Abstract
A targeted discovery effort is required to identify low frequency single nucleotide polymorphisms (SNPs) in human coding and regulatory regions. We here describe combining mismatch repair detection (MRD) with dideoxy terminator sequencing to detect SNPs in pooled DNA samples. MRD enriches for variant alleles in the pooled sample, and sequencing determines the nature of the variants. By using a genomic DNA pool as a template, ∼100 fragments were amplified and subsequently combined and subjected en masse to the MRD procedure. The variant-enriched pool from this one MRD reaction is enriched for the population variants of all the tested fragments. Each fragment was amplified from the variant-enriched pool and sequenced, allowing the discovery of alleles with frequencies as low as 1% in the initial population. Our results support that MRD-based SNP discovery can be used for large-scale discovery of SNPs at low frequencies in a population.
Random sequencing approaches have led to the identification of a tremendous number of single nucleotide polymorphisms (SNPs) in the human genome. Through the work of The SNP Consortium, 1.4 million SNPs have been identified (Sachidanandam et al. 2001). This has been followed by other public projects, leading to the presence of ∼3 million SNPs with some level of validation. These SNPs provide researchers with a wealth of candidate SNPs in their desired candidate regions. Unfortunately, only a fraction of the disease-causing variations in regulatory and coding regions (cSNP) are identified through this approach (Kruglyak and Nickerson 2001; Carlson et al. 2003). The identification of low frequency cSNPs requires a targeted discovery effort. An extensive targeted effort to identify common cSNPs has been advocated before (Johnson et al. 2001) and partially implemented (Haga et al. 2002). Cost has been a major drawback for the targeted approach. To have 95% confidence of identifying an allele with a 2% frequency, 75 individuals need to be sequenced due to the Poisson statistics of chromosome sampling. In addition, detection of these alleles assumes the ability to detect heterozygote peaks in a sequencing trace with good accuracy.
We present our utilization of mismatch repair detection (MRD; Faham et al. 2001) to enrich fragments amplified from pooled genomic DNA samples for variant alleles that are then subjected to standard dideoxy sequencing. MRD has been described before as a method for multiplex variation scanning (Faham et al. 2001). Here we describe its use in combination with standard dideoxy terminator sequencing to discover variant alleles in pooled genomic DNA. MRD detects variants using the mismatch repair system of Escherichia coli (Modrich 1991). A specific strain (mutation sorter) is engineered to sort a mixture of transformed fragments into two pools: those carrying a variation and those that do not.
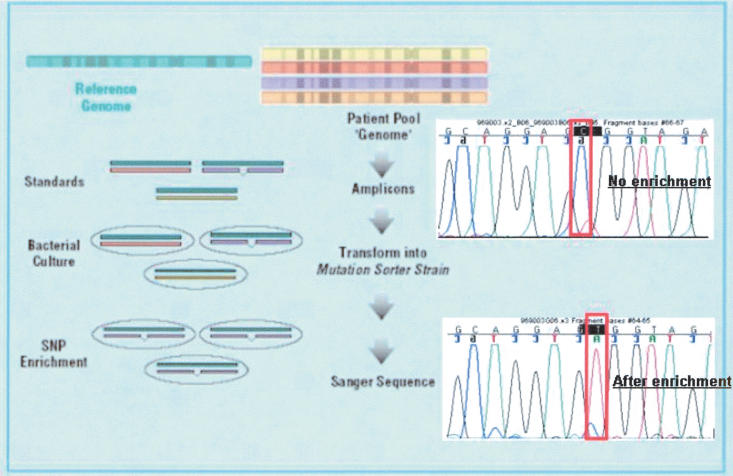
The basic approach is shown schematically in Figure 1. Sanger sequencing does not have sufficient sensitivity to detect rare alleles from genomic pools, as demonstrated in Figure 1, top trace, in which the PCR product from the pooled sample is sequenced directly. Instead, individual PCR reactions using pooled genomic DNA as a template are, in turn, pooled together and hybridized to PCR fragments from a single homozygous source (standard). These heteroduplexes are transformed into the mutation sorter strain, generating a pool of colonies enriched for variant alleles (compared with the standard). One amplification reaction from the variant-enriched pool is done for each amplicon, followed by a sequencing reaction to identify variant alleles in the population examined. The end result of this process is that the necessity of amplifying and sequencing many individuals is replaced with a pooled enrichment process that is carried out for hundreds or thousands of amplicons in a multiplexed fashion. The sequencing effort is thus reduced to the task of sequencing a standard and the variant-enriched pool.
Figure 1.
A schematic of the MRD SNP discovery process. Genomic DNA samples are pooled together, and PCR amplicons are generated by using the pooled genomic DNA as a template. If this PCR product was simply sequenced, the SNP shown would be lost in the noise of sequencing (top trace). The various amplicons (each amplified by using the genomic pool sample as a template) are pooled and hybridized to standards and transformed into the specifically designed mutation sorter strain. The fragments containing mismatches between the test sequence and the standard grow preferentially. This variant-enriched pool is then used as a template for PCR amplification and sequencing of the various amplicons. As seen when comparing the top and bottom traces, the minor allele becomes dominant after enrichment. Of note is that the frequency of this SNP is 15% in the tested population, as estimated by peak height as previously described (Kwok et al. 1994).
