Abstract
Fragile sites are gaps and breaks in metaphase chromosomes generated by specific culture conditions. Fragile site FRA3B is the most unstable site and is directly involved in the breakpoints of deletion and translocation in a wide spectrum of cancers. To learn about the general characteristics of common fragile sites, we investigated the chromatin structure of the FRA3B site. Because FRA3B spans several hundred kilobases, we focused our study on two breakpoint clusters found in FRA3B. Using various nucleases, we demonstrated that these two regions contain phased nucleosomes, regardless of treatment with aphidicolin. Because these regions are located in intron 4 of the FHIT gene, it is very interesting to observe phased nucleosomes over these regions, which are several hundred kilobases downstream from the promoter. Further, by using nucleosome assembly assays, we demonstrate that these two regions do not contain strong nucleosome positioning elements. These results suggest that other factors appear to cooperate with the DNA sequence of these regions to impart nucleosome phasing. This study provides the first information on the chromatin structure of breakpoint regions in a common fragile site. The observation of phased nucleosomes over these breakpoint regions could offer a foundation to understand the expression of fragile sites.
Fragile sites are DNA loci that are especially sensitive to the formation of gaps or breaks on metaphase chromosomes when cultured with aphidicolin, 5-bromodeoxyuridine, or under other stress conditions (Richards 2001). Instability of fragile sites has been implicated in the incidence of certain cancers and other severe disorders. On the basis of their frequency in the human population, fragile sites are typically classified into two major groups, rare and common (Sutherland 1991). All rare fragile sites sequenced to date, such as FRAXA, a site directly linked to fragile X syndrome, contain expanded repeating sequences. On the other hand, common sites are found in all individuals and do not contain those repeating sequences. Common fragile sites have been shown to be involved in development of numerous cancers through the mechanism of large-scale chromosomal abnormalities, such as deletions and rearrangements. One of these, FRA3B, is the most inducible fragile region in the human genome and is located within the FHIT gene, spanning about 300 kb between exon 4 and the centromeric end of intron 5 (Fig. 1; Ohta et al. 1996; Boldog et al. 1997). Abnormal expression of the FHIT gene has been observed in various cancers, such as digestive tract, lung, and breast cancers. From tumor cell lines and aphidicolininduced hybrid cells, many deletions and rearrangements of the FHIT gene have been mapped to the FRA3B site, suggesting that instability of FRA3B contributes to the development of various cancers.
Figure 1.
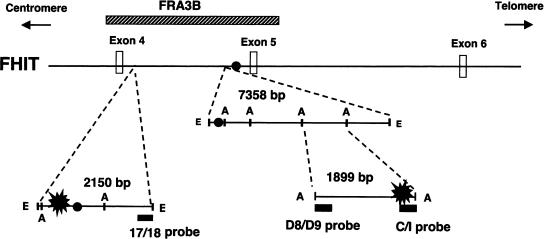
Genomic organization of the FHIT/FRA3B region. The FHIT sequence is shown from intron 3 through intron 6. The position of the unstable FRA3B region is indicated by a hatched bar. The major breakpoint clusters are marked by black blast symbols within the 2150-bp EcoRI fragment of the proximal region and the 1899-bp AflIII fragment of the distal region. Solid circles represent the locations of Alu elements. (E) EcoRI; (A) AflIII.
The instability of common fragile sites has been elegantly demonstrated to be governed by the S-phase and G2/M checkpoint protein, Ataxia-Telangiectasia, and Rad3-related protein kinase (ATR; Casper et al. 2002). In ATR-deficient cells, fragile site expression increases up to 20-fold with aphidicolin induction and, even in the absence of aphidicolin, is present at a similar level as for aphidicolin-treated control cells. Delayed replication has been observed in several fragile sites; FRA3B (Le Beau et al. 1998; Wang et al. 1999), FRAXA (Subramanian et al. 1996; Hansen et al. 1997), FRA7H (Hellman et al. 2000), and FRA7G (Hellman et al. 2002). Therefore, fragile sites are proposed to be unreplicated regions, possibly caused by a stalled replication fork, which the ATR replication checkpoint fails to repair. However, it is not clear why fragile sites are more susceptible to delayed replication than other regions.
DNA sequence itself and/or other factors, such as chromatin structure, might cause difficulties for passage of the replication fork. The CGG-repeating sequence derived from the rare fragile site FRAXA has been shown in vitro to block DNA synthesis (Usdin and Woodford 1995). However, common sites do not contain such repeating sequence, but they often exhibit high DNA flexibility. For example, high-flexibility DNA sequences have been found near the aphidicolin-induced breakpoint regions in FRA3B, FRA7H, and a partial sequence of FRA7G (Mishmar et al. 1998; Mimori et al. 1999). In addition, high-flexibility DNA has been shown to affect the affinity of interaction between DNA and protein, such as histones and transcriptional activators (Sarai et al. 1989; Travers and Drew 1997). Therefore, it is important to examine the chromatin structure of common fragile sites in order to understand the mechanistic details that underlie fragile site expression. Studies of chromatin structure of the FRAXA site have revealed a heterochromatin-like structure (Eberhart and Warren 1996; Coffee et al. 2002). Whereas in vitro binding studies have shown that CGG repeating sequences derived from FRAXA display a low affinity to bind histone octamers (Godde et al. 1996; Wang and Griffith 1996; Wang et al. 1996) and that DNA from another rare site (FRA16B) excludes nucleosomes in the presence of the fragile site inducer distamycin (Hsu and Wang 2002). Kerem and colleagues used FISH analysis to demonstrate that induction of fragile sites could lead to a perturbed chromatin organization in the FRA7G site (Hellman et al. 2002). However, no studies have been carried out to investigate the chromatin structure of common fragile sites in detail.
