Abstract
Single nucleotide polymorphisms (SNPs) are the most common source of genetic variation in populations and are thus most likely to account for the majority of phenotypic and behavioral differences between individuals or strains. Although the rat is extensively studied for the latter, data on naturally occurring polymorphisms are mostly lacking. We have used publicly available sequences consisting of whole-genome shotgun (WGS), expressed sequence tag (EST), and mRNA data as a source for the in silico identification of SNPs in gene-coding regions and have identified a large collection of 33,305 high-quality candidate SNPs. Experimental verification of 471 candidate SNPs using a limited set of rat isolates revealed a confirmation rate of ∼50%. Although the majority of SNPs were identified between Sprague-Dawley (EST data) and Brown Norway (WGS data) strains, we found that 66% of the verified variations are common among different rat strains. All SNPs were extensively annotated, including chromosomal and genetic map information, and nonsynonymous SNPs were analyzed by SIFT and PolyPhen prediction programs for their potential deleterious effect on protein function. Interestingly, we retrieved three SNPs from the database that result in the introduction of a premature stop codon and that could be confirmed experimentally. Two of these “in silico-identified knockouts” reside in interesting QTL regions. Data are publicly available via a Web interface (http://cascad.niob.knaw.nl), allowing simple and advanced search queries.
Single nucleotide polymorphisms (SNPs) are the most common type of DNA variation and the main source of phenotypic differences between individuals. Furthermore, SNPs have proven to be highly stable within populations and very useful as genetic markers in several applications including physical mapping, evolutionary studies, and association genetics. Although many experimental techniques for high-throughput SNP mining have been developed (for review, see Vignal et al. 2002), the generation of dense polymorphism maps is still a laborious and time-consuming procedure. The alternative, in silico identification of SNPs using large data sets generated by sequencing projects, is a quick and cost-efficient approach for the generation of impressive amounts of candidate SNPs.
Several candidate SNP detection pipelines based on EST data analysis have been described previously (Buetow et al. 1999; Marth et al. 1999; Picoult-Newberg et al. 1999; Brett et al. 2000). Application of computationally intensive BLAST search and/or Phrap fragment assembler, which are common first steps in most pipelines for candidate SNPs discovery, does not allow incorporation of large amounts of sequence data, as, for example, produced by whole-genome shotgun sequencing (WGS) projects. Although a genome assembly, if available, could be used in addition to ESTs (Schmid et al. 2003), the WGS data partition could be extremely useful for SNP mining because it represents a genome-wide redundant collection of sequences and contains base-calling quality information not provided by genome assemblies.
Currently, there is only a very limited amount of data available on polymorphisms in the rat (Rattus norvegicus), a major model for understanding basic biology and human health and disease. In contrast, there are several thousands of mRNA sequences available from GenBank as well as millions of traces from two high-throughput EST and genome sequencing projects. We have implemented an SNP discovery pipeline based on fast sequence search algorithm (SSAHA; Ning et al. 2001) for building a candidate SNP database using different types of public domain data as an input. The database, named CASCAD (CASCAD SNP CAndidates Database), stores in silico discovered polymorphisms that are primarily associated with gene-coding regions. It can be queried through a Web interface that is tightly linked to other public databases and contains predictions of the potential effect on protein function for nonsynonymous SNPs. Experimental validation studies were performed to address the quality of the candidate SNP collection.
RESULTS AND DISCUSSION
SNP Discovery Pipeline and CASCAD Database
We have developed an SSAHA-based pipeline for rapid discovery of candidate SNPs (see Supplemental Fig. S1 for outline). The increased performance of the SSAHA search algorithm allowed us to compare raw WGS sequence data (∼13 × 109 bp) against mRNA and EST sequences. There are currently two major sequencing projects in R. norvegicus: the University of Iowa EST project (http://ratEST.uiowa.edu) for the outbred Sprague-Dawley (SD) strain (Scheetz et al. 2001) and the rat whole-genome shotgun (WGS) project (http://www.hgsc.bcm.tmc.edu/projects/rat) using the inbred Brown Norway (BN) strain. The WGS data used in our study have an advantage over the genome assembly as it allows incorporation of unassembled sequences, base-calling qualities, and coverage information. This additional information is used to filter for high-quality candidate SNPs. Besides the data from these large-scale projects, all rat-derived data (mRNA) from the NCBI database are exploited. This data partition has the disadvantage that no sequence quality information is available.
