Abstract
A second-generation 5000 rad radiation hybrid (RH) map of the cattle genome was constructed primarily using cattle ESTs that were targeted to gaps in the existing cattle–human comparative map, as well as to sparsely populated map intervals. A total of 870 targeted markers were added, bringing the number of markers mapped on the RH5000 panel to 1913. Of these, 1463 have significant BLASTN hits (E < e–5) against the human genome sequence. A cattle–human comparative map was created using human genome sequence coordinates of the paired orthologs. One-hundred and ninety-five conserved segments (defined by two or more genes) were identified between the cattle and human genomes, of which 31 are newly discovered and 34 were extended singletons on the first-generation map. The new map represents an improvement of 20% genome-wide comparative coverage compared with the first-generation map. Analysis of gene content within human genome regions where there are gaps in the comparative map revealed gaps with both significantly greater and significantly lower gene content. The new, more detailed cattle–human comparative map provides an improved resource for the analysis of mammalian chromosome evolution, the identification of candidate genes for economically important traits, and for proper alignment of sequence contigs on cattle chromosomes.
The Cetartiodactyla include a phenotypically diverse group of relatively large mammals, including the Cetacea (e.g., whales, dolphins, and porpoises), Ruminantia (e.g., cattle, goats, and giraffes), Suiformes (e.g., pigs, wart hogs, and peccaries), Tylopoda (e.g., camels, lamas, and vicunas), and Hippopotamidae (e.g., hippopotamus). The Cetartiodactyla diverged from a common ancestor of the Rodentia and Primates approximately 94 million years ago at the Laurasiatheria-Euarchontoglires split (Springer et al. 2003). The deep divergence time of Cetartiodactyla relative to Primates has important consequences for comparative genomic analysis. A representative Cetartiodactyl genome will be useful for annotating the human genome for genes and conserved non-coding regulatory elements (Thomas and Touchman 2002). In addition, the cumulative adaptive phenotypes within the Cetartiodactyla make them ideal for exploring the molecular bases for adaptive evolution. For example, ruminants such as cattle have a distinctive digestive system (a four-chambered stomach), possess a cotyledonary, synepitheliochorial placenta, and exhibit variable structures in their immune system (e.g., hemal lymph nodes and high frequency of γ/δ T cells) compared to human primates. These features provide a strong justification for using cattle in comparative evolutionary studies and were essential criteria used for assigning a high priority to sequencing the cattle genome (http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/BovineSEQ; Bovine Genomic Sequencing Initiative 2002). Furthermore, the importance of cattle to sustainable agriculture in the developed and developing world imparts a special significance to improving genomic resources that can be used for the molecular dissection of complex traits of cattle, such as resistance to infectious diseases, growth rate, lactation, and fecundity.
We described a radiation hybrid (RH)-based, whole-genome cattle–human comparative map consisting of 768 genes spanning 9330 cR (Band et al. 2000). This “first-generation” whole-genome map, as well as earlier chromosome-specific maps, revealed significant features of mammalian genome evolution and facilitated the identification of genes responsible for double-muscling (Grobet et al. 1997; Kambadur et al. 1997), chondrodysplasia (Takeda et al. 2002), and a major QTL for milk production (Grisart et al. 2002). However, neither of the whole-genome cattle RH maps (Band et al. 2000; Williams et al. 2002), are adequate for assembly of the cattle genome sequence or detailed evolutionary studies. For example, the first-generation cattle–human RH5000 map (Band et al. 2000) had less than 60% comparative coverage. Our goal for the second-generation whole-genome cattle RH map was to fill in existing gaps in the cattle–human comparative map as well as segments of the map with large distances between markers. To clarify these ambiguous regions of the cattle–human comparative map, the COMPASS bioinformatics strategy (Ma et al. 1998; Rebeiz and Lewin 2000; Larkin et al. 2003) was employed for targeted selection of expressed sequence tags (ESTs) for RH mapping. In addition, human genome sequence coordinates were used to construct the comparative chromosome maps. This approach significantly increased the number of genes on the cattle RH map, thereby improving comparative coverage of the cattle genome and providing additional insights into mammalian chromosome evolution.
RESULTS
A Second-Generation Cattle RH5000 Map
The second-generation RH5000 map has 870 new assignments, bringing the total number of mapped markers to 1913. Among these markers, 1564 are genes and ESTs (Type I markers) and 349 are microsatellites (Type II markers) that facilitated proper orientation of the linkage groups (Supplemental Table 1). The map contains 86 linkage groups assigned to all 29 autosomes and the X chromosome (Table 1); 667 are framework markers (Fig. 1). Relatively few markers are unlinked (n = 58), or linked with ambiguous placement (n = 63). Failure of these markers to be placed in the map may be the result of amplification of paralogous sequences (resulting in higher than expected retention frequencies), genotyping errors, and map position outside terminal framework markers or within gaps. The higher LOD score threshold used for linkage analysis (LOD > 7.0) resulted in removal of 44 markers found on the first-generation map.
Table 1.
Summary Statistics of Cattle RH5000 Map
|
Markersa
|
|||||||
|---|---|---|---|---|---|---|---|
| BTA | Type I | Type II | Total | Framework | Retention frequency | Length (cR5000) | Linkage Groups |
| 1 | 73 | 17 | 90 | 20 | 0.27 | 256 | 3 |
| 2 | 77 | 11 | 88 | 33 | 0.19 | 502 | 5 |
| 3 | 74 | 14 | 88 | 14 | 0.19 | 303 | 2 |
| 4 | 51 | 13 | 64 | 24 | 0.20 | 431 | 5 |
| 5 | 69 | 34 | 103 | 25 | 0.18 | 508 | 1 |
| 6 | 58 | 14 | 72 | 26 | 0.16 | 521 | 3 |
| 7 | 67 | 14 | 81 | 20 | 0.16 | 547 | 3 |
| 8 | 52 | 16 | 68 | 27 | 0.19 | 513 | 5 |
| 9 | 39 | 11 | 50 | 14 | 0.13 | 345 | 2 |
| 10 | 58 | 8 | 66 | 30 | 0.23 | 466 | 1 |
| 11 | 78 | 8 | 86 | 33 | 0.26 | 557 | 3 |
| 12 | 28 | 10 | 38 | 11 | 0.17 | 226 | 5 |
| 13 | 62 | 8 | 70 | 22 | 0.19 | 384 | 2 |
| 14 | 44 | 21 | 65 | 27 | 0.24 | 361 | 6 |
| 15 | 47 | 8 | 55 | 24 | 0.18 | 452 | 3 |
| 16 | 58 | 9 | 67 | 26 | 0.28 | 563 | 4 |
| 17 | 52 | 14 | 66 | 27 | 0.21 | 440 | 3 |
| 18 | 110 | 11 | 121 | 44 | 0.24 | 549 | 4 |
| 19 | 71 | 5 | 76 | 26 | 0.43 | 513 | 5 |
| 20 | 20 | 9 | 29 | 10 | 0.19 | 150 | 3 |
| 21 | 38 | 12 | 50 | 18 | 0.18 | 316 | 2 |
| 22 | 41 | 5 | 46 | 16 | 0.26 | 291 | 3 |
| 23 | 59 | 13 | 72 | 28 | 0.27 | 462 | 1 |
| 24 | 37 | 5 | 42 | 11 | 0.23 | 219 | 1 |
| 25 | 48 | 7 | 55 | 21 | 0.24 | 286 | 2 |
| 26 | 34 | 5 | 39 | 17 | 0.22 | 333 | 1 |
| 27 | 18 | 14 | 32 | 8 | 0.14 | 175 | 2 |
| 28 | 16 | 7 | 23 | 14 | 0.30 | 170 | 1 |
| 29 | 38 | 5 | 43 | 23 | 0.30 | 469 | 1 |
| X | 47 | 21 | 68 | 28 | 0.16 | 533 | 4 |
| Total | 1,564 | 349 | 1,913 | 667 | 0.22 | 11,841 | 86 |
BTA; Bos taurus chromosome; Type I markers are located within the 5′ and 3′ boundaries of known or predicted genes; Type II markers are all microsatellites.