To learn whether an unusual chromatin structure is present at common fragile sites, which could be one of the factors causing a delayed replication fork, we investigated the chromatin structure of the FRA3B breakpoint regions in this report. Because large regions (over hundreds of kilobases) found in five common fragile sites characterized so far have shown breaks or gaps in metaphase chromosomes (Huebner and Croce 2001), we focused our study on two breakpoint clusters found in FRA3B on the basis of the following reasons. First, by examining aphidicolin-induced hybrid cells, these regions are two major clusters of aphidicolin-induced breakpoints in FRA3B (Wang et al. 1997). Both are located within intron 4 of the FHIT gene. The proximal breakpoint cluster (∼200 bp) is located at the centromeric end of the 2.2-kb EcoRI fragment, and the distal cluster (∼350 bp) is near the telomeric end of the 1.9-kb AflIII fragment (Fig. 1). Second, the EcoRI and AflIII fragments are highly AT-rich, and contain the vertebrate topoisomerase II consensus sequences and the core autonomously replicating sequences (Wang et al. 1997). Third, these two major breakpoint regions have been shown to be located very close to high-flexibility DNA sequences (Mimori et al. 1999; Corbin et al. 2002). In fact, a high-flexibility DNA sequence, which is included in the 1.9-kb AflIII fragment, is only 0.35 kb centrometric of the distal breakpoint region. In total, these features suggest that possible replication-related elements are located in these two regions and prompt us to examine their chromatin structure.
Using restriction enzymes as well as DNase I and micrococcal nuclease (MNase), we demonstrated that these two regions of FRA3B contain phased nucleosomes, regardless of treatment with aphidicolin. They do not form a heterochromatin-like structure as found in the FRAXA site (Eberhart and Warren 1996; Coffee et al. 2002), and no DNase I hypersensitive sites are observed in these regions. Further, using in vitro nucleosome assembly and MNase analysis, we examined whether the DNA sequence is the cause of the nucleosome phasing that we observed in these regions. We found that DNA sequences derived from these two major breakpoint regions do not form phased nucleosomes in vitro, suggesting that other factors, not DNA sequences of these two regions alone contribute to this nucleosome phasing phenomenon in cells.
RESULTS
The Chromatin Structure of FRA3B Breakpoint Regions Is Not Heterochromatinized
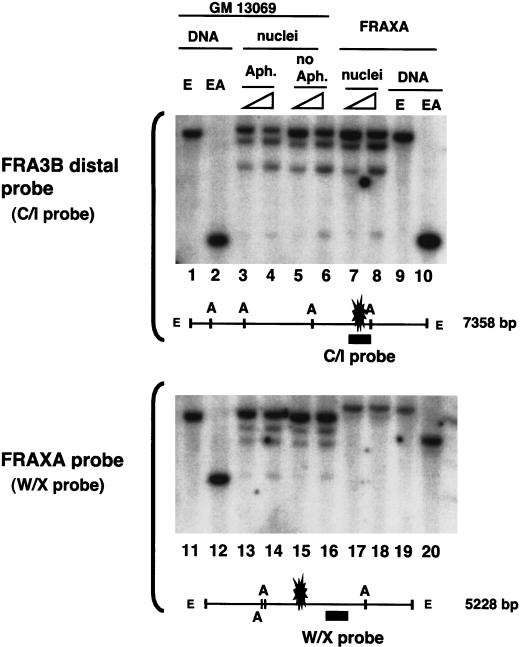
To evaluate the chromatin packaging of the FRA3B breakpoint regions, the restriction enzyme accessibility was used to compare the chromatin structure between the FRA3B and FRAXA sites. The normal lymphoblastoid cell line GM13069 was determined by cytogenetic analysis to contain the FRA3B at the 3p14.2 position of the metaphase chromosome when treated with 0.4 μM aphidicolin, a concentration that has been used in many studies for observing the breakpoints in FRA3B. Therefore, nuclei isolated from aphidicolin-treated and untreated GM 13069 cells and from the FRAXA cells were incubated with increasing amounts of AflIII, and the purified DNA was further digested to generate a parental DNA fragment with EcoRI. By using a FRA3B distal breakpoint specific probe (Fig. 2, top), cleavage at the AflIII sites flanking the distal breakpoint region within the 7.4-kb EcoRI fragment was observed with no apparent hypersensitive or resistant sites. Aphidicolin treatment has no significant effect on the chromatin structure. On the other hand, probing with FRAXA site-specific DNA (Fig. 2, bottom), all three AflIII sites within the EcoRI fragment containing the FRAXA breakpoint sites in the FRAXA cells (Fig. 2, lanes 17,18) were much less accessible than that of GM13069, which does not contain the FRAXA site (Fig. 2, lanes 13–16), indicating a condensed chromatin structure present in the FRAXA site as shown previously (Eberhart and Warren 1996; Coffee et al. 2002). The FRA3B proximal breakpoint region was also examined with a specific probe, and cleavage at the AflIII site was observed to be similar to the distal region (data not shown). This suggests that the FRA3B breakpoint regions are not in a condensed chromatin and, therefore, are different from the heterochromatin-like structure seen in the FRAXA site.