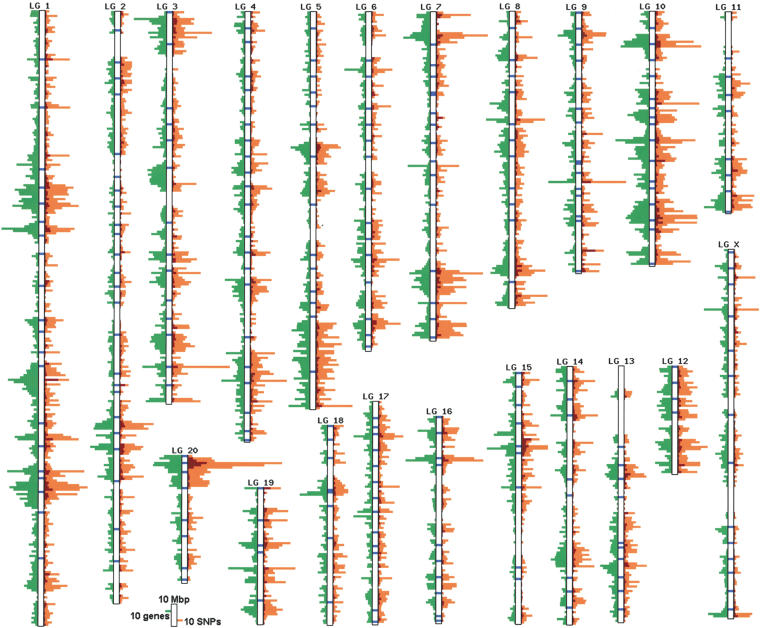
Although an initial set of ∼600,000 polymorphisms was identified by SSAHA comparison of all input sequences (Table 1), thorough filtering and clustering resulted in 33,305 unique high-quality candidate SNPs (Table 1). All polymorphisms are stored in the CASCAD database that was constructed to incorporate annotations and characteristics related to polymorphisms, links to public databases, and the results of severity prediction for nonsynonymous SNPs. We have mapped the candidate SNPs to the latest rat genome assembly (RGSC 3.1, February 2004), demonstrating a good correlation with the location of expressed sequences (r = +0.77, P < 0.0001; Fig. 1). We found that 77% of the SNPs can be mapped to a unique genomic location, but for ∼11% of the SNPs, two or more matches were found when using the same search conditions and criteria as used in the original candidate SNP identification pipeline. Furthermore, ∼12% of the SNPs could not be found in the current genome assembly. The first class of unmapped SNPs may be due to pseudogenes, closely related gene family members, or genome assembly errors, whereas the latter class may reflect the proportion of the rat genome that is not represented in the current genome assembly. Using the Web interface (http://cascad.niob.knaw.nl), the candidate SNP database can be searched using nucleotide sequences or arbitrary combinations of accessions (GenBank, UniGene, LocusLink, Ensembl gene, etc.), location (radiation hybrid maps, position in genome assembly), and SNP characteristics (e.g., substitution class, base-calling quality) as input.
Table 1.
Database Statistics
| Input sequence dataa | |
| mRNA | 25,634 |
| EST | 244,518 |
| WGS | 19,813,313 |
| SNPs predicted | |
| Total | 33,305 |
| Synonymous | 3842 (11.5%) |
| Nonsynonymous | 3708 (11.1%) |
| Nonsense | 162 (0.5%) |
| At CpG site | 8028 (24.1%) |
| Transition/Transversion ratio | 1.76 |
| Database coverage | |
| LocusLink IDs | 2547 |
| Unigene IDs | 15015 |
| Ensembl genes | 7558 |
| Nonsynonymous variations predicted to be damaging | |
| SIFT | 1069/2316b |
| PolyPhen | 1086/2855b |
| Both programs | 597/2075b |
Number of sequence reads.
