Abstract
Homologous recombination (HR) is an indispensable tool to modify the genome of yeast and mammals. More recently HR is also being used for gene targeting in Drosophila. Here we show that HR can be used efficiently to engineer chromosomal rearrangements such as pericentric and paracentric inversions and translocations in Drosophila. Two chromosomal double-strand breaks (DSBs), introduced by the rare-cutting I-SceI endonuclease on two different mobile elements sharing homologous sequences, are sufficient to promote rearrangements at a frequency of 1% to 4%. Such rearrangements, once generated by HR, can be reverted by Cre recombinase. However, Cre-mediated recombination efficiency drops with increasing distance between recombination sites, unlike HR. We therefore speculate that physical constraints on chromosomal movement are modulated during DSB repair, to facilitate the homology search throughout the genome.
The use of chromosomal rearrangements has a long tradition in Drosophila genetics. For example, chromosomal inversions are indispensable in balancing lethal mutations, deficiencies have been important for mapping recessive mutations, and translocations have been used to analyze chromosome segregation during meiosis. Most of these inversions, translocations, and deletions were induced by random mutagenesis with ionizing radiation (Lindsley and Zimm 1992). Despite a large collection of random chromosomal rearrangements in Drosophila, there is a need for precisely defined deletions, inversions, and other molecular lesions, because in many cases it has been difficult to separate phenotypic effects intrinsic to the rearrangement from gene mutations at the breakpoint, or from secondary mutations linked to the rearrangement. Recently, a method was introduced to generate specific chromosomal rearrangements in Drosophila. It relies on two P-elements located at different positions in the same genome, each of which carry a recognition site for FLP recombinase in opposite directionality. Two partial white (w) genes in the two P-elements become functionally joined only after FLP-mediated recombination, thus permitting the selection of the chromosomal rearrangement (Golic and Golic 1996; Beumer et al. 1998). This method is currently being used to generate isogenic deficiencies covering most of the Drosophila genome (Drosdel Drosophila isogenic deficiency kit; http://www.drosdel.org.uk/; Parks et al. 2004). Mouse chromosomes can be engineered in a similar way: Cre recombinase inverts or deletes a sequence between two loxP sites, restoring the function of a selective marker (Yu and Bradley 2001). Cre recombinase, well known from its applications in the mouse, is also being used in Drosophila as an alternative system to the FLP recombinase to delete or invert sequences within a transgene (Siegal and Hartl 1996).
Chromosomal double-strand breaks (DSBs) are dangerous lesions for cells; they can result in the loss of chromosomes or chromosome segments or in the rearrangement of genetic information. Any of these outcomes are potentially lethal. DSB repair occurs by homology-dependent and homology-independent recombination mechanisms (van Gent et al. 2001). Homology-independent recombination relies on the direct joining of DNA ends and is referred to as nonhomologous end joining (NHEJ). This process is imprecise and often results in the deletion or insertion of a few nucleotides. Repair by homologous recombination (HR) relies on identical sequence stretches serving as templates and therefore usually restores the original sequence. A remarkable capacity of the cell is that a DSB can find a repair partner anywhere in the genome. The template may be on the sister chromatid, on the homologous chromosome, or ectopic: at a nonallelic position or even on a plasmid. At least in yeast and in mammalian cells, the preferred template for repair seems to be the sister chromatid (Kadyk and Hartwell 1992; Johnson and Jasin 2001). Recombination with an ectopic repair template is readily observed in yeast, but is rare in mammals (Loidl and Nairz 1997; Richardson et al. 1998; Inbar and Kupiec 1999). The choice of the repair template is of profound consequence for the organism. Most likely, the preference for the sister chromatid is to ensure that the original sequence is restored and to prevent unwanted recombination with either an allele on the homologous chromosome, leading to loss of heterozygosity, or with a gene of the same family, potentially leading to loss of the functions of the gene and to chromosomal rearrangements. This might be especially important for organisms with more complex genomes, in which interspersed repetitive sequences provide ample possibilities for ectopic recombination. Indeed, HR between the highly abundant Alu elements has been reported to contribute to cancer, genome instability, and several diseases (Deininger and Batzer 1999; Deininger and Batzer 2002; Kolomietz et al. 2002). However, in some systems the recombination with an ectopic template and a change of the original sequence is required, for example, in yeast mating type switching or in the generation of chicken antibody diversity (Nasmyth 1982; Thompson 1992).
Although the preferred repair templates for Drosophila are the sister chromatid and the homologous chromosome (Rong and Golic 2003), several examples of ectopic recombination have also been reported. DSB ends generated by the mobilization of a P-element transposon may recombine with a homologous sequence at an ectopic position on the same or on another chromosome, or even with a homologous sequence offered on a plasmid (Gloor et al. 1991; Engels et al. 1994; Keeler et al. 1996; Kolomietz et al. 2002). The ability of Drosophila to undergo ectopic recombination is successfully exploited for gene targeting: The method relies on the recombination of an episomal DNA fragment with its homologous target gene (Rong and Golic 2000, 2001; Rong et al. 2002; Seum et al. 2002; Egli et al. 2003; Gong and Golic 2003;). It is not well understood how a DSB end can find its homologous partner sequence at an ectopic position with high efficiency. Drosophila chromosomes are organized in territories and undergo specific contacts with the nuclear envelope, and at least in Drosophila embryos, telomeres and centromeres are located at different poles of the nucleus (Vazquez et al. 2001; Marshall 2002). This organization constrains the motion of chromosomes and thus has a strong impact on interactions between chromosomal loci. The efficiency of recombination by Cre recombinase or FLP is thought to reflect those interactions and thus chromosomal organization (Burgess and Kleckner 1999). Nuclear organization is likely to also restrict the ability of a DSB end to find a homologous repair template. A direct comparison of the efficiency of recombination at loxP sites and HR of a DSB may give an indication about the mechanism of the search for a homologous repair partner.
Several models have been suggested to explain DSB repair by HR in meiotic cells (for review see, Paques and Haber 1999). Single-strand annealing (SSA) can occur when a DSB appears within two direct repeats. Resection of the DSB ends produces two complementary single strands that are annealed and ligation restores two continuous strands, a process that reduces the repeat to a single copy. SSA has been shown to be a very efficient DSB repair pathway in Drosophila (Rong and Golic 2000; Preston et al. 2002). The synthesis-dependent strand annealing model (SDSA) proposed by Nassif et al. (1994) explains gene conversion in mitotic cells: The 3′ end of the DSB invades an intact homologous template and acts as a primer for new DNA synthesis. The newly synthesized strands are displaced from the template and returned to the broken molecule, allowing them to anneal to each other. This model readily explains the lack of crossover and the directionality of information flow from the intact to the broken strand during gap repair in mitotic cells.
Chromosomal rearrangements by HR at ectopic positions have been experimentally demonstrated in yeast and in mammalian cells, showing that the whole genome is efficiently scanned for repair partners during HR (Haber and Leung 1996; Richardson and Jasin 2000). Here we demonstrate that Drosophila cells are very efficient at generating inversions and translocations by HR. Some of those rearranged chromosomes could be reverted by Cre recombinase. This establishes HR and Cre recombinase as tools to engineer the Drosophila genome, similar to the FLP recombinase, but with HR being up to two or more orders of magnitude more efficient than is Cre or FLP recombinase. A comparison of the efficiency of Cre recombinase and HR to recombine homologous sequence pairs at different positions along the chromosome suggests that the search for a homologous repair partner during DSB repair is not under the same constraints as Cre-mediated recombination: Unlike Cre-mediated recombination, the efficiency of DSB repair did not decrease with increasing distance. Finally, we compare the efficiency of a chromosomal DSB with an episomal DSB to find its repair partner. We make use of the fact that gene targeting is enhanced by an I-SceI–induced DSB in the target gene (see also Smih et al. 1995). Surprisingly, we find the two processes, gene targeting and chromosomal inversion, are similarly efficient.