RESULTS
Scheme for SNP Discovery in Pooled Samples
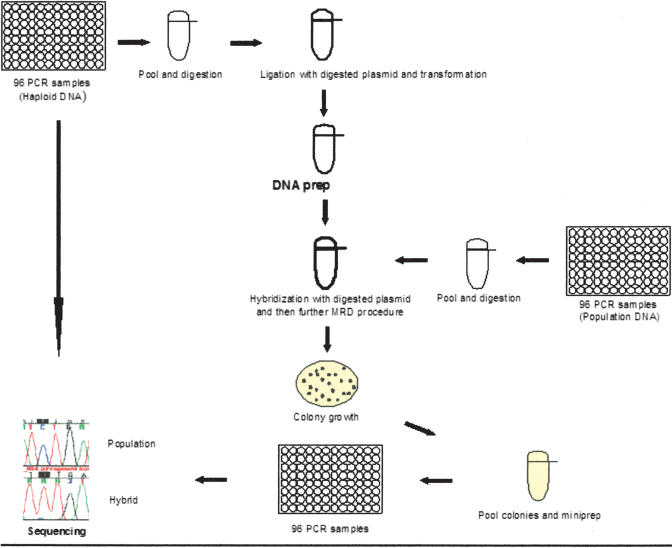
The basic MRD protocol and the mechanism of the fragment sorting based on the presence or absence of a mismatch (variation) by the mutation sorter have been described before (Faham et al. 2001). The experimental procedure for SNP discovery by MRD is depicted in Figure 2. At first, sequences of interest are amplified by PCR using the homozygous hydatidiform mole DNA as a template. These PCR products are pooled and cloned en masse in a specific vector, producing a standard plasmid library that is used to compare the test PCR products. To identify SNPs in the fragments of interest in the population under examination, individual PCR reactions amplified by using the pooled sample as a template are, in turn, pooled together and hybridized to DNA from the standards. These heteroduplex molecules are transformed into the mutation sorter strain. Two features of this strain allows for the enrichment of the variant allele in the initial population. First, bacteria transformed with a fragment containing a mismatch (between the test PCR product and the corresponding sequence in the standard) repair the mismatch in such a way as to preserve the sequence on the test (not the standard) strand. Second, a specific selective medium selects for bacteria transformed with fragments carrying a mismatch. These bacteria selected to have a mismatch repair event are collected, and the pool of plasmid DNAs enriched for population-specific variant alleles is purified. Then two PCR reactions for each sequence of interest are produced: one from standard plasmid library and another from variant-enriched plasmid pool. Each PCR product is sequenced, and the traces from variant-enriched pool and standard plasmid library are compared. Sequencing PCR fragments from a pool of colonies and not from individual clones alleviates the problem of erroneously detecting PCR-induced mutations. Because PCR-induced mutations are expected to occur in many positions and each fragment is represented by many colonies, it is unlikely that a specific PCR-induced mutation would dominate the pool and be detected in the sequencing trace.
Figure 2.
Scheme for laboratory process for the MRD-based SNP discovery. Individual PCR reactions from a homozygous source are pooled and cloned en masse to vector DNA to construct standards. Individual PCR reactions from the population of interest sample are pooled, hybridized to the standards' DNA, and transformed into the mutation sorter strain that enriches for the variant fragments. The resultant colonies are pooled, and the pool plasmid DNA is prepared followed by individual PCR and sequencing reactions for each amplicon. Sequencing traces from the enriched pool are compared with those of the homozygous source in order to detect SNP sites.
The enrichment procedure, with the exception of the PCR and sequencing steps, allows simultaneous processing of hundreds or thousands of sequences in one reaction. These multiplex steps replace a large number of PCR and sequencing reactions that would have been required in the traditional targeted SNP discovery procedure.
SNP Discovery in 126 Amplicons on Human Chromosome 21
The availability of an extensive SNP discovery study on human chromosome 21, using chip wafer technology by Perlegen Sciences (Mountain View), allowed us to design an experiment to test our method of SNP discovery (Patil et al. 2001). We designed primers to amplify 126 amplicons containing exons and some flanking intron sequences. To get a good estimate on the false-negative rate of our method, we increased the number of SNPs that should be detected by selecting 24 of 126 amplicons on the basis that they have SNPs discovered by Perlegen Sciences chip wafer technology. These 24 amplicons as well as the other 102 randomly chosen amplicons were processed and analyzed together in the same fashion.
To construct a set of standards, we performed 126 PCR reactions using as a template a genomic DNA purified from mouse–human hybrid carrying one copy of human chromosome 21. The use of the hybrid ensured the presence of only one allele in the standards. PCR products were pooled, and a library of the cloned standards was produced. The bacterial clones were pooled, and DNA extracted from this pool was used as the standard that was compared with PCR products from the population of interest.
We used a pool of genomic DNA from 100 whites or 100 African Americans as a template for the PCR amplification. PCR products for each population were pooled and subjected to an MRD reaction, producing a variant pool of colonies enriched for alleles that differ from the standards. Plasmid DNA was isolated from the variant pool and used as a template for PCR reaction for each amplicon that was then sequenced by forward and reverse primers.
We obtained sequence information on 105 and 102 of 126 amplicons in each of the white and African American populations, respectively. No sequence information was obtained for 15 amplicons in either population, and 60% of these failures was due to failure of the PCR amplification from genomic DNA. Seven of 111 sequenced amplicons showed many “variants” as a result of amplifying paralogous sequences and they were removed from the subsequent analysis,4 reducing the total number of fragments analyzed to 104 fragments. For each of these 104 fragments, the sequence traces (forward and reverse) from the enriched pool(s) were compared with the traces of the standard, and SNPs were called. The list of SNPs identified by our method was then compared with the SNPs detected by the wafer technology.
In the 104 products that succeeded in at least one population, 44 SNPs were previously identified by using the wafer technology. We identified 42 of 44 of these SNPs. SNPs identified by Patil et al. (2001) generally have minor allele frequency of ≥10%. The high sensitivity of our method to detect SNPs that were previously identified by Patil et al. is consistent with an excellent sensitivity for alleles with minor frequency of ≥10%. It is of note that among randomly chosen fragments, we identified 36 SNPs.