Figure 2.
The FRA3B distal breakpoint region does not have a condensed chromatin structure. Nuclei isolated from aphidicolin-treated (lanes 3,4,13,14), untreated GM 13069 cells (lanes 5,6,15,16) or from FRAXA cells (lanes 7,8,17,18) were incubated with 25 or 100 U of AflIII (indicated by triangles), and purified DNA was completely digested with EcoRI. Genomic DNA from the GM 13069 and FRAXA cells was completely digested with EcoRI (lanes 1,9,11,19) or EcoRI and AflIII (lanes 2,10,12,20). Southern blot analysis was carried out first with the FRA3B distal region specific probe (the C/I probe, top), then reprobing with the W/X probe, specific for the FRAXA site (bottom). The EcoRI fragment of the FRAXA site is 5.2 kb in GM13069 cells, whereas due to CGG expansion, it is 6.8 kb in FRAXA expressing cells. The positions of the distal breakpoint cluster of the FRA3B site and the breakpoint region (the CGG repeating sequence) of the FRAXA site are indicated by blasts. (E) EcoRI; (A) AflIII.
FRA3B Breakpoint Regions Contain No Detectable DNase I Hypersensitive Sites
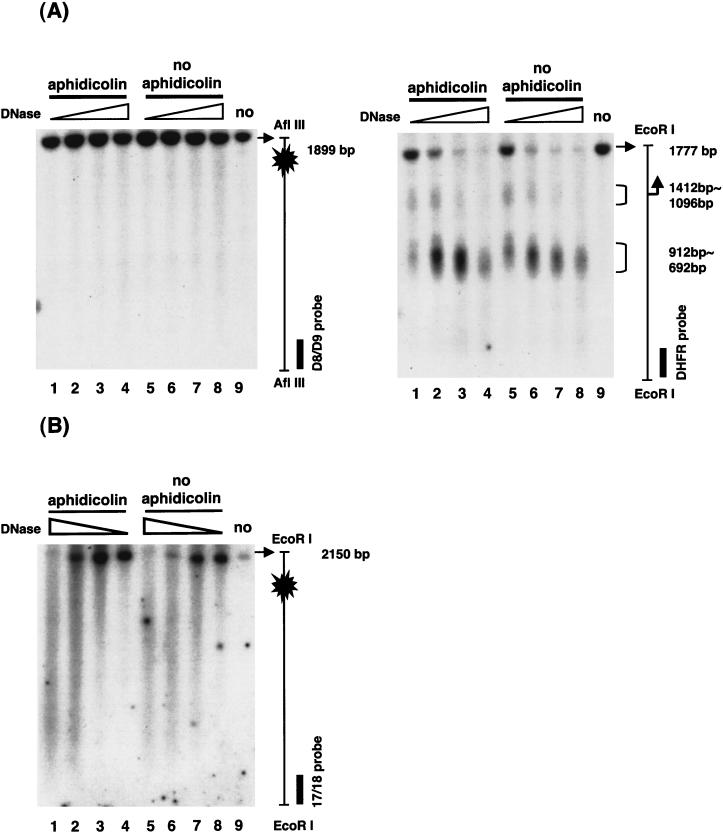
We next examined whether these two breakpoint regions contain any DNase I hypersensitive sites that usually represent nucleosome-free regions. Nuclei isolated from aphidicolin-treated and untreated cells were incubated with increasing amounts of DNase I, and the purified DNA was further digested and analyzed by Southern blotting and indirect end labeling to examine the pattern of DNase I sensitivity. As a control to insure the condition of DNase I digestion, the promoter region of the DHFR gene was examined. As expected from Shimada et al. (1986), two major DNase I hypersensitive sites were observed, one spanning the TATA box, and the other located at the adjacent upstream promoter region (Fig. 3A, right). However, when the same blots were probed with FRA3B-specific probes, no distinct DNase I hypersensitive sites were found in either breakpoint region of FRA3B (Fig. 3A, left, distal region, and B, proximal region). Cells treated with aphidicolin showed no significant difference in DNase I sensitivity from untreated cells. This result suggests that both FRA3B major breakpoint sites do not contain nucleosome-free regions.
Figure 3.
