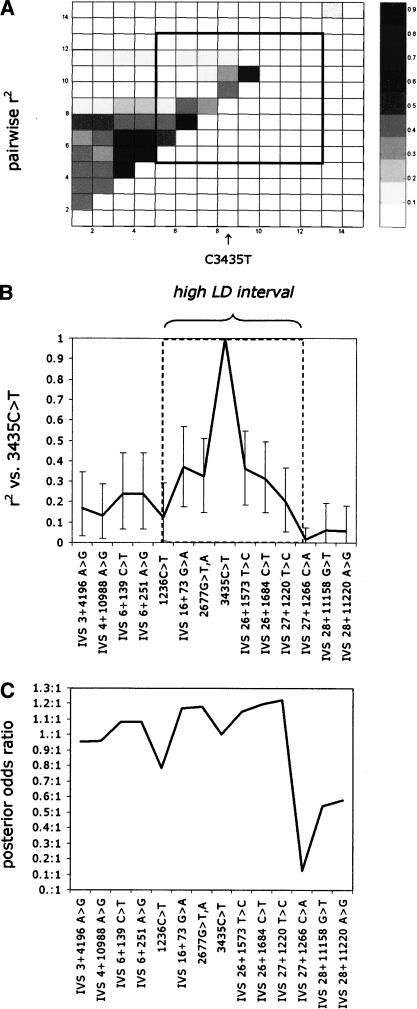
Resequencing of 12 amplicons distributed along the length of the ABCB1 gene and corresponding to a total of 4.1 kb identified 17 SNP loci in 24 CEPH trios (Table 1). Three loci had a low minor allele frequency (<6%) and were therefore excluded from further analysis, because it is unlikely that these low-frequency variants could be responsible for the observed association between PGP activity and the common 3435C>T polymorphism. We used the 14 SNPs with high minor allele frequency to assess the LD pattern throughout the gene. In our sample, we detected significant evidence of LD (Fisher's exact test is significant at the 0.05 level) between intron 3 and intron 27 (Fig. 1). We do not know how far LD extends upstream of intron 3, as the polymorphism at the 5′ end of the gene was too low in frequency for reliable LD inference. r2 values drop in the region between the two polymorphisms in intron 27, about 4.6 kb downstream of the 3435C>T site (Fig. 1, upper panels).
Table 1.
SNPs Used for LD Analysis of ABCB1 in 24 CEPH Trios
| SNP ID | Positiona | dbSNP ID | Ancestral alleles | Locationb | A1c | A2,3c | f(1)d | f(2)d | f(3)d | f(1/1)e | f(1/2)e | f(2/2)e | f(2/3)e | χ2f | 6Pf | r2vs. 3435C>Tg |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 12504716 | rs2157926 | A | 5′ UTR | A | T | 1 | 0 | 1 | 0 | 0 | — | — | — | ||
| 2 | 12455102 | rs3789243 | — | IVS 3+4196 A>G | A | G | 0.489 | 0.511 | 0.250 | 0.477 | 0.273 | 0.089 | 0.766 | 0.17 | ||
| 3 | 12438056 | rs1202180 | A | IVS 4+10988 A>G | A | G | 0.689 | 0.311 | 0.489 | 0.400 | 0.111 | 0.201 | 0.654 | 0.13 | ||
| 4 | 12430178 | rs1202168 | C | IVS 6+139 C>T | C | T | 0.536 | 0.464 | 0.214 | 0.643 | 0.143 | 3.589 | 0.058 | 0.24 | ||
| 5 | 12430066 | rs1202169 | A | IVS 6+251 A>G | A | G | 0.536 | 0.464 | 0.214 | 0.643 | 0.143 | 3.589 | 0.058 | 0.24 | ||
| 6 | 12413817 | rs1128503 | C | 1236C>T | C | T | 0.544 | 0.457 | 0.239 | 0.609 | 0.152 | 2.363 | 0.124 | 0.13 | ||
| 7 | 12413659 | rs2032588 | C | IVS 12+44 C>T | C | T | 0.940 | 0.060 | 0.900 | 0.080 | 0.020 | 4.228 | 0.040 | — | ||
| 8 | 12408282 | rs2235046 | G | IVS 16+73 G>A | G | A | 0.467 | 0.533 | 0.152 | 0.630 | 0.217 | 3.261 | 0.071 | 0.37 | ||
| 9 | 12402243 | rs3789246 | C | IVS 19+557 C>T | C | T | 0.958 | 0.042 | 0.917 | 0.083 | 0.000 | 0.091 | 0.763 | — | ||
| 10 | 12394834 | rs2032582 | G | 2677G>T,A | G | T,A | 0.472 | 0.493 | 0.0347 | 0.208 | 0.528 | 0.194 | 0.0694 | 0.804 | 0.370 | 0.32 |
| 11 | 12372861 | rs1045642 | C | 3435C>T | C | T | 0.396 | 0.604 | 0.104 | 0.583 | 0.313 | 2.315 | 0.128 | — | ||
| 12 | 12371234 | rs1882478 | T | IVS 26+1573 T>C | C | T | 0.750 | 0.250 | 0.546 | 0.409 | 0.046 | 0.364 | 0.547 | 0.36 | ||
| 13 | 12371123 | — | C | IVS 26+1684 C>T | C | T | 0.773 | 0.227 | 0.591 | 0.364 | 0.046 | 0.055 | 0.815 | 0.31 | ||
| 14 | 12368209 | rs1186746 | T | IVS 27+1220 T>C | C | T | 0.192 | 0.809 | 0.043 | 0.298 | 0.660 | 0.068 | 0.794 | 0.20 | ||
| 15 | 12368163 | rs1186745 | C | IVS 27+1266 C>A | A | C | 0.117 | 0.883 | 0.000 | 0.234 | 0.766 | 0.826 | 0.364 | 0.02 | ||
| 16 | 12356238 | rs2178658 | G | IVS 28+11158 G>T | G | T | 0.702 | 0.298 | 0.447 | 0.511 | 0.043 | 2.291 | 0.130 | 0.06 | ||
| 17 | 12356176 | — | A | IVS 28+11220 A>G | A | G | 0.198 | 0.802 | 0.042 | 0.313 | 0.646 | 0.012 | 0.913 | 0.06 |
SNP position within the reference contig (GenBank acc. no. NT_007933). bDerived from sequencing in chimp, calternative alleles of one locus, dallelic frequencies in unrelated CEPH individuals, egenotypic frequencies, fχ2 and P-values for test of fit to Hardy-Weinberg equilibrium, gpairwise LD values against 3435C>T.
Figure 1.
(A) Graph of pairwise r2 measures of LD among the 14 high-frequency (>6%) ABCB1 SNP loci described in Table 1. (B) r2 for 3435C>T against each of the remainder SNPs, together with Bayesian 95% credible intervals calculated by the method described in the Supplemental materials. The dashed box identifies the high-LD interval around 3435C>T. All sites upstream of IVS 27+1220 show association with 3435C>T, significant at the 0.001 level (Fisher's exact test). At the loci downstream of this site, we observe no significant association (P > 0.05) with 3435C>T. Throughout the study we used r2 to measure association, to take into account the dependency of LD on allelic frequencies. (C) Graph of posterior odds of each SNP being causal relative to 3435C>T being causal, based on the combined LD data and case control data on 3435C>T (Siddiqui et al. 2003), using the method described in the Supplemental materials.
We used a Bayesian method (described in Methods and in Supplemental material) to assess the support for each SNP in turn being causal relative to the alternative model that 3435C>T was causal. We found that the boundaries of the associated interval (Fig. 1) were not adequately described when the Bayesian method was applied to the LD data (from the 24 CEPH trios) combined with clinical case-control data for 3435C>T on anti-epileptic drug response (Siddiqui et al. 2003). We also found that data on intermediate phenotype association for 3435C>T did not delimit the associated interval either (data not shown). Further investigations using simulated data suggest that the association with clinical phenotype is too weak to constrain the interval well, whereas available data on intermediate phenotype suffers from measurement error and low sample sizes. Although the formal assessment of the associated interval does not appear to provide useful guidance in this case for prioritizing which part of the gene should be further studied, it does provide a framework for making judgement about the cost effectiveness of fine localization strategies. For example, in this case, it appears that a larger and more accurate assessment of the intermediate phenotype would provide more discrimination among markers throughout the gene.