RESULTS
HR Between Two P-Element Transgenes
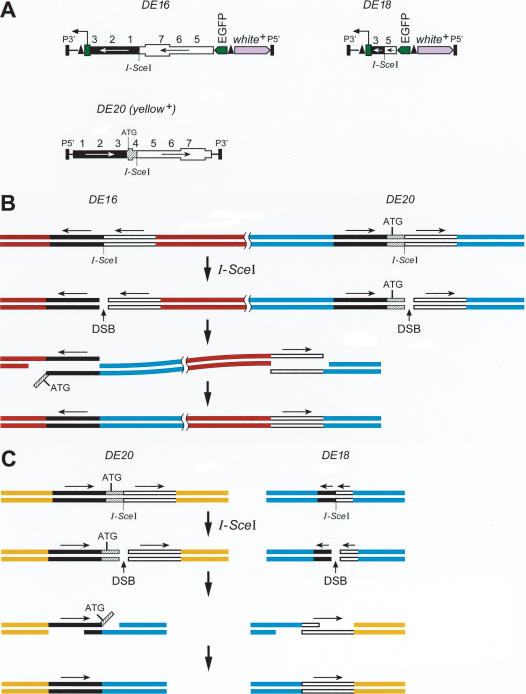
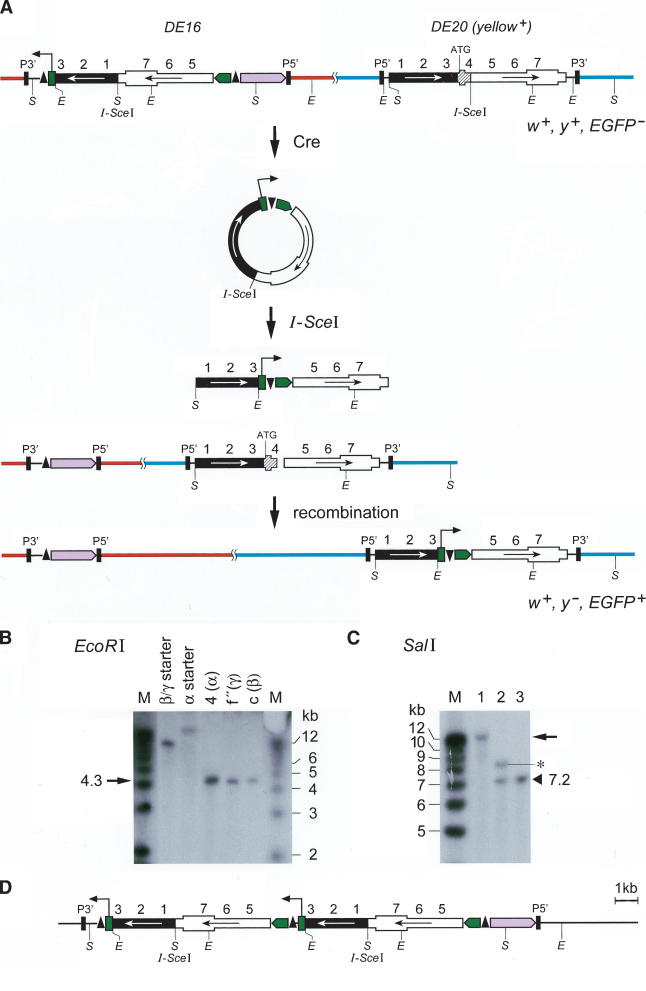
We established several fly strains termed α, β, γ, all carrying two homologous P-element constructs on the third chromosome, and a fly strain δε with one P-element on the second and the other on the third chromosome (Fig. 1A, Table 1). DSBs can be induced by expression of the I-SceI endonuclease that recognizes an 18-bp restriction site present within DE20 and DE16, or DE18 transgenes. DSB repair by recombination between two P-elements would result in an inversion or a deletion or a duplication or in a translocation, depending on their cytological location and their relative orientation. Inversions, translocations, and deletions should all result in the elimination of the functional yellow gene (y+) on the P-element DE20 (Fig. 1B,C,D). On average, 50% of DE20–DE16 or DE20–DE18 pairs should lie in the correct relative orientation to produce inversions or translocations by HR. We expected to recover inversions and translocations only, because deletions of that size are not expected to be viable. DSB repair at DE16 or DE18 and DE20 may lead not only to HR-mediated chromosomal rearrangements but also to DSB repair by NHEJ or gene conversion using any yellow sequence in the genome, such as the identical sequence on the sister chromatid in the G2 phase of the cell cycle or the endogenous mutant y1 gene, which is inactivated by a point mutation in the ATG start codon. Repair using the sister chromatid as a template restores the original sequence. DE20, carrying a y+ gene with an I-SceI site in the intron, may acquire the y1 mutation from the endogenous y1 locus and thereby lose the y+ gene function (Fig. 1E).
Figure 1.
Possible chromosomal rearrangements following two DNA double-strand breaks (DSBs) and repair by single-strand annealing (SSA). Two transgenes (DE16 or DE18 and DE20) that share homologous sequences of the yellow (y) gene undergo homologous recombination (HR) upon cleavage by I-SceI. (A) DE20 is a P-element carrying a full-length y+ gene and a recognition site for the rare cutting endonuclease I-SceI in the intron. This y+ gene is functional and confers a dark body color to the fly. DE16 and DE18 carry the same sequences in reverse orientation (the 3′ part of y is placed upstream of the 5′ part), and the first exon of y is removed rendering this incomplete y gene nonfunctional. The recognition site for I-SceI is located at the junction of the two inverted sequences. In addition, DE16 and DE18 carry a white+ (w+) gene serving as a transformation marker. (B, C) Mechanism of SSA as described in the Introduction, leading to chromosomal inversion (B) or to chromosomal translocation (C). Sequences shared between the two recombining elements are reduced to single copy, and the y+ function within DE20 is thereby lost. (D) Recombination of DE20 with DE16 or DE18 results in two hybrid elements composed of DE16 or DE18 and DE20. (E) Possibilities of DSB repair leading to the loss of y gene function. Indicated are only elements undergoing a change in the respective case. (F) Cre-mediated excision from DE16 joins the 3xP3 promoter to the EGFP coding region, resulting in a transient EGFP expression from an episome. (Boxed key) An explanation of all symbols used in this and the other figures is as follows: The y gene sequences are shown as either two bars to represent the DNA double-strand or as a single box to illustrate the structure of the gene. Arrows indicate the arrangement of the homologous regions of the y gene shared between the P-elements. Sequences other than y are not drawn to scale. Numbers (one to seven) denote kilobase intervals on the y gene. In some figures, the P-elements DE16, DE18, and DE20 are simplified to emphasize the components involved in the depicted process.
Table 1.
Frequency of Chromosomal Rearrangements by HR
| A. Frequency of Chromosal Inversions | |||||||||
| Strain | P-element DE16 insertion | P-element DE20 insertion | Inversion size (Mbp) | Yellow loss (w+y- / w+y- + w+y+) | Episomal EGFP- | Rearrangement by SSA (according to Southern) | Inversions | ||
| α | 16da (3L;79A;mub)b | 20z (3R;100D;ttk) | 29 | 230/1789 (12.8%) | 31/94; 33% | 26 of 26 | 4.12% (17)c | ||
| β | 16c (3R;83C;CG2017) | 20h (3L; 62E;CG32306) | 22.6 | 40/466 (8.6%) | 9/19; 47% | 8 of 9 | 3.9% (3) | ||
| γ | 16c (3R;83C;CG2017) | 20y (3R; 86E; CG14709) | 5.7 | 27/836 (3.2%) | ND | 1 | ∼0.8% (1) | ||
| B. Frequency of Chromosomal Translocations | |||||||||
| Strain | P-element DE18 insertion | P-element DE20 insertion | Yellow loss | Other loss of y events | Translocations | ||||
| δε | 18ca (2R;45C)b | 20d (3R;89A) | 46/955 (4.8%) | 3% (8; 4 NHEJ, 4 ND) | 1.8% (5)c | ||||
The frequency of chromosomal rearrangement is not equivalent to the number of events since the heat shock is performed at larval stages, and thus, the frequency is also influenced by the expansion of germ cells during development. Inversion frequencies for individual starter strains were calculated from the number of w+y- flies without episomal EGFP expression and corrected by a factor 0.97 because one out of 35 inversions was a false-positive event according to Southern data. For strain γ the frequency was calculated according to the Southern data only, where 25% of the w+y- had an inverted chromosome. ND indicates not determined.
