Abstract
Gitelman’s syndrome is an autosomal recessive salt losing nephropathy caused by inactivated mutations of the SLC12A3 gene, encoding the NaCl cotransporter of the distal convoluted tubule. We report a French family with five affected members over two generations suggesting a dominant transmission. After SLC12A3 sequencing of seven individuals, four mutations were detected. Pseudo-dominant transmission was explained by the union of a compound heterozygous woman (two mutations on one allele and one mutation on the other) with a heterozygous healthy man. This study shows the importance of complete genetic analysis of families with unusual presentation.
Keywords: Gitelman syndrome, mutation in cis, mutation in trans, pseudo-dominant inheritance
Background
Gitelman’s syndrome (GS; OMIM 263800) is an autosomal recessive disease characterized by a defect in renal tubular sodium reabsorption with secondary hyperaldosteronism, renal hypokalaemia metabolic alkalosis, hypomagnesaemia and hypocalciuria [1]. GS results from inactivated mutations of the SLC12A3 gene, encoding the NaCl cotransporter of the distal convoluted tubule [2]. Most subjects carry two different heterozygous mutations inherited from each parent. Estimated prevalence of GS and of heterozygous carriers is ∼1/40 000 and 1/100 in the Caucasian population [3]. We report phenotypic and genotypic data of a French family with two unusual features: five members affected over two generations and the presence of four different mutations.
Case report
A 20-year-old man (Patient II.2, Figure 1) was referred for severe hypokalaemia (2 mmol/L) with hypomagnesaemia discovered 2 months before. He reported cramps, episodes of tetany and weakness. He had no past medical history, particularly, no prematurity or growth retardation. He was treated with potassium and magnesium supplementation. Normal blood pressure, orthostatic tachycardia, hyper-reninaemic hyperaldosteronism, hypokalaemia, hypomagnesaemia, metabolic alkalosis and high urinary excretion of NaCl, K and Mg were confirmed. There was no polyuria or hypercalciuria.
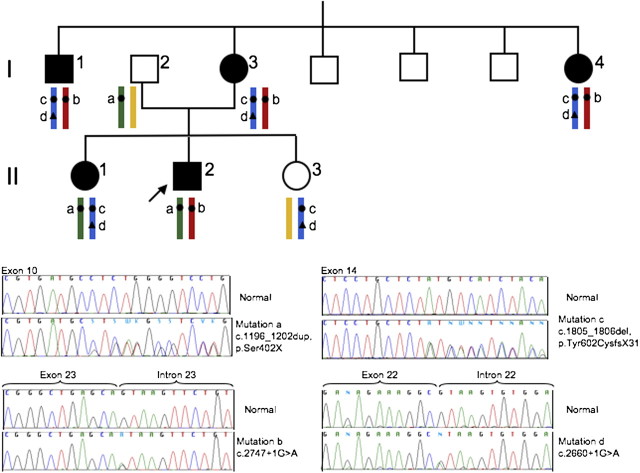
Fig. 1.
Family tree illustrating the transmission of GS over two generations. Sequencing chromatograms and segregation of the four detected mutations in the SLC12A3 gene are shown. Affected subjects are represented by dark symbols. The index patient is II.2. Mutations c and d are on the same allele.
Familial history of hypokalaemia was present in his mother, his maternal aunt and one of his maternal uncles. We first investigated his mother and then five other family members. Clinical and biochemical data are summarized in Table 1.
Table 1.
Clinical and biological data of the five GS patients and the two heterozygous carriers
| Standards | II-2 (proband) | II-1 (affected sister) | II-3 (unaffected sister) | I-1 (affected uncle) | I-2 (father) | I-3 (mother) | I-4 (affected aunt) | |
| Age, years | 20 | 24 | 19 | 52 | 52 | 51 | 41 | |
| Age at hypokalaemiadetection, years | 20 | 24 | 42 | 47 | 21 | |||
| Medical history and symptoms | Cramps, tetany and weakness | Tetany and weakness | Faintness and tetany at 42 | Hypertension | Pre-term, generalized seizures at 31 and 41, transient right hemiparesis at 47. Normal pregnancies | Generalized seizures | ||
| Treatment (dose/day) | ||||||||
| Potassium | 50 mmol | 90 mmol | 100 mmol | 70 mmol | 40 mmol | |||
| Magnesium | 15 mmol | 10 mmol | 5.5 mmol | |||||
| Spironolactone | 100 mg | |||||||
| Recumbent blood pressure, mmHg (cardiac frequency, bpm) | 112/55 (46) | 105/53 (52) | 119/68 (55) | 132/78 (47) | 114/60 (59) | 110/60 (72) | ||
| Standing blood pressure, mmHg (cardiac frequency, b.p.m.) | 110/54 (115) | 118/63 (93) | 87/43 (66) | 136/84 (52) | 116/63 (79) | 63/37 (106) | ||
| Plasma parameters | ||||||||
| Sodium, mmol/L | 135–145 | 140 | 140 | 137 | 138 | 140 | 139 | 137 |
| Potassium, mmol/L | 3.5–4.5 | 2.6 | 2.4 | 3.4a | 2.5 | 4.3 | 2.2 | 2.5 |
| Chloride, mmol/L | 95–105 | 94 | 99 | 101 | 92 | 103 | 95 | 94 |
| Bicarbonate, mmol/L | 22–27 | 33 | 29 | 26 | 37 | 29 | 35 | 36 |
| Magnesium, mmol/L | 0.64–0.90 | 0.52 | 0.62 | 0.90 | 0.49 | 0.87 | 0.58 | 0.64 |
| Standing renin, mU/L | 15–50 | 272 | 197 | 133 | 124 | 104 | ||