One expects that comparing the sequence against itself would generate no variants. So in additional experiment, we substituted 49 PCR products from genomic DNA pool with PCR product from hybrid DNA. We did not detect any variation, suggesting that the false-positive rate for our method is low.
Two-Round MRD Enrichment
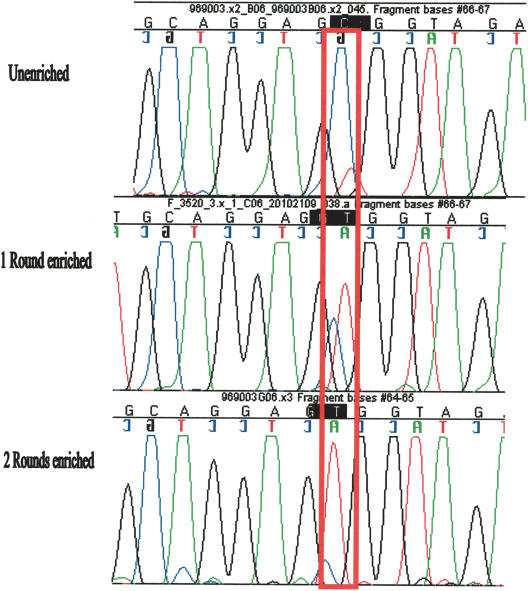
We reasoned that another round of MRD enrichment can improve the sensitivity of the assay. A more robust enrichment would make the SNP detection easier and more automated and allows for the detection of less frequent SNPs. We used the variant-enriched pool obtained above as a template to amplify 49 different amplicons that were hybridized to the standard library and subjected to another round of MRD reaction. Each of the 49 amplicons was amplified again from the twice enriched pool and sequenced. Indeed, we found the variant alleles are further enriched in the second MRD round (Fig. 3).
Figure 3.
Sequencing traces assessing enrichment after one and two MRD rounds. Sequencing traces of products that had undergone zero (top), one (middle) or two (bottom) MRD rounds. The red box indicates the site of variation (C/T SNP). The reference has the C, and the variant base is the T. The bases have the following colors: A, green; G, black; T, red; and C, blue. The top trace is a sequence of the PCR amplified from 100 whites. The T/C polymorphism is evident after the first round of enrichment, and after the second round, the T becomes the only major peak. Of note is that this SNP is the same as that in Figure 1.
SNP Discovery in BRCA1 and BRCA2 Genes
The above experiment with chromosome 21 markers showed that our method has an excellent sensitivity for alleles of ≥10%. However, it did not clearly define the lower limit of the sensitivity. This is especially true because we implemented multiple significant improvements to the process, including the two rounds of enrichment. We designed an experiment to investigate the sensitivity of the method by testing multiple variants at different frequencies.
We used 94 samples that had already been sequenced for all the coding exons of BRCA1 gene (R. Kroiss, T.M.U. Wagner, D. Muhr, D. Richards, P. Shen, M. Schreiber, E. Fleischmann, G. Longbauer, E. Kubista, M. Kubista, et al., in prep.). There were 10 known SNPs in the BRCA1 exons. To assess our sensitivity at different allele frequency levels, we wanted to construct genomic pools to test each of these SNPs at frequencies ranging from 1% to 30%. We designed 95 amplicons that encompass all the exons of BRCA1 and BRCA2 genes. We used homozygous DNA from hydatidiform mole as a template for PCR. The PCR products were pooled and cloned en masse to generate the standard DNA.
We constructed five different pools. The first pool was an equimolar ratio of all 94 samples. The other four pools were constructed by using five genomic samples and the homozygous mole DNA. The five genomic samples were very carefully quantitated and mixed in equal amounts. This DNA pool, the five-genome pool, was again carefully quantitated and mixed with four different ratios of excess mole DNA. The four pools had one part of the five-genome pool to 6, 13, 34, or 69 parts of the mole DNA. The mole DNA obviously had no variation to itself, and so it effectively acted as a diluent of variant alleles in the other samples. The frequency of an allele in a pool was then the frequency of that allele among the five individuals divided by the dilution factor. For example, an allele that is present in seven of 10 chromosomes among the five individuals has the final frequency of 10%, 5%, 2%, and 1% in the four pools.
We used these four pools as well as the pool of all 94 samples as a template for 95 PCR reactions amplifying BRCA1 and BRCA2. The 95 PCR reactions from each of the five genomic mixtures were pooled and subjected to two rounds of MRD enrichment as described above. Eighty-nine out of 95 fragments yielded sequencing results from at least one of the five MRD reactions, and the failure in four of six cases was in the initial genomic PCR reaction. The sequencing traces from the standard and those from each MRD reaction were independently compared, and SNPs were called. A list of the SNPs detected in each pool was then compiled and compared with the known SNPs.
False Negatives
Two of the 10 SNPs were not detected in any of the pools. One of these SNPs was on an amplicon that failed the initial PCR step. The other SNP was missed even when present at 30% frequency. Further investigation of the sequencing data showed that another SNP that is perfectly correlated with this missing SNP was present one nucleotide away from the 3′ end of one of the primers. This secondary SNP caused allele-specific amplification, resulting in the absence of the primary SNP from the amplicon. Given that these two nondetected SNPs (one for amplification failure and the other because of allelic drop-out due to secondary SNPs) would be missed in all PCR-based methods, we did not include them in the further analysis.