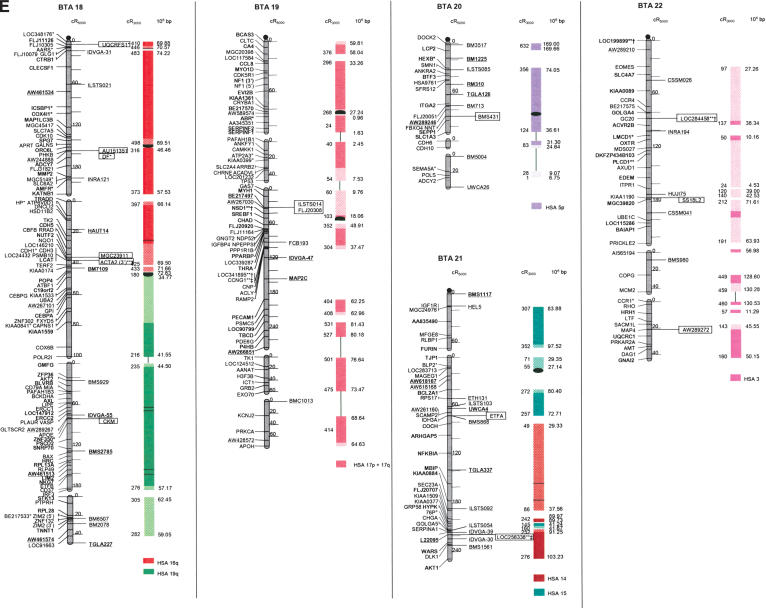
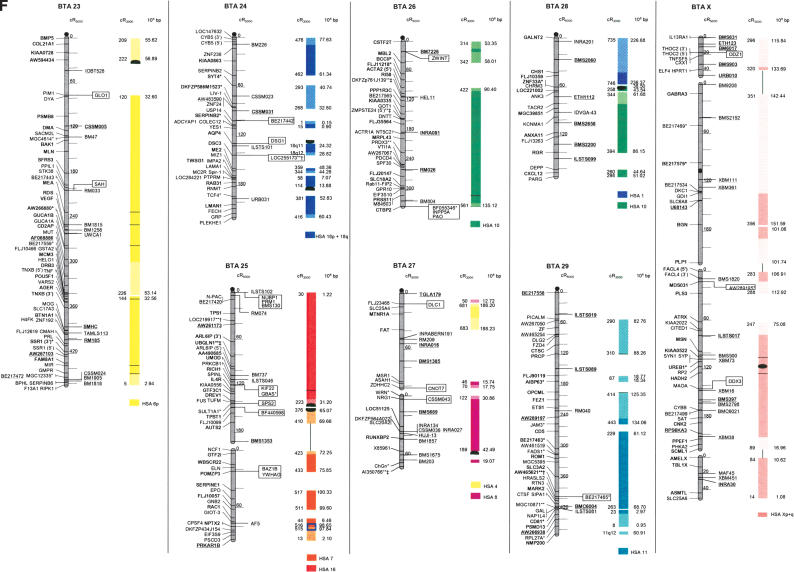
Figure 1.
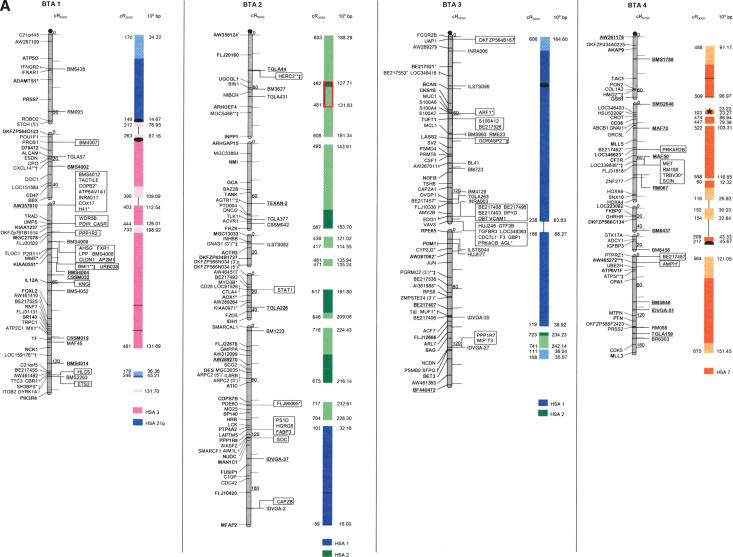
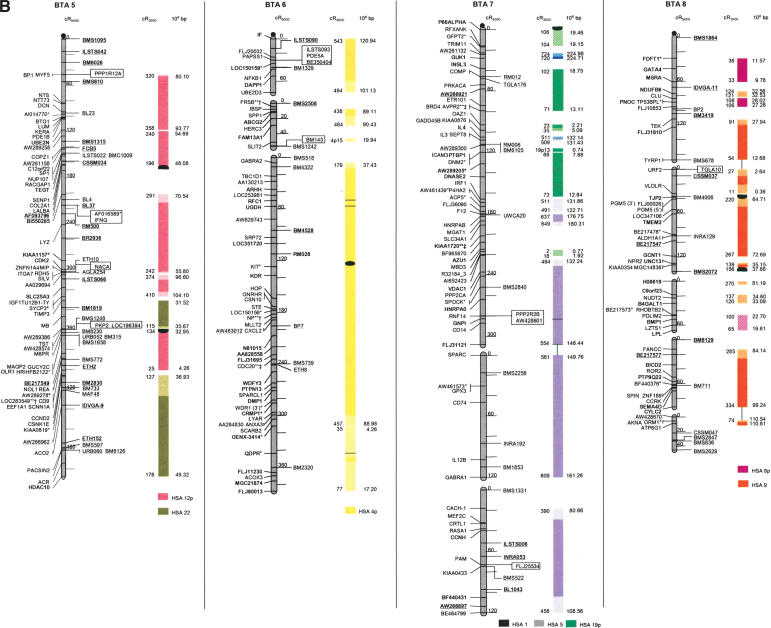
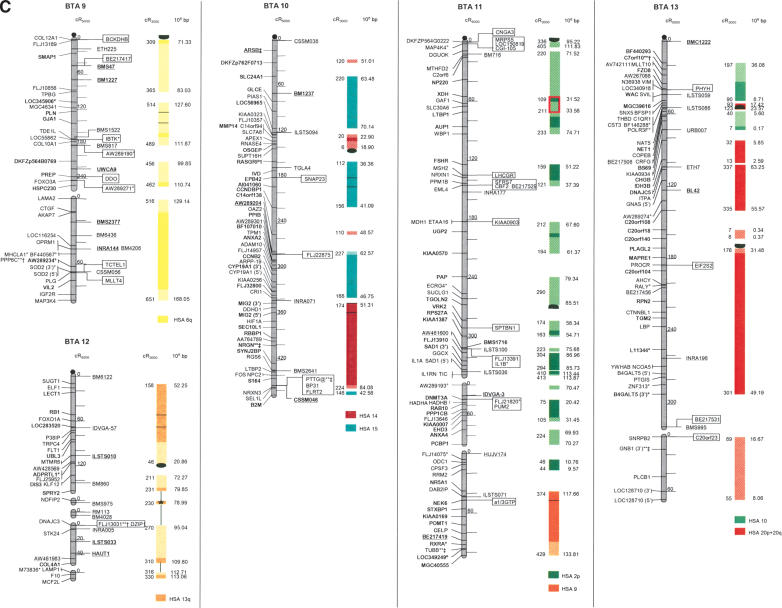
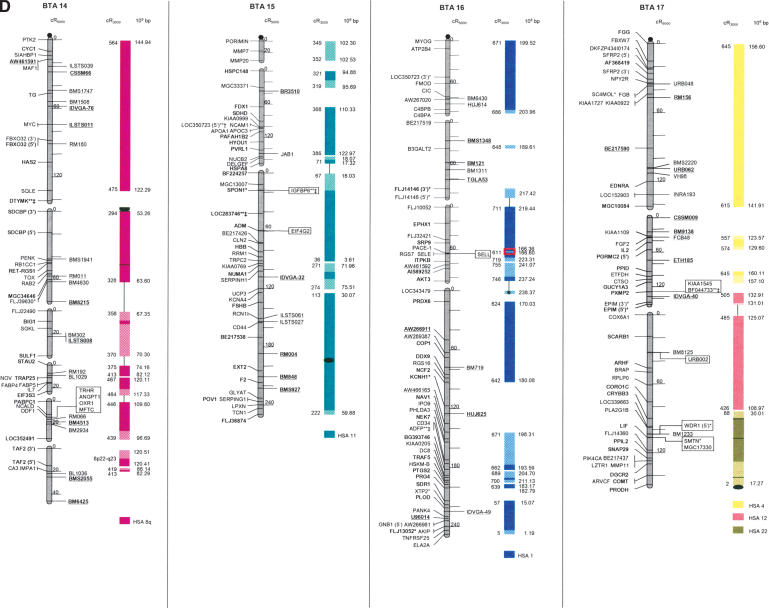
The second-generation cattle RH5000 and cattle–human whole-genome comparative chromosome maps. Cattle chromosomes are indicated by vertical shaded gray bars. Linkage groups on chromosomes are separated by blank spaces, and markers that are placed on the map on the basis of their two-point linkage data are binned to the right of the chromosomes. Cattle orthologs of human genes are named with the human gene symbol. Homologous human chromosome segments are color-coded, with segments defined on the first-generation map (Band et al. 2000) indicated as solid bars and improvements in coverage indicated by shading. GenBank accession nos. are used for cattle-specific sequences and for significant hits to human sequences with no gene symbol. Orthologs identified with sensitive BLAST options are indicated by the symbol “°”. Framework markers are shown in boldface type. Solid black lines running through a segment (e.g., the fifth conserved segment on BTA1) indicate framework markers that are not in the same order as in the human map. The comparative maps were constructed as described in Methods. Markers for which there is no map position based on the human genome sequence are underlined. An asterisk (*) indicates a single gene at a position that is inconsistent with the expected gene order based on the human sequence-based map. Single genes that map to chromosomes that are inconsistent with COMPASS predictions based on human sequence coordinates have a double asterisk (**). Single genes that have inconsistent COMPASS predictions based on human sequence coordinates but COMPASS predictions consistent with UniGene are indicated with a double asterisk and double dagger (**‡). A double asterisk and single dagger (**†) are used when there is inconsistent mapping information based on human genome sequence information and no UniGene GB4 information for comparison. Lines connect human segments that span gaps in the cattle RH map and have no evidence of chromosome rearrangement. Small regions on BTA2, BTA11, BTA16, and BTA25 that have gene orders that produced problems in creating distinct comparative segments are indicated by solid lines (colored for visibility) forming a box within the corresponding