For this reason, we have decided to concentrate here only on a region of particularly high LD within the overall associated interval, on the assumption that this core region would be included even in a more tightly defined associated interval following, for example, a larger and more accurate assessment of the association between 3435C>T and the intermediate phenotype. Our approach here therefore cannot be viewed as completely exhaustive, but rather is an appropriate first step giving the costs of exhaustive resequencing in genes as large as ABCB1. This high LD interval is defined in Figure 1.
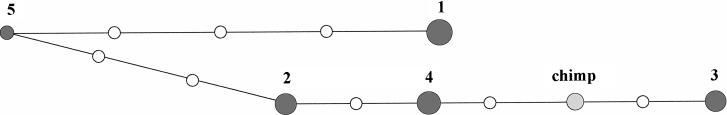
In the region between IVS 6+139 and IVS 26+1684 we identify four major haplotypes in Europeans (Table 2). In the two most frequent haplotypes (35%, haplotype 1; 15%, haplotype 4), the derived T and ancestral C variants at 3435C>T are associated with haplotypes corresponding respectively to the derived and ancestral state at the loci upstream of 3435C>T. The haplotype network for this region of the ABCB1 gene suggests a deep split in the gene genealogy in Europeans (Fig. 2). We selected two individuals for sequencing based on the following two criteria: (1) they were, respectively, homozygous for the C and T alleles at 3435C>T, and (2) their haplotypes were homozygous and non-recombinant at all loci in the high-LD interval. This directed genotyping strategy will preferentially identify SNPs with high r2 values with 3435C>T.
Table 2.
ABCB1 Haplotypes for High-Frequency Loci Between IVS 6+139 and IVS 26+1684
| Id | Haplotypea | Freqb | Nb |
|---|---|---|---|
| 1 | TGTTTTCC | 0.35 | 34 |
| 2 | CACCGTCC | 0.11 | 11 |
| 3 | CACCGCTT | 0.12 | 12 |
| 4 | CACCGCCC | 0.15 | 14 |
| 5 | CACTTTCC | 0.05 | 5 |
| 6 | TGTTTCTT | 0.03 | 3 |
| 7 | TGCTTTCC | 0.03 | 3 |
| 8 | TGTTGCTT | 0.03 | 3 |
| Ancestral | CACCGCTC |
Derived by EM inference from CEPH trio data on the following eight loci (details in Table 1): IVS 6+139 C>T, IVS 6+251 A>G, 1236C>T, IVS 16+73 G>A, 2677G>T,A, 3435C>T, IVS 26+1573 T>C, IVS 26+1684 C>T.
Haplotype numbers and frequencies are relative to parental chromosomes only. The 3435C>T variant is bold underlined.
Figure 2.
Reduced-median haplotype network of ABCB1 haplotypes found with frequencies >5% in the set of 24 CEPH trios, as described in Table 2. Loci include IVS 6+139C>T, IVS 6+251A>G, 1236C>T, IVS 16+73G>A, 2677G>T,A, 3435C>T, IVS 26+1573T>C, and IVS 26+1684 of Table 1. Node size is proportional to haplotype frequency. Mutated positions are indicated by white dots. The ancestral haplotype was determined by resequencing of a chimpanzee sample (gray).
Screening for Candidate Polymorphic Sites Within the High-LD Interval
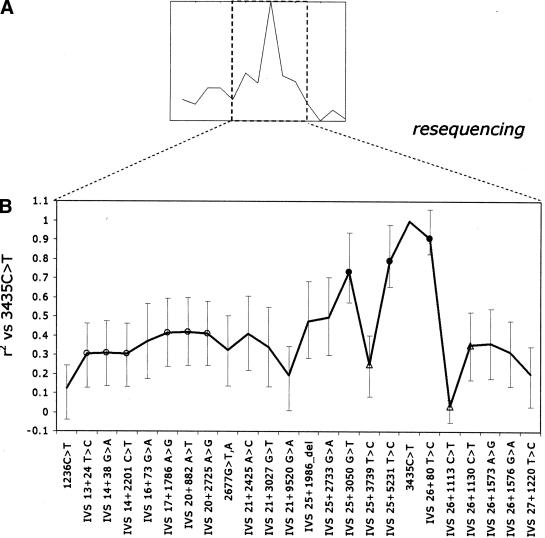
In these two individuals (CEPH ID 1420-02 and 1333-01, Table 3), the 1236C>T–IVS 27+1266 interval was sequenced entirely except for gaps corresponding to less than 3 kb, where the presence of low-complexity regions prevented sequencing. A total of 53 additional polymorphisms were found in the 42 kb that was resequenced, 32 of which were previously unknown (Table 3). Fourteen of these were homozygous (and different) in the two individuals, and 39 were heterozygous in at least one of them. In general, these are all potential candidate causal sites. However, for reasons given above we concentrated on homozygous sites, as they are more likely to have high or complete LD with the 3435C>T polymorphism. To assess these new polymorphisms, we calculated LD with 3435C>T by resequencing CEPH trios at each of the 14 polymorphisms that showed homozygous differences. Typing of the new homozygous variants revealed that, within the high-LD interval, a smaller region downstream of the IVS 25+3050 G>T site had the highest r2 values. Therefore, within this region of elevated LD, we also calculated LD with 3435C>T for all the sites that were heterozygous in at least one of the sequenced individuals.
Table 3.
List of Polymorphic Sites Identified by Representing CEPH 1420-02 and 1333-01 in the High LD Interval Surrounding 3435C>T
| Namea | Location | dbSNPIDb | Position (bp)c | CEPH 142002 | CEPH 133301 | Target sequence | MAF in CEPHse | r2 vs 3435C>Tf |
|---|---|---|---|---|---|---|---|---|
| 1236C>T | Exon 12 | rs1128503 | 12413817 | CC | TT | TCTTGAAGGGyCTGAACCTGA | 0.47 | 0.13 |
| IVS 13+24 T>C | Intron 13 | rs2235033 | 12413359 | CC | TT | GCCCTTTGCCyTTCTAGAGGT | 0.45 | 0.30 |
| IVS 13+81 C>T | Intron 13 | rs2235035 | 12413302 | CT | CC | TAGGAAACTAyTATAAATCGG | ||
| IVS 14+38 G>A | Intron 14 | rs2235013 | 12412842 | AA | GG | TGATTTATAArCATAAGAACA | 0.45 | 0.31 |
| IVS 14+2201 C>T | Intron 14 | ss20398881 | 12410679 | CC | TT | TTGCTAGTTAyTAGAGAGACA | 0.44 | 0.30 |
| IVS 14+3038 G>A | Intron 14 | ss20398882 | 12409842 | AG | GG | CACCACGCCCrGCCCAAGCCC | ||
| IVS 15+675 G>A | Intron 15 | rs2091766 | 12408720 | AG | GG | GGTGGTTTTCrTTTTCAAATA | ||
| IVS 16+73 G>A | Intron 16 | rs2235046 | 12408282 | GG | AA | CTAGGGCTACrGTAGGAGTGG | 0.49 | 0.37 |
| IVS 17+1786 A>Gd | Intron 17 | rs4728700 | 12405875 | GG | AA | TTCTGGAGATrGTTGCCAGGA | 0.46 | 0.42 |
| IVS 17+1918 A>G | Intron 17 | rs10276603 | 12405743 | AG | AA | CTCTTCAAAArTCCTTGTTGT | ||
| IVS 17+2062 T>C | Intron 17 | rs4148736 | 12405599 | CT | CC | GAATGGTTATyCTCTGTGTTC | ||