Letters arbitrarily denote the isolated insertion.
The cytological position together with the gene next to the insertion site is given in parentheses.
The minimal number of independent events is given in parentheses.
Two DSBs Are Sufficient To Generate Frequent Chromosomal Inversions and Translocations by HR
We screened for the loss of y+ upon DSB generation by I-SceI, the expression of which was induced by heat-shocking Drosophila larvae. We observed clonal inactivation of y in heat-shocked adult flies carrying DE16 and DE20 together with the heat inducible I-SceI transgene (data not shown). Offspring of those flies carrying a w+ y– chromosome have inactivated their yellow gene by DSB repair and are candidates for chromosomal rearrangements (Fig. 1B–E).
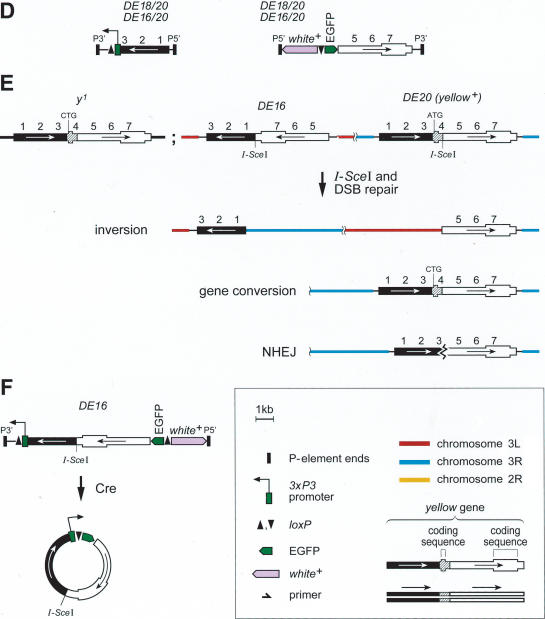
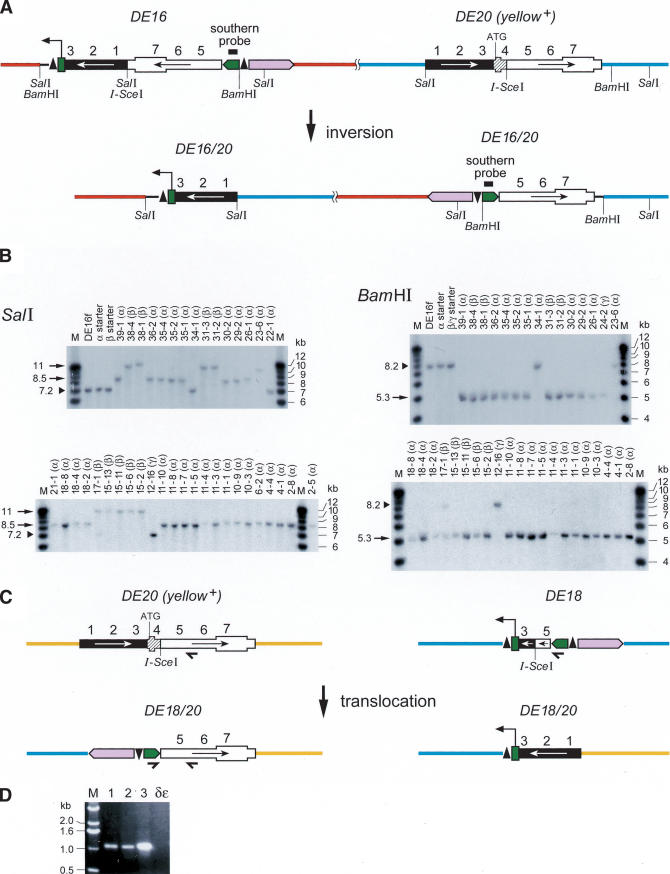
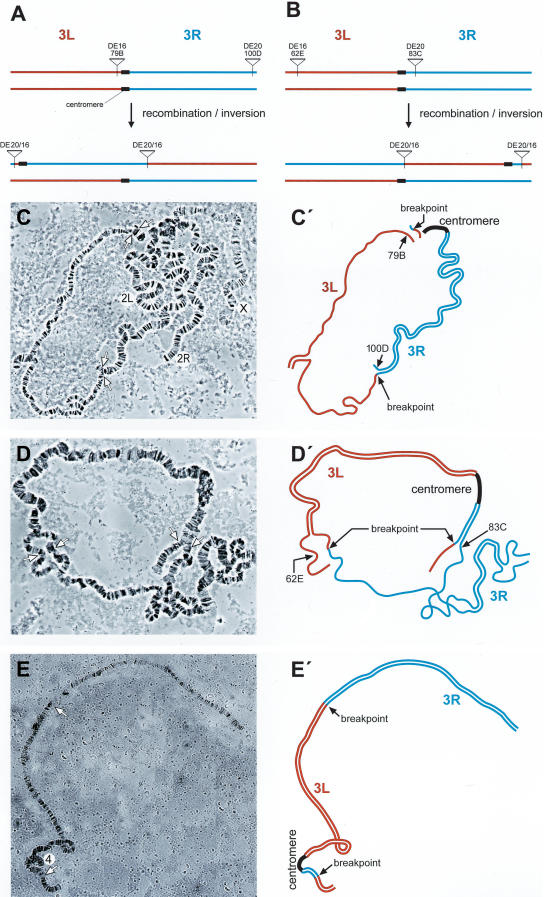
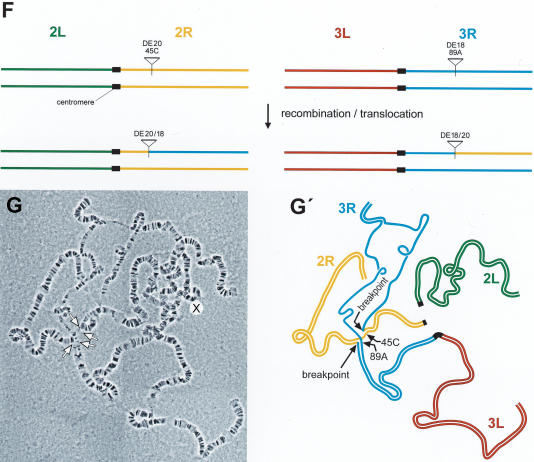
From flies carrying DE16 and DE20 on chromosome 3, we recovered 297 w+ y– flies out of 3091 inheriting the chromosome carrying the P-elements (Table 1A). To determine whether the chromosome had been inverted, we screened for flies, which had lost the ability to transiently express EGFP from an episome excised from the DE16 original construct. Cre recombinase action on DE16 joins the artificial 3xP3 promoter, driving strong expression in larval tissues and the adult eye, to the EGFP coding sequence by generating an episome (Fig. 1F). However, chromosomal inversions lose the ability to generate an episome. Among 113 originally recovered w+ y– flies, 40 (∼35%) lacked episomal EGFP expression. For the molecular verification of the inversion, we performed Southern blotting by using the two different enzymes BamHI and SalI (Fig. 2A,B). Indeed, 34 out of 35 of the selected events tested showed bands at a position consistent with an inversion by precise elimination of the duplicated sequences, suggesting a SSA-like mechanism as shown in Figure 1B. The screen for inversions with Cre recombinase gives thus a good estimate of the number of chromosomal inversions with only one false-positive event out of 35 (Table 1A). In addition, we performed Southern analysis of four w+ y– events (derived from strain γ with the P-element combination DE20y [3R; 86E] DE16c [3R; 83C]) without prescreening for absence of episomal EGFP expression, one of which showed the expected pattern of an inversion (Fig. 2B, lane 24-2). Figure 3, A through E, shows the cytological analysis of the inversions. The unequal length of the inverted chromosomal arms is obvious. Inversions In (3R) DE20y-86E; DE16c-83C and In (3LR) DE20h-62E; DE16c-83C are homozygous viable and fertile, whereas In (3LR) DE16d-79A; DE20z-100D is homozygous lethal, possibly due to disruption of ttk and/or mub function, where the two elements are inserted (DE20z is inserted 330 bp in front of the ttk transcription start and DE16d 25 bp in front of the transcription start of mub). All starter strains are homozygous viable. The lethality of inversion In (3LR) DE16d-79A; DE20z-100D is therefore a genetic confirmation of the chromosomal rearrangement. The rearrangements appear to be stable, because they were preserved for >1 year, corresponding to >25 generations, after their recovery. Taken together, on average about one-third of w+ y– flies and ∼3% of total flies had the chromosome inverted by SSA. Frequencies of inversions for the individual starter strains are indicated in Table 1.