No DNase I hypersensitive sites in either breakpoint region of FRA3B. Nuclei were isolated from aphidicolin-treated and untreated cells and incubated with DNase I at 10, 25, 50, and 100 U/mL at 37°C for 5 min. Reactions were stopped by a solution containing EDTA and EGTA and DNA was extracted. (A) For probing the FRA3B distal breakpoint region, DNA was further digested with EcoRI and AflIII and hybridized with the D8/D9 probe to visualize a 1899-bp AflIII fragment of the distal breakpoint region (the 1899-bp AflIII fragment of FRA3B does not contain an internal EcoRI site). The blot was also hybridized with the C/I probe (see Fig. 1) to examine the same fragment from the other end (data not shown). As a control to insure the condition of DNase I digestion, the same blot was hybridized with the DHFR probe to visualize a 1777-bp EcoRI fragment of the DHFR promoter region (the 1777-bp EcoRI fragment of the DHFR gene does not contain an internal AflIII site). (B) For probing the proximal breakpoint region, DNase I-treated DNA was further digested with EcoRI and hybridized with the 17/18 probe to visualize a 2150-bp EcoRI fragment of the FRA3B proximal breakpoint region. The same blot was hybridized with the DHFR probe to visualize a 1777-bp EcoRI fragment of the DHFR promoter region (data not shown). The major breakpoint clusters are marked by blasts within the 1899-bp AflIII fragment of the distal region and the 2150-bp EcoRI fragment of the proximal region. The transcription initiation site of the DHFR gene is indicated by a right angle arrow.
Nucleosomes Are Phased on DNA of The FRA3B Breakpoint Clusters
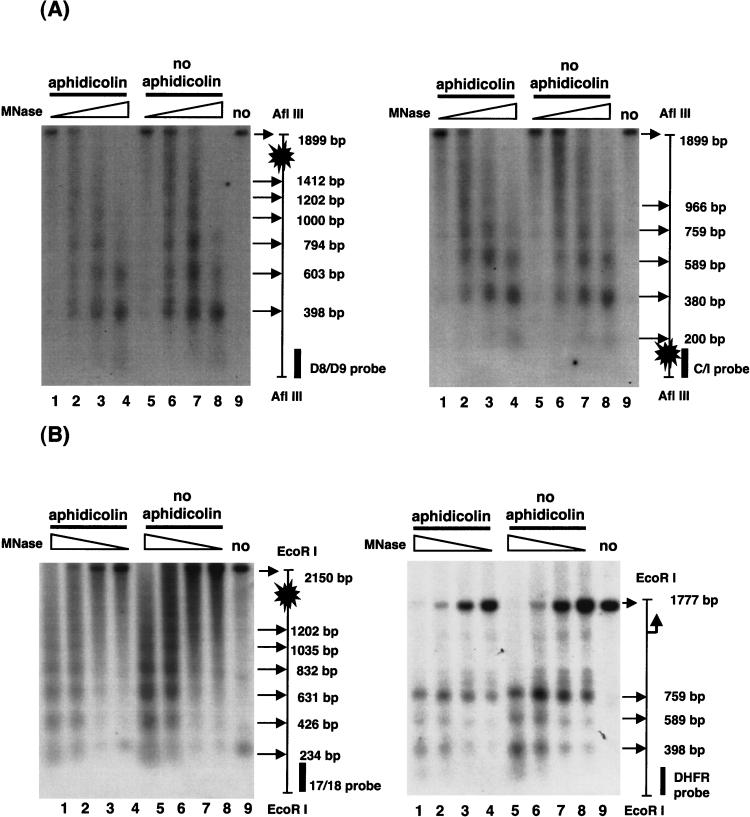
We then examined the arrangement of nucleosomes in the FRA3B breakpoint regions by digestion with MNase. Nuclei isolated from aphidicolin-treated and untreated cells were incubated with increasing amounts of MNase, and the purified DNA was further digested and analyzed by Southern blotting and indirect end labeling. An ∼200 bp periodicity in MNase cleavage was observed over the FRA3B distal breakpoint region (Fig. 4A, left), and when the same blot was hybridized with the C/I probe (Fig. 1) to the other end of the same fragment, similar periodicity was observed (Fig. 4A, right). The same pattern was also found in the proximal region (Fig. 4B, left), indicating that nucleosomes are present in fixed and contiguous positions, not randomly distributed, over both FRA3B breakpoint regions. However, when the blot of Figure 4B was examined with the DHFR probe (Fig. 4B, right), nucleosome-free regions were present around the TATA box and adjacent upstream promoter region, and three positioned nucleosomes were found in the upstream region of the DHFR gene (agreeing with the DNase I mapping in Fig. 3A and previously published results in Shimada et al. 1986). This suggests that the FRA3B breakpoint regions contain contiguously phased nucleosomes.
Figure 4.
Both major breakpoint regions of FRA3B contain contiguously phased nucleosomes. Nuclei were isolated from aphidicolin-treated and untreated cells and incubated with MNase at 0.00375, 0.0125, 0.0375, and 0.125 U/mL at 37°C for 5 min. Reactions were stopped and DNA was extracted. (A) The Southern blot probing strategy was the same as described in Fig. 3. For probing the FRA3B distal breakpoint region, MNase-treated DNA was further digested with EcoRI and AflIII and hybridized with the D8/D9 probe (left) and the C/I probe (right). The same blot was hybridized with the DHFR probe (data not shown) (B) For probing the proximal breakpoint region, MNase-treated DNA was further digested with EcoRI and hybridized with the 17/18 probe. The same blot was hybridized with the DHFR probe to examine the DHFR promoter region. Symbols are the same as in Figure 3.