human segments. Segments on BTA4, BTA12, and BTA16 that appear to be out of place on the comparative map (i.e., segments that should be contained within other segments but appear as distinct) are indicated by a large asterisk within the segment. Human genome sequence coordinates are shown on the right side and GB4 cR3000 distances are on the left side of the conserved segments. Human centromeres are represented as full black circles if the position is within a conserved segment or on an acrocentric chromosome located at a segment boundary, or half circles if on a metacentric chromosome located at a segment boundary.
Seven cattle chromosomes are represented by one contiguous RH linkage group: BTA5, BTA10, BTA23, BTA24, BTA26, BTA28, and BTA29. The most fragmented chromosome, BTA14, has six linkage groups containing 65 markers (Table 1). BTA18 has the largest number of mapped markers (n = 121), whereas BTA28 has the fewest (n = 23). Relative differences in marker distribution are due in part to targeting of specific chromosomes for high-density mapping (e.g., BTA18; Goldammer et al. 2002) as well as chromosome size. The average chromosome length is 397 cR5000, ranging from 563 cR for BTA16 to 150 cR for BTA20 (Table 1). Total length of the map is 11,841 cR5000, with an approximate genome-wide ratio of 4 cR:1 cM. The total map length is underestimated due to the presence of 56 remaining gaps between RH linkage groups on 23/30 chromosomes. The number of gaps represents an increase compared to the first-generation map (Band et al. 2000) and can be explained by the more stringent threshold value used for marker placement, that is, 40 cR from the closest framework marker in the present study compared to 50 cR in the first-generation map. In contrast, the use of increased stringency for map assembly resulted in fewer unlinked markers (58 as opposed to 113) and fewer linked markers that were ambiguously placed (63 as opposed to 114).
The average retention frequency (RF) of the mapped markers is 22%, ranging from 43% for BTA19, which contains the selectable marker thymidine kinase, to 13% for BTA9 (Table 1). The relatively low RF for markers on BTAX (16%) was due to the fact that X-chromosome genes are present in the RH cell lines in the hemizygous state (the cattle parental cell line was created from a male). The large variation in RF resulted in widely different numbers and densities of framework markers among the chromosomes (Table 1).