| IVS 17+2293 A>G | Intron 17 | rs4148737 | 12405368 | AA | AG | TTTTCCCCAGrCACCTTGGGA | ||
| IVS 18+656 G>A | Intron 18 | ss20398883 | 12404233 | AG | GG | TTGGGAGGCCrAGGTGAGTGG | ||
| IVS 18+971 T>G | Intron 18 | ss20398884 | 12403918 | TT | GT | AAGTGCTCCTkGTACCTGTTT | ||
| IVS 18+1004 A>G | Intron 18 | rs10268314 | 12403885 | AG | AA | CCTTGGGCACrTAAGTAAACT | ||
| IVS 18+1317 A>G | Intron 18 | ss20398885 | 12403572 | AA | AG | CCTCACTGGGrAATTTGACCT | ||
| IVS 20+788 T>C | Intron 20 | rs10248420 | 12399202 | CT | TT | AATGAGGAAAyACATAGAGTT | ||
| IVS 20+882 A>Td | Intron 20 | rs10234411 | 12399108 | TT | AA | AGCTCCTCTTwGTAATTGTTG | 0.49 | 0.42 |
| IVS 20+1545 A>G | Intron 20 | ss20398887 | 12398445 | AG | AA | TTTTTTAAAGrGACAGGGTCT | ||
| IVS 20+2725 A>G | Intron 20 | rs4148738 | 12397265 | AA | GG | TTTGGCTGACrGGTTTTAGTT | 0.47 | 0.41 |
| IVS 20+2758 A>G | Intron 20 | ss20398888 | 12397232 | AG | AA | CCTAGTCTTTrCAATGAAAAT | ||
| IVS 20+4254 A>G | Intron 20 | ss20398889 | 12395736 | AG | AA | GATGTGCTGGrCTAAATTATA | ||
| 2677G>T,A | Exon 21 | rs2032582 | 12394834 | GG | TT | ACTAGAAGGTkCTGGGAAGGT | 0.47 | 0.32 |
| IVS 21+49 T>C | Intron 21 | rs2032583 | 12394777 | CT | TT | TAAAGTATTCyAATCAGTGTT | ||
| IVS 21+597 C>G | Intron 21 | ss20398890 | 12394229 | CC | CG | ATGGAAAAATsCTAGGGGTAT | ||
| IVS 21+614 T>A | Intron 21 | ss20398891 | 12394212 | TT | AT | GTATTTATTAwATCTGTTTTA | ||
| IVS 21+1574 C>A | Intron 21 | ss20398861 | 12393252 | AC | CC | AGAAAGCATTmTAGTTAACAG | ||
| IVS 21+2425 A>C | Intron 21 | ss20398862 | 12392401 | AA | CC | ATTGCCTCTGmTCTTCTCTTG | 0.50 | 0.41 |
| IVS 21+2964 A>T | Intron 21 | ss20399352 | 12391862 | AA | AT | ACACAAACCTwTAATTAAAAA | ||
| IVS 21+3027 G>Td | Intron 21 | rs2373586 | 12391799 | GG | TT | ACCATATAAGkCACCATTCAC | 0.47 | 0.34 |
| IVS 21+5964 T>C | Intron 21 | ss20398863 | 12388862 | CT | TT | TAAACTTGTTyATTTGAGGGT | ||
| IVS 21+7026 T>G | Intron 21 | ss20398864 | 12387800 | GT | GT | AAGTATCTTGkGGTAAACATA | ||
| IVS 21+7394 A>G | Intron 21 | ss20398865 | 12387432 | AG | GG | AATTCTGGAArTTATTTCTCT | ||
| IVS 21+7473 A>G | Intron 21 | ss20398866 | 12387353 | AG | AA | CAAATGAGGArAATGAGACAG | ||
| IVS 21+7724 G>A | Intron 21 | ss20398867 | 12387102 | AG | GG | ACGCCATCTTrACTCACTGCA | ||
| IVS 21+7987 A>T | Intron 21 | ss20398868 | 12386839 | AT | AA | TATGTGCCACwTTCCTTAAAA | ||
| IVS 21+8099 C>T | Intron 21 | ss20398869 | 12386727 | CT | CC | GTGTGTGAGGyTGCAGTGAGC | ||
| IVS 21+8507 T>C | Intron 21 | rs4148740 | 12386319 | TT | CT | AAAAAAACAAyATGGAAATGT | ||
| IVS 21+8565 T>C | Intron 21 | rs4148742 | 12386261 | CT | TT | TATTCAGCATyATGATCAGAC | ||
| IVS 21+8952 G>A | Intron 21 | rs2373589 | 12385874 | AG | GG | TCATGGTTTGrCAAAGTACTG | ||
| IVS 21+9086 C>G | Intron 21 | rs1882477 | 12385740 | CG | GG | TTGCTTCCATsATTACCAAAT | ||
| IVS 21+9520 G>A | Intron 21 | rs4148743 | 12385306 | AA | GG | TATCTTTTTCrCAGTTGGGTG | 0.45 | 0.19 |
| IVS 23+314 G>A | Intron 23 | ss20398870 | 12382544 | GG | AG | CCTGAGCAAGrAGTCTGACTG | ||
| IVS 25+1986_1991_del | Intron 25 | ss20398871 | 12376777 | +/+ | -/- | AGCCTCC[[TTTTC]/-]TTTCACT | 0.48 | 0.48 |
| IVS 25+2733 G>A | Intron 25 | ss20398872 | 12376030 | GG | AA | TTGACCTGAArTGGTGGTTCT | 0.49 | 0.50 |
| IVS 25+3050 G>T | Intron 25 | ss20398873 | 12375713 | GG | TT | (A)12(T)9kAATGCAAAAT | 0.40 | 0.73 |
| IVS 25+3739 T>C | Intron 25 | ss20398874 | 12375024 | CT | TT | AATTATTATTyCACAGTAAAT | 0.19 | 0.25 |
| IVS 25+4091 C>T | Intron 25 | ss20398875 | 12374672 | CT | CT | GTTTGCAATTyTAGGGTATTA | ||
| IVS 25+4983 T>G | Intron 25 | ss20398876 | 12373780 | GT | GT | TCCATGCTAAkCCTGGGCACT | ||
| IVS 25+5231 T>C | Intron 25 | ss20398880 | 12373532 | CC | TT | TGATCTGTTTyCTTGCTTGTG | 0.42 | 0.79 |
| 3435C>T | Exon 26 | rs1045642 | 12372861 | CC | TT | AGGAAGAGATyGTGAGGGCAG | 0.42 | 1.00 |
| IVS 26+80 T>C | Intron 26 | rs2235048 | 12372727 | TT | CC | AGGGGCTGGTyTCCCAGAAGT | 0.43 | 0.91 |
| IVS 26+1113 C>T | Intron 26 | ss20398877 | 12371694 | CT | CC | TATTAAAAGTyCAAAATTAGA | 0.05 | 0.03 |
| IVS 26+1130 C>T | Intron 26 | ss20398878 | 12371677 | CT | CC | TAGATTTTTTyCAACCTTTAT | 0.23 | 0.35 |
| IVS 26+1573 A>G | Intron 26 | rs1882478 | 12371234 | AG | GG | TCAACCCGGCrGGGAAGACAG | 0.25 | 0.36 |
| IVS 26+1576 G>A | Intron 26 | ss20398879 | 12371231 | AG | GG | ACCCGGCGGGrAAGACAGTTT | 0.22 | 0.31 |
For exonic SNPs, the locus name refers to the base position in the ABCB1 cDNA (GenBank acc. no. M14758), with the first base of the ATG start set to 1. Intronic SNPs were named as follows: the intron number was followed by a number indicating the distance from the G of the donor site invariant GT immediately upstream of the SNP. bSNP position within the reference contig (GenBank acc. no. NT_007933). The ancestral state of the polymorphism was determined either by sequencing a chimpanzee sample, or assuming that the ancestral state was the most frequent allele in our sample. For SNPs marked with dthis information is missing. cThe new variants were deposited in the GenBank database (ss ID above). eMAF (minor allele frequency), and fr2 (posterior mean) against 3435C>T were calculated in unrelated CEPH chromosomes only for the newly identified homozygous sites, and for the heterozygous sites located in the region downstream IVS 25+3050 G>T.