Figure 2.
Molecular verification of chromosomal rearrangements. (A) Scheme of a chromosome before and after inversion. (B) Molecular verification of chromosomal inversions by genomic Southern-blotting with SalI and BamHI digested DNA. The Southern probe detects the EGFP sequence in DE16. The size of the SalI fragment is dependent on the chromosomal insertion site of the P-element DE20, whereas the BamHI fragment is independent of the genomic context of DE20 because both BamHI sites are located within the P-element. Arrows mark bands characteristic for an inversion, and arrowheads mark bands representing the original construct. Numbers above the lanes denote arbitrary isolate numbers: The first number denotes flies from different tubes and thus different events, and the second number represents the number of an individual fly taken from a particular tube. The lines from which the events are derived are indicated in parentheses above the lanes: α, DE16d*-DE20z; β, DE16c-DE20h; and γ, DE16c-DE20y. The latter two share the same DE16 starter P-element insertion. Lowercase letters arbitrarily denote the isolated P-element insertion. (C) Scheme of a chromosome 2 (left) and chromosome 3 (right) before the translocation and of the rearranged chromosomes. Small arrows indicate primers for the verification of the translocation. (D) PCR of three different translocation events (1–3) and of the δε starter strain.
Figure 3.
Cytology of chromosome 3 inversions (A–E) and chromosome 2; three translocations (F, G). Scheme of the generation of inversions In(3LR)DE16d*-79A;DE20z -100D derived from stock α (A) and of In(3LR) DE20h-62E; DE16c-83C derived from stock β (B). (C, C′) Polytene chromosomes and schematic representation from In(3LR)DE16d-79A;DE20z -100D/+ heterozygotes. (D, D′) Polytene chromosomes and schematic representation from In(3LR) DE20h-62E; DE16c-83C/+ heterozygotes, and (E, E′) from In(3LR) DE20h-62E; DE16c-83C homozygotes. Note the unequal length of chromosomal arms of the inverted chromosomes. (F) Scheme of the generation of chromosome 2; three translocations. (G, G′) Polytene chromosomes and schematic representation from T(2; 3) DE18c-45C; DE20d-89A/+ heterozygotes. Lowercase letters arbitrarily denote the isolated P-element insertion.
Chromosomal translocations were selected by screening for thelossof y+ upon DSB induction by I-SceI and for pseudolinkage of the third and second chromosome, distinguishing translocations from other y– events. The P-element combination used for generating translocations (DE18c [2R; 45C]-DE20d [3R; 89A]; strain δε) carries DE18 instead of DE16 (Fig. 1A,C; Table 1). DE18 is derived from DE16 but shares only 1.4-kb homology instead of 6.88-kb homology with DE20. Putative translocations were further characterized by PCR and sequencing, revealing that, as for inversions, the duplicated sequences had been precisely eliminated by SSA (Figs. 2C,D, 1C). The translocation frequency was calculated to be 1.8% of total flies (Table 1B). Figure 3, F through G, shows the cytology of the translocation.
NHEJ and Gene Conversion Events
Our analysis also revealed some events other than a chromosomal rearrangement (Fig. 1E). Because the I-SceI site is only 250 bp away from the first y exon, we expected to also recover flies with an inactivated y gene as a result of exonuclease resection and subsequent NHEJ. Sequencing of the defective y gene of four independent events derived from the strain δε revealed junctions typical for NHEJ without apparent homology at the junction point, resembling the junctions previously observed by Hagmann et al. (1998; Table 2). Event “H” has a deletion of 0.88 kb and an insertion of 54 bp at the junction, the sequence of which resembles an I-SceI site. Lane 34-1 in the Southern blot of Figure 2B does not show a pattern of inversion at DE-16, but the y+ gene in DE20 has been converted to y–. Sequencing revealed that the ATG of the y+ gene in the P-element has been converted to CTG as present in the y1 locus. This event is best explained by resection of the DSB end followed by SDSA using the y1 locus on the X chromosome as repair template. Lane 23-6 of Figure 2B represents an NHEJ event at DE16, whereby the SalI site together with the adjacent I-SceI site in DE16 is eliminated, resulting in a longer SalI fragment but not changing the BamHI fragment size.
Table 2.
NHEJ Events Deleting the First Exon of the Yellow Locus in DE20
| Ea: 5′-ATTCGATAGGGTAATACTAGT-3′ | Δ 0.88 kb, +1 bp |
| C: 5′-GATTTAACAGGGTAATACTAGT-3′ | Δ 1.3 kb |
| Y: 5′-CTAAAGTATT*ATAACAGGGTAATACTAGT-3′ | Δ 0.37 kb, +4 bp |
| H: 5′-GTATGGGTATAACAGGTATAACCCTGTACAGGTATATACCTGTACCCTGTACCCTGTACCCTACTAG-3′ | Δ 0.88 kb, +54 bp |
Events are derived from strain δε. Nucleotides derived from the I-Scel site are underlined and the site of cleavage is indicated with an asterisk. The size of the deletion (Δ) is given on the right. Nucleotides, which have been inserted during the process of NHEJ are indicated in bold letters, and the number is given on the right (+).
Letters arbitrarily denote the isolated event.
Cre Recombinase Can Revert loxP Containing Chromosomal Rearrangements
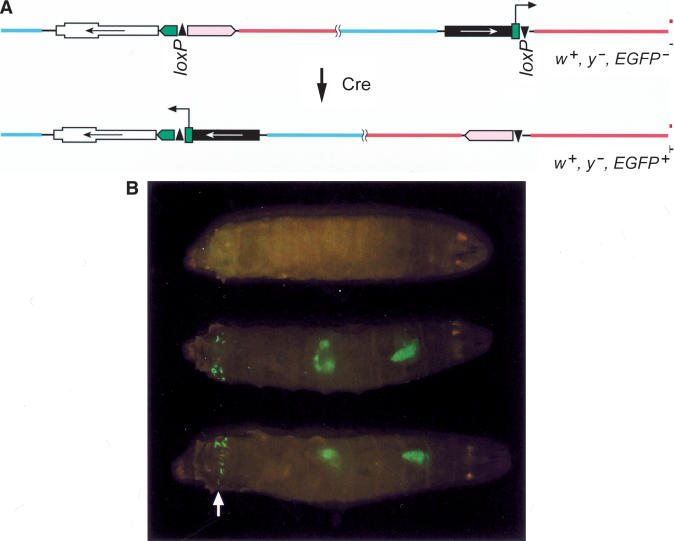
Cre recombinase has been used to generate chromosomal rearrangements in the mouse (Van Deursen et al. 1995; Zheng et al. 1999, 2000). Inversions and deletions of several megabases were recovered at a rate of 10–1 to 10–6 per cell with exponentially decreasing efficiency at increasing distances between the loxP sites. Our chromosomal inversion generated by HR carries two loxP sites in opposite orientation, which offered a convenient opportunity to also test for the ability of Cre recombinase to revert the inverted chromosome region to its original orientation, thereby joining the synthetic 3xP3 promoter to the EGFP coding sequence (Fig. 4A). Indeed, we observed mosaic EGFP expression in the larval brain, the gut, the anal plates, and the eye of adult flies in heat-shocked larvae carrying a hsCre transgene-together with the inversion (Fig. 4B; data not shown). This expression pattern conforms exactly to the one reported by Horn et al. (2000) showing the specificity of the inversion. Yet the number of clones was not equally frequent for the three inversions tested. For In (3R) DE20y-86E; DE16c-83C, spanning 5.7 Mbp, we observed extensive clonal EGFP expression, but for In (3LR) DE16d-79A; DE20z-100D and In (3LR) DE20h-62E; DE16c-83C, spanning 29.5 and 22.6 Mbp, respectively, EGFP spots were rare (Table 3).