A Whole-Genome Cattle–Human Comparative Map
The construction of individual comparative chromosome maps utilized the UniGene database (NCBI build 148) for human GB4 map coordinates and the human genome sequence (NCBI build 33) for precisely positioning the location of the mapped amplicons. Among the 1564 Type I markers on the cattle RH map, 1352 (86.4%) represented putative human orthologs identified by default BLASTN parameters against the human reference sequence; the remaining 212 (13.6%) were comprised of either ESTs or database sequences that had no human hit with E < e–5 using default BLASTN parameters. For those 212 ESTs, BLASTN was rerun using “sensitive” options. This resulted in 111 additional hits against the human sequence, all of which were consistent with their expected position on the comparative map (Fig. 1).
For construction of the cattle–human comparative chromosome maps, human chromosome coordinates of genes mapped in both species were used to align human chromosome segments along the cattle chromosomes (Fig. 1). Despite the limited resolution of marker order in the cattle RH5000 map, the location of conserved segment boundaries and their relative orientation were unambiguously determined for most segments. In total, 195 conserved regions containing two or more genes were identified, of which 31 are newly defined (shown as shaded bars in Fig. 1). Furthermore, 34 putative segments identified by singletons in the first-generation map were extended by at least one gene. The largest new segment is 15.73 human-Mbp, which is syntenic to a region on BTA9. The chromosome with the greatest number of new syntenies (n = 7) is BTA11. Among the 31 singletons defining putative segments, 19 originate from the marker set used to create the new map, and 12 remain from the first-generation map. Thus, the second-generation RH5000 map provides evidence for 226 chromosome segments conserved between the cattle and human genomes.
On the basis of human genome sequence coordinates, comparative coverage of the human genome on the second-generation cattle RH5000 map is 65.8% (Table 2). Comparative coverage for each human chromosome was estimated from the first telomeric gene on the human p-arm to the last gene proximal to the centromere, and from the first gene proximal to the centromere on the q-arm to the last telomeric gene. The current estimate of comparative coverage excluded telomeric, centromeric, and heterochromatic regions, because we assumed that these regions are, for the most part, devoid of genes. For the first-generation map (shown as solid colors in Fig. 1), the new estimate of comparative coverage using the human genome sequence as a reference (rather than GB4 map definitions) is 45.9% (Table 2), somewhat less than the previous estimate of 50% based on GB4 map definitions.
Table 2.
Comparative Coverage of the Cattle-Human Comparative Map, Listed by Human Chromosomes
|
Comparative coverage
|
|||||
|---|---|---|---|---|---|
|
First-generation mapa
|
Second-generation map
|
||||
| HSA | Total human comparative length (Mbp)b | (Mbp) | (%) | (Mbp) | (%) |
| 1 | 224 (245) | 135 | 60.2 | 164 | 73.2 |
| 2 | 240 (243) | 52 | 21.9 | 159 | 66.2 |
| 3 | 194 (199) | 98 | 50.6 | 132 | 68.1 |
| 4 | 188 (192) | 83 | 44.2 | 109 | 57.7 |
| 5 | 177 (181) | 91 | 51.6 | 109 | 61.2 |
| 6 | 167 (171) | 76 | 45.3 | 129 | 77.1 |
| 7 | 154 (158) | 50 | 32.5 | 91 | 59.0 |
| 8 | 142 (146) | 64 | 44.8 | 83 | 58.6 |
| 9 | 117 (135) | 44 | 37.7 | 61 | 52.3 |
| 10 | 132 (135) | 77 | 58.3 | 115 | 86.9 |
| 11 | 132 (135) | 60 | 45.8 | 81 | 61.7 |
| 12 | 130 (133) | 82 | 63.4 | 89 | 68.6 |
| 13 | 97 (114) | 25 | 25.7 | 54 | 55.8 |
| 14 | 87 (105) | 41 | 46.8 | 57 | 65.7 |
| 15 | 82 (100) | 45 | 55.4 | 51 | 62.9 |
| 16 | 80 (90) | 56 | 70.0 | 62 | 77.0 |
| 17 | 78 (82) | 32 | 40.6 | 42 | 54.1 |
| 18 | 75 (78) | 28 | 37.7 | 48 | 64.3 |
| 19 | 55 (64) | 22 | 40.2 | 38 | 68.8 |
| 20 | 60 (64) | 34 | 56.6 | 43 | 71.8 |
| 21 | 37 (47) | 20 | 52.9 | 28 | 76.7 |
| 22 | 35 (49) | 23 | 65.7 | 22 | 61.9 |
| X | 149 (153) | 60 | 40.2 | 97 | 65.2 |
| Total | 2,834 (3,020) | 1,300 | 45.9 | 1,865 | 65.8 |
Comparative length of chromosome is the total length minus area of telomere, centromere, and heterochromatin regions. The full length of the chromosome is given in parentheses.
There are 44 single genes or ESTs on the comparative map with nonhomologous positions in the human and cattle genomes (Fig. 1). These may represent as yet unidentified paralogs (including pseudogenes) or technical errors. Among these 44 genes, only one (PLCD3 on BTA22) has a position in the human genome sequence that is inconsistent but GB4 mapping information that is consistent with the surrounding markers within the conserved segment. Twenty-six genes on the comparative map have nonhomologous human genome sequence positions as well as nonhomologous GB4 map positions (possible technical errors or novel rearrangements). The remaining 17 genes have nonhomologous comparative genome positions and no similarity to any sequence mapped on the GB4 panel, and thus map inconsistencies could not be further explored. Among the 43 genes with contradictory human reference sequence positions and either no mapping information in the GB4 database or a contradictory GB4 map location, 35 were mapped within conserved segments, and the remaining eight genes mapped outside conserved segments (Fig. 1). In addition, among these 43 genes, 29 had sequences with less significant BLASTN hits (not the top hit) that could be further examined for putative orthologs. Among them only five additional putative orthologs were identified with human map positions that are consistent with their location in the cattle genome: PTTG@ (BTA10), TUBB (BTA11), ZMPSTE24_5 (BTA26), LOC284458 (BTA22), and LOC219917 (BTA25). The less significant BLASTN hits identified the likely orthologs of these genes to be PCNX, LOC286222, WDR11, LAMR1, and LOC283880, respectively.
Consistent with the first-generation RH5000 map, four cattle chromosomes show complete conservation of synteny with their four human homologs: BTA12 and HSA13, BTA19 and HSA17, BTA24 and HSA18, and BTAX and HSAX. However, multiple internal rearrangements have occurred, most likely representing recent inversions that date after the divergence of the most recent ancestor of the Primates and Cetartiodactyls. By examination of the conserved segments, 41 putative translocations leading to the present organization of the human and cattle chromosomes can be identified (Fig. 1), an addition of two new translocations compared to the information derived from the first-generation map. The two new translocations are small segments of homology between BTA7 and HSA1, and between BTA3 and HSA2. Fifteen cattle autosomes appear to be comprised of genes found on only one human chromosome (consistent with the first-generation map), and 12 cattle chromosomes were comprised of genes on an entire conserved human chromosome arm: BTA11-HSA2p, BTA6-HSA4p, BTA20-HSA5p, BTA23-HSA6p, BTA8-HSA9p, BTA5-HSA12p, BTA25-HSA16p, BTA7-HSA19p, BTA9-HSA6q, BTA14-HSA8q, BTA18-HSA16q, and BTA18-HSA19q (Fig. 1).