Number of predicted damaging SNPs per total number of predictions by the specified program.
Figure 1.
Distribution of candidate SNPs on the rat linkage group map. The density (per 1-Mb window) of synonymous (orange) and nonsynonymous (red) candidate SNPs is plotted against gene density (green) as annotated in rat genome build 3.1 (February 2004). Loci with confirmed polymorphisms are indicated by closed blue boxes.
Prediction of the Effect of Nonsynonymous SNP Candidates on Protein Function
We used two programs, SIFT (Ng and Henikoff 2003) and Poly-Phen (Ramensky et al. 2002), to predict a potential effect on protein function for the 3708 nonsynonymous cSNPs in our database. Although SIFT was found to predict ∼70% of polymorphisms that are annotated to be involved in disease as damaging (Ng and Henikoff 2002), it should be mentioned that the performance of these programs strongly depends on the availability of orthologous/homologous sequences and structural protein information (in case of PolyPhen), and, therefore, predictions should be used with care. More than 3000 candidate SNPs (3096) were scored by at least one program, with 2075 being scored by both programs (Table 1). Although the programs disagreed in 28.7% of the predictions, 597 variations were classified as intolerant/damaging by both algorithms and, therefore, are likely to affect protein function and thus may account for phenotypic differences between strains.
Confirmation of Candidate SNPs
To get an indication about the quality of the CASCAD database, we addressed the verification rate for different subsets of candidate SNPs. Initially, we checked 68 candidate SNPs by resequencing in 10 widely used rat strains (BN, BUF, COP, DA, F344, LEW, PVG, SHR, SD, and WIST) and were able to confirm six out of 15 synonymous SNPs, eight out of 18 nonsynonymous SNPs, and 20 out of 35 SNPs that are localized in UTRs or regions not annotated for an open reading frame. When data were sorted by resource type (i.e., mRNA, EST), SNPs based on mRNA sequences were found to have the lowest reliability, with only 13 of 32 candidates confirmed. As no sequencing quality information is available for mRNA sequences that are deposited in public databases, it is most likely that the low verification rate for this category reflects the frequent occurrence of sequencing errors in (mainly the older) mRNA database entries. However, if we only take into account the data with sequencing quality information (EST and WGS), the confirmation rate can be as high as 74% (e.g., 20 of 27 SNPs can be confirmed; this partition contributes 63% of the database entries). Applying even more stringent parameters to the data by filtering for SNPs for which each polymorphic state is supported by at least two sequencing reads, restricting the total number of candidates to 3346, resulted in the confirmation of all five candidates in our verification set that meet these criteria. The verification of polymorphisms within an outbred strain (i.e., Sprague-Dawley EST vs. EST comparison) is problematic, as only three out of 14 variants could be confirmed when screening only two independent animals. Additional sampling of 20 randomly selected animals from a Sprague-Dawley breeding colony did not increase this number. However, it should be mentioned that the observed genetic variation in this breeding population was very limited, and therefore it might be necessary to genotype Sprague-Dawley animals from colonies around the world to increase the validation rate for this category. Taking into account the contribution of each subset to the database (Table 1) and the observed confirmation rate from our initial verification study, we estimate the average confirmation rate for the whole data set to be ∼59%.
To extend the set of verified SNPs, we designed a second data set of 340 candidates that are evenly distributed on the 21 rat chromosomes and thus could result in a genome-wide SNP-based mapping panel. By resequencing in five laboratory rat strains (BN, F344, SHR, SD, and WIST) and two wild rat samples, we confirmed 171 candidate SNPs (50.3%). Although the wild rat samples were essential for the confirmation of only nine SNPs, up to 99 SNPs confirmed in the other strains were also observed in the wild rat, suggesting that wild rat isolates are well suited for initial rat SNP discovery.