Other Factors, Not DNA Sequences of the FRA3B Breakpoint Regions Alone, Contribute to the Nucleosome Phasing in Cells
To examine the possibility of strong nucleosome positioning sequence(s) residing in the distal and proximal breakpoint regions, in vitro chromatin assembly was used with plasmids containing the 1899-bp AflIII fragment of the distal breakpoint region and the 2150-bp EcoRI fragment of the proximal breakpoint region. (Note: the 1899-bp AflIII fragment also contains a high-flexibility DNA sequence [Fig. 5; Mimori et al. 1999]).
Figure 5.
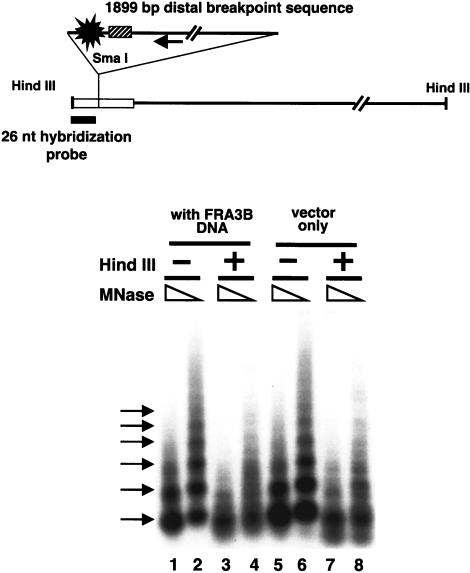
The formation of nucleosome arrays on a plasmid containing a 1899-bp FRA3B distal breakpoint sequence. A diagram of the plasmid with a 1899-bp FRA3B distal breakpoint sequence inserted at the SmaI site of the polylinker region (white bar) of the pGEM3zf(+) vector is shown. The single arrow indicates the direction of the FHIT gene. The position of the distal breakpoint cluster of the FRA3B site is indicated by a blast. The position of the high-flexibility DNA sequence is indicated by a hatched bar. (E) EcoRI; (A) AflIII. RSF-assembled plasmids (lanes 1–4, the plasmid containing the FRA3B sequence; lanes 5–8, the parental plasmid) were partially digested by two different concentrations of MNase. The MNase-treated DNA was purified, and half of the DNA was used directly in lanes 1, 2, 5, and 6. The other half of the DNA samples was further digested by HindIII (lanes 3,4,7,8), and the MNase digestion patterns were analyzed by Southern blots. The 26-nt hybridization probe is located between the HindIII and SmaI sites. The multiple arrows to the left indicate the nucleosome ladder.
Human histones from HeLa cells were mixed with these pGEM3zf(+)-based plasmids to assemble nucleosome arrays with RSF, and the assembled plasmids were partially digested by MNase. Half of the MNase-treated DNA was further digested by HindIII to determine the positions of MNase cleavage sites relative to the HindIII site within the polylinker region of the plasmid. The MNase digestion pattern of all DNA samples was analyzed by Southern blotting and indirect end-labeling (Fig. 5). Without HindIII digestion, the distal breakpoint regions showed an ∼180 bp ladder (Fig. 5, lanes 1 and 2) similar to the parental plasmid (Fig. 5 lanes 5 and 6), indicating that both sequences formed regularly spaced nucleosome arrays. As expected, the nucleosome ladders were lost for the parental plasmid upon HindIII digestion (Fig. 5 lanes 7 and 8), because the parental vector does not contain strong nucleosome positioning sequences. Interestingly, the nucleosome ladders were also lost for the FRA3B distal breakpoint sequence upon HindIII digestion (Fig. 5 lanes 3 and 4). These results suggest that the distal breakpoint sequence cannot form positioned nucleosomes in the same manner on each molecule in the reconstituted arrays, in contrast to the strong positioning of nucleosomes on the same sequence in cells (Fig. 4). The plasmid containing the 2150-bp EcoRI fragment of the proximal breakpoint region displayed a MNase digestion pattern (data not shown) similar to the distal region, implying that other factors beside the DNA sequences of the two breakpoint regions are responsible for the nucleosome phasing observed in cells.
DISCUSSION
In this study, the chromatin structure of the two major aphidicolin-induced breakpoint regions of FRA3B, the most inducible common fragile site, were examined. By restriction enzyme accessibility, we found that the chromatin structure of these regions is not as condensed as that in the FRAXA site (Fig. 2; Eberhart and Warren 1996; Coffee et al. 2002). From DNase I and MNase mapping, no hypersensitive sites were observed in these regions, and nucleosomes are phased over both regions. In vitro binding assays did not show DNA sequences derived from these regions to be strong nulceosome positioning elements, indicating that other factors cooperate with the DNA sequence of these regions to impart nucleosome phasing.