Sensitivity
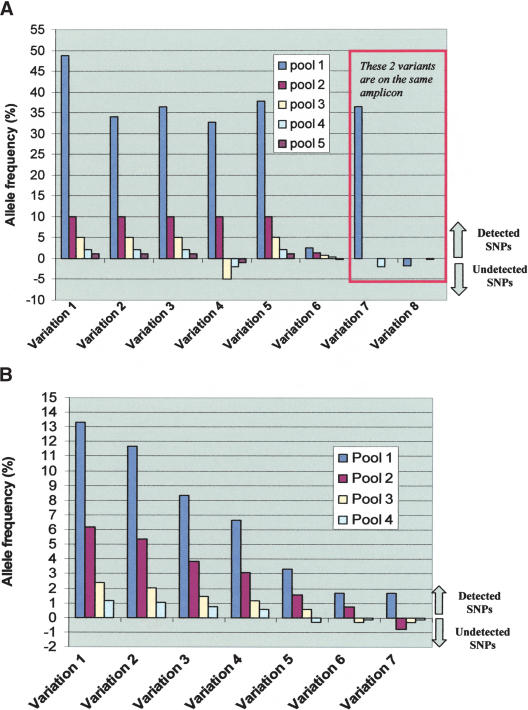
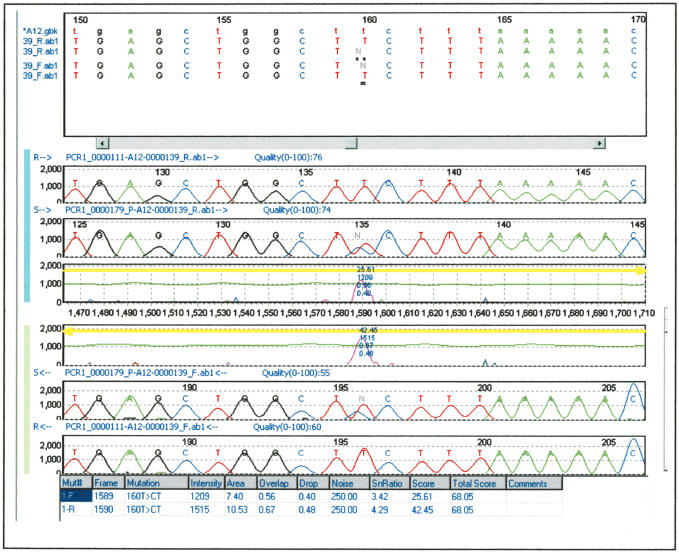
As can be seen in Figure 4A, all the variants were detected when their frequency was at least 10%. The discovery rate decreases when the allele frequency is lower with most of the variants still detected when present at 1% frequency. An example of a SNP detected at the 1% frequency is shown in Figure 5. Variant 8 is in the same amplicon as variant 7 and is not detected underscoring the difficulty with this technology to detect two SNPs with different frequencies on the same amplicon.
Figure 4.
Detection of the various SNPs in the study. (A) Detection of known SNPs. This panel depicts the allele frequency (y-axis) of the seven detected variations in the four constructed genomic mixtures (x-axis) with data from each pool represented with a different color. Pool 1 is where all 94 samples are mixed. The other four pools are different mole DNA dilutions of the five-genome pool. To distinguish SNPs that were detected from those that were not detected in a particular genomic mixture in the MRD-based SNP discovery, we depict the undetected SNPs with negative allele frequency. Variations 7 and 8 are in the same amplicon. Sequence data for this amplicon was obtained for only two pools. Data from pool 1 of this amplicon demonstrates how the presence of a common variant (variation 7) masks other rare variant(s) (variant 8) in the same amplicon. (B) Detection of unknown SNPs. This panel depicts the allele frequency (y-axis) of the seven detected variations in the four constructed genomic mixtures (x-axis) with data from each pool represented with a different color. The four pools shown are for the different mole DNA dilutions of the five-genome pool. To distinguish SNPs that were detected from those that were not detected in a particular genomic mixture in the MRD-based SNP discovery, we depict the undetected SNPs with negative allele frequency. Some variations are detected when present at frequency as low as 0.5%.
Figure 5.
Sequencing traces allowing detection of 1% allele. This is an example of the enrichment of 1% allele detected by the mutation surveyor software (variation 1 in Fig. 4A). The top two traces are the forward traces from the standard and the variant-enriched pool, and the bottom two traces are the reverse traces. The letters R and S in the left side of each trace denote the standard and variant-enriched pool, respectively. The middle traces are the software graphical representation of the “difference” between the traces. At the top, the sequence of the traces (uppercase letters) are aligned with the public database sequence (lowercase letters), allowing the localization of the variation.
False Positives
In addition to the already known SNPs, we found eight new variants (seven of them in BRCA2 and one in BRCA1) in the four pools constructed from pooling the five individuals. We sequenced these eight amplicons in the five individuals that made up the pools and detected seven of eight variants in at least one individual. The frequency of each SNP in the initial genomic mixture was calculated and is depicted in Figure 4B. As seen in Figure 4B, the detection of SNPs at 1% frequency is robust, and some variants were detected at a frequency as low as 0.5%.
The last SNP that was detected in one of the pools could not be seen in any of the individuals. This estimates that this method generates a false positive in ∼0.25% of the amplicons (one false positive in ∼400 amplicons in the four pools) and that the proportion of false-positive SNPs is ∼2% of all SNPs detected (in the four pools). It is likely that this false positive is secondary to PCR-induced mutation that happened to be unusually enriched in one of the four pools. Of note is that this false-positive variation had a very modest enrichment, and one could potentially reduce the false-positive results by implementing more rigorous rules about the extent of enrichment for accepting a variation. Given the traces seen in this study, such rules would likely decrease the sensitivity of detecting alleles at <1% frequency but have minimal effect for alleles that are >1% that are generally robustly enriched.