To obtain parameters for selecting candidate SNPs with the highest likelihood to be confirmed, we used all verification data to correlate validation of SNPs with characteristics present in the CASCAD database. Six significant correlations were found (P < 0.005): showing higher validation rates for candidates (1) located at hypervariable CpG dinucleotides, (2) caused by transitions, (3) supported by multiple reads, or (4) predicted in EST versus WGS subset comparison; and lower validation rates for candidates that are (5) nonsynonymous or (6) polymorphisms predicted within the EST data subset. Based on these correlations, we introduced an arbitrary SNP score (from 1 to 10), allowing database users to restrict their search to subsets of SNPs with a higher validation rate (see Supplemental Tables S1 and S2 for correlation analysis and SNP category details). No correlation between the confirmation of candidates and the quality of the sequencing reads was found, indicating that the initial filtering by using a cutoff phred score of 20 is sufficiently stringent to eliminate the majority of sequencing errors. Although the overall confirmation rate of the candidate SNPs in this study is only ∼50%, it is similar to verification rates for other SNP mining pipelines where public domain data were used (63%, based on 18 individuals [Picoult-Newberg et al. 1999]; 71%, based on 24 individuals [Irizarry et al. 2000]). In contrast, confirmation rates can also be as high as 96% as shown by Schmid et al. (2003) for Arabidopsis SNPs. However, in this study, primary data generation (EST sequencing) and the confirmation analysis were performed on the same source material. The unavailability of the original DNA samples and the genetic heterogeneity between individuals from outbred rat strains and substrains, in combination with the limited number of strains and individuals tested, may explain the relatively low confirmation rate for our data set. In addition, we cannot exclude a negative effect of repetitive elements or pseudogenes on our prediction method, although only 11% of the final set of candidate SNPs was found to map to multiple genomic loci.
Nucleotide Diversity
In addition to the 208 SNPs confirmed in our verification experiments (34 from the first set, 171 from the second set, and three nonsense mutations; see below) we discovered 287 novel SNPs and 10 indels (1–14 bp, all intronic). Of these novel SNPs, 65 can be mapped to exons, but were not predicted by our in silico identification approach. The remaining 222 SNPs were unlikely to be predicted by our approach, as they could not be assigned to any annotated exon in the Ensembl database and are thus most likely to be intronic polymorphisms. In total, we have screened ∼112,000 bp, resulting in a frequency of 1 SNP per 226 bp (or 1 per 252 when only considering the laboratory rat strains). The calculated nucleotide diversity (θ = 1.25 × 10–3) is higher than estimates obtained for human (θ = 3–8 × 10–4; Deutsch et al. 2001). However, the SNP frequency is similar to that observed for STS-derived polymorphisms in comparison between mouse subspecies, for example, Mus mus castaneus and Mus mus domesticus (Wade et al. 2002).
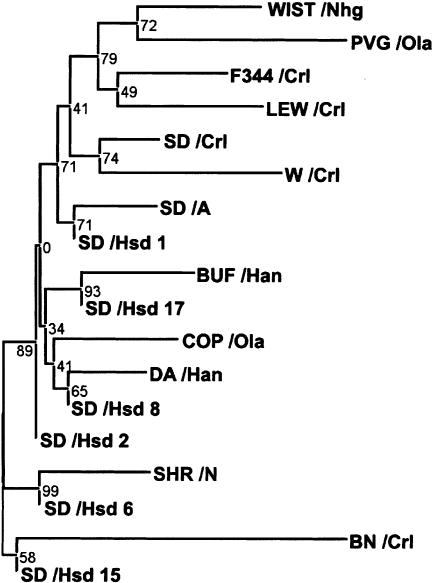
Two-thirds of the polymorphisms (318) were common with minor allele frequency >20%. Of the uncommon SNPs, 27 discriminate BN from all other strains tested (the closest competitors SHR/N and WIST/Crl have eight), suggesting that Brown Norway is the most divergent among laboratory rat strains. We have used the polymorphic positions observed in the first verification experiment as a multilocus marker set to build a population tree of 18 individuals from 13 rat isolates (Fig. 2). As expected, the Brown Norway strain occupies the basal position, whereas Sprague-Dawley isolates from the breeding population tend to outgroup other inbred strains indicating that this strain may retain the genetic variation of the common ancestor that was used as a source for generating most of the inbred strains (only three alleles were specific to the Sprague-Dawley strain for 82 polymorphic loci studied). In contrast, the two Wistar individuals that were obtained from completely different sources (WIST/Crl and WIST/Nhg) seem to represent only a limited collection of the inbred strains rather than a common mixed genetic background.