Centromere Repositioning
All human centromeres, including those on the eight human chromosomes or chromosome arms that had an unplaced centromere position on the first-generation comparative map (HSA2p, HSA7q, HSA8, HSA5, HSA19q, and HSA22), were accounted for on the second-generation comparative map. Seven human chromosomes (HSA1, HSA3, HSA4, HSA5, HSA10, HSA11, and HSAX) have repositioned centromeres in the cattle genome. These centromeres are located within large conserved syntenic blocks without apparent gene rearrangement. Six human chromosomes or chromosome arms (HSA2p, HSA7, HSA8p, HSA8q, HSA16, and HSA19) have repositioned centromeres that have predicted locations on the cattle map that lie at evolutionary breakpoints. Seemingly, the centromere of acrocentric HSA22 has been repositioned from the telomere of BTA17. The centromeres of HSA7 and HSA16 lie precisely at the junction of homologous segment boundaries on BTA25. Three entire human chromosome arms appear to have undergone either fission or fusion in relation to the cattle chromosomes: HSA19p, HSA2p, and HSA6q. The centromere positions on these human chromosomes have apparently been conserved in the cattle genome on BTA7, BTA11, and BTA9, respectively.
Gap Size Distribution
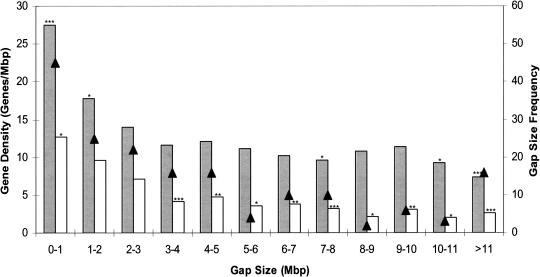
Half of the 175 gaps between conserved segments resulting from translocations and inversions are resolved at less than 2.88 Mbp, with 45 (25.7%) less than 1 Mbp (Fig. 2). Gap sizes in the comparative map were corrected for gaps in the human reference sequence resulting from noninformative telomeric, centromeric, and heterochromatic regions. The human gene density within the gaps was then compared for all gaps (Fig. 2). The mean gene density across the entire human genome is 5.8 RefSeq genes/Mbp and 12.3 predicted and known genes/Mbp. A lower gene density was correlated with an increase in gap size as determined by analysis of variance (P < 0.0001 for both known and predicted genes, and RefSeq genes). When gaps of different sizes were compared, gaps less than 2 Mbp had a higher density of known and predicted genes, whereas gaps greater than 10 Mbp had significantly lower density of known and predicted genes than the genome-wide average. Similar results were obtained for RefSeq genes, although the lower gene density was significant for all comparisons of gaps greater than 3 Mbp. Among the 56 gaps between linkage groups in the cattle RH map, 25 occurred at sites of ancient inversion or translocation breakpoints.
Figure 2.
Gap size distribution and gene density within gaps on the cattle–human comparative map. The frequency of gaps of different sizes is indicated on the figure by “▴”. Gene density was determined as described in Methods. Shaded bars indicate known and predicted genes (NCBI build 33); clear bars are RefSeq genes (NCBI release 1). Asterisks above the bars indicate the significance level of the comparison of gene density to the genome-wide average (*, P < 0.05; **, P < 0.001; ***, P < 0.0001). Gap sizes include those gaps in the comparative map that correspond to chromosome rearrangements, excluding boundaries at conserved telomeres and around centromere insertions.
DISCUSSION
Radiation-hybrid mapping was used in conjunction with COMPASS, an in silico predictive mapping tool, to produce a second-generation cattle–human whole-genome comparative map. The COMPASS strategy (Ma et al. 1998; Rebeiz and Lewin 2000; Larkin et al. 2003) was used to target more than 900 new cattle genes or ESTs to gaps in the first-generation RH and comparative maps (Band et al. 2000). Although there are still 56 gaps in cattle chromosome coverage, due in part to the increased stringency used for map construction, gene density on the map was more than doubled compared to the first-generation RH5000 map. Moreover, the number of framework markers was increased from 468 to 667, substantially improving the detail and accuracy of the map.
The new RH5000 map proved to be a powerful resource for increasing the resolution of the cattle–human comparative map, thus permitting a detailed analysis of comparative chromosome organization between the two species. For the first time, the human genome sequence coordinates were used to construct the cattle–human comparative chromosome maps, replacing the GB4-based coordinates used for the first-generation map. This resulted in a more accurate definition of segment boundaries and estimation of comparative chromosome coverage. The comparative coverage of the human genome on the new cattle RH map (based on the human genome reference sequence) was increased from ∼46% to ∼66%, representing a 20% overall increase coverage and a 43.4% increase compared to the first-generation map. Using the human genome sequence as a standard allowed for a more precise prediction of coverage on the human map, because the gene annotation available in the reference sequence is greater than the comparable information in UniGene. In addition, use of the human genome sequence to construct the comparative map resulted in correct positioning of 26/44 problem genes (all singletons) that did not map where expected based on the human map locations in UniGene (Fig. 1). These discrepancies may be the result of mapping errors or small unaccounted-for conserved segments. Because genes for the present map were selected for mapping on the basis of cattle genes/ESTs having human orthologs with GB4 (human RH) mapping information, the remaining gaps in the comparative map are due, in part, to gene-poor regions around centromeres and telomeres. Among the 56 gaps in the cattle RH linkage map, 25 are at inversion and translocation breakpoints, which correspond to the position of gaps in the cattle–human comparative map (Fig. 1). These data suggest that difficulties in obtaining contiguous RH linkage groups may be related to preferential breakage of the cattle chromosomes at radiation-induced fragile sites, many of which may be devoid of genes (see below).
We identified 195 conserved segments between the human and cattle genomes containing two or more genes, and a potential for 226 total segments if singletons are included. Of these, 65 are newly discovered, 123 are extended relative to the first-generation map, and the remainder are unchanged in length. It is noteworthy that there are many changes not only in the number of conserved segments on the new map, but also that several conserved segments on the first-generation map have been reoriented or even moved to different positions on a chromosome. This was expected as the density of markers on the map increased. An example is on BTA16, where the HSA1 170–180-Mbp segment has changed place with the HSA1 1–15-Mbp segment.
The total number of conserved segments is substantially higher than the number (n = 105) identified in the first-generation map using GB4 data (Band et al. 2000). Reanalysis of the first-generation map using the human genome reference sequence revealed 130 conserved segments, which is still substantially less than the new total. These data are generally consistent with the notion that the cattle genome is less rearranged than the mouse genome, which has 342 conserved syntenic segments with the human genome (Waterston et al. 2002). However, as map resolution increases, it is possible that the number of conserved segments could approach that found for mice. According to the random breakage model of chromosome evolution (Nadeau and Taylor 1984), one would expect to find more small conserved segments than large conserved segments. Indeed, the average size of the 31 newly discovered conserved segments defined by at least two genes/ESTs is 3.59 Mbp, compared to the overall average of 9.58 Mbp. Thirty of the comparative segments are smaller than 1 Mbp. The boundaries of at least some of these small segments may represent regions where chromosome breakages reoccur during evolution (Larkin et al. 2003; Pevzner and Tesler 2003). The occurrence of reuse boundaries in mammalian chromosome evolution is an important issue that will be addressed in a systematic way as map resolution increases for additional species.