| Standing aldosterone, pmol/L | 208–1000 | 943 | 1961 | 1488 | 596 | 917 | ||
| Urine parameters | ||||||||
| Urine volume, L/24 h | 2.01 | 0.76 | 1.93 | 2.00 | 1.99 | |||
| Sodium, mmol/24 h | <10 if NaCl depletion | 258 | 123 | 245 | 168 | 247 | ||
| Potassium, mmol/24 h | <20 if K depletion | 119 | 87 | 179 | 132 | 127 | ||
| Chloride, mmol/24 h | <10 if NaCl depletion | 286 | 106 | 345 | 251 | 324 | ||
| Magnesium, mmol/24 h | <1 if Mg depletion | 2.9 | 0.7 | 6.2 | 3.8 | 2.1 | ||
| Calcium, mmol/24 h | Man <7.5, woman <6.25 | 2.5 | 0.1 | 5.8 | 3.8 | 3.4 | ||
| Creatinine mmol/kg/day | Men 0.17–0.23, women 0.12–0.19 | 0.28 | 0.17 | 0.17 | 0.12 | 0.11 | ||
Plasma potassium concentration 1 year before was normal.
The phenotypes of Patients I.1, I.3, I.4, II.1 and II.2 were similar and strongly suggested GS. Genetic investigations were performed after patients’ informed consent was obtained. DNA was extracted from blood leucocytes by standard procedures. Mutation analysis was performed by polymerase chain reaction amplification and direct sequencing of the SLC12A3 gene, as previously described [4]. Molecular genetic analysis showed the following results (Figure 1).
The proband (Patient II.2) was compound heterozygous for one frameshift mutation in exon 10 (c.1196_1202dup, p.Ser402X, mutation a) and one splice-site mutation in intron 23 (c.2747 + 1G > A, mutation b). For his mother (Patient I.3), we first analysed Exons 10 and 23 but only mutation b was detected. Analysis of the remaining exons revealed two additional mutations: one frameshift mutation in Exon 14 (c.1805_1806del, p.Tyr602CysfsX31, mutation c) and another splice mutation in intron 22 (c.2660 + 1G > A, mutation d). Patients I.1 and I.4 carried the same three mutations as Patient I.3. Thus, Patient II.1 carried heterozygous mutations a, c and d. The healthy father (Patient I.2) was heterozygous for mutation a, and the healthy sister (Patient II.3) was heterozygous for mutations c and d. From these results, we deduced that mutations c and d were on the same allele (i.e. in cis) (Figure 1). Mutations a and c have been previously described [5, 6]. Mutation b has been detected only in the proband of this family in our cohort [4] and finally, this is the first description of mutation d. We did not perform RNA analysis, but mutation d was not detected in 200 normal chromosomes and four different bioinformatic methods integrated in the Alamut V.2 software predict a loss of donor splice-site (Interactive Biosoftware, Rouen, France; http://www.interactivebiosoftware.com/).
Discussion
We report a French non-consanguineous family with apparent autosomal-dominant transmission of GS with five cases over two generations. Direct SLC12A3 sequencing has allowed us to show that the dominant-like transmission of the disease was explained by the union of a compound heterozygous woman, carrying two mutations on one allele and another mutation on the other, with a healthy man heterozygous for the mutation p.Ser402X. Consequently, the proband and his affected sister differed by the allele transmitted by their mother. An apparent dominant transmission of GS has been previously reported. Bettinelli et al. [7] in 1995 hypothesized that two different genetic transmissions of GS exist, autosomal recessive or autosomal dominant. Subsequently, the molecular analysis of the family described by Bettinelli et al. and another similar family showed that the affected parent was compound heterozygous with two distinct mutations and the other parent was heterozygous for a third mutation [5, 8]. It is well established that GS is transmitted as an autosomal recessive trait, and patients presenting typical GS phenotype are homozygous or compound heterozygous (i.e. bearing two pathogenic mutations in trans). The estimated prevalence of SLC12A3 heterozygous carriers is ∼1% [3]. Recently, this prevalence was estimated to 0.48% in unrelated subjects of the Framingham Heart study population [9]. Hence, in non-consanguineous families, the theoretical probability to have an affected child when one of the parents has GS is at least 1/200 and for a couple of one heterozygous carrier and one homozygous or compound heterozygous GS patient, as in the family described here, the probability to have an affected child is 50%.