Figure 2.
Phylogenetic tree of 18 rat individuals based on a set of 82 polymorphic loci as obtained in the first verification experiment. Coefficients represent interior branch test support values for tree nodes.
The observed average heterozygote frequency at the verified polymorphic loci ranges from 7.6% to 9.2% for animals from inbred strains, from 13.7% to 22.1% for animals from outbred strains, and from 25.5% to 28.5% for wild rat isolates (Supplemental Table S3). Although the degree of heterozygosity increases from inbred to outbred and wild animals as expected, animals from inbred strains are expected to be >98% isogenic (Beck et al. 2000). Our results suggest that the inbred animals that we used are less inbred than statistically expected, although it should be mentioned that only a single animal per strain was analyzed. However, these results are in line with the observed genetic variation (between 2% and 20%) between colonies of the same inbred strain (Smits et al. 2004), which may well reflect the residual genetic variation in a common ancestor.
CASCAD Applications
This study presents the first large-scale SNP discovery effort in the rat. Using data from public databases and large-scale genome sequencing projects, it was possible to build a representative and well-annotated database on genetic variations. The CASCAD database potentially contains a major part of variation associated with rat expressed sequences and can be exploited in many ways. For example, one can search for SNPs between specified strains in specific chromosomal regions that could serve as polymorphic markers for mapping or association studies. In the verification experiments, we have genotyped 495 SNPs (208 confirmed cSNPs and 287 newly discovered SNPs), 487 of which can be mapped to 261 loci on the rat genome assembly (Fig. 1). This collection of randomly distributed validated polymorphisms represents a first-generation SNP map for the rat genome. The CASCAD database in combination with the verification parameters provided here can now be used to refine this map and develop a rat SNP data set for use on any high-throughput SNP typing platform. For smaller-scale studies, one can also extract a dense RFLP mapping panel, based on the use of selected restriction enzymes. Alternatively, the database can be used for retrieving nonsynonymous SNPs that are annotated to be deleterious for protein function by the SIFT and/or PolyPhen programs and that are located in a specific genomic region, for example, where a specific quantitative trait locus (QTL) is mapped. Finally, it is also possible to search for known rat polymorphisms in custom nucleotide sequences.
We have explored the possibilities for identifying gene knockouts by in silico analysis and mined the CASCAD database for cSNPs that result in the introduction of premature stop codons. In total, 162 such polymorphisms are in the database. We selected 38 of these nonsense candidates for verification and were able to retrieve three knockouts by experimental confirmation. Although the relatively low confirmation rate for this class of SNPs may be affected by a potential low frequency of null alleles in outbred SD populations, selection for high rates of sequencing errors in this class is probably more likely. Unfortunately, no DNA is available from the animals that were used to generate the EST libraries to confirm this assumption. The knockout polymorphisms that were confirmed (Table 2) are located in the fibroblast growth factor receptor-1-like (Fgfr1-like) gene (GenBank: U58466; CASCAD: RS011240), the endo-α-mannosidase gene (GenBank: NM_080785; CASCAD: RS017460), and an uncharacterized predicted Ensembl gene (ENSRNOG00000010341; CASCAD: RS018213). Interestingly, Fgfr1-like is located in a QTL for uveitis severity (severe eye disease) on Chromosome 4 and may be a good candidate for this disease as the closest paralogous gene. Fgfr1 is known to be up-regulated in stressed retinas (Valter et al. 2002). Likewise, endo-α-mannosidase is located on Chromosome 9 in a region where two blood-pressure QTLs are mapped. Fascinatingly, the “knockout” allele is present in the widely studied Spontaneously Hypertensive Rat (SHR) strain that is an important genetic model for studying high blood pressure.