Figure 4.
Cre-mediated reversion of inverted chromosomes. (A) Schematic view of the Cre-mediated reversion joining the 3xP3 promoter to the EGFP coding sequence resulting in EGFP expression. Note the inverted orientation of the two loxP sites in the two P-elements. w+ y– EGFP–, and w+ y– EGFP+ indicates the phenotype of the flies carrying the respective chromosomes. (B) Larvae with the genotype In (3R) 16c-83C; 20y-86E (top) and In(3R) 16c-83C;20y-86E/hsCre (the two larvae below) under UV light. Note the clonal nature of the inversion visualized by EGFP fluorescence, which can be particularly well observed at the anal plates (arrow). The expression pattern of 3xP3 in the anal plates, the gut, and the brain is as reported by Horn et al. (2000).
Table 3.
Frequency of Chromosomal Rearrangements by Cre Recombinase
| Genotype | Derived from strain | Size of inversion | Clonal GFPa | Reversion | Reversion % | Inversions | Inversion % |
|---|---|---|---|---|---|---|---|
| In (3R) DE20y-86E; DE16c-83C | γ | 5.7 | 22.25 (178/8)a | 10/450 (4)b | 2.25 | ||
| Reversion γ | In (3R) DE20y-86E; DE16c-83C | 5.7 | 6/2179 (3) | 0.28 | |||
| In (3LR) DE16d-79A; DE20z-100D | α | 29.5 | 0.1 (3/30) | 0/4500 | <0.02 | ||
| In (3LR) DE20h-62E; DE16c-83C | β | 22.6 | 0.5 (5/10) | 0/1100 | <0.09 | ||
| T(2;3) DE18c-45C; DE20d-89A | δε | ND | 2/1500 (2) | 0.13 |
The total number of larvae has divided the number of EGFP+ clones in the anal plates of third instar larvae. ND indicates not determined.
The minimal number of independent events is given in parentheses.
This difference of frequency was also reflected in the germ line. Heat-shocked parents that carry a hsCre recombinase transgene together with the inversion In (3R) DE20y-86E; DE16c-83C or together with the translocation T(2;3) DE18c-45C; DE20d-89A gave rise to offspring that showed the full expression pattern of 3xP3-EGFP and thus had the chromosome reverted, but no reversion was recovered from In (3LR) DE20h-62E; DE16c-83C or from In (3LR) DE16d-79A; DE20z-100D. The reverted chromosome derived from In (3R) DE20y-86E; DE16c-83C could also be inverted again by Cre recombinase, but with eightfold lower efficiency than the reversion as indicated in Table 3. At first sight, one might expect similar frequencies for Cre-mediated inversion and reversion. However, a similar difference in efficiency was also found for FLP-mediated inversion and reversion (Golic and Golic 1996). The observed difference might be explained by the pairing of homologs in mitotic cells of Drosophila, which would result in a greater proximity of sites for the reversion. In conclusion, Cre recombinase is also able to rearrange a chromosome, but at a much lower frequency, and unlike inversion by HR, the frequency is strongly dependent on the location of the loxP sites on the chromosome.
Gene Targeting of a Linearized Episome to a DSB Is Similarly Efficient to Chromosomal Inversion
In a further experiment, we specifically targeted the y+ gene in the DE20 P-element by a homologous mutant y sequence located in another P-element (DE16; Fig. 5). The y sequence was liberated and circularized from DE16 by Cre recombinase. In the resulting episome, the synthetic 3xP3 promoter is joined to the EGFP coding sequence. The release of the episome in the larva can be followed by transient expression of EGFP. I-SceI cuts the episome, creating recombinogenic ends. Recombination with DE20 inactivates y+ by inserting the 3xP3-EGFP marker and deleting the first exon. Gene targeting to a DSB is essentially identical to the chromosomal inversion or translocation, except that in this case recombination takes place between a chromosomal and an episomal DSB-end rather than between two chromosomal DSB ends. Indeed, out of 4600 flies from three different starter lines, we recovered 26 w+, y–, EGFP+ flies, representing at least 14 independent events (Table 4). Southern blot analysis of targeting events derived from the three starter strains α, β, and γ showed the expected band pattern for integration into DE20: The EGFP probe detected an EcoRI fragment of 4.2 kb instead of a fragment characteristic of a DE16 starter element, the size of which depends on the insertion site (Fig. 5B). We did not recover any targeted alleles to the endogenous y locus on the X chromosome, which shares an extensive homology segment but lacks an I-SceI cleavage site, nor did we have any indication for an unspecific NHEJ-mediated insertion anywhere else in the genome. In a control experiment without the presence of DE20 (w1118 background), we were unable to recover any EGFP-positive line from ∼6000 flies using the insertion DE16d.
Figure 5.
Gene targeting to an I-SceI–induced DSB. (A) Schematic view of the ends-out targeting strategy. Cre recombinase excises an episome from DE16, which is subsequently cleaved by I-SceI to create a linear molecule able to undergo HR with the second transgene DE20, which is also cleaved by I-SceI. Cre-mediated recombination joins the 3xP3 promoter to the EGFP coding sequence. E indicates EcoRI; S, SalI. w+ y– EGFP– and w+ y– EGFP+ indicate the phenotypes of the flies carrying the corresponding chromosomes. (B, C) Verification of targeting by genomic Southern blotting. The arrows mark bands characteristic for targeting. Letters above the lanes denote arbitrary isolate numbers. The starter lines from which the events are derived are indicated in parentheses above the lanes: α, DE16d-DE20z; β, DE16c-DE20h; and γ, DE16c-DE20y. β and γ share the same DE16 starter P-element insertion DE16c. (B) EcoRI digestion. (C) SalI digestion of a targeted event with a novel band at ∼12 kb (lane 1), of a Cre-mediated duplication with two bands at 7.2 kb and 8.5 kb (lane 2) and of DE16 with a single band at 7.2 kb (lane 3). The asterisk marks the band generated by Cre-mediated duplication. (D) Scheme of a Cre-mediated duplication leading to EGFP expression.
Table 4.
Frequency of Gene Targeting to a DSB
| Strain | P-element DE16 insertion | P-element DE20 insertion | EGFP+ w+ y- | w+ y+ | Frequency |
|---|---|---|---|---|---|
| α | 16da (3L;79A;mub)b | 20z (3R;100D;ttk) | 12 (6)c | 2250 | 0.53% |
| β | 16c (3R;83C;CG2017) | 20h (3L; 62E;CG32306) | 10 (5) | 1300 | 0.77% |
| γ | 16c (3R;86C;CG2017) | 20y (3R; 86E;CG14709) | 4 (3) | 1050 | 0.38% |
| Total | 26 (14) | 4600 | 0.56% |
Letters arbitrarily denote the isolated insertion.
The cytological position together with the gene next to the insertion site is given in parentheses.
The minimal number of independent events is given in parentheses.
As an unexpected side product of the targeting, we observed Cre-mediated duplications of DE16 placing the 3xP3 promoter in front of EGFP (Fig. 5D). These flies express EGFP but retain a functional y gene in DE20. As expected, the EGFP expression could be eliminated by reducing the duplication to a single copy by another round of treatment with Cre recombinase. These duplications probably arise by unequal sister chromatid exchange similar to FLP-mediated duplications observed by Golic and Lindquist (1989). Southern blot analysis of genomic DNA digested with SalI and a labeled EGFP probe shows that the EGFP sequence in these lines had not integrated into DE20 but rather duplicated at the original position of DE16, whereas gene targeting results in the movement of EGFP to the genomic location of DE20 (Fig. 5B,C). On average, we recovered targeted flies at a frequency of ∼0.5%. Similar results were obtained by Bibikova et al. (2003), who found a strongly increased targeting frequency, up to 1.5%, by specifically cleaving the gene of interest with a designer-made zinc finger nuclease.