Reproducibility
As a first indication of reproducibility, we noted that variation number 5 was present in another overlapping amplicon. This variation was detected in all five pools in the two amplicons (data not shown). To further assess reproducibility, we did the MRD enrichment in duplicates for the four pools that were constructed from the five-genome pool and the mole DNA. The results were essentially identical for all the duplicates with two exceptions: missing one variation at 0.7% frequency and the absence of the false-positive SNP that was described above. So all the variations with an allele frequency of ≥1% that were detected in the first experiment were also detected in the replication experiment. This underscores the general reproducibility of these results as well as the fact that the false-positive result is due to random rather than systematic error consistent with an enrichment of a PCR-induced mutation.
Extent of Enrichment
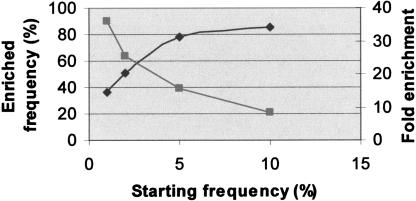
We have estimated from the sequencing traces, the frequency of the SNPs in the variant pool as described previously (Kwok et al. 1994). Because the starting frequency of each SNP was known, we were able to estimate the level of enrichment for each SNP. Figure 6 shows the average level of enrichment for variants at different allele frequencies. Rarer alleles have higher fold enrichment than do common alleles. The incomplete enrichment stems from the background of the system that is mainly a reflection of PCR error as described in the Discussion section.
Figure 6.
Extent of enrichment. This figure shows the average enrichment for detected SNPs with starting allele frequencies of 1%, 2%, 5%, and 10%. To generate this graph, we obtained the average frequency in the variant pool of the detected SNPs that were initially at 1%, 2%, 5%, or 10%. The estimation of the enriched frequency was done as previously described (Kwok et al. 1994). As expected, the rarer alleles have higher-fold enrichment but continue to have lower frequency in the enriched pool. For example 1% and 10% alleles get enriched 36- and 8.5-fold, respectively, to achieve in the variant pool 36% and 85% frequencies, respectively.
DISCUSSION
Dideoxy terminator sequencing is the standard method to determine the nature of a variation. However, to identify a relatively infrequent allele, a large number of sequencing reactions need to be performed. The premise of our method is to combine a highly multiplexed assay to provide a variation-enriched sample that can be analyzed with a much smaller number of sequencing reactions. At least 1000 fragments can be processed in parallel by MRD without loss of sensitivity (H. Fakhrai-Rad, E. Namsaraev, and M. Faham, unpubl.), making it an ideal method for generating the variant-enriched sample (we have compared the discovery rate when the experiment was done in 200 plex and 950 plex and obtained identical sensitivity). In this proof of principle, we demonstrated that SNPs with frequency as low as 1% can be detected with high sensitivity.
The sensitivity threshold of 1% is largely defined by the PCR error rate. By using pfu ultra, we have determined that ∼1% of the fragments contain a PCR error in some position (J. Zheng and M. Faham, unpubl.). These PCR errors that occur at many sites are enriched in the variant pool. The occurrence of the errors in many sites ensures that no single error is dominant enough to be detected by sequencing the pool of colonies. Although these errors are not detected by sequencing, they are enriched in the variant pool, creating a minimum allele frequency threshold of ∼1% for a SNP to be detected. For example, after enrichment of fragments carrying SNPs at 0.1%, 1%, and 10% frequencies, the proportion of the SNP in the sequencing traces is expected to be ∼10%, 50%, and 90%, respectively, and the proportion of colonies with PCR errors is expected to be ∼90%, 50%, and 10%, respectively. The observed enrichment (Fig. 6) is somewhat lower than these expected numbers because of other sources of background such as mutations in the oligonucleotides. Because the dominant background is due to PCR-induced error, improvement of the sensitivity requires the utilization of a polymerase with lower error rate.
This work describes the first methodology that uses pooled genomic DNA to detect previously unknown variations in many fragments. Methodologies that study genetic variations in pooled genomic DNA can be divided into three classes. The first class of methods is focused on the estimation of the frequency of a known SNP in a specific population (Krook et al. 1992; Kwok et al. 1994; Werner et al. 2002; http://brie2.cshl.org). The second class targets the identification of a specific allele in vast excess of the other alleles (Parsons and Heflich 1998; McKinzie and Parsons 2002). The third class attempts to identify unknown variations in a pooled sample (Amos et al. 2000; Wolford et al. 2000). Unlike the first two classes, our MRD-based SNP discovery detects variations that are previously unknown. Our method differs from the third class because the enrichment step it uses is highly multiplexed.
Multiple applications can be considered for this technology, including the identification of somatic mutations in which only a fraction of the cells carry mutant alleles, or the cataloguing of mutations in many genes in a pool of mutagenized animals. We believe an important application of this SNP discovery platform is the large-scale discovery of coding and regulatory SNPs in human populations.
One limitation for MRD-based SNP discovery is that multiple SNPs can occur on a particular sequencing fragment. If this occurs with the two SNPs having very different frequencies, the SNP with the higher frequency will tend to dominate the enriched pool, suppressing the signal of the rarer SNP. This effect can be mitigated in several ways. The first is to use fairly small PCR fragments to minimize the chances of the presence in the tested population of more than one SNP within a single fragment (we use fragments with average size of ∼300 bp). For application of SNP discovery in human exons, this technical limitation does not create a large burden because the average size of human exons is 150 to 160 bp. Second, in cases in which common SNPs are known to occur, PCR primers can be designed to exclude these SNPs. The a priori knowledge of SNPs can be obtained from databases or the use of information from the MRD-based SNP discovery. In this latter scheme, all amplicons for which a SNP is found can be retested using primers that avoid the SNP discovered in the first round. This approach of performing two cycles of discovery is certainly more comprehensive than relying on the databases, but it doubles the time needed for the experiment.