Assessing Candidate Polymorphisms
The 14 polymorphisms that were homozygous in the two individuals, and the three polymorphisms that were heterozygous in the region of greatest LD with 3435C>T, were typed in two sets of 24 CEPH trios, and LD with 3435C>T was calculated (Fig. 3; not all of the polymorphisms were sequenced in the same individuals, but in all cases the 3435C>T polymorphism was typed to allow assessment of pairwise r2). All 14 homozygous polymorphisms are intronic; six are novel. Eleven of these 14 homozygous sites have intermediate to low values of r2 against 3435C>T (0.19–0.50, Table 3), and will not be considered further. The three other originally homozygous sites have high or complete LD with 3435C>T in the 24 CEPH trios (Fig. 2). The IVS 26+80 T>C polymorphism (corresponding to dbSNP rs2235048) is located 134 bp downstream of 3435C>T in intron 26. This SNP is in near-complete LD with 3435C>T in the CEPH trios (r2 = 0.91, Fisher's exact test P < 0.001). Two other variants showing high LD with 3435C>T, IVS 25+3050 G>T (r2 = 0.73, Fisher's exact test P < 0.001) and IVS 25+5231 T>C (r2 = 0.79, P < 0.001), represent new SNPs, and are both located in intron 25 (Table 3). IVS 25+3050 G>T is associated with a polymorphic AnT9 repeat, which itself could not be accurately genotyped by resequencing in the CEPH trios. Among the variants showing heterozygous differences between the two sequenced individuals, r2 never exceeds 0.35. This suggests that in sequencing through a high-LD interval in representative derived and ancestral chromosomes at the associated polymorphism, it may be sufficient to subsequently characterize only the polymorphisms showing homozygous differences, in order to identify high-LD variants.
Figure 3.
The high-LD interval (A, dashed box) was fully sequenced in two chromosomes, leading to the identification of candidate causal sites; resequencing of these variants in CEPH trios was used to resolve LD structure within this interval (B). Of all homozygous variants, those that are in high or complete r2 with 3435C>T (•) are the most likely candidates to be causal; heterozygous mutations (▵) result in decreased r2. (○) The cases where a new set of CEPH trios was used to calculate r2 compared to the rest of the study. Bars indicate the Bayesian 95% credible intervals calculated by the method described in Supplemental materials.
Association With Drug Response in Patients With Epilepsy
We reanalyzed 3435C>T in a cohort of 286 drug-resistant and 135 drug-responsive patients with epilepsy, partially overlapping with the cohort analyzed by Siddiqui et al. (2003). We confirmed the significant association between 3435C>T and response to anti-epileptic drugs reported by Siddiqui et al. (2003). The C allele (high activity) was significantly overrepresented in the drug-resistant group for both allelic and genotypic intergroup comparisons (χ2 = 4.509, P = 0.034 and χ2 = 6.855, P = 0.032; Table 4).
Table 4.
Association of ABCB1 Variants and Drug Resistance in Epileptic Patients
| Variant | Phenotype | Alleles | Χ2a | Pb | Genotypes | Χ2a | Pb | r2 vs 3435C>T | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | A | G | AT | GA | TT | GT | GG | |||||||
| 2667G>T,A | Drug-resistant | 184 | 8 | 266 | 0.66 ± 0.0048c | 4 | 4 | 39 | 102 | 80 | 0.87 ± 0.0019c | 0.21 | ||
| Drug-responsive | 86 | 3 | 107 | 2 | 1 | 18 | 48 | 29 | ||||||
| G | T | GG | GT | TT | ||||||||||
| IVS 25+3050 G>T | Drug-resistant | 286 | 284 | 2.108 | 0.147 | 69 | 148 | 68 | 2.840 | 0.242 | 0.84 | |||
| Drug-responsive | 121 | 149 | 23 | 75 | 37 | |||||||||
| C | T | CC | CT | TT | ||||||||||
| IVS 25+5231 T>C | Drug-resistant | 259 | 291 | 0.766 | 0.381 | 57 | 145 | 73 | 1.560 | 0.458 | 0.80 | |||
| Drug-responsive | 113 | 145 | 20 | 73 | 36 | |||||||||
| C | T | CC | CT | TT | ||||||||||
| 3435C>T | Drug-resistant | 291 | 269 | 4.509 | 0.034 | 73 | 145 | 62 | 6.855 | 0.032 | 1 | |||
| Drug-responsive | 120 | 152 | 20 | 80 | 36 | |||||||||
| T | C | TT | CT | CC | ||||||||||
| IVS 26+80 T>C | Drug-resistant | 285 | 271 | 3.782 | 0.052 | 70 | 145 | 63 | 5.629 | 0.060 | 0.98 | |||
| Drug-responsive | 118 | 150 | 20 | 78 | 36 | |||||||||
Χ2 and P-values for Pearson's Χ2 test of association with one (alleles) and two (genotypes) d.f.
Fisher's exact test P and s.e.
In addition, we genotyped the three newly identified SNPs having high r2 with 3435C>T, and the well characterized exon 21 SNP (2677G>T,A) that has been proposed to be a functional variant (Kim et al. 2001). Among the three intronic variants, the T allele at IVS 26+80 T>C also showed borderline-significant association with drug response for both allelic and genotypic intergroup comparisons (χ2 = 3.782, P = 0.052 and χ2 = 5.629, P = 0.060). At the three other variants investigated, we could not detect a significant association with drug resistance for either alleles and genotypes: IVS 25+3050 G>T (χ2 = 2.108, P = 0.147 and χ2 = 2.840, P = 0.242), IVS 25+5231 T>C (χ2 = 0.766, P = 0.381 and χ2 = 1.560, P = 0.458), and 2677G>T,A (Fisher's Exact test P = 0.660 and P = 0.870).
Analysis of two-locus haplotypes, representing all possible combinations of two of the five loci, did not reveal a significant association with drug resistance (data not shown).
To assess whether 3435C>T provided a significantly better association with phenotype than the other four SNPs, we added 3435C>T genotype as an additional explanatory factor to each of four logistic regression models (one for each other SNP) in which the other SNP genotype had already been entered as an explanatory factor (to allow fair comparison and make all the SNPs diallelic, the 11 people with the third A allele at 2677G>T were excluded). In one case, adding the 3435C>T genotype improved association significantly (2677G>T: P = 0.047), whereas in the other three cases there was no significant improvement (IVS 25+3050 G>T: P = 0.135; IVS 25+5231 T>C: P = 0.067; IVS 26+80 T>C: P = 0.387). These results suggest that among the proposed candidate SNPs, only one could be functionally causal, with the others associated through LD alone. It is of interest to consider the relative probabilities of these five SNPs being causal, under the assumption that only one of them is in fact causal. We assigned equal prior weight to the five alternatives, then restricted the data set to those patients with no missing data for any SNP (n = 260) and used Laplace's method of approximation for the Bayes factors (Kass and Raftery 1995). The posterior probabilities for each alternative model were estimated to be 0.002 that 2677G>T is the causal SNP, 0.036 that IVS 25+3050 G>T is causal, 0.024 that IVS 25+5231 T>C is causal, 0.559 that 3435C>T is causal, and 0.379 that IVS 26+80 T>C is causal. These data suggest that 3435C>T is 236 times more likely to be the causal SNP than 2677G>T, between 15 and 24 times more likely to be the causal SNP than IVS 25+3050 G>T or IVS 25+5231 T>C, but only 1.5 times more likely to be the causal SNP than IVS 26+80 T>C.
These results indicate that IVS 26+80 T>C and 3435C>T are almost equally strong candidates for explaining resistance to anti-epileptic drugs due to PGP activity, and that IVS 25+3050 G>T and IVS 25+5231 T>C are less likely, though still possible. IVS 26+80 T>C and 3435C>T are less than 200 bp apart and in near-complete LD in the patients (pairwise r2 = 0.98). These results are consistent with the two hypotheses that either one, or both, of these variants are directly causal to the phenotype. If only one of the two variants were causal, the other then displays significant association with the phenotype because of LD between the variants. Note that although the two other variants are also in high LD with 3435C>T in the patients (r2 = 0.84 and r2 = 0.80), they are not significantly associated with the drug-resistant phenotype, and based on the current evidence are less likely candidates. Finally, we note that when these four candidate SNPs plus 3435C>T were assessed directly by typing all of these in cases and controls, a positive relationship between association with the phenotype and association with 3435C>T (measured by r2) was found. Although this can only be taken as suggestive, as only a small number of SNPs were typed, it may indicate in this case that the causal SNP is indeed one of those in high LD with 3435C>T.