DISCUSSION
The efficiency of genome rearrangements in Drosophila by HR is striking. We were able to recover translocations and paracentric and pericentric inversions spanning several megabases at a frequency of 1% to 4%. Rearrangements were specific, and the products were compatible with a SSA mechanism. NHEJ did not contribute to the observed translocations or inversions. Rearrangement by SSA is competing with several other possible repair pathways for the repair of the lesion, among them NHEJ and gene conversion, for example, with the sister chromatid (Fig. 1). Inversion by SSA requires DSBs in both transgenes at the same time (see also Richardson and Jasin 2000). Other mechanisms leading to inversions, such as DSB repair with an associated crossover, are highly unlikely because mitotic crossovers are suppressed in several organisms, including Drosophila (Nassif et al. 1994; Paques and Haber 1999). The ease to recover inversions and translocations at high frequency leads us to assume that deletions and duplications could also be recovered readily. This would allow for any desired rearrangement of the Drosophila genome, for example, to study the effects of chromosome size and chromosome number on the organism.
Cre recombinase is also an efficient tool to generate chromosomal rearrangements. We observed somatic reversion with a translocation and all three inversions, but the efficiency was much greater for the inversion spanning 5.7 Mbp than with the two inversions spanning 22.6 or 29.5 Mbp, and only the smallest of the three inversions could be reverted and also reinverted in the germline. The different efficiencies of Cre-mediated recombination are most likely due to the difference in interlocus distance or due to structural constraints in the nucleus. Cre-mediated recombination is known to be strongly dependent on the distance between the loxP sites in mammalian cells (Zheng et al. 2000). The same holds true for FLP in Drosophila. FLP-mediated inversions have been recovered at a frequency of 0.03% to 0.3%, depending on the distance of FRT sites (Golic and Golic 1996). The Cre/loxP system seems to be at least as efficient for rearranging a chromosome as FLP. A disadvantage of the Cre/loxP system is that Cre recombinase, especially if strongly expressed, also recognizes cryptic loxP sites in the genome, which can lead to sterility and lethality (Sauer 1992; Schmidt et al. 2000; Siegal and Hartl 2000). This hampers some practical applications of Cre recombinase but demonstrates its potency to generate genomic rearrangements. We have demonstrated here that Cre recombinase can also be used as a tool in manipulating the Drosophila genome as a convenient complement to FLP recombinase. Yet HR is by far the most efficient tool to rearrange Drosophila chromosomes, being up to two orders of magnitude more efficient than either FLP or Cre recombinase. This advantage, in Drosophila, of HR over Cre- or FLP-mediated rearrangements recommends this technique for application in other species.
We observed ends-out gene targeting to an I-SceI induced DSB at a frequency of ∼0.5%, with less than a twofold variation between the three different P-element combinations. Because we have used the very same insertions for targeting as for the chromosomal inversions, the main difference between the two experiments is that in the first case the repair occurs between two DSBs in the chromosome, whereas in the second it occurs between the chromosome and a linearized episome. As with the chromosomal inversion, gene targeting to a DSB is apparently mediated by SSA. Surprisingly, the efficiency of gene targeting was slightly lower than was chromosomal inversion, even though an episome is expected to diffuse more readily than a chromosome and is not anchored within the nucleus, and thus might be expected to find the homologous repair partner more easily. However, it might be argued that the simultaneous induction of Cre recombinase and I-SceI is not optimal to ensure that circularization/excision takes place before linearization, and might therefore not be directly compared with chromosomal inversion. Because we did not recover any targeted event at the endogenous y locus, we conclude that the DSB at the target gene strongly stimulated targeting. Other groups using either zinc finger nucleases in Drosophila, or I-SceI in mammalian cell culture, obtained similar results (Smih et al. 1995; Bibikova et al. 2003).
Our study suggests that DSB repair by SSA is not impaired by chromosomal organization or by the interlocus distance on a chromosome. The efficiency of chromosomal rearrangements and gene targeting to a DSB only varied within a factor of two to five, whereas Cre-mediated recombination varied by more than a factor of 100 between different P-element combinations. In the strain α, the recombining P-elements are separated by ∼29.5 Mbp, with one P-element close to the telomere and the other one close to the centromere, and in strain γ they are separated by ∼5.7 Mbp. Whereas HR was slightly more efficient to rearrange the elements separated by ∼29.5 Mbp than those separated by ∼5.7 Mbp, Cre recombinase acts with at least two orders of magnitude lower efficiency on the elements separated by ∼29.5 Mbp than on those separated by only ∼5.7 Mbp. This suggests that for HR, in contrast to Cre recombinase, there is no correlation between the efficiency and the distance of the recombination partners within a chromosome. Even though there is a variation of the efficiency of HR depending on the location on the chromosome, a clear dependence on the genetic distance of recombining elements could not be established (Engels et al. 1994; Haber and Leung 1996; Rong et al. 2002). Engels et al. (1994) found intrachromosomal gene conversions by SDSA were more frequent than interchromosomal ones, but intrachromosomal events exhibited no dependence on the interlocus distance. The published results and our own data therefore argue for a relaxed chromosomal organization and against the existence of rigid chromosomal domains for DSB repair, even though some level of chromosomal organization is most probably maintained. Our data could also be explained by a mechanism, which actively directs the search for homologous sequences. Evidence for such a mechanism might come from yeast mating type switching. In this example, DSB repair is selectively directed toward one of the silent donor loci, depending on the mating type of the cell (Wu et al. 1996). In mammalian cells and in yeast, chromosomal regions containing a DSB are juxtaposed (Lisby et al. 2003; Aten et al. 2004). It is very likely that this mechanism is also operative in Drosophila, and thus, it could be responsible for the high rate of recombination between homologous DNA segments containing a DSB.
The high frequency of inversions by HR in our study suggests a role of HR in genomic rearrangements. Ectopic recombination between transposable elements and repetitive sequences is most likely the cause of many naturally occurring inversions and chromosomal rearrangements in Drosophila and probably also in other species (Lim and Simmons 1994; Strout et al. 1998; Caceres et al. 1999). Ectopic recombination may thus be an important determinant for the rate at which the structure of the Drosophila genome changes. Interestingly, the frequency of ectopic recombination of Drosophila is higher than in mammals but lower than in yeast. Although it is difficult to directly compare the studies in different organisms, the frequency of ectopic recombination tends to decrease with the size and complexity of the genome. In Drosophila ectopicrepairofaDSB liestobeinthe range of 10–1 to 10–3 (this study and Engels et al. 1994); in mammals it is in the range of 10–4 to 10–6 (Richardson et al. 1998; Richardson and Jasin 2000), whereas in yeast the frequency is high, between 0.2 and 0.9 (Haber and Leung 1996; Wu et al. 1997; Inbar et al. 2000). One might speculate that these differences reflect solely the difference in genome size, which makes it less likely for a DSB to meet its repair partner anywhere in the huge genome of mammalian cells. Alternatively, ectopic recombination is specifically inhibited in more complex organisms because the higher amount of interspersed repetitive DNA sequences would threaten genome stability if ectopic recombination was efficient.
This study, to our knowledge, is the first one providing a direct comparison of DSB repair of a chromosome with Cre-mediated recombination and with DSB repair of an episome. Taken together, the methods for genome engineering described in this article will be useful tools for further studies on DSB repair and genome maintenance.