FRA3B is located within an actively transcribed gene, FHIT. It has been shown that aphidicolin induction of FRA3B does not change FHIT transcription (Michael and Rajewsky 2001). For an active gene, well-positioned nucleosomes are often found adjacent to nucleosome-free regions, which typically contain promoters and activator binding sites. Nonhistone proteins bind to these sites and allow nucleosomes on either side of the sites to arrange at regular intervals. For example, the DHFR gene used in our study contains three phased nucleosomes upstream of the hypersensitive promoter region (Fig. 4B). The two major breakpoint regions of FRA3B, proximal and distal, are about 500 and 750 kb, respectively, downstream of the transcription start site (Ohta et al. 1996; Corbin et al. 2002). Because of variations in the length of the linker DNA of the chromatin, the effects of nucleosome phasing gradually diminish when it is increasingly far away from the nonhistone binding sites. Regions remotely downstream from the promoter, such as the location of the two breakpoint regions, are expected to have randomly positioned nucleosomes. Therefore, it is very interesting to reveal nucleosome phasing over these regions, which suggests an opportunity for gene regulation (see below).
Because these breakpoint regions do not contain strong nucleosome positioning sequences, we suspect that other factors and/or other DNA sequences outside the regions could serve as boundaries to cause the nucleosome phasing over these two breakpoint regions. Examining DNA sequences around the breakpoint sites, we found two partial Alu elements, each about 2 and 3 kb from the 1.9-kb distal region, and one Alu element within the 2.2-kb proximal region (Fig. 1). Human Alu elements have been shown to be associated with phased nucleosomes (Englander et al. 1993; Englander Howard 1995), and could possibly direct the nucleosome positioning that we observe. However, the 2.2-kb fragment fails to form phased nucleosomes by in vitro assembly assay, suggesting that this Alu element cannot position nucleosomes. In addition, the sequences of all three Alu elements do not contain the box A and box B promoters, which are important for nucleosome positioning (Englander et al. 1993). Further, the primers used in the study of Englander and Howard (1995) to observe phased nucleosomes in vivo do not anneal to these three Alu sequences. Therefore, the nucleosome positioning that we observed at the breakpoint clusters is not caused by the Alu repeats.
The inherent nature of phased nucleosomes can restrict certain DNA sequences to be always either wrapped within nucleosomes or be in the linker region. The 2.2-kb EcoRI and the 1.9-kb AflIII fragments that we examined in this study have properties consistent with origins of replication (Wang et al. 1997). Therefore, we suggest that when the replication origin sequences are continuously wrapped in nucleosomes, it could serve as a mechanism for delay in replication initiation and cause the replication fork to stall. This mechanism, combined with the presence of aphidicolin, an inhibitor of DNA polymerases, would severely slow down the replication fork through the FRA3B site. Le Beau et al (1998) have shown that replication of FRA3B occurs late in S phase and is delayed further upon aphidicolin induction. However, it is not clear whether late replication found in FRA3B is due to delayed replication initiation, slowed replication fork progress, or other factors.
In summary, the results from this study provide the first insights about chromatin structure of breakpoint regions in a common fragile site. Whether or not phased nucleosomes are a feature of the FRA3B site as a whole, and whether such nucleosomes are responsible for the chromosome fragility remains to be seen.
METHODS
DNAs and Proteins
Genomic DNA for endonuclease digestion and Southern blot was isolated from the cells by a procedure described in Miller et al. (1988). The 2150-bp EcoRI fragment of the proximal breakpoint region of the FRA3B site (Fig. 1) is located from nucleotides 134736 to 136886 of the GenBank accession no. AF152363 sequence (Mimori et al. 1999) in the antisense orientation, and the 1899-bp AflIII distal breakpoint region is from nucleotides 38228 to 40127 of the GenBank accession no. U66722 sequence (Boldog et al. 1997) in the antisense orientation. The pUC 18-based plasmid containing the 2150-bp EcoRI fragment was kindly provided by D.I. Smith (Mayo Foundation; Wang et al. 1997). The plasmid containing the 1899-bp AflIII fragment was constructed with the pGEM3zf(+) vector by inserting a fragment generated by PCR of human genomic DNA. The proximal probe (17/18, nucleotides 134736∼134949 of AF152363) and the distal probes (D8/D9 and C/I, nucleotides 39896∼40080, and nucleotides 38228∼38641 of U66722, respectively), were obtained by PCR of the above respective plasmids and isolated by agarose gel electrophoresis. The dihydrofolate reductase (DHFR) probe contains the 5′ flanking region of the human DHFR gene from the EcoRI site at –1252 to the RsaI site at –994 (Shimada et al. 1986). The FRAXA probe is a 448-bp fragment isolated from plasmid pE5.1 (Pieretti et al. 1991) and located 121 bp downstream from the CGG repeat block. HeLa cell histone octamers were isolated as previously described (Hsu and Wang 2002). RSF was kindly provided by D. Reinberg (Robert Wood Johnson Medical School).
Cell Culture and Cytogenetic Analysis
The normal lymphoblastoid cell line GM13069 transformed by Epstein-Barr-virus was obtained from Coriell Cell Repositories. For aphidicolin treatment, GM13069 cells were grown to 105 cells/mL and treated with 0.4 μM aphidicolin (Sigma) for 24 h. For fragile site analysis, cells were analyzed in the University Diagnostic Laboratories (RWJMS-UMDNJ) by the GTG bending method. FRAXA cell line GM3200A was kindly provided by S. Warren (Emory University).