The four mutations detected in this family are two frameshift and two splice-site mutations probably resulting in unstable/abnormal messenger RNAs or truncated proteins. Interestingly, two of them are located on the same allele. In GS and other recessive diseases such as cystic fibrosis, two mutations on the same allele have been described; they are often missense mutations, which could be either a frequent not pathogenic variant or be functional and contribute to the phenotype severity [10–12]. Here, the two mutations detected on the same allele are pathogenic and rare. Indeed, in our cohort of 396 GS patients harbouring SLC12A3 mutations [4], mutation c was detected in only one other proband and for mutation d, it is the first ever description. This finding of triple pathogenic mutations appears to be a rare phenomenon (0.7% in our cohort) but illustrates the importance of sequencing all the exons for probands and parents to check the segregation and to determine if the mutations are located in cis or in trans. Otherwise, individuals with two mutations on the same allele, as sister II.3 in this family, could be wrongly considered as affected.
In summary, complete phenotypic and genotypic characterization are critical to define autosomal recessive diseases with vertical transmission. The probability of a union between a GS-affected subject and a healthy heterozygous subject must be considered to provide accurate genetic advice.
Acknowledgments
We would like to thank all the members of this family for their participation. Funding. The study was made possible by funds from The French Ministry of Health (Plan Maladies Rares) and the European Community FP7 (EUNEFRON, 201590).
Conflict of interest statement. None declared.
References
- 1.Gitelman HJ, Graham JB, Welt LG. A new familial disorder characterized by hypokalemia and hypomagnesemia. Trans Assoc Am Physicians. 1966;79:221–235. [PubMed] [Google Scholar]
- 2.Simon DB, Nelson-Williams C, Bia MJ, et al. Gitelman's variant of Bartter's syndrome, inherited hypokalaemic alkalosis, is caused by mutations in the thiazide-sensitive Na-Cl cotransporter. Nat Genet. 1996;12:24–30. doi: 10.1038/ng0196-24. [DOI] [PubMed] [Google Scholar]
- 3.Knoers NV, Levtchenko EN. Gitelman syndrome. Orphanet J Rare Dis. 2008;30:3–22. doi: 10.1186/1750-1172-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vargas-Poussou R, Dahan K, Kahila D, et al. Spectrum of mutations in Gitelman syndrome. J Am Soc Nephrol. 2011;22:693–703. doi: 10.1681/ASN.2010090907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mastroianni N, Bettinelli A, Bianchetti M, et al. Novel molecular variants of the Na-Cl cotransporter gene are responsible for Gitelman syndrome. Am J Hum Genet. 1996;59:1019–1026. [PMC free article] [PubMed] [Google Scholar]
- 6.Maki N, Komatsuda A, Wakui H, et al. Four novel mutations in the thiazide-sensitive Na-Cl co-transporter gene in Japanese patients with Gitelman's syndrome. Nephrol Dial Transplant. 2004;19:1761–1766. doi: 10.1093/ndt/gfh239. [DOI] [PubMed] [Google Scholar]
- 7.Bettinelli A, Bianchetti MG, Borella P, et al. Genetic heterogeneity in tubular hypomagnesemia-hypokalemia with hypocalcuria (Gitelman's syndrome) Kidney Int. 1995;47:547–551. doi: 10.1038/ki.1995.68. [DOI] [PubMed] [Google Scholar]
- 8.Lemmink HH, van den Heuvel LP, van Dijk HA, et al. Linkage of Gitelman syndrome to the thiazide-sensitive sodium-chloride cotransporter gene with identification of mutations in Dutch families. Pediatr Nephrol. 1996;10:403–407. doi: 10.1007/s004670050129. [DOI] [PubMed] [Google Scholar]
- 9.Ji W, Foo JN, O'Roak BJ, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claustres M, Altiéri JP, Guittard C, et al. Are p.I148T, p.R74W and p.D1270N cystic fibrosis causing mutations? BMC Med Genet. 2004;5:19. doi: 10.1186/1471-2350-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clain J, Lehmann-Che J, Girodon E, et al. A neutral variant involved in a complex CFTR allele contributes to a severe cystic fibrosis phenotype. Hum Genet. 2005;116:454–456. doi: 10.1007/s00439-004-1246-z. [DOI] [PubMed] [Google Scholar]
- 12.Lin SH, Shiang JC, Huang CC, et al. Phenotype and genotype analysis in Chinese patients with Gitelman's syndrome. J Clin Endocrinol Metab. 2005;90:2500–2507. doi: 10.1210/jc.2004-1905. [DOI] [PubMed] [Google Scholar]