This limitation is to be weighed against the high costs of sequencing and analysis of many individuals in the traditional sequencing approach. Reducing the number of individuals sequenced in the classical manner reduces coverage by introducing Poisson noise in the choice of a small population. For example, by sequencing 15 individuals there is 40% chance of missing a 3% allele and a 37% chance of seeing the allele once. When an allele is seen only once, it is difficult to distinguish this allele from other private alleles that are present in the sequenced individuals. This is a distinct advantage for the MRD-based SNP discovery, which is insensitive to private variations. For example, by using 300 individuals, a 2% allele is represented on average 12 times and can be readily enriched, whereas a private allele at frequency of 0.16% would not be sufficiently enriched to be detected.
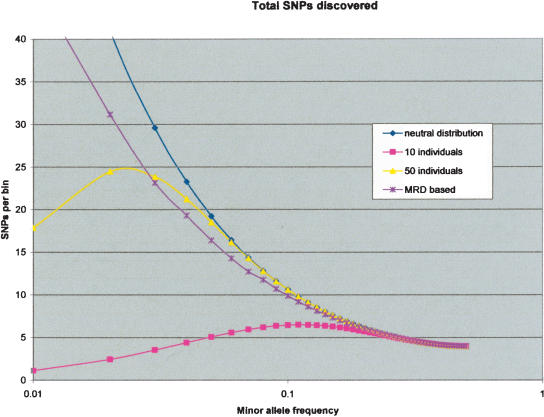
We have modeled the expected performance of MRD-based SNP discovery and compared it to the performance of traditional sequencing. As is shown in Figure 7, the sensitivity of the MRD-based SNP discovery is comparable to that obtained from sequencing 50 individuals. In this model, we assumed that for the MRD-based SNP discovery we would design our amplicons in such a way as to avoid validated SNPs in the public databases (to ameliorate the effect of having two SNPs in one amplicon). A better performance would be obtained by doing another cycle of SNP discovery, avoiding all SNPs detected in the first cycle.
Figure 7.
Sensitivity of MRD-based and traditional SNP discovery. We have modeled the proportion of SNPs that would be detected by various approaches. The y-axis is a relative measure of the number of SNPs, and the x-axis is the logarithm of the minor allele frequency. The model assumes a variation distribution consistent with neutral theory, and the total number of SNPs is shown in the blue curve. A perfect SNP discovery approach would have essentially the same curve. This neutral theory assumption is an approximation for at least three reasons. First, SNPs aggregate close together, more than expected randomly (Reich et al. 2002). Second, this aggregation is often generally due to variants on the same chromosome (Reich et al. 2002). Third, the variation distribution in exons is shifted toward the rarer alleles (Cargill et al. 1999; Halushka et al. 1999). The aggregation of SNPs would make the problem of two SNPs in the same amplicon more pronounced. This is balanced by the presence of LD and the occurrence of variants on the same chromosome, allowing for the enrichment of both SNPs together. The shift toward rare allele frequencies in the exons also should decrease the effect of the two SNPs in the same amplicon. Performance of traditional sequencing assumes perfect technical detection but takes into account the sampling error described above and requires two observations of the allele. We show the performance for sequencing 10 and 50 people (in pink and yellow, respectively). For the MRD-based SNP discovery (purple curve), we have taken into account the sensitivity determined from the BRCA experiment (Fig. 4). We also assumed that we would design our amplicons in such a way as to avoid validated SNPs in the public databases (to ameliorate the two SNPs in one amplicon effect). The proportion of SNP detected in the model has taken into account the expected times two SNPs are present on the same amplicon after avoiding database-validated SNPs in the amplicon design phase.
The use of MRD to enrich for variant alleles can cut the cost and effort involved in sequencing. With MRD enrichment for each amplicon, two samples need to be sequenced: the variant pool and the standard compared with 50 individuals in the traditional approach. Therefore, MRD enrichment can lead to 25-fold reduction in sequencing. The cost and effort of performing the MRD reaction are amortized over hundreds of distinct fragments that can be processed in one reaction. With 1000 fragments processed simultaneously, each MRD reaction replaces ∼96,000 sequencing reactions (48 forward and reverse sequencing reaction saved per amplicon multiplied by 1000 amplicons) and the associated trace analysis overhead.
Much effort is being spent to develop linkage disequilibrium maps for the human genome to be used in later association studies (Dawson et al. 2002; Gabriel et al. 2002; International Hap-Map Consortium 2003). Integration of coding and regulatory SNPs in these maps can be of great use for the identification of the basis of human common disease. We believe MRD-based SNP discovery can be an enabling technology to generate this resource.