Table 2.
Characteristics of In Silico Identified Gene Knockouts
| CASCAD ID | Description | Location | RGD QTL ID | Variation | BN | BUF | COP | DA | F344 | LEW | PVG | SD | SHR | WIST |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RS011240 | Fgfr1-like | 4q24 | 61330a | Y127X | KO | WT | WT | WT | WT | WT | WT | WT | KO | WT |
| RS017460 | Enman | 9q31 | 61352b | Y8X | KO | WT | ndd | ndd | KO | WT | KO | WT | KO | KO |
| 70218c | ||||||||||||||
| RS018213 | LOC362986 | 7q34 | — | Q215X | WT | KO | KO | KO | KO | WT | WT | KO | KO | KO |
Ratmap locus ID 45703.
Ratmap locus ID 39687.
Ratmap locus ID 45644.
Not determined.
METHODS
SNP Discovery
The sequence data used in this study were downloaded on November 8, 2002 from the NCBI GenBank (http://www.ncbi.nlm.nih.gov/Genbank, mRNA and EST subsets) and the rat Ensembl trace archive (http://trace.ensembl.org, WGS subset; Table 1). EST and mRNA sequences were masked for rodent- and mammalian-specific repeats, low-complexity regions, rat mitochondrial DNA, and hypervariable and high-copy immune receptor genes using RepeatMasker. Local SSAHA search was performed to collect hits with nearly exact homology containing a single mismatch in the mRNA/EST subset and remote search (using Ensembl SSAHA search server) in case of mRNA/EST versus WGS comparison. Only hits with a high-quality mismatch (phred score >20 for both reads) within a sequence stretch of >80 bp identity were retained. The mRNA subset that is not annotated for base-calling quality data was treated as having a reliable overall quality. Hits were clustered to represent unique variations and stored in an MySQL database.
Perl scripts were developed to extend the candidate SNP database to incorporate annotation information. Translation tags stored in GenBank sequences were used to add information on amino acid variants. For sequences lacking translation information, similar information was obtained from search against all known proteins in Ensembl rat genome build 2 using BLASTX (expectation cutoff = 1E – 10). Using the same genome build, mRNA + EST sequences and candidate SNPs were mapped to the whole genome assembly by SSAHA search (parameters: -wl 10 -mp 80 -mg 10). A Web interface was built using Perl with CGI and Mysql modules and Bioperl toolkit (Stajich et al. 2002).
SNPs and haplotypes obtained in this study were submitted to dbSNP (accession numbers: ss12535137–ss12568440, ss12588074–ss12588105). The database is publicly available at http://cascad.niob.knaw.nl; all scripts are freely available upon request from the authors.
Prediction of Effect of Nonsynonymous Candidate SNPs on Protein Function
Protein sequences and predicted amino acid variants were extracted from the CASCAD database and submitted to the publicly available SIFT (Ng and Henikoff 2003) and PolyPhen (Ramensky et al. 2002) prediction services. We used the stand-alone version of SIFT and queried the PolyPhen server remotely. To get comparable results, the same BLAST parameters (expectation cutoff = 1E – 04) and the same database of protein sequences were chosen (SWISS-PROT, SWISS-PROT-NEW, TrEMBL, TrEMBL_NEW, and PIR dated from April 9, 2003 and downloaded from ftp://ftp.expasy.org) for both programs.