Evolutionary Conservation of ABCB1 Intronic Sites and Possible Effect of the Newly Discovered Polymorphisms
Comparative genomics approaches can help to prioritize associated polymorphisms by identifying evolutionarily conserved sequences with a potential functional role, for example in intronic or promoter regions of genes. In particular, the phylogenetic shadowing method (Boffelli et al. 2003) can be applied to the comparison of multiple species closely related to humans, allowing detecting functional regions that evolved late in mammalian evolution.
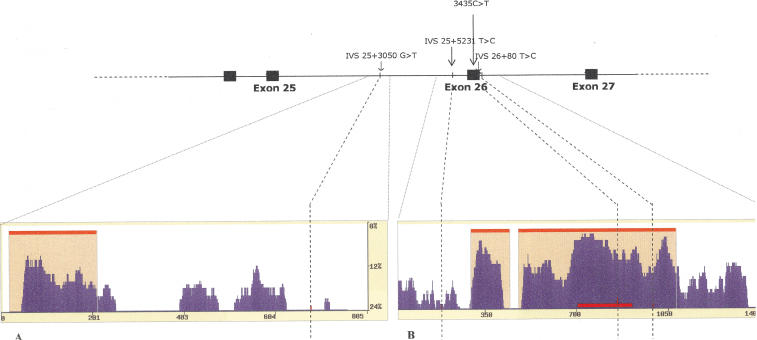
Phylogenetic shadowing exploits a two-rate nucleotide divergence model (slow for constrained evolution and fast for unconstrained), where these two states are calibrated on the divergence of a known functional region for a given group of species. These are then used to calculate the relative likelihood that any given nucleotide site within a region of interest is subjected to a slower or faster rate of accumulation of variation, related to functional constraints imposed on each site (Boffelli et al. 2003). We implemented this method by sequencing approximately 0.8 kb and 1.5 kb around the IVS 25+3050 G>T and 3435C>T sites in 15–17 species spanning the primate phylogeny (details in Methods). The divergence between exon 26, contained within the larger amplicon, and the flanking intronic sequences was used to calibrate the parameters of the two-rate divergence model for the set of sequences (eS 0.9227, eF 0.6986, T 0.0013).
The plots of cumulative divergence obtained by phylogenetic shadowing are shown in Figure 4. The block of significant conservation (orange shading) between nucleotides 477 and 1078 of the reference human sequence in Figure 4A comprises the entire exon 26 and the flanking intronic sequences. It is interesting to note that the IVS 26+80 T>C variant is included within this block of significant conservation. Conversely, both the IVS 25+3050 G>T and IVS 25+5231 T>C variants in intron 25 are within stretches of low or no conservation (Fig. 4A,B). This analysis suggests that, among the three intronic sites, IVS 26+80 T>C is the most likely to be functional (in addition to 3435C>T). The proximity of IVS 26+80 T>C to exon 26 however makes it difficult to confirm whether the variant itself is functional, or its conservation is due to its physical proximity to exon 26.
Figure 4.
Position of the three candidate causal mutations within ABCB1, showing the position of the three new candidate variants in high LD with 3435C>T. In the boxes are the two regions further investigated by phylogenetic shadowing. Plots of cumulative nucleotide divergence were generated using the eShadow tool by amplifying the region surrounding IVS 25+3050 G>T (A) and 3435C>T (B) in 17 and 15 primate species, respectively. Orange shading indicates the region of significant conservation; the horizontal red bar at nucleotides 715–921 of the alignment in B indicates the position of exon 26. The position of the three intronic sites is marked by a vertical dashed line: IVS 25+3050 G>T at nucleotide 710 (A), and IVS 25+5231 T>C, 3435C>T, and IVS 26+80 T>C sites at 184, 867, and 1001 bp of the alignment (B).
A possible effect for the intronic variants could be through either RNA splicing or transcriptional regulation. For instance, a polymorphism in a region mediating the binding of the spliceosome to the pre-mRNA, associated with the low-activity T allele at 3435C>T, may act to reduce ACBC1 splicing efficiency. This would result in a lower rate of production of the correctly spliced RNA, and hence of PGP, in low-activity TT homozygotes (Hoffmeyer et al. 2000).
Another possibility is that one of the variants sits in one as yet undiscovered transcriptional regulator of ABCB1. We used a bioinformatic approach to infer the presence of putative transcription factor binding sites that may be affected by one of the three polymorphisms. This analysis showed for instance that at the most likely (non-3435C>T) candidate causal site IVS 26+80 T>C, the T to C transition, associated with the low-activity T allele at 3435C>T, may lead to the loss of a binding site for Sp1, a known regulator of ABCB1 (Cornwell and Smith 1993). Similarly, the G to T mutation at IVS 25+3050 G>T may generate a new putative TATA binding protein (TBP) site, which may inhibit ABCB1 transcription through p53 (Truant et al. 1993; Nguyen et al. 1994). At IVS 25+5231 T>C, a T to C substitution creates a putative NF-ATp binding site associated with the high-activity CC at 3435C>T. Although intronic transcriptional enhancers/silencers are sometimes found at the 3′ end of genes, these results should be taken with caution, as bioinformatic predictions of transcription factor binding sites are expected to generate a large number of false positives. No published evidence suggests the presence of enhancers near the exon 26 of ABCB1.
It is clear that a correct evaluation of these or other possible biological effects will only be obtained with the functional characterization of these putative candidate causal SNPs, including in vitro splicing experiments and/or promoter analysis.
Design of a Set of Haplotype Tagging SNPs (tSNPs) for ABCB1
The analysis of haplotype tagging SNPs (tSNPs) was carried out on the set of 14 high-frequency SNPs (Table 1). Tagging SNPs were identified that provide a coefficient of determination of at least 0.85 in predicting all the known SNPs (haplotype r2 criterion as defined in TagIT [Weale and Goldstein 2003; Goldstein et al. 2003]). One set of SNPs satisfying the criterion corresponds to SNPs 2, 6, 8, 12 (or 11), 14, and 15 of Table 1. Full details on the tag design are given in the Supplemental material.
Conclusions
This study illustrates the importance of following up reported associations with detailed genetic analysis including, where necessary, deep resequencing of the genomic region within which the causal variant may reside (i.e., the associated interval, as defined in Goldstein 2003). We emphasize however that in general, the boundaries of the associated interval will be difficult to define. One reason for this is uneven decay of LD with physical distances. Another is because the uncertainties attached to the phenotypic data (both in terms of low odds ratios and small sample sizes) may be such that even SNPs with a low r2 value to an initial candidate (in this case, 3435C>T) cannot be excluded.
Our formalization of the associated interval suggests that when the initial association between a variant and a phenotype is weak, the extent of associated interval will be extremely large, even if LD is only modest in the region. In these situations, there would be a considerable advantage to the use of intermediate phenotypes, where available, to refine the interval. The method described in the Supplemental material would allow prior evaluation of how additional sorts of data (more accurate estimation of LD pattern in controls, more accurate assessment of the clinical association, or intermediate phenotypes) might be used to help constrain the associated interval.
This approach therefore can help to determine the most cost-effective strategies for follow-up of first associations, as opposed to simply carrying out deep resequencing through very large associated intervals. We suspect that in this case refinement of the intermediate phenotype could lead to an identification of the associated interval commensurate with the high-LD interval described in Figure 1, and within this region we have identified, a reasonably complete set of candidate causal variants can be identified with modest experimental effort. It is also worth noting that failure to identify any functional variants in well defined associated intervals would have to lower confidence in the original reported association (Lohmueller et al 2003).
The application of our approach may be particularly effective to screen intronic regions of genes in cases where candidate coding variants have not been discovered, because deep resequencing through introns can be extremely costly, and all possible sources of information should be appropriately combined to identify the associated interval. In pharmacogenomics, for instance, variation in gene expression appears to underlie at least in part differences in drug response among individuals. As most studies to date have focused on the resequencing of exons, core promoters, and intron-exon boundaries, the bulk of intronic and other regulatory variation is likely to have been systematically undetected. A future challenge for genetic association studies will be to identify intronic polymorphisms of medical relevance.