METHODS
DNA Constructs
DE16 was constructed by using a Bluescript (Stratagene)-based vector containing two loxP sites (a gift from K. Basler, University of Zurich, Zurich, Switzerland). XhoI and XbaI sites flank the two loxP sites, and they themselves flank the sites used for cloning mentioned below. One of the loxP sites is shortened by two nucleotides at the 5′ end, which does not affect the efficiency of Cre recombinase to act on it, because excision is close to 100% (data not shown). The 3xP3 promoter was cloned as an EcoRI-SalI fragment into the EcoRI-SalI opened vector and EGFP as a SalI fragment blunted by Klenow treatment into the vectors NotI site. Both fragments are derived from pMos [3xP3-EGFPafm], a gift from E. Wimmer (University of Bayreuth, Bayreuth, Germany). The 3xP3 promoter contains three Pax6 binding sites and drives expression in the larval gut, brain, and anal plates and in the eye of the adult fly (Berghammer et al. 1999; Horn et al. 2000). The poly A site of HSV-TK was cloned as a PCR fragment digested with MunI-EcoRI into the EcoRI site. The PCR for HSV-TK was done using the primer pair 5′-AGGAATTCGGGAGGCTAACTGAAA CACG-3′ and 5′-TATCCAATTGAGTAACCTGAGGCTATGGCA-3′ and pEGFP-1 from Clontech as a template. SalI and SpeI sites were eliminated by digestion and Klenow treatment. Oligonucleotides 5′-CGCGTACTAGTTCTAAGATCTATTACCCT GTTATCCCTAGTCGACATTAAACGTTGTCTTCG-3′ and 5′-AAT TCGAAGACAACGTTTAATGTCGACTAGGGATAACA GGGTAATAGATCTTAGAACTAGTA-3′ were cloned into MluI-EcoRI–digested vector to clone the y gene in reverse arrangement and lacking the first exon as BbsI (–190)-SalI (–2900) and SpeI (662)-BglII (4756) fragments released from a y containing vector. Numbers in parentheses indicate the position relative to the transcription start of y. The whole cassette between the loxP sites was released from the Bluescript vector backbone by XhoI (blunted by Klenow) and XbaI and inserted into pCasper4 opened by XbaI and EcoRI (blunted by Klenow).
DE18 is derived from DE16 by digestion with SphI and Eco47III and insertion of the oligonucleotides 5′-GCTAG GGATAACAGGGTAATGCATG-3′ and 5′-CATTACCCTGTTATC CCTAGC-3′ containing an I-SceI site.
DE20 is based on a P-element vector containing a y+ gene from SalI (–2900) to BglII (4756), a gift from K. Basler. The I-SceI site was cloned into the SpeI (662) site of the first intron using the oligonucleotides 5′-CTAGTATTACCCTGTTATCCCTA-3′ and 5′-CTAGTAGGGATAACAGGGTAATA-3′. DE20 contains two I-SceI sites in the SpeI site with head to head arrangement.
Fly Stocks and Genetics
The stock y1 w; P[ry+, 70FLP]4 P[v+, 70I-SceI]2B Sco/S2 CyO were kindly provided by Y. Rong (National Cancer Institute, Bethesda, Maryland) and K. Golic (University of Utah, Utah), the stock y1 w; +; P[hsCre 666.10]/TM3y+ by K. Basler and the stock y1 w; CyO, P[hsCre*, y+]/Sco by Mark L. Siegal (Stanford University, Stanford, California; Siegal and Hartl 2000). Cre* carries an uncharacterized mutation resulting in reduced activity and eliminates toxicity of Cre following a heat shock. The mutation y1 is an ATG-to-CTG change in the start codon. Combinations of the P-elements DE20 and DE16 or DE18 used in this study are indicated in Table 1. For chromosomal inversions and gene targeting, the following stock was constructed: y1 w; P[ry+, 70FLP]4 P[v+, 70I-SceI]2B Sco/S2 CyO; P[hsCre666.10]/(TM2, y+). The FLP does not play any role in this process because there are no FRT sites it could act on. Three heat shocks for 60 min in a 38°C water bath were performed during larval development. Cre recombinase expression is toxic to the fly, causing lethality and sterility, possibly by recombination between cryptic loxP sites (Sauer 1992; Schmidt et al. 2000; Siegal and Hartl 2000). Therefore, only a small number of offspring could be recovered from heat-shocked flies carrying hsCre. Targeting experiments were therefore repeated with hsCre*, and similar results were obtained. Cre recombinase–mediated reversion and inversion was done by using the stock y1 w; +; hsCre 666.10/(TM3, y+) with a single 15-min 38°C heat shock at larval stages, which does not lead to lethality or reduced fertility. The male and female germ line was not discriminated in this study.
Photographs of EGFP expression were taken with a Leica MZ FLIII fluorescence stereomicroscope and a Nikon COOLPIX950 digital camera.
Cytology and Localization of P-Elements
Polytene chromosomes were prepared as described (Sullivan et al. 2000) and examined with a Zeiss Axiophot microscope. Mapping of P-elements was done by in situ hybridization to polytene chromosomes as described (Sullivan et al. 2000). Labeling of a y probe was done by using the DIG DNA Labeling and Detection Kit (Roche 1093657). Inverse PCR was done according to http://www.fruitfly.org/, using primer pairs Plac1–Plac4 and Pry1–Pry2 and sequenced with primer P*.
Molecular Characterization of DSB Repair
Southern blots were performed with a probe derived from a SalI fragment released from pMos [3xP3-EGFPafm]. PCR to determine the junction of the event 23-6 in the P-element DE16 was done with the primer pair 5′-TGTGGATCGCGATGACGTTG-3′ and 5′-CAAAACAACATCAAGCGGAGC-3′ and sequencing with primer 5′-TCAGCTCGATGCGGTTCACCA-3′. The sequence of the y start codon and the NHEJ junction in DE20 was determined with the primers Plac4 and 5′-ACTGAAGTATTTGTATGCGCATC-3′ for PCR and primer 5′-TGTCATATTTATTCCTCTGCCAA-3′ for sequencing. The translocation junction was sequenced by using primers 5′-TTTCCAATTGAGCCCAGCAT-3′ and 5′-CTGCA TTCTAGTTGTGGTTTG-3′ for PCR and primer 5′-AGGGCC GCCACATAGAAATGG-3′ for sequencing.
Acknowledgments
We are grateful to Christoph Hugentobler, Mike Hirsch, Patrick Faller, and Bruno Schmid for technical assistance; to Kent Golic (University of Utah) and Yikang Rong (NCI) for the generous gift of Drosophila strains; to Konrad Basler (Universität Zürich) for plasmids, fly strains, and valuable advice; to Ernst Wimmer (Universität Bayreuth) for the 3xP3 promoter plasmid; to Mark L. Siegal (Stanford University) for fly strains; and to Fritz Ochsenbein for the preparation of figures. We thank Oleg Georgiev for important discussion; we also thank O. Georgiev, George Hausmann, Renjie Jiao, and two anonymous reviewers for comments on the manuscript. D.E. is grateful to Inspiration. This work was supported by the Kanton Zürich and by the Swiss National Science Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2279804. Article published online before print in June 2004.
Footnotes
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: K. Golic, Y. Rong, K. Basler, E. Wimmer, and M.L. Siegal.]