The recent improvement in genome maps of other mammalian species permits a more detailed analysis of comparative genome organization and its evolutionary significance (Larkin et al. 2003; Murphy et al. 2003; Pevzner and Tesler 2003). Interesting examples include the conserved homologs BTA12/HSA13, BTA19/HSA17, BTA24/HSA18, and BTAX/HSAX. HSA17 is a particularly interesting chromosome as it is fully conserved in most mammals studied to date: SSC12 in pig, E1 in cat, MMU11 in mouse, ECA11 in horse, and the h-arm of hm in shrew (Larkin et al. 2000; Rink et al. 2002; Waterston et al. 2002; Chowdhary et al. 2003; Menotti-Raymond et al. 2003). The dog genome shows conservation of the distal part of HSA17p on CFA5, and the remainder of HSA17 on CFA9 (Guyon et al. 2003). On a genome-wide scale, 20/29 cattle autosomes have either complete homology with a human chromosome, homology with a human p- or q-chromosome arm, and/or genes from a single human chromosome. Although the functional significance of these observations is difficult to prove, maintenance of synteny (not necessarily gene order) involving thousands of genes over more than 90 million years of evolution suggests that conservation of many large chromosome segments may be necessary for supporting normal mammalian development and physiology.
The second-generation comparative map provides greater detail for the analysis of centromere repositioning (Montefalcone et al. 1999; Ventura et al. 2001) in mammalian evolution (all cattle chromosomes except BTAX are acrocentric). Three main categories of centromere repositioning can be deduced from the comparative maps: (1) complete repositioning, (2) centromere insertions at evolutionary breakpoints, and (3) centromere fusion/fission. Distinction of repositioning events requires comparison to a putative ancestral chromosome. A comprehensive reconstruction of the mammalian ancestral karyotype was recently made by Murphy et al. (2003), permitting development of hypothetical models of centromere repositioning events in mammalian evolution. For example, BTA25 represents the putative ancestral chromosome for conserved segments found on HSA7p and the entire p-arm HSA16. Centromeres of HSA7p and HSA16p are located at the point of the junction of conserved segments on BTA25, which represents the putative ancestral arrangement. Thus, the location of the centromeres on these conserved segments in the human genome has likely resulted from a fission of the ancestral chromosome and centromere insertion. Simple fission/fusion events and more complex fission/fusion events followed by internal rearrangements can also be discerned, such as on HSA6 (ancestral) and BTA9 and BTA23. Such observations contribute significantly to our understanding of karyotypic evolution.
An analysis of the size distribution of the gaps between conserved segments, using the human genome as a reference, demonstrated the power of the COMPASS strategy for producing high-resolution comparative maps. The majority of the 175 remaining gaps in the cattle–human comparative map are less than 3 Mbp. Many of these gaps can be closed with further marker targeting using comparatively anchored cattle ESTs or BAC-ends as a resource (Larkin et al. 2003). Targeted marker selection based on comparative mapping information is therefore a very efficient means of defining the boundaries of conserved segments and is extensible to any species if the primer sets used for mapping are judiciously chosen for amplification in other species. An interesting biological question that arises from the analysis of size distribution of the gaps is the gene content within these regions. The gene content within the gaps is important to our understanding of chromosome evolution and the role of rearrangements in speciation and adaptation (Navarro and Barton 2003), especially given the recent evidence for breakpoint reuse (Larkin et al. 2003; Pevzner and Tesler 2003). Although many of the larger gaps result simply by chance, other gaps may be due to the absence of orthologous sequences within these intervals. We attempted to address this question by comparing the number of genes in the gaps of different sizes, using either RefSeq (NCBI release 1) or all predicted and known genes (NCBI build 33) as standards. The analysis revealed gaps of three types: (1) tight gaps of less than 1 Mbp with high gene density, (2) larger gaps with approximately average gene density, and (3) very large gaps with significantly lower gene density than that found in the human genome on average. If one accepts the theoretical arguments concerning the role of chromosomal rearrangements in speciation (Navarro and Barton 2003) and the mounting evidence for reuse breakpoints (Larkin et al. 2003; Pevzner and Tesler 2003), then the gene content in the comparative map gaps suggests that the patterns of segment conservation observed in extant mammals is related to the function(s) and distribution of genes at or near breakpoint sites. For example, breakpoints in gene deserts (and without regulatory elements) would be compatible with a model of neutral evolution, whereas breakpoints at gene-rich sites could produce novel genes and regulatory elements by shuffling exons/regulatory elements that may be acted upon by selection. Breakpoints within duplicated regions may also create new gene families by separation of paralogs. Multispecies comparative mapping and DNA sequence information will lead to improved definition of segment boundaries and gene content within the boundary regions, which in turn will result in greater understanding of the biological significance of the differences in gene density along the chromosomes.
The initial promise of comparative mapping, dating back to the maps produced by linkage analysis and by somatic cell (synteny) mapping (O'Brien 1987) is now being realized from the rapid progress in physical mapping techniques and high-throughput DNA sequencing. The new cattle–human comparative map described in this report, the most detailed for any non-sequenced mammalian species, will facilitate multispecies comparisons of segment boundaries, centromere repositioning, and gene distribution in regions of known breakpoints. Furthermore, the new map will be an essential tool for the proper alignment of sequence contigs on the cattle chromosomes as the cattle genome sequencing initiative begins.
METHODS
Marker Selection and Mapping Strategy
Markers were selected for mapping using the COMPASS strategy (Ma et al. 1998; Band et al. 2000; Rebeiz and Lewin 2000; Larkin et al. 2003). All of the 47,787 cattle ESTs in the April 2000 GenBank release were used as a resource for filling gaps in the first-generation comparative map (Band et al. 2000). COMPASS was then implemented using a series of PERL scripts. Briefly, BLASTN was used to identify mapped human homologs in UniGene (NCBI build 148) represented by the cattle ESTs, using default BLASTN parameters and an expectation value (E-value) threshold of E = e–5 (Rebeiz and Lewin 2000). Cattle chromosome positions were then predicted using the first-generation RH5000 basis map (Band et al. 2000). Genes that were predicted to fall into comparative gaps or regions of low gene density on the first-generation cattle RH map (Band et al. 2000) were selected for mapping. Using the COMPASS procedure, a total of 947 genes were selected for subsequent mapping on a 5000-rad RH panel (Womack et al. 1997) and for analysis.
Oligonucleotide primers were designed from cattle EST sequences using Primer Designer (Version 3.0, Scientific & Educational Software) and Vector NTI v7.0 software (InforMax), targeting regions of low similarity between bovine and murine sequences. Primers were designed within the 3′ UTR whenever possible to avoid amplification of introns. Primer sequences and annealing temperatures used are listed in Supplemental Table 1. Primers for 349 additional microsatellite markers used to anchor the RH map to the linkage map were obtained from published sources (Suppl. Table 1; Barendse et al. 1994, 1997; Bishop et al. 1994; Ma et al. 1996; Kappes et al. 1997). Optimization of the primers and PCR amplification using the RH5000 panel was performed as described (Womack et al. 1997; Band et al. 1998; Band et al. 2000).