The application of the proposed method to the analysis of the ABCB1 has led to the identification of three intronic polymorphisms that represent, in addition to the well characterized 3435C>T, alternative well supported candidates for the well known and clinically relevant polymorphism affecting PGP activity. Based on our additional analyses we could not rule out any of these three sites as a possible candidate, although a test of association of these three variants with drug-resistant epilepsy, and evolutionary conservation, show IVS 26+80 T>C to be the strongest candidate. It is now important to assess experimentally whether any of these polymorphisms has a functional effect. If none of the three well supported non-3435C>T polymorphisms do, this would either reduce confidence that the original associations between 3435C>T are real, or indicate that a SNP with low r2 to 3435C>T was responsible.
Assuming that many of the originally reported associations are in fact real, the identification of the causal variant in the case of ABCB1 is of considerable importance. The discrepancies highlighted by the large number of clinical studies addressing the effect of 3435C>T on ABCB1 expression and function, and its pharmacokinetic and pharmacodynamic properties, call for a more thorough analysis of the regions surrounding 3435C>T and the corresponding haplotypes (Sakaeda et al 2003; Sparreboom et al. 2003).
If the new polymorphisms were confirmed to influence enhancer or splicing activity, this could be of relevance in a range of clinical settings. ABCB1 expression is finely regulated through a complex network of transcription factors and posttranscriptional mechanisms, in response to a variety of different stresses (Kantharidis et al. 2000; Labialle et al. 2002). Despite extensive efforts, studies have failed to describe an unequivocal correlation between core promoter elements and stress-inducible PGP overexpression, suggesting that additional undiscovered cis-acting regulatory elements may exist (Labialle et al 2002). The identification of novel genomic regions with enhancer function in ABCB1 would help elucidate the mechanisms underlying both constitutive and inducible PGP overexpression.
METHODS
SNP Genotyping and Resequencing
Seventeen SNPs distributed along the length of the gene were genotyped in 24 CEPH (Centre d'Etude du Polymorphisme Humain) trios following Siddiqui et al. (2003; Table 1). Amplified PCR products were sequenced using the Big Dye Terminator cycle sequencing kit (Applied Biosystems) in the presence of 10 pmoles forward or reverse primer. Sequencing was carried out using a 3700 ABI automated sequencer. Sequence analysis and contig assembly were done using Sequencher software (v. 4.0.5, Genecodes). Sequence traces were scored twice, and the genotyping data were checked by random resequencing of 10% of the samples. The estimated error rate was 0.015. The ancestral state of each allele was inferred by sequencing one chimpanzee.
Two individuals who were homozygous at assayed SNPs in the region between IVS 6+139 and 3435C>T, and representing respectively two ancestral (CEPH1422-02) and two derived (CEPH1333-01) chromosomes at 3435C>T were selected for sequencing. Resequencing of the region between 1236C>T–IVS 27+1266 C>A (∼45 kb) in these two samples was done using 51 pairs of primers designed to amplify partially overlapping amplicons (available at http://popgen.biol.ucl.ac.uk/supdata.html). The new variants were named as described in Table 3.
Data Analysis
Bayesian Assessment of Associated Interval
Here we formalize the idea of an associated interval as defined in Goldstein (2003). The idea applies to a situation in which a polymorphism has been associated with a phenotype of interest in one data set (“phenotyped” data set). This polymorphism is referred to as the “associated variant,” M. In a separate data set (“LD” data set) there is information about the pattern of association between a set of linked SNPs and the “associated variant.” These two sources of information are combined to assess which markers, and the sequence stretch that they delimit, should be considered as candidates for causing the original genotype-phenotype association. This stretch of sequence is the “associated interval” and should be exhaustively searched for causal variants. We note that given the imminent availability of a relatively dense set of LD genotype data stemming from the HapMap project, this situation will soon be very common. A variant will be associated in a “phenotyped” data set, and typing it in the same individuals used in the HapMap project will provide data on the association between the associated variant and linked polymorphisms. The approaches described here will allow estimation of the associated interval in this context.
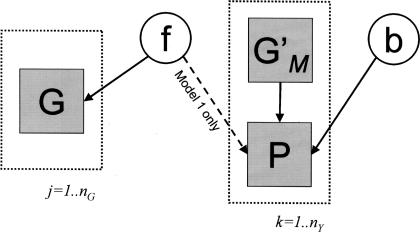
Figure 5 describes the two Bayesian models, which are applied separately to each SNP i in the LD data set. Model 0 proposes that SNP M, the “associated variant,” is causal. Model 1 proposes that SNP i is causal. Under Model 0, the phenotypic data P in a set of nY phenotyped individuals is explained directly by the known genotype information, G′M, regarding SNP M, via an association model described by parameters in b (call this set b0 for Model 0). Under Model 1, P is explained indirectly by linkage of M to SNP i (determined by f, the vector of four two-locus haplotype frequencies) and then by the association model between SNP i and phenotype described by parameters in b1. Information on f is obtained from the LD data set, G, which contains genotype information on M and SNP i in a sample of nG individuals. G is allowed to contain phase-resolved information on heterozygotes, if available from, for example, typing family members.
Figure 5.
Description of Bayesian models for assessment of the associated interval (see text). The link between f and P is dashed to indicate that it is present in Model 1 but not Model 0.
Many different forms of the association model, described by the parameters in b, are possible (see Supplemental material). One such model, used to generate Figure 1C, is a logistic regression model appropriate for case-control data. It is assumed that the genotypic odds ratio is greatest between the two homozygous genotypes—denoted by parameter θ. The odds ratio between the heterozygous genotype and the reference homozygote is given by θc, where c is a dominance modifier taking a value between 0 and 1 (½ indicates no dominance). We gave c an uninformative uniform prior, but we chose to apply an informative prior to θ, based on the emerging picture of the strength of clinical association found for variants implicated in complex diseases. To date the strength of association, even for variants of strong effects, does not usually exceed θ = 5. We therefore applied a Normal prior to loge(θ), centred on zero, with 2.5% and 97.5% percentiles of ±loge(5).
We used WinBUGs (Spiegelhalter et al. 2003) to find joint posterior distributions of f and b via Markov chain Monte Carlo approximation. We assessed the associated interval by estimating the posterior odds ratio: P(Model 1|Data) / P(Model 0|Data). We assumed equal prior weight to each model, and so estimated the posterior odds as the Bayes Factor: P(Data|Model 1) / P(Data|Model 0). For further details see Supplemental material.
Linkage Disequilibrium (LD)
Haplotypes were inferred in the 24 CEPH trios using an Expectation-Maximization algorithm developed for trio data and implemented in the TagIT software package (Weale and Goldstein 2003; http://popgen.biol.ucl.ac.uk/software.html) tests for Hardy-Weinberg equilibrium, and pairwise r2 measures of LD, were calculated on the inferred haplotypes with routines available in the TagIT package, and using a Bayesian approach as detailed in the Supplemental material. In the unrelated patients, haplotypes were inferred using the Partition-Ligation algorithm implemented by PL-EM (Qin et al. 2002), and the haplotypes and frequencies were imported into TagIT for analysis of LD patterns. Design of haplotype tagging SNPs was done following Goldstein et al. (2003), as described in the Supplemental material.
Tests of Association
Associations of allelic and genotypic frequencies and multidrug resistance at 3435C>T and the three candidate causal variants identified in this study were assessed in a cohort of 286 drug-resistant and 135 drug-responsive epileptic patients. Genotypes were obtained by direct sequencing using primers available upon request from the authors. Appropriate details on the patients are given in Siddiqui et al. (2003). All subjects gave written informed consent for the study. The study of patient data was approved by the joint research ethics committee of the National Hospital for Neurology and Neurosurgery and the UCL Institute of Neurology. Allelic and genotypic frequencies were tested for significant association with the drug response phenotype using χ2 and Fisher's exact tests. All reported values are two-sided. Significance was set at 0.05. Haplotypic association was assessed using the log-likelihood ratio method implemented in the EH and PM programs (Zhao et al 2000; http://web1.iop.kcl.ac.uk/IoP/Departments/PsychMed/GEpiBSt/software.shtml). This involves calculating log-likelihoods of estimated haplotype frequencies for each of cases, controls, and cases and controls combined. The test statistic 2*(ln(Lcase)+ln(Lcontrol)–ln(Lcase/Lcontrol)) is then applied, giving a χ2 value with n–1 degrees of freedom (where n = number of haplotypes). The evidence of causation for each of the four polymorphisms described above, beneath the heading “Association With Drug Response in Patients With Epilepsy,” was compared using a log linear model.