References
- Aten, J.A., Stap, J., Krawczyk, P.M., van Oven, C.H., Hoebe, R.A., Essers, J., and Kanaar, R. 2004. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303: 92–95. [DOI] [PubMed] [Google Scholar]
- Berghammer, A.J., Klingler, M., and Wimmer, E.A. 1999. A universal marker for transgenic insects. Nature 402: 370–371. [DOI] [PubMed] [Google Scholar]
- Beumer, K.J., Pimpinelli, S., and Golic, K.G. 1998. Induced chromosomal exchange directs the segregation of recombinant chromatids in mitosis of Drosophila. Genetics 150: 173–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova, M., Beumer, K., Trautman, J.K., Carroll, D., Britt, A.B., and May, G.D. 2003. Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. [DOI] [PubMed] [Google Scholar]
- Burgess, S.M. and Kleckner, N. 1999. Collisions between yeast chromosomal loci in vivo are governed by three layers of organization. Genes Dev. 13: 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres, M., Ranz, J.M., Barbadilla, A., Long, M., and Ruiz, A. 1999. Generation of a widespread Drosophila inversion by a transposable element. Science 285: 415–418. [DOI] [PubMed] [Google Scholar]
- Deininger, P.L. and Batzer, M.A. 1999. Alu repeats and human disease. Mol. Genet. Metab. 67: 183–193. [DOI] [PubMed] [Google Scholar]
- Deininger, P.L. and Batzer, M.A. 2002. Mammalian retroelements. Genome Res. 12: 1455–1465. [DOI] [PubMed] [Google Scholar]
- Egli, D., Selvaraj, A., Yepiskoposyan, H., Zhang, B., Hafen, E., Georgiev, O., and Schaffner, W. 2003. Knockout of “metal-responsive transcription factor” MTF-1 in Drosophila by homologous recombination reveals its central role in heavy metal homeostasis. EMBO J. 22: 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels, W.R., Preston, C.R., and Johnson-Schlitz, D.M. 1994. Long-range cis preference in DNA homology search over the length of a Drosophila chromosome. Science 263: 1623–1625. [DOI] [PubMed] [Google Scholar]
- Gloor, G.B., Nassif, N.A., Johnson-Schlitz, D.M., Preston, C.R., and Engels, W.R. 1991. Targeted gene replacement in Drosophila via P element–induced gap repair. Science 253: 1110–1117. [DOI] [PubMed] [Google Scholar]
- Golic, K.G. and Golic, M.M. 1996. Engineering the Drosophila genome: Chromosome rearrangements by design. Genetics 144: 1693–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic, K.G. and Lindquist, S. 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59: 499–509. [DOI] [PubMed] [Google Scholar]
- Gong, W.J. and Golic, K.G. 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. 100: 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J.E. and Leung, W.Y. 1996. Lack of chromosome territoriality in yeast: Promiscuous rejoining of broken chromosome ends. Proc. Natl. Acad. Sci. 93: 13949–13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann, M., Bruggmann, R., Xue, L., Georgiev, O., Schaffner, W., Rungger, D., Spaniol, P., and Gerster, T. 1998. Homologous recombination and DNA-end joining reactions in zygotes and early embryos of zebrafish (Danio rerio) and Drosophila melanogaster. Biol. Chem. 379: 673–681. [DOI] [PubMed] [Google Scholar]
- Horn, C., Jaunich, B., and Wimmer, E.A. 2000. Highly sensitive, fluorescent transformation marker for Drosophila transgenesis. Dev. Genes Evol. 210: 623–629. [DOI] [PubMed] [Google Scholar]
- Inbar, O. and Kupiec, M. 1999. Homology search and choice of homologous partner during mitotic recombination. Mol. Cell. Biol. 19: 4134–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar, O., Liefshitz, B., Bitan, G., and Kupiec, M. 2000. The relationship between homology length and crossing over during the repair of a broken chromosome. J. Biol. Chem. 275: 30833–30838. [DOI] [PubMed] [Google Scholar]
- Johnson, R.D. and Jasin, M. 2001. Double-strand-break–induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 29: 196–201. [DOI] [PubMed] [Google Scholar]
- Kadyk, L.C. and Hartwell, L.H. 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeler, K.J., Dray, T., Penney, J.E., and Gloor, G.B. 1996. Gene targeting of a plasmid-borne sequence to a double-strand DNA break in Drosophila melanogaster. Mol. Cell. Biol. 16: 522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolomietz, E., Meyn, M.S., Pandita, A., and Squire, J.A. 2002. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Gene Chromosomes Cancer 35: 97–112. [DOI] [PubMed] [Google Scholar]
- Lim, J.K. and Simmons, M.J. 1994. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. Bioessays 16: 269–275. [DOI] [PubMed] [Google Scholar]
- Lindsley, D.L. and Zimm, G.G. 1992. The genome of Drosophila melanogaster. Academic Press, San Diego, CA.
- Lisby, M., Mortensen, U.H., and Rothstein, R. 2003. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell. Biol. 5: 572–577. [DOI] [PubMed] [Google Scholar]
- Loidl, J. and Nairz, K. 1997. Karyotype variability in yeast caused by nonallelic recombination in haploid meiosis. Genetics 146: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W.F. 2002. Order and disorder in the nucleus. Curr. Biol. 12: R185–R192. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K.A. 1982. Molecular genetics of yeast mating type. Annu. Rev. Genet. 16: 439–500. [DOI] [PubMed] [Google Scholar]
- Nassif, N., Penney, J., Pal, S., Engels, W.R., and Gloor, G.B. 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element–induced gap repair. Mol. Cell. Biol. 14: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F. and Haber, J.E. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, A.L., Cook, K.R., Belvin, M., Dompe, N.A., Fawcett, R., Huppert, K., Tan, L.R., Winter, C.G., Bogart, K.P., Deal, J.E., et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- Preston, C.R., Engels, W., and Flores, C. 2002. Efficient repair of DNA breaks in Drosophila: Evidence for single-strand annealing and competition with other repair pathways. Genetics 161: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, C. and Jasin, M. 2000. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 405: 697–700. [DOI] [PubMed] [Google Scholar]
- Richardson, C., Moynahan, M.E., and Jasin, M. 1998. Double-strand break repair by interchromosomal recombination: Suppression of chromosomal translocations. Genes & Dev. 12: 3831–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y.S. and Golic, K.G. 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rong, Y.S. and Golic, K.G. 2001. A targeted gene knockout in Drosophila. Genetics 157: 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y.S. and Golic, K.G. 2003. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y.S., Titen, S.W., Xie, H.B., Golic, M.M., Bastiani, M., Bandyopadhyay, P., Olivera, B.M., Brodsky, M., Rubin, G.M., and Golic, K.G. 2002. Targeted mutagenesis by homologous recombination in Drosophila melanogaster. Genes & Dev. 16: 1568–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, B. 1992. Identification of cryptic lox sites in the yeast genome by selection for Cre-mediated chromosome translocations that confer multiple drug resistance. J. Mol. Biol. 223: 911–928. [DOI] [PubMed] [Google Scholar]
- Schmidt, E.E., Taylor, D.S., Prigge, J.R., Barnett, S., and Capecchi, M.R. 2000. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc. Natl. Acad. Sci. 97: 13702–13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum, C., Pauli, D., Delattre, M., Jaquet, Y., Spierer, A., and Spierer, P. 2002. Isolation of Su(var)3-7 mutations by homologous recombination in Drosophila melanogaster. Genetics 161: 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal, M.L. and Hartl, D.L. 1996. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144: 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal, M.L. and Hartl, D.L. 2000. Application of Cre/loxP in Drosophila: Site-specific recombination and transgene coplacement. Methods Mol. Biol. 136: 487–495. [DOI] [PubMed] [Google Scholar]
- Smih, F., Rouet, P., Romanienko, P.J., and Jasin, M. 1995. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 23: 5012–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strout, M.P., Marcucci, G., Bloomfield, C.D., and Caligiuri, M.A. 1998. The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proc. Natl. Acad. Sci. 95: 2390–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, W., Ashburner, M., and Hawley, R.S. 2000. Drosophila protocols. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Thompson, C.B. 1992. Creation of immunoglobulin diversity by intrachromosomal gene conversion. Trends Genet. 8: 416–422. [DOI] [PubMed] [Google Scholar]
- Van Deursen, J., Fornerod, M., Van Rees, B., and Grosveld, G. 1995. Cre-mediated site-specific translocation between nonhomologous mouse chromosomes. Proc. Natl. Acad. Sci. 92: 7376–7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent, D.C., Hoeijmakers, J.H., and Kanaar, R. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2: 196–206. [DOI] [PubMed] [Google Scholar]
- Vazquez, J., Belmont, A.S., and Sedat, J.W. 2001. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr. Biol. 11: 1227–1239. [DOI] [PubMed] [Google Scholar]
- Wu, X., Moore, J.K., and Haber, J.E. 1996. Mechanism of MAT α donor preference during mating-type switching of Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Wu, C., and Haber, J.E. 1997. Rules of donor preference in Saccharomyces mating-type gene switching revealed by a competition assay involving two types of recombination. Genetics 147: 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. and Bradley, A. 2001. Engineering chromosomal rearrangements in mice. Nat. Rev. Genet. 2: 780–790. [DOI] [PubMed] [Google Scholar]
- Zheng, B., Sage, M., Cai, W.W., Thompson, D.M., Tavsanli, B.C., Cheah, Y.C., and Bradley, A. 1999. Engineering a mouse balancer chromosome. Nat. Genet. 22: 375–378. [DOI] [PubMed] [Google Scholar]
- Zheng, B., Sage, M., Sheppeard, E.A., Jurecic, V., and Bradley, A. 2000. Engineering mouse chromosomes with Cre-loxP: Range, efficiency, and somatic applications. Mol. Cell. Biol. 20: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCE
- http://www.fruitfly.org/; Berkeley Drosophila Genome Project home page.
- http://www.drosdel.org.uk/; Department of Genetics, University of Cambridge.