Validation Experiments
For the first verification experiment, 13 genomic DNA samples representing 10 different rat strains (inbred: BN/Crl, BUF/Han, COP/OlaHsd, DA/Han, F344/Crl, LEW/Crl, PVG/OlaHsdCpb, SHR/N [Bender et al. 1984]; outbred: SD/Crl, SD/A, SD/Hsd, WIST/Crl, WIST/Nhg) were kindly provided by L.F.M. van Zutphen (Department of Laboratory Animal Science, Faculty of Veterinary Medicine, Utrecht University, The Netherlands) and B. Smits (Hubrecht Laboratorium, Utrecht, The Netherlands). Twenty Sprague-Dawley (SD/Hsd 1–20) samples randomly taken from a breeding colony were obtained from Harlan Netherlands, and DNA isolation was done using the protocol described in Bender et al. (1984). Local genome assemblies were constructed using GENOTRACE (Berezikov et al. 2002) and verified with rat genome assembly by BLASTN search.
For the second verification experiment, seven laboratory isolates were used (inbred: BN/Crl, F344/Crl, SHR/N; outbred: SD/Crl, SD/A, SD/Hsd, WIST/Crl) as well as two wild rat isolates. The genomic context of each candidate SNP was obtained from rat genome assembly using the corresponding genome position field in the CASCAD database.
Primers for PCR amplification and sequencing of the genomic region were designed using a customized Web interface (http://primers.niob.knaw.nl) to the Primer3 program (http://www-genome.wi.mit.edu/genome_software/other/primer3.html). The touchdown PCR amplification consisted of 30 cycles starting with reannealing temperature 65°C and ending with 53°C. The sequencing of the amplicons was performed with PCR primers using an ABI 3700 automated sequencer (Applied Biosystems) and Dyenamic ET terminator (Amersham Biosciences) as recommended by the manufacturer. The obtained sequences were scored for SNPs using PolyPhred (Nickerson et al. 1997).
Phylogenetic Reconstruction
Sequence alignment for 82 observed variable positions was used as an input for the MEGA 2.1 program (Kumar et al. 2001). The phylogenetic tree was built with Neighbor-joining algorithm using p-distances. Support for each node was determined by interior-branch test.
Acknowledgments
This work was supported by the Dutch Ministry of Economic Affairs through the Innovation Oriented Research Program on Genomics, grant #IGE01017. We thank M.B. Soares, T. Scheetz (The University of Iowa), and the Rat Genome Sequencing Consortium for providing rat EST and genomic sequences, respectively, including corresponding quality files; and B. Smits (Hubrecht Laboratory) and L.F.M. van Zutphen (Utrecht University) for providing rat genomic DNA.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2154304.
Footnotes
[Supplemental material is available online at www.genome.org. The SNPs identified in this study can be found in the National Center of Biotechnology Information (NCBI) SNP database under submitter handle FGG_NIOB (ss12535137–ss12568440, ss12588074–ss12588105). The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: L.F.M. van Zutphen, B. Smits, M.B. Soares, T. Scheetz, and the Rat Genome Sequencing Consortium.]