METHODS
Construction of Standards
All enzymes used were from New England Biolabs (NEB) unless otherwise specified. Amplicons were designed to amplify exons and flanking intron sequences. Primer selection was done through a batch version of PRIMER3 (Rozen and Skaletsky 1996, 1997, 1998). The amplicon sizes were designed to have approximately the same size, 100 to 500 bp. Restriction sites were added to the 5′ ends of all the primers; the sites were Cla I and Sac II sites for the chromosome 21 study and Cla I and Asc I for the BRCA study. PCR was performed by using 0.8 U pfu turbo hotstart (or pfu ultra in the BRCA experiment) polymerase (Stratagene) with the standard buffer, dNTP concentration in 20 μL reaction, and a human-mouse hybrid containing one copy of chromosome 21 (Corriell) as a template or hydatidiform mole (Corriell) for the BRCA study. The cycling conditions were 95°C 30 sec, 57°C 1 min, and 72°C 1 min for 35 cycles. The PCR products were run on a 1% agarose gel, and equal mass were pooled and purified through gel electrophoresis. The product pool was digested with Cla I and Sac II (or Cla I and Asc I) at 37°C for 2 h. One hundred nanograms of the digested products were ligated to 100 ng digested and dephosphorylated plasmid pMRD300 carrying the active Cre gene. pMRD300 is an improvement over the previously published vector pMRD100, which in combination with a new Mutation Sorter strain (MS3) can reduce the background by as much as fourfold (M. Faham and R.W. Davis, unpubl.). After overnight ligation at 16°C, a QiaQuick purification column (Qiagen) was performed to purify the product from the ligase and salts. Transformation is then done to a standard cloning strain such as DH5α, and selection for transformants was done in liquid by adding 100μg/mL carbenicillin. DNA was prepared and transformed into GM2929. The two step transformation is because dam– strains have low efficiency of transformation. DNA obtained from this transformation was used in later steps.
MRD Protocol
For the chromosome 21 study, equal amount genomic DNA from 100 whites and 100 African Americans (Corriell) were pooled. For the BRCA study, different pools were generated. In one pool, equal amounts of each of 94 samples were mixed. In the other four pools, five genomic DNA samples were mixed after picogreen and real-time PCR quantitation. The pool along with DNA from a hydatidiform mole were carefully quantitated again by using picogreen and real-time PCR. The pool and the mole DNA were then mixed at four different ratios.
PCR reactions using the genomic pool as a template was performed by using pfu turbo hotstart (or pfu utra for the BRCA experiment) polymerase using a similar protocol as described above. The PCR products from each population were pooled, and a purification column QiaQuick (Qiagen) was performed. Methylation of the PCR products was carried out by addition of Tris (ph 7.6) to a final concentration of 50 mM, as well as SAM (NEB) to a final concentration of 80 μM and 8 U dam methylase (NEB) at 37°C for 1 or 2 h. The PCR pool was then digested with Cla I and Sac II h at 37°C for 1 to 2.
For each MRD reaction, 2 μg of the above PCR product pool was mixed with 2 μg of the pool of the unmethylated standard DNA and 2 μg of digested vector carrying the inactive Cre gene pMRD400. pMRD400 is the same as pMRD300 except for a 5-bp deletion in the Cre gene. The three components were concentrated to 10 μL by using a QiaQuick minielute column (Qiagen); 0.5 μL of 0.5 M EDTA, 0.5 μL of 200 mM Tris (ph 7.6), 0.5 μL 20× SSC, and 1.25 μL of freshly diluted 1 M NaOH was added, and incubation for 15 min at room temperature followed. Then 1.25 μL of 2 M Tris (ph 7.2) and 12.5 μL formamide were then added, and reannealing was allowed to occur overnight at 42°C. The hybridization mixture was desalted by using a column (Edge Biosystems). Three microliters Taq Ligase buffer and 5 U Mbo I was added and incubated for 15 min followed by addition of 40 U Taq Ligase (NEB), and further incubation followed for 30 min at 65°C. Fifty units of exonuclease III (USB) and 20 U of T7 exonuclease (USB) were added and incubated for 30 min at 37°C. Ten microliters of SOPE Resin (Edge Biosystems) was added to eliminate single-stranded DNA, and a QiaQuick cleanup (Qiagen) was done before transformation.
Transformation of the MS3 strain was done by electroporation (Micropulser, BioRad). The electrocompetent MS3 cells preparation and the electroporation procedure were done as recommended (Ausubel et al. 1999). During the 1-h recovery phase, 1 μL 0.1 M IPTG was added into 1 mL SOC medium (Invitrogen). The culture was plated onto a plate supplemented with carbenicillin (75 μg/mL) and tetracycline (3.25 μg/mL) to select for colonies with variant alleles. The next day, all the colonies from the plate were collected into a tube with 1 mL LB. DNA from the cells obtained after the overnight growth with the selective media was minipreped by using the QIAPrep columns (Qiagen) as recommended by the manufacturer.
Sanger Sequencing and Sequence Analysis
By using DNA from the variant pool of the two electroporations, we performed PCR reactions followed by bidirectional sequencing for each amplicon. For the chromosome 21 project, the sequencing traces were analyzed by using Sequencher software and compared with the genome database by using BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat?command=start). SNPs were called when the dominant base in the enriched pool was different from the hybrid or when mixed peaks were seen in both strands of the enriched pool. For the BRCA study, mutation surveyor software (SoftGenetics) was used. SNPs automatically called in both strands were accepted directly. Variations called in only one strand were then inspected manually.
Acknowledgments
We thank Dr. Martin G. Marinus for the generous gift of the dam– strain GM2929 and Dr. Peidong Shen at the Stanford Genome Technology Center for providing the BRCA1 sequence. We also thank the members of the Stanford Genome Technology Center and ParAllele Bioscience for their constant support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2373904.
Footnotes
In order to ameliorate this problem, amplification primers specific for one member of the family need to be designed; we have implemented software that would accomplish that by performing BLAST on the primer sequences and changing primer pairs that were not unique.
References
- Amos, C.I., Frazier, M.L., and Wang, W. 2000. DNA pooling in mutation detection with reference to sequence analysis. Am. J. Hum. Genet. 66: 1689–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. 1999. Current protocols in molecular biology. John Wiley and Sons, New York.
