Abstract
Snakes provide a unique and valuable model system for studying the extremes of physiological remodeling because of the ability of some species to rapidly upregulate organ form and function upon feeding. The predominant model species used to study such extreme responses has been the Burmese python because of the extreme nature of postfeeding response in this species. We analyzed the Burmese python intestine across a time series, before, during, and after feeding to understand the patterns and timing of changes in gene expression and their relationship to changes in intestinal form and function upon feeding. Our results indicate that >2,000 genes show significant changes in expression in the small intestine following feeding, including genes involved in intestinal morphology and function (e.g., hydrolases, microvillus proteins, trafficking and transport proteins), as well as genes involved in cell division and apoptosis. Extensive changes in gene expression occur surprisingly rapidly, within the first 6 h of feeding, coincide with changes in intestinal morphology, and effectively return to prefeeding levels within 10 days. Collectively, our results provide an unprecedented portrait of parallel changes in gene expression and intestinal morphology and physiology on a scale that is extreme both in the magnitude of changes, as well as in the incredibly short time frame of these changes, with up- and downregulation of expression and function occurring in the span of 10 days. Our results also identify conserved vertebrate signaling pathways that modulate these responses, which may suggest pathways for therapeutic modulation of intestinal function in humans.
Keywords: hyperplasia, organ remodeling, physiological remodeling, small intestine, RNA-Seq
snakes represent an emerging model system for studying extreme vertebrate phenotypes, including extreme examples of modulation of physiological form and function (1, 13, 19, 23, 31, 37, 69, 70). Studying such extreme examples of vertebrate phenotypes and physiology may provide novel insight into vertebrate biology and how snakes have evolutionarily manipulated vertebrate pathways to achieve such phenotypes. Furthermore, such studies have the potential to demonstrate how conserved vertebrate pathways may be modulated to achieve desired therapeutic phenotypes in other vertebrates, such as humans. A particularly interesting feature to study is the ability of certain snake species to rapidly and reversibly remodel major organ systems, including growth of new organ tissue, in response to feeding. Multiple species of snakes appear to have evolved the ability to massively downregulate metabolic and physiological functions during periods of fasting, including the atrophy of organs, such as the heart, liver, kidney, and intestine (44, 51, 58, 59). Upon feeding, their metabolism, along with the size and function of these major organ systems, is massively and rapidly upregulated to accommodate the digestion of prey. While some snake species represent the most extreme exemplars of such remodeling, complex cyclic physiological remodeling has been documented in other ectotherms including frogs and fishes and is proposed to have evolved as a mechanism to reduce daily energy needs while enduring long periods (months or years) of fasting (10, 51, 52, 58, 59).
Among snake species that experience such large fluctuations in physiology with each meal, the Burmese python (Python molurus bivittatus) is the most well studied (2, 10, 15, 45, 51, 55, 56, 60). Within 48 h of feeding, Burmese pythons experience major shifts in systemic physiology, including as much as 44-fold increases in metabolic rate and 160-fold increases in plasma triglyceride content (56, 58). Major organ-specific changes also occur within 72 h of feeding, including 40–100% increases in the mass of the heart, liver, pancreas, kidneys, and small intestine (15, 34, 51, 55, 64).
Evolutionary processes have driven improvement of anatomical and physiological traits of the digestive system to ensure that organisms can effectively digest and assimilate large meals. The extreme phenotypic plasticity of the Burmese python small intestine has been well documented, with results indicating major fluctuations in form and function of this organ between fasted and postfed animals. Within just 24 h after feeding, the wet mass of the small intestine doubles, including a doubling of the mucosal enterocyte volume and a sixfold increase in microvillus length (34). Within 48 h, intestinal nutrient transport capacity increases 20-fold compared with fasted levels (15, 44). The pH of the stomach also drops by as much as 4.6 (from 7.5 to 2.9) within 12 h of feeding, indicating that every 3 h there is a 10-fold increase in intragastric hydrogen ions [H+] (53). Digestion is completed within 6–10 days, and this completion coincides with the downregulation of intestinal form and function to fasted levels (15, 55). These extreme cycles of physiological fluctuations with feeding suggest tightly regulated gene expression patterns that coordinate massive reconstructions of the anatomy and physiology of the python digestive system.
Recent studies on the Burmese python have demonstrated that rapid, massive changes in gene expression do indeed broadly coincide with physiological remodeling phenotypes and that these differentially expressed genes involved in remodeling include those known to be involved in human development, metabolism, and disease (10). Previous studies have, however, only surveyed two postfeeding time-points during this postfeeding response, 1 day postfeeding (1 DPF; when phenotypes are continuing to increase) and 4 DPF (when phenotypes have just begun to regress), and lacked any substantial analyses linking changes in gene expression with corresponding changes in phenotypes and physiological function (10). Therefore, major gaps in our understanding remain, including how patterns of gene expression precisely underlie shifts in intestinal phenotype and function throughout the intestinal remodeling process, how rapidly gene expression responses occur postfeeding, and how different temporal phases of gene expression direct temporal phases of intestinal phenotypic and functional change.