Map Construction
The cattle RH maps were constructed using RHMAP (Version 3.0, Boehnke 1992) and RHMAPPER (Version 1.22, Slonim et al. 1997) following the strategy used for the first-generation cattle RH map (Band et al. 2000), with the following adjustments. Using two-point analysis in RHMAPPER, new markers were assigned to the different cattle chromosomes on the basis of anchored markers from the first-generation cattle RH5000 map and a threshold of LOD > 9.0. To create linkage groups for each chromosome, data were loaded into RHMAP using a minimum LOD threshold of 7.0 as a first pass. Framework markers within each linkage group were identified using RHMAP, accepting 1:1000 as the threshold and then cross-checked with RHMAPPER using the “grow_framework” option. RHMAPPER was used to place all other markers relative to the framework map, using 40 cR as a maximum distance from a framework marker to be included in the linkage group. Comparisons were then made of the order of microsatellites in the RH and reference linkage maps (Barendse et al. 1994, 1997; Bishop et al. 1994; Ma et al. 1996; Kappes et al. 1997) in order to ensure consistency of the maps. If the order of microsatellites in the linkage map and the RH map was consistent (LOD ≥ 7.0 for the RH linkage groups), the RH framework map(s) was accepted. If the order was inconsistent, the LOD threshold was increased step-wise to a maximum value of 10 until a consistent framework order between the RH map and linkage map was obtained. If increasing the LOD threshold resulted in breakage of the RH linkage group into smaller groups with consistent microsatellite orders, the smaller linkage groups were merged manually and reanalyzed with RHMAPPER. If the merged linkage groups retained a microsatellite order that was consistent with the genetic linkage map, the merged group was accepted. If the order was inconsistent, then the linkage groups were represented separately on the map, in an orientation that is consistent with the genetic map. If the final placement of a marker was not between two framework markers (i.e., it was terminal to a linkage group) and the two-point data contradicted this placement, the marker was placed in a bin adjacent to the most strongly linked marker in the RH linkage group (LOD > 7.0). Binning was also done for markers not placed by RHMAPPER due to the 40-cR threshold.
Comparative Mapping Strategy
Following the completion of marker genotyping on the RH panel, all genes/ESTs, including those on the first-generation cattle RH map, were re-annotated using BLASTN with default parameters against an updated version of human UniGene (NCBI build 148) and the human genome reference sequence (NCBI build 33). For those ESTs having a BLASTN hit with E > e–5, “sensitive” BLASTN options were used with the following parameters: –W 7 –r 17 –q –21 –f 280 –G 29 –E 22 –X 240 –e 0.00000001 (http://sapiens.wustl.edu/∼ikorf/mmhs/index.html). Human gene nomenclature was used for all putative cattle orthologs identified by BLASTN. A minimum of 50 contiguous bases of alignment was necessary for a hit to be considered a putative ortholog. If the BLASTN hit was segmented within a gene(s), the highest-scoring hit was used to define the putative ortholog. If there was no human ortholog of a known cattle gene, the cattle gene symbol was used (e.g., DYA). GenBank accession numbers were used for BLASTN hits to unannotated human ESTs and for cattle ESTs with no significant BLASTN hit using default or sensitive parameters (underlined accession no., Fig. 1). Consistency between COMPASS predictions, GB4 mapping information, and location in the human genome reference sequence was checked and indicated on the chromosome maps for all exceptions.
The order of genes on the cattle RH map and their corresponding positions in the human genome were used to define the boundaries and gene content of conserved segments, similar to the process used previously (Band et al. 2000). The human genome position of each cattle ortholog was defined by the start and end coordinates for the locus given in LocusLink. If there was no locus identification in human UniGene for a BLASTN hit, the start and end coordinates in the human genome of the region of similarity with the cattle EST were used. Segments were oriented according to the position of framework markers within the segments. Segment boundaries were defined by the starting and ending coordinates of the most distant intrasegmental orthologs in the human genome, regardless of the marker order within the homologous cattle genome segment. Conserved segments were broken, indicating an internal rearrangement or translocation, only if supported by more than one framework marker on the cattle RH map. If there was no framework marker within a conserved segment, the most parsimonious interpretation of segment orientation was made by minimizing the number of rearrangements required to produce a given orientation of adjacent segments. The number of putative translocations between the cattle and human genomes was calculated as the number of syntenies, corrected for possible fissions (e.g., HSA6, BTA9, and BTA23) and for entire chromosomes that have apparently not been disrupted (e.g., HSA17 and BTA19). The location of human centromeres in conserved segments was determined using the human genome sequence (NCBI build 33).
Analysis of Gene Density in Map Gaps
Gap sizes in the comparative map were determined using the human genome sequence as a reference (NCBI build 33). Gap size was calculated as the distance in base pairs in the human genome between terminal markers on two adjacent conserved segments. Only those gaps between clearly identified chromosome rearrangements between cattle and humans were counted, thus excluding boundaries at conserved telomeres and centromere insertions. Gene density within a gap was determined by counting all RefSeq (NCBI release 1) genes within the gap or by counting all known and predicted genes (NCBI build 33) and dividing by the total size of the gap as defined in the human genome. A general linear models procedure (PROC GLM, Version 8.02, SAS) was used to test the hypothesis that gene density is different in gaps of different sizes. The gene density within gaps compared to the gene density on a genome-wide basis was compared using Student' s t-test (UNIVARIATE procedure in SAS).
Acknowledgments
This work was made possible in part by a grant to H.A.L. and J.E.W. from the U.S. Dept. of Agriculture, National Research Initiative, Project No. AG99-35205-8534.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2554404.
Footnotes
[Supplemental material is available online at www.genome.org, including all relevant information on mapped genes and markers.]