Restriction Enzyme, DNAse I, and MNase Accessibility Analysis
Nuclei were isolated from aphidicolin-treated and control cells by lysing cell pellets in chilled homogenization buffer (10 mM Tris-HCl at pH 7.4, 1 mM EDTA, 15 mM NaCl, 60 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 0.2% Nonident P-40, 5% sucrose, 0.1 mM PMSF) and centrifuging the homogenate through a cushion of 10% sucrose in homogenization buffer for 20 min at 1600g. Isolated nuclei were suspended in restriction enzyme digestion buffers (as specified by the suppliers) for restriction enzyme digestion, in a buffer containing 40 mM Tris-HCl (pH 7.5), 10 mM MgSO4, 1 mM CaCl2 for DNase I digestion, and in a buffer containing 50 mM Tris-HCl (pH 7.4), 60 mM KCl, 3 mM CaCl2, 0.34 M sucrose for MNase digestion. The amounts of endonucleases used are indicated in the figure legends. DNA was purified by proteinase K and RNase A treatment, restricted to completion with appropriate enzymes, and analyzed by Southern blot hybridization.
Assembly and Analysis of Nucleosome Arrays
Nucleosome assembly reactions were performed as described in Loyola et al. (2001) with 1 μg of plasmid DNA, 0.9 μg of histones, 0.15 μg of RSF, 75 μg of bovine serum albumin, 3 mM ATP, 30 mM phosphocreatine, 0.2 μg of phosphocreatine kinase, 5 mM MgCl2, 60 mM KCl, 10 mM Hepes (pH 7.6), 0.2 mM EDTA, and 5% glycerol. Reactions were incubated at 30°C for 16 h. The reaction mixtures were then partially digested with two different concentrations of MNase (0.0125 and 0.125 units), and the MNase-treated DNA was purified. Half of each DNA sample was further digested by HindIII, and the MNase digestion patterns were analyzed by Southern blots.
Acknowledgments
We thank Danny Reinberg for RSF protein, David. I. Smith for the pUC 18-based plasmid containing the 2150-bp EcoRI fragment from the proximal breakpoint region of the FRA3B site, and Hailin Wang, Ying Ying Hsu, and Hussein Mastafa for technical support. This work was supported by Public Health Service grant CA85826 from the National Cancer Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2304404.
Footnotes
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: D. Reinberg, S. Warren, and D.I. Smith.]
References
- Boldog, F., Gemmill, R.M., West, J., Robinson, M., Robinson, L., Li, E., Roche, J., Todd, S., Waggoner, B., Lundstrom, R., et al. 1997. Chromosome 3p14 homozygous deletions and sequence analysis of FRA3B. Hum. Mol. Genet. 6: 193–203. [DOI] [PubMed] [Google Scholar]
- Casper, A.M., Nghiem, P., Arlt, M.F., and Glover, T.W. 2002. ATR regulates fragile site stability. Cell 111: 779–789. [DOI] [PubMed] [Google Scholar]
- Coffee, B., Zhang, F., Ceman, S., Warren, S.T., and Reines, D. 2002. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. Am. J. Hum. Genet. 71: 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin, S., Neilly, M.E., Espinosa III, R., Davis, E.M., McKeithan, T.W., and Le Beau, M.M. 2002. Identification of unstable sequences within the common fragile site at 3p14.2: Implications for the mechanism of deletions within fragile histidine triad gene/common fragile site at 3p14.2 in tumors. Cancer Res. 62: 3477–3484. [PubMed] [Google Scholar]
- Eberhart, D.E. and Warren, S.T. 1996. Nuclease sensitivity of permeabilized cells confirms altered chromatin formation at the fragile X locus. Somat. Cell Mol. Genet. 22: 435–441. [DOI] [PubMed] [Google Scholar]
- Englander, E.W. and Howard, B.H. 1995. Nucleosome positioning by human Alu elements in chromatin. J. Biol. Chem. 270: 10091–10096. [DOI] [PubMed] [Google Scholar]
- Englander, E.W., Wolffe, A.P., and Howard, B.H. 1993. Nucleosome interactions with a human Alu element. Transcriptional repression and effects of template methylation. J. Biol. Chem. 268: 19565–19573. [PubMed] [Google Scholar]
- Godde, J.S., Kass, S.U., Hirst, M.C., and Wolffe, A.P. 1996. Nucleosome assembly on methylated CGG triplet repeats in fragile X mental retardation gene 1 promoter. J. Biol. Chem. 271: 24325–24328. [DOI] [PubMed] [Google Scholar]
- Hansen, R.S., Canfield, T.K., Fjeld, A.D., Mumm, S., Laird, C.D., and Gartler, S.M. 1997. A variable domain of delayed replication in FRAXA fragile X chromosomes: X inactivation-like spread of late replication. Proc. Natl. Acad. Sci. 94: 4587–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman, A., Rahat, A., Scherer, S.W., Darvasi, A., Tsui, L.C., and Kerem, B. 2000. Replication delay along FRA7H, a common fragile site on human chromosome 7, leads to chromosomal instability. Mol. Cell. Biol. 20: 4420–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman, A., Zlotorynski, E., Scherer, S.W., Cheung, J., Vincent, J.B., Smith, D., Trakhtenbrot, L., and Kerem, B. 2002. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell 1: 89–97. [DOI] [PubMed] [Google Scholar]