In this study we address gaps in our understanding of how the python intestine undergoes such transformations of form and function by substantially increasing sampling of gene expression data to include more biological replicates and postfeeding time-points. Though of a distinct body form, the Burmese python shares with humans and other vertebrates the same structure and function of organs at the tissue and cellular levels, including the intestine, making them a useful system for investigating the regulation of vertebrate organ structure and function (55, 58, 60). This expanded sampling allows us to link shifts in gene expression through time with the phenotypic and functional shifts that co-occur in the intestine by linking particular gene expression programs to cellular and functional physiological processes that they underlie. Our integrated analyses of gene expression and intestinal physiology provide new perspectives on the surprising scales associated with this response and new insight into the molecular mechanisms and foundations driving these extensive shifts in phenotype and physiology.
MATERIALS AND METHODS
Feeding experiments.
Burmese pythons were obtained as hatchlings from commercial breeders and maintained on a weekly to biweekly diet of prekilled rodents. All animal care and experiments were conducted under approved protocols of the University of Alabama Institutional Animal Care and Use Committee. Two- to three-year-old Burmese pythons (mean mass = 910 g, range = 415-5,776 g) were maintained at 30°C and studied fasted (30 days since last meal) as well as at 6 h postfeeding (6 HPF), 12 HPF, 1 DPF, 4 DPF, and 10 DPF following the consumption of meals (prekilled rats) equaling at least 25% of their body mass (Table 1). The sampling times were chosen to represent episodes of clear shifts in tissue physiology and gene expression (10, 11, 15, 51). Each time-point was sampled with multiple biological replicates (multiple animals). We humanely euthanized the snakes by severing the spinal cord immediately behind the head, and, following a midventral incision, organs were immediately extracted and weighed (44). For fed snakes, elements of the digestive tract were emptied of their contents and reweighed. Segments of the anterior third of the small intestine were placed in ice-cold reptilian Ringer's solution (for nutrient uptake), fixed in reptilian Ringer's-buffered 10% formalin solution (for light microscopy), fixed in reptilian Ringer's-buffered 2.5% gluteraldehyde solution (for electron microscopy), snap-frozen in liquid N2, and stored at −80°C (for enzyme assays and gene expression analyses).
Table 1.
Sampling design and number of mapped reads per sample
| Sample | Mass, g | Tissue | Time-point | Mapped Reads |
|---|---|---|---|---|
| AI6 | 462.5 | SI | 0 h | 1,511,751 |
| AJ6 | 445.8 | SI | 0 h | 3,965,717 |
| Ai8 | 414.5 | SI | 0 h | 800,799 |
| U25 | 5,800 | SI | 0 h | 372,003 |
| AF2 | 581 | SI | 0 h | 950,773 |
| S19 | 868.4 | Muc | 0 h | 203,389 |
| AJ7 | 610 | SI | 6 h | 881,249 |
| AJ7 | 610 | Muc | 6 h | 1,492,138 |
| Ak25 | 725 | SI | 6 h | 1,626,270 |
| Ak25 | 725 | Muc | 6 h | 403,645 |
| R27 | 815.7 | Muc | 6 h | 590,892 |
| W11 | 788.7 | Muc | 6 h | 532,607 |
| Ak4 | 635 | SI | 12 h | 936,987 |
| Ak4 | 635 | Muc | 12 h | 928,321 |
| Ak10 | 747 | Muc | 12 h | 936,420 |
| U7 | 796.3 | Muc | 12 h | 797,731 |
| U13 | 774.8 | Muc | 12 h | 654,142 |
| Z14 | 467 | SI | 24 h | 3,420,055 |
| V43 | 782 | SI | 24 h | 678,000 |
| Z18 | 525 | SI | 24 h | 596,198 |
| W20 | 797.8 | SI | 24 h | 1,280,360 |
| S6 | 809 | SI | 24 h | 1,281,658 |
| S6 | 809 | Muc | 24 h | 974,889 |
| Y5 | 839 | SI | 96 h | 1,911,844 |
| Y18 | 971 | SI | 96 h | 2,099,417 |
| Y23 | 802 | SI | 96 h | 1,016,261 |
| Y24 | 957 | SI | 96 h | 871,196 |
| V40 | 867.5 | SI | 96 h | 503,975 |
| W2 | 807.7 | Muc | 96 h | 235,595 |
| W22 | 764 | SI | 10 d | 481,585 |
| W22 | 764 | Muc | 10 d | 923,344 |
| U1 | 767.9 | Muc | 10 d | 532,607 |
| V29 | 825 | Muc | 10 d | 2,688,732 |
| W12 | 981.6 | Muc | 10 d | 799,404 |
The number of reads mapped to at least one transcript from the Burmese python transcriptome. Each row represents a single replicate (individual animal), which are grouped by time-point. Mapped reads are composed of RNA-Seq reads generated from cross-sectional (SI) or mucosal (Muc) samples of intestinal tissue or from both types of sampling. d, Days.