References
- Band, M., Larson, J.H., Womack, J.E., and Lewin, H.A. 1998. A radiation hybrid map of BTA23: Identification of a chromosomal rearrangement leading to separation of the cattle MHC class II subregions. Genomics 53: 269–275. [DOI] [PubMed] [Google Scholar]
- Band, M.R., Larson, J.H., Rebeiz, M., Green, C.A., Heyen, D.W., Donovan, J., Windish, R., Steining, C., Mahyuddin, P., Womack, J.E., et al. 2000. An ordered comparative map of the cattle and human genomes. Genome Res. 10: 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barendse, W., Armitage, S.M., Kossarek, L.M., Shalom, A., Kirkpatrick, B.W., Ryan, A.M., Clayton, D., Li, L., Neibergs, H.L., and Zhang, N. 1994. A genetic linkage map of the bovine genome. Nat. Genet. 6: 227–235. [DOI] [PubMed] [Google Scholar]
- Barendse, W., Vaiman, D., Kemp, S.J., Sugimoto, Y., Armitage, S.M., Williams, J.L., Sun, H.S., Eggen, A., Agaba, M., Aleyasin, S.A., et al. 1997. A medium-density genetic linkage map of the bovine genome. Mamm. Genome 8: 21–28. [DOI] [PubMed] [Google Scholar]
- Bishop, M.D., Kappes, S.M., Keele, J.W., Stone, R.T., Sunden, S.L., Hawkins, G.A., Toldo, S.S., Fries, R., Grosz, M.D., and Yoo, J. 1994. A genetic linkage map for cattle. Genetics 136: 619–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehnke, M. 1992. Multipoint analysis for radiation hybrid mapping. Ann. Med. 24: 383–386. [DOI] [PubMed] [Google Scholar]
- Chowdhary, B.P., Raudsepp, T., Kata, S.R., Goh, G., Millon, L.V., Allan, V., Piumi, F., Guérin, G., Swinburne, J., Binns, M., et al. 2003. The first-generation whole-genome radiation hybrid map in the horse identifies conserved segments in human and mouse genomes. Genome Res. 13: 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldammer, T., Kata, S.R., Brunner, R.M., Dorroch, U., Sanftleben, H., Schwerin, M., and Womack, J.E. 2002. A comparative radiation hybrid map of bovine chromosome 18 and homologous chromosomes in human and mice. Proc. Natl. Acad. Sci. 99: 2106–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisart, B., Coppieters, W., Farnir, F., Karim, L., Ford, C., Berzi, P., Cambisano, N., Mni, M., Reid, S., Simon, P., et al. 2002. Positional candidate cloning of a QTL in dairy cattle: Identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 12: 222–231. [DOI] [PubMed] [Google Scholar]
- Grobet, L., Martin, L.J., Poncelet, D., Pirottin, D., Brouwers, B., Riquet, J., Schoeberlein, A., Dunner, S., Menissier, F., Massabanda, J., et al. 1997. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 17: 71–74. [DOI] [PubMed] [Google Scholar]
- Guyon, R., Lorentzen, T.D., Hitte, C., Kim, L., Cadieu, E., Parker, H.G., Quignon, P., Lowe, J.K., Renier, C., Gelfenbeyn, B., et al. 2003. A 1-Mb resolution radiation hybrid map of the canine genome. Proc. Natl. Acad. Sci. 100: 5296–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambadur, R., Sharma, M., Smith, T.P., and Bass, J.J. 1997. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 7: 910–916. [DOI] [PubMed] [Google Scholar]
- Kappes, S.M., Keele, J.W., Stone, R.T., McGraw, R.A., Sonstegard, T.S., Smith, T.P., Lopez-Corrales, N.L., and Beattie, C.W. 1997. A second-generation linkage map of the bovine genome. Genome Res. 7: 235–249. [DOI] [PubMed] [Google Scholar]
- Larkin, D.M., Serov, O.L., Borodin, P.M., Zhdanova, N.S., and Searle, J.B. 2000. Comparative genome mapping in mammals: The shrew map. Acta Theriologica 2000 45: 131–142. [Google Scholar]
- Larkin, D.M., Everts-van der Wind, A., Rebeiz, M., Schweitzer, P.A., Bachman, S., Green, C.A., Wright, C.L., Campos, E.J., Benson, L.D., Edwards, J., et al. 2003. A cattle–human comparative map built with cattle BAC-ends and human genome sequence. Genome Res. 13: 1966–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, R.Z., Beever, J.E., Da, Y., Green, C.A., Russ, I., Park, C., Heyen, D.W., Everts, R.E., Fisher, S.R., Overton, K.M., et al. 1996. A male linkage map of the cattle (Bos taurus) genome. J. Hered. 87: 261–271. [DOI] [PubMed] [Google Scholar]
- Ma, R.Z., van Eijk, M.J., Beever, J.E., Guérin, G., Mummery, C.L., and Lewin, H.A. 1998. Comparative analysis of 82 expressed sequence tags from a cattle ovary cDNA library. Mamm. Genome 9: 545–549. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond, M., David, V.A., Chen, Z.Q., Menotti, K.A., Sun, S., Schäffer, A.A., Agarwala, R., Tomlin, J.F., O'Brien, S.J., and Murphy, W.J. 2003. Second-generation integrated genetic linkage/radiation hybrid maps of the domestic cat (Felis catus). J. Hered. 94: 95–106. [DOI] [PubMed] [Google Scholar]
- Montefalcone, G., Tempesta, S., Rocchi, M., and Archidiacono, N. 1999. Centromere repositioning. Genome Res. 9: 1184–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W.J., Bourque, G., Tesler, G., Pevzner, P., and O'Brien, S.J. 2003. Reconstructing the genomic architecture of mammalian ancestors using multispecies comparative maps. Human Genomics 1: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau, J.H. and Taylor, B.A. 1984. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc. Natl. Acad. Sci. 81: 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, A. and Barton, N.H. 2003. Chromosomal speciation and molecular divergence—Accelerated evolution in rearranged chromosomes. Science 300: 321–324. [DOI] [PubMed] [Google Scholar]
- O'Brien, S.J. 1987. Genetic Maps 1987: A compilation of linkage and restriction maps of genetically studied organisms. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Pevzner, P. and Tesler, G. 2003. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. 100: 7672–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz, M. and Lewin, H.A. 2000. Compass of 47,787 cattle ESTs. Anim. Biotechnol. 11: 75–241. [DOI] [PubMed] [Google Scholar]
- Rink, A., Santschi, E.M., Eyer, K.M., Roelofs, B., Hess, M., Godfrey, M., Karajusuf, E.K., Yerle, M., Milan, D., and Beattie, C.W. 2002. A first-generation EST RH comparative map of the porcine and human genome. Mamm. Genome 13: 578–587. [DOI] [PubMed] [Google Scholar]
- Slonim, D., Kruglyak, L., Stein, L., and Lander, E. 1997. Building human genome maps with radiation hybrids. J. Comput. Biol. 4: 487–504. [DOI] [PubMed] [Google Scholar]
- Springer, M.S., Murphy, W.J., Eizirik, E., and O'Brien, S.J. 2003. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. 100: 1056–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, H., Takami, M., Oguni, T., Tsuji, T., Yoneda, K., Sato, H., Ihara, N., Itoh, T., Kata, S.R., Mishina, Y., et al. 2002. Positional cloning of the gene LIMBIN responsible for bovine chondrodysplastic dwarfism. Proc. Natl. Acad. Sci. 99: 10549–10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J.W. and Touchman, J.W. 2002. Vertebrate genome sequencing: Building a backbone for comparative genomics. Trends Genet. 18: 104–108. [DOI] [PubMed] [Google Scholar]
- Ventura, M., Archidiacono, N., and Rocchi, M. 2001. Centromere emergence in evolution. Genome Res. 11: 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Williams, J.L., Eggen, A., Ferretti, L., Farr, C.J., Gautier, M., Amati, G., Ball, G., Caramorr, T., Critcher, R., Costa, S., et al. 2002. A bovine whole-genome radiation hybrid panel and outline map. Mamm. Genome 13: 469–474. [DOI] [PubMed] [Google Scholar]
- Womack, J.E., Johnson, J.S., Owens, E.K., Rexroad III, C.E., Schläpfer, J., and Yang, Y.P. 1997. A whole-genome radiation hybrid panel for bovine gene mapping. Mamm. Genome 8: 854–856. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://sapiens.wustl.edu/∼ikorf/mmhs/index.html; Mouse–Human Experiment.
- http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/BovineSEQ; Bovine Genomic Sequencing Initiative.