References
- Beck, J.A., Lloyd, S., Hafezparast, M., Lennon-Pierce, M., Eppig, J.T., Festing, M.F., and Fisher, E.M. 2000. Genealogies of mouse inbred strains. Nat. Genet. 24: 23–25. [DOI] [PubMed] [Google Scholar]
- Bender, K., Adams, M., Baverstock, P.R., den Bieman, M., Bissbort, S., Brdicka, R., Butcher, G.W., Cramer, D.V., von Deimling, O., Festing, M.F., et al. 1984. Biochemical markers in inbred strains of the rat (Rattus norvegicus). Immunogenetics 19: 257–266. [DOI] [PubMed] [Google Scholar]
- Berezikov, E., Plasterk, R., and Cuppen, E. 2002. GENOTRACE: cDNA-based local GENOme assembly from TRACE archives. Bioinformatics 18: 1396–1397. [DOI] [PubMed] [Google Scholar]
- Brett, D., Lehmann, G., Hanke, J., Gross, S., Reich, J., and Bork, P. 2000. EST analysis online: WWW tools for detection of SNPs and alternative splice forms. Trends Genet. 16: 416–418. [DOI] [PubMed] [Google Scholar]
- Buetow, K.H., Edmonson, M.H., and Cassidy, A.B. 1999. Reliable identification of large numbers of candidate SNPs from public EST data. Nat. Genet. 21: 323–325. [DOI] [PubMed] [Google Scholar]
- Deutsch, S., Iseli, C., Bucher, P., Antonarakis, S.E., and Scott, H.S. 2001. A cSNP map and database for human chromosome 21. Genome Res. 11: 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, K., Kustanovich, V., Li, C., Brown, N., Nelson, S., Wing, W., and Lee, C.J. 2000. Genome-wide analysis of single-nucleotide polymorphisms in human expressed sequences. Nat. Genet. 26: 233–236. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jacobsen, I.B., and Nei, M. 2001. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Marth, G.T., Korf, I., Yandell, M.D., Yeh, R.T., Gu, Z., Zakeri, H., Stitziel, N.O., Hillier, L., Kwok, P.Y., and Gish, W.R. 1999. A general approach to single-nucleotide polymorphism discovery. Nat. Genet. 23: 452–456. [DOI] [PubMed] [Google Scholar]
- Ng, P.C. and Henikoff, S. 2002. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 12: 436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, P.C. and Henikoff, S. 2003. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, D.A., Tobe, V.O., and Taylor, S.L. 1997. Polyphred: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 25: 2745–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, Z., Cox, A.J., and Mullikin, J.C. 2001. SSAHA: A fast search method for large DNA databases. Genome Res. 11: 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picoult-Newberg, L., Ideker, T.E., Pohl, M.G., Taylor, S.L., Donaldson, M.A., Nickerson, D.A., and Boyce-Jacino, M. 1999. Mining SNPs from EST databases. Genome Res. 9: 167–174. [PMC free article] [PubMed] [Google Scholar]
- Ramensky, V., Bork, P., and Sunyaev, S. 2002. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 30: 3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz, T.E., Raymond, M.R., Nishimura, D.Y., McClain, A., Roberts, C., Birkett, C., Gardiner, J., Zhang, J., Butters, N., Sun, C., et al. 2001. Generation of a high-density rat EST map. Genome Res. 11: 497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K.J., Sörensen, T.R., Stracke, R., Törjek, O., Altmann, T., Mitchell-Olds, T., and Weisshaar, B. 2003. Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res. 13: 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits, B.M.G., van Zutphen, B.F.M., Plasterk, R.H.A., and Cuppen, E. 2004. Genetic variation in coding regions between and within commonly used inbred rat strains. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Stajich, J.E., Block, D., Boulez, K., Brenner, S.E., Chervitz, S.A., Dagdigian, C., Fuellen, G., Gilbert, J.G., Korf, I., Lapp, H., et al. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12: 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valter, K., van Driel D., Bisti, S., and Stone, J. 2002. FGFR1 expression and FGFR1-FGF2 colocalisation in rat retina: Sites of FGF-2 action on rat photoreceptors. Growth Factors 20: 177–188. [DOI] [PubMed] [Google Scholar]
- Vignal, A., Milan, D., San Cristobal, M., and Eggen, A. 2002. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 34: 275–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, C.M., Kulbokas III, E.J., Kirby, A.W., Zody, M.C., Mullikin, J.C., Lander, E.S., Lindblad-Toh, K., and Daly, M.J. 2002. The mosaic structure of variation in the laboratory mouse genome. Nature 420: 574–578. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://cascad.niob.knaw.nl; candidate SNP database.
- http://primers.niob.knaw.nl; primer design Web interface.
- http://ratEST.uiowa.edu; Rat Gene Discovery and Mapping Project.
- http://trace.ensembl.org; Ensembl trace archive.
- http://www.hgsc.bcm.tmc.edu/projects/rat; Rat Genome Project.
- http://www.ncbi.nlm.nih.gov/Genbank; NCBI GenBank.
- http://www-genome.wi.mit.edu/genome_software/other/primer3.html; Primer3.