Prediction of Transcription Factor Binding Sites
The ABCB1 sequences were compared with published sequence nucleotide and dbSNP divisions of the NCBI database using the BLASTN algorithm (Altschul et al. 1990). To assess whether the identified polymorphisms affected binding to transcription factors, for each of the three sites we submitted 40 base pair DNA stretches containing each of the two alleles to the TESS algorithm (Schug and Overton 1997), which predicts transcription factor binding sites (http://www.cbil.upenn.edu/tess). The algorithm was set at 12 minimum log-likelihood ratio score, 6 minimum string length, 0.75 minimum core similarity, and 0.85 minimum matrix similarity.
Phylogenetic Shadowing
We amplified 850 nucleotides around the IVS 25+3050 G>T site in 17 primate species (C. neglectus, S. entellus, C. olivaceus, G. gorilla, C. guereza, M. sphinx, T. obscurus, C. polykomos, P. nemaeus, C. mitis, M. mulatta, H. leucogenis, P. hamadryas, M. arctoides, M. fascicularis, H. sapiens, P. troglodytes), and 1.6 kb around the 3435C>T site, containing the entire exon 26 (C. aetiops, C. guereza, C. mitis, C. neglectus, C. polykomos, P. troglodytes, G. gorilla, H. lar, H. sapiens, M. fascicularis, M. mulatta, M. sphinx, P. abelii, P. hamadryas, S. entellus). Evolutionary conservation was analyzed using phylogenetic shadowing (Boffelli et al. 2003) implemented using the eShadow program (I. Ovcharenko, pers. comm.; http://eshadow.dcode.org). Alignment gaps were included in the analysis. Optimization of parameters for phylogenetic shadowing was obtained under the HMM Islands model with Maximum Likelihood option. Emission probabilities for nucleotide substitution being in slow- (eS) or fast-mutation state (eF), and the transition probability T from one state to another, were determined experimentally by training the algorithm on the 207 nucleotides of exon 26, included within the larger amplicon. The parameters so obtained (eS 0.9227, eF 0.6986, T 0.0013) were applied for the analysis of (1) the region surrounding the IVS 25+3050 G>T site (Fig. 4A), and (2) the intronic sequences surrounding the IVS 25+5231 T>C and IVS 26+80 T>C sites (Fig. 4B), using the HMM Islands model no-parameter optimization. All analyses were implemented using eShadow (http://eshadow.dcode.org).
Acknowledgments
We thank Alice Smith and Mari Wyn Burley for technical support, Prof. E. Shepherd (Dept. of Biochemistry, UCL) for useful advice, the German Primate Centre for supplying primate samples, Dr. I. Ovcharenko (Lawrence Livermore National Laboratory, CA) for help with the implementation of phylogenetic shadowing and for sharing unpublished material, and three anonymous referees for their comments on a previous version of the manuscript. This research was supported by a Royal Society/Wolfson Research Merit Award and by the Leverhulme Trust. D.B.G. is a Royal Society/Wolfson Research Merit Award holder.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1965304. Article published online before print in June 2004.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: I. Ovcharenko.]
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bellamy, W.T. 1996. P-glycoproteins and multidrug resistance. Annu. Rev. Pharmacol. Toxicol. 36: 161–183. [DOI] [PubMed] [Google Scholar]
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391–1394. [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo, C., O'Brien, J.P., Casals, D., Rittman-Grauer, L., Biedler, J.L., Melamed, M.R., and Bertino, J.R. 1989. Multidrug-resistance gene (P-glycoprotein) is expressed by endothelial cells at blood-brain barrier sites. Proc. Natl. Acad. Sci. 86: 695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornwell, M.M. and Smith, D.E. 1993. SP1 activates the MDR1 promoter through one of two distinct G-rich regions that modulate promoter activity. J. Biol. Chem. 268: 19505–19511. [PubMed] [Google Scholar]
- de Lannoy, I.A.M. and Silverman, M. 1992. The MDR1 gene product, P-glycoprotein, mediates the transport of the cardiac glycoside, digoxin. Biochem. Biophys. Res. Commun. 189: 551–557. [DOI] [PubMed] [Google Scholar]
- Fellay, J., Marzolini, C., Meaden, E.R., Back, D.J., Buclin, T., Chave, J.P., Decosterd, L.A., Furrer, H., Opravil, M., Pantaleo, G., et al. 2002. Response to antiretroviral treatment in HIV-1-infected individuals with allelic variants of the multidrug resistance transporter 1: A pharmacogenetics study. Lancet 359: 30–36. [DOI] [PubMed] [Google Scholar]
- Fromm, M.F. 2002. The influence of MDR1 polymorphisms on P-glycoprotein expression and function in humans. Adv. Drug Deliv. Rev. 54: 1295–1310. [DOI] [PubMed] [Google Scholar]
- Goldstein, D.B. 2003. Pharmacogenetics in the laboratory and the clinic. New Engl. J. Med. 348: 553–556. [DOI] [PubMed] [Google Scholar]
- Goldstein, D.B., Ahmadi, K.R., Weale, M.E., and Wood, N.W. 2003. Genome scans and candidate gene. Approaches in the study of common diseases and variable drug responses. Trends Genet. 19: 615–622. [DOI] [PubMed] [Google Scholar]
- Gottesman, M.M., Fojo, T., and Bates, S.E. 2002. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer 2: 48–58. [DOI] [PubMed] [Google Scholar]
- Hitzl, M., Drescher, S., van der Kuip, H., Schaffeler, E., Fischer, J., Schwab, M., Eichelbaum, M., and Fromm, M.F. 2001. The C3435T mutation in the human MDR1 gene is associated with altered efflux of the P-glycoprotein substrate rhodamine 123 from CD56+ natural killer cells. Pharmacogenomics 11: 293–298. [DOI] [PubMed] [Google Scholar]
- Hoffmeyer, S., Burk, O., von Richter, O., Arnold, H.P., Brockmoller, J., Johne, A., Cascorbi, I., Gerloff, T., Roots, I., Eichelbaum, M., et al. 2000. Functional polymorphisms of the human multidrug-resistance gene: Multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc. Natl. Acad. Sci. 97: 3473–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, S., Koren, G., and Harper, P.A. 1992. Energy-dependent transport of digoxin across renal tubular cell monolayers (LLC-PK1). Can. J. Physiol. Pharmacol. 71: 40–47. [DOI] [PubMed] [Google Scholar]
- Johne, A., Kopke, K., Gerloff, T., Mai, I., Rietbrock, S., Meisel, C., Hoffmeyer, S., Kerb, R., Fromm, M.F., Brinkmann, U., et al. 2002. Modulation of steady-state kinetics of digoxin by haplotypes of the P-glycoprotein MDR1 gene. Clin. Pharmacol. Ther. 72: 584–594. [DOI] [PubMed] [Google Scholar]
- Kantharidis, P., El-Osta, S., Silva, M., Lee, G., Hu, X.F., and Zalcberg, J. 2000. Regulation of MDR1 gene expression: Emerging concepts. Drug Resist. Updat. 3: 99–108. [DOI] [PubMed] [Google Scholar]
- Kass, R.E. and Raftery, A.E. 1995. “Bayes factors.” J. Am. Stat. Assoc. 90: 773–795. [Google Scholar]