- Hsu, Y.-Y. and Wang, Y.-H. 2002. Human fragile site FRA16B DNA excludes nucleosomes in the presence of distamycin. J. Biol. Chem. 277: 17315–17319. [DOI] [PubMed] [Google Scholar]
- Huebner, K. and Croce, C.M. 2001. FRA3B and other common fragile sites: The weakest links. Nat. Rev. Cancer 1: 214–221. [DOI] [PubMed] [Google Scholar]
- Le Beau, M.M., Rassool, F.V., Neilly, M.E., Espinosa III, R., Glover, T.W., Smith, D.I., and McKeithan, T.W. 1998. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: Implications for the mechanism of fragile site induction. Hum. Mol. Genet. 7: 755–761. [DOI] [PubMed] [Google Scholar]
- Loyola, A., LeRoy, G., Wang, Y.-H., and Reinberg, D. 2001. Reconstitution of recombination chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes & Dev. 15: 2837–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, D. and Rajewsky, M.F. 2001. Induction of the common fragile site FRA3B does not affect FHIT expression. Oncogene. 20: 1798–1801. [DOI] [PubMed] [Google Scholar]
- Miller, S.A., Dykes, D.D., and Polesky, H.F. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori, K., Druck, T., Inoue, H., Alder, H., Berk, L., Mori, M., Huebner, K., and Croce, C.M. 1999. Cancer-specific chromosome alterations in the constitutive fragile region FRA3B. Proc. Natl. Acad. Sci. 96: 7456–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar, D., Rahat, A., Scherer, S.W., Nyakatura, G., Hinzmann, B., Kohwi, Y., Mandel-Gutfroind, Y., Lee, J.R., Drescher, B., Sas, D.E., et al. 1998. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc. Natl. Acad. Sci. 95: 8141–8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, M., Inoue, H., Cotticelli, M.G., Kastury, K., Baffa, R., Palazzo, J., Siprashvili, Z., Mori, M., McCue, P., Druck, T., et al. 1996. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell 84: 587–597. [DOI] [PubMed] [Google Scholar]
- Pieretti, M., Zhang, F., Fu, Y.H., Warren, S.T., Oostra, B.A., Caskey, C.T., and Nelson, D.L. 1991. Absence of expression of the FMR1 gene in fragile X syndrome. Cell 66: 817–822. [DOI] [PubMed] [Google Scholar]
- Richards, R.I. 2001. Fragile and unstable chromosomes in cancer: Causes and consequences. Trends Genet. 17: 339–345. [DOI] [PubMed] [Google Scholar]
- Sarai, A., Mazur, J., Nussinov, R., and Jernigan, R.L. 1989. Sequence dependence of DNA conformational flexibility. Biochemistry 28: 7842–7849. [DOI] [PubMed] [Google Scholar]
- Shimada, T., Inokuchi, K., and Nienhuis, A.W. 1986. Chromatin structure of the human dihydrofolate reductase gene promoter. Multiple protein-binding sites. J. Biol. Chem. 261: 1445–1452. [PubMed] [Google Scholar]
- Subramanian, P.S., Nelson, D.L., and Chinault, A.C. 1996. Large domains of apparent delayed replication timing associated with triplet repeat expansion at FRAXA and FRAXE. Am. J. Hum. Genet. 59: 407–416. [PMC free article] [PubMed] [Google Scholar]
- Sutherland, G.R. 1991. Chromosomal fragile sites. Genet. Anal. Tech. Appl. 8: 161–166. [DOI] [PubMed] [Google Scholar]
- Travers, A. and Drew, H. 1997. DNA recognition and nucleosome organization. Biopolymers 44: 423–433. [DOI] [PubMed] [Google Scholar]
- Usdin, K. and Woodford, K.J. 1995. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 23: 4202–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Paradee, W., Mullins, C., Shridhar, R., Rosati, R., Wilke, C.M., Glover, T.W., and Smith, D.I. 1997. Aphidicolin-induced FRA3B breakpoints cluster in two distinct regions. Genomics 41: 485–488. [DOI] [PubMed] [Google Scholar]
- Wang, L., Darling, J., Zhang, J.S., Huang, H., Liu, W., and Smith, D.I. 1999. Allele-specific late replication and fragility of the most active common fragile site, FRA3B. Hum. Mol. Genet. 8: 431–437. [DOI] [PubMed] [Google Scholar]
- Wang, Y.-H. and Griffith, J.D. 1996. Methylation of expanded CCG triplet repeat DNA from fragile X syndrome patients enhances nucleosome exclusion J. Biol. Chem. 271: 22937–22940. [PubMed] [Google Scholar]
- Wang, Y.-H., Gellibolian, R., Shimizu, M., Wells, R.D. and Griffith, J.D. 1996. Long CCG triplet repeat blocks exclude nucleosomes: A possible mechanism for the nature of fragile sites in chromosomes. J. Mol. Biol. 263: 511–516. [DOI] [PubMed] [Google Scholar]