- Cargill, M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., Shaw, N., Lane, C.R., Lim, E.P., Kalyanaraman, N., et al. 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22: 231–238. [DOI] [PubMed] [Google Scholar]
- Carlson, C.S., Eberle, M.A., Rieder, M.J., Smith, J.D., Kruglyak, L., and Nickerson, D.A. 2003. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat. Genet. 33: 518–521. [DOI] [PubMed] [Google Scholar]
- Dawson, E., Abecasis, C.R., Bumpstead, S., Chen, Y., Hunt, S., Beare, D.M., Pabial, J., Dibling, T., Tinsley, E., Kirby, S., et al. 2002. A first-generation linkage disequilibrium map of human chromosome 22. Nature 418: 544–548. [DOI] [PubMed] [Google Scholar]
- Faham, M., Baharloo, S., Tomitaka, S., DeYoung, J., and Freimer N. 2001. Mismatch repair detection (MRD): High throughput scanning for DNA variations. Hum. Mol. Genet. 10: 1657–1664. [DOI] [PubMed] [Google Scholar]
- Gabriel, S., Schaffner, S.F., Nguyen, H., Moore, J.M., Roy, J., Blumenstiel, B., Higgins, J., DeFelice, M., Lochner, A., Faggart, M., et al. 2002. The structure of haplotype blocks in the human genome. Science 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- Haga, H., Yamada, R., Ohnishi, Y., Nakamura, Y., and Tanaka, T. 2002. Gene-based SNP discovery as part of the Japanese Millennium Genome Project 2002: Identification of 190,562 genetic variations in the human genome. J. Hum. Genet. 47: 605–610. [DOI] [PubMed] [Google Scholar]
- Halushka, M.K., Fan, J.B., Bentley, K., Hsie, L., Shen, N., Weder, A., Cooper, R., Lipshutz, R., and Chakravarti, A. 1999. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet. 22: 239–247. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. 2003. The international HapMap project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- Johnson, G.C.L., Esposito, L., Barratt, B.J., Smith, A.N., Heward, J., Di Genova, G., Ueda, H., Cordell, H.J., Eaves, I.A., Dudbridge, F., et al. 2001. Haplotype tagging for the identification of common disease genes. Nat. Genet. 29: 233–237. [DOI] [PubMed] [Google Scholar]
- Krook, A., Stratton, I.M., and O'Rahilly, S. 1992. Rapid and simultaneous detection of multiple mutations by pooled and multiplex single nucleotide primer extension: Application to the study of insulin-responsive glucose transporter and insulin receptor mutations in non-insulin dependent diabetes. Hum. Mol. Genet. 1: 391–395. [DOI] [PubMed] [Google Scholar]
- Kruglyak, L. and Nickerson, D.A. 2001. Variation is the spice of life. Nat. Genet. 27: 234–236. [DOI] [PubMed] [Google Scholar]
- Kwok, P-Y., Carlson, C., Yager, T.D., Ankener, W., and Nickerson, D.A. 1994. Comparative analysis of human DNA variations by fluorescence-based sequencing of PCR products. Genomics 23: 138–144. [DOI] [PubMed] [Google Scholar]
- McKinzie, P.B and Parsons, B.L. 2002. Detection of rare K-ras codon 12 mutations using allele-specific competitive blocker PCR. Mutat. Res. 27: 209–220. [DOI] [PubMed] [Google Scholar]
- Modrich, P. 1991. Mismatch repair. Ann. Rev. Genet. 25: 229–248. [DOI] [PubMed] [Google Scholar]
- Parsons, B.L. and Heflich, R.H. 1998. Detection of basepair substitution mutation at a frequency of 1 × 10– 7 by combining two genotypic selection methods, MutEx enrichment and allele-specific competitive blocker PCR. Environ. Mol. Mutagen. 32: 200–211. [PubMed] [Google Scholar]
- Patil, N., Berno, A.J., Hinds, D.A., Barrett, W.A., Doshi, J.M., Hacker, C.R., Kautzer, C.R., Lee, D.H., Marjoribanks, C., McDonough, D.P., et al. 2001. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294: 1719–1723. [DOI] [PubMed] [Google Scholar]
- Reich, D.E., Schaffner, S.F., Daly, M.J., McVean, G., Mullikin, J.C., Higgins, J.M., Richter, D.J., Lander, E.S., and Altshuler, D. 2002. Human genome sequence variation and the influence of gene history, mutation and recombination. Nat. Genet. 32: 135–142. [DOI] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H.J. 1996, 1997, and 1998. Primer3. Code available at: http://www-genome.wi.mit.edu/genome_software/other/primer3.html.
- Sachidanandam, R., Weissman, D., Schmidt, S.C., Kakol, J.M., Stein, L.D., Marth, G., Sherry, S., Mullikin, J.C., Mortimore, B.J., Willey, D.L., et al. 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928–933. [DOI] [PubMed] [Google Scholar]
- Werner, M., Sych, M., Herbon, N., Illig, T., Konig, I.R., and Wjst, M. 2002. Large-scale determination of SNP allele frequencies in DNA pools using MALDI-TOF mass spectrometry. Hum. Mutat. 20: 57–64. [DOI] [PubMed] [Google Scholar]
- Wolford, J.K., Blunt, D., Ballecer, C., and Prochazka, M. 2000. High-throughput SNP detection by using DNA pooling and denaturing high performance liquid chromatography (DHPLC). Hum. Genet. 107: 483–487. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://brie2.cshl.org/; The SNP Consortium Web site.
- http://genome.ucsc.edu/cgi-bin/hgBlat?command=start; UCSC Genome Bioinformatics.