- Kim, R.B., Fromm, M.F., Wandel, C., Leake, B., Wood, A.J., Roden, D.M., and Wilkinson, G.R. 1998. The drug transporter P-glycoprotein limits oral absorption and brain entry of HIV-1 protease inhibitors. J. Clin. Invest. 101: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, R.B., Leake, B.F., Choo, E.F., Dresser, G.K., Kubba, S.V., Schwarz, U.I., Taylor, A., Xie, H.G., McKinsey, J., Zhou, S., et al. 2001. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin. Pharmacol. Ther. 70: 189–199. [DOI] [PubMed] [Google Scholar]
- Labialle, S., Gayet, L., Marthinet, E., Rigal, D., and Baggetto, L.G. 2002. Transcriptional regulators of the human multidrug resistance 1 gene: Recent views. Biochem. Pharmacol. 64: 943–948. [DOI] [PubMed] [Google Scholar]
- Lohmueller, K.E., Pearce, C.L., Pike, M., Lander, E.S., and Hirschhorn, J.N. 2003. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 33: 177–182. [DOI] [PubMed] [Google Scholar]
- Lown, K.S., Mayo, R.R., Leichtman, A.B., Hsiao, H.L., Turgeon, D.K., Schmiedlin-Ren, P., Brown, M.B., Guo, W., Rossi, S.J., Benet, L.Z., et al. 1997. Role of intestinal P-glycoprotein (mdr1) in interpatient variation in the oral bioavailability of cyclosporine. Clin. Pharmacol. Ther. 62: 248–260. [DOI] [PubMed] [Google Scholar]
- Morita, N., Yasumori, T., and Nakayama, K. 2003. Human MDR1 polymorphism: G2677T/A and C3435T have no effect on MDR1 transport activities. Biochem. Pharmacol. 65: 1843–1852. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Sakaeda, T., Horinouchi, M., Tamura, T., Aoyama, N., Shirakawa, T., Matsuo, M., Kasuga, M., and Okumura, K. 2002. Effect of the mutation (C3435T) at exon 26 of the MDR1 gene on expression level of MDR1 messenger ribonucleic acid in duodenal enterocytes of healthy Japanese subjects. Clin. Pharmacol. Ther. 71: 297–303. [DOI] [PubMed] [Google Scholar]
- Nguyen, K.T., Liu, B., Ueda, K., Gottesman, M.M., Pastan, I., and Chin, K.V. 1994. Transactivation of the human multidrug resistance (MDR1) gene promoter by p53 mutants. Oncol. Res. 6: 71–77. [PubMed] [Google Scholar]
- Oselin, K., Nowakowski-Gashaw, I., Mrozikiewicz, P.M., Wolbergs, D., Pahkla, R., and Roots, I. 2003a. Quantitative determination of MDR1 mRNA expression in peripheral blood lymphocytes: A possible role of genetic polymorphisms in the MDR1 gene. Eur. J. Clin. Invest. 33: 261–267. [DOI] [PubMed] [Google Scholar]
- Oselin, K., Gerloff, T., Mrozikiewicz, P.M., Pahkla, R., and Roots, I. 2003b. MDR1 polymorphisms G2677T in exon 21 and C3435T in exon 26 fail to affect rhodamine 123 efflux in peripheral blood lymphocytes. Fundam. Clin. Pharmacol. 17: 463–469. [DOI] [PubMed] [Google Scholar]
- Polli, J.W., Jarrett, J.L., Studenberg, S.D., Humphreys, J.E., Dennis, S.W., Brouwer, K.R., and Woolley, J.L. 1999. Role of P-glycoprotein on the CNS disposition of amprenavir (141W94), an HIV protease inhibitor. Pharm. Res. 16: 1206–1212. [DOI] [PubMed] [Google Scholar]
- Sakaeda, T., Nakamura, T., and Okumura, K. 2002. MDR1 genotype-related pharmacokinetics and pharmacodynamics. Biol. Pharm. Bull. 25: 1391–1400. [DOI] [PubMed] [Google Scholar]
- Sakaeda, T., Nakamura, T., and Okumura, K. 2003. Pharmacogenetics of MDR1 and its impact on the pharmacokinetics and pharmacodynamics of drugs. Pharmacogenomics 4: 397–410. [DOI] [PubMed] [Google Scholar]
- Schinkel, A.H. 2001. The roles of P-glycoprotein and MRP1 in the blood-brain and blood-cerebrospinal fluid barriers. Adv. Exp. Med. Biol. 500: 365–372. [DOI] [PubMed] [Google Scholar]
- Schug, J. and Overton, G.C. 1997. Modeling transcription factor binding sites with Gibbs Sampling and Minimum Description Length encoding. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5: 268–271. [PubMed] [Google Scholar]
- Schwab, M., Schaeffeler, E., Marx, C., Fromm, M.F., Kaskas, B., Metzler, J., Stange, E., Herfarth, H., Schoelmerich, J., Gregor, M., et al. 2003. Association between the C3435T MDR1 gene polymorphism and susceptibility for ulcerative colitis. Gastroenterology 124: 26–33. [DOI] [PubMed] [Google Scholar]
- Siddiqui, A., Kerb, R., Weale, M.E., Brinkmann, U., Smith, A., Goldstein, D.B., Wood, N.W., and Sisodiya, S.M. 2003. Association of multidrug resistance in epilepsy with a polymorphism in ABCB1 New Engl. J. Med. 348: 1442–1448. [DOI] [PubMed] [Google Scholar]
- Slatkin, M. 1994. Linkage disequilibrium in growing and stable populations. Genetics 137: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn-Langguth, H., Baktir, G., Radschuweit, A., Okyar, A., Terhaag, B., Ader, P., Hanafy, A., and Langguth, P. 1998. P-glycoprotein transporters and the gastrointestinal tract: Evaluation of the potential in vivo relevance of in vitro data employing talinolol as model compound. Int. J. Clin. Pharmacol. Ther. 36: 16–24. [PubMed] [Google Scholar]
- Sparreboom, A., Danesi, R., Ando, Y., Chan, J., and Figg, W.D. 2003. Pharmacogenomics of ABC transporters and its role in cancer chemotherapy. Drug Resist. Updat. 6: 71–84. [DOI] [PubMed] [Google Scholar]
- Spiegelhalter, D., Thomas, A., and Best, N. 2003. WinBUGS Version 1.4, User Manual. MRC Biostatistics Unit, Cambridge, UK.
- Tang, K., Ngoi, S.M., Gwee, P.C., Chua, J.M., Lee, E.J., Chong, S.S., and Lee, C.G. 2002. Distinct haplotype profiles and strong linkage disequilibrium at the MDR1 multidrug transporter gene locus in three ethnic Asian populations. Pharmacogenetics 12: 437–450. [DOI] [PubMed] [Google Scholar]
- Thiebaut, F., Tsuruo, T., Hamada, H., Gottesman, M.M., Pastan, I., and Willingham, M.C. 1987. Cellular localization of the multidrug-resistance Proc. Natl. Acad. Sci. 84: 7735–7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant, R., Xiao, H., Ingles, C.J., and Greenblatt, J. 1993. Direct interaction between the transcriptional activation domain of human p53 and the TATA box-binding protein. J. Biol. Chem. 268: 2284–2287. [PubMed] [Google Scholar]
- Wandel, C., Kim, R., Wood, M., and Wood, A. 2002. Interaction of morphine, fentanyl, sufentanil, alfentanil, and loperamide with the efflux drug transporter P-glycoprotein. Anesthesiology 96: 913–920. [DOI] [PubMed] [Google Scholar]
- Weale, M.E. and Goldstein, D.B. 2003. TagIT User Guide Version 1.14 (02 January 2003). http://popgen.biol.ucl.ac.uk/software.html.
- Zhao, J.H., Curtis, D., and Sham, P.C. 2000. Model-free analysis and permutation tests for allelic associations. Hum. Hered. 50: 133–139. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://popgen.biol.ucl.ac.uk/supdata.html; primer information.
- http://popgen.biol.ucl.ac.uk/software.html; TagIT and routines for the Bayesian analysis of the associated interval.
- http://www.cbil.upenn.edu/tess; TESS algorithm.
- http://eshadow.dcode.org/; eShadow.
- http://web1.iop.kcl.ac.uk/IoP/Departments/PsychMed/GEpiBSt/software.shtml; EH and PM programs.