Analysis of tissue and cellular structure.
We used light microscopy to examine postprandial changes in intestinal tissue thickness and enterocyte size. Formalin-fixed intestinal samples were embedded in paraffin, cross-sectioned (6 μm), and stained with hematoxylin and eosin on glass slides. Samples were viewed with a light microscope linked to a computer loaded with image-analysis software (Motic Image Plus, Richmond, British Columbia, Canada). From each cross section, we measured mucosal/submucosal thickness at 10 locations along with muscularis/serosa thickness and enterocyte height and width. We calculated enterocyte volume according to the formula for a cylinder. We assessed ultrastructural changes to the intestinal brush-border membrane by transmission electron microscopy. Small pieces of intestinal mucosa were fixed in 2.5% gluteraldehyde, postfixed in 1% osmium tetroxide, dehydrated in a graded series of ethanol, and embedded in Spurr's epoxy resin. Ultrathin sections (∼80 nm) were cut and placed on copper mesh grids and examined under a Hitachi (H-7650) transmission electron microscope. Sections of microvillus membrane were viewed with the scope's high-definition camera (Orca) and image analysis software (Advance Microscopy Techniques). For each sample, we measured the height and width of 100 individual microvilli and calculated microvillus surface area with the equation width × π × height.
Measuring intestinal function.
We quantified intestinal nutrient uptake and hydrolase activities to determine the magnitude that intestinal function is modulated with feeding and fasting. We used the everted-sleeve technique to measure the brush-border uptake of the amino acids l-leucine and l-proline and of the sugar d-glucose (28, 63). Sleeves (1 cm) of everted intestine were preincubated in reptile Ringer's solution for 5 min at 30°C and incubated for 2 min at 30°C in reptile Ringer's solution containing both an unlabeled and radiolabeled nutrient (3H-l-leucine, 3H-l-proline, or 14C-d-glucose) and a radiolabeled adherent fluid marker (14C-polyethylene glycol for the amino acids or 3H-l-glucose for d-glucose). We quantified total uptake (carrier-mediated and passive) of each amino acid, and carrier-mediated uptake of d-glucose, as nanomoles of nutrient per minute per milligram of tissue (29, 63). We used colorimetric methods to quantify (μmol·min−1·mg−1 protein) the activities of the membrane-bound intestinal aminopeptidase N (APN) (15).
Transcriptome library generation.
Total RNA was extracted from snap-frozen intestinal cross sections and intestinal mucosa with Trizol reagent (Invitrogen). The details of the sampling design and the numbers of replicates per time-point are outlined in Table 1. Illumina mRNA-Seq barcoded libraries were constructed with either the Illumina TruSeq RNA-Seq kit or the NEB Next RNA-Seq kit, both of which include poly-A selection, RNA fragmentation, cDNA synthesis, and indexed Illumina adapter ligation. Completed RNA-Seq libraries were quantified on a BioAnalyzer (Agilent), pooled in various multiplex arrangements, and sequenced on either an Illumina GAIIx or Illumina HiSeq2000.
Analysis of gene expression.
Raw Illumina RNA-Seq reads were quality filtered, trimmed, and mapped to the complete annotated transcript set of the Burmese python genome (10). The numbers of mapped reads per sample are provided in Table 1. Based on preliminary analyses, we found intestinal mucosa and intestinal cross section samples to be extremely similar in terms of gene expression (Fig. 1). Mucosal and intestinal sections were therefore analyzed together, and where both samples existed for a single specimen (same animal), these reads were combined for final analysis. In instances where replicates of a sample were available (e.g., same library sequenced in multiple different runs), replicated samples were combined and mapped for each individual. Mapping of reads to the reference transcriptome of the Burmese python (10) was conducted in CLC Genomics Workbench using the following parameters: maximum number of mismatches = 2, minimum length fraction = 0.8, minimum similarity fraction = 0.8, and maximum number of hits for a read = 10. We determined expression by counting the number of unique gene reads that mapped to a particular annotated transcript, while excluding reads that mapped to multiple positions. Combined with data previously published (10), new data are accessioned in the National Center for Biotechnology Information Short Read Archive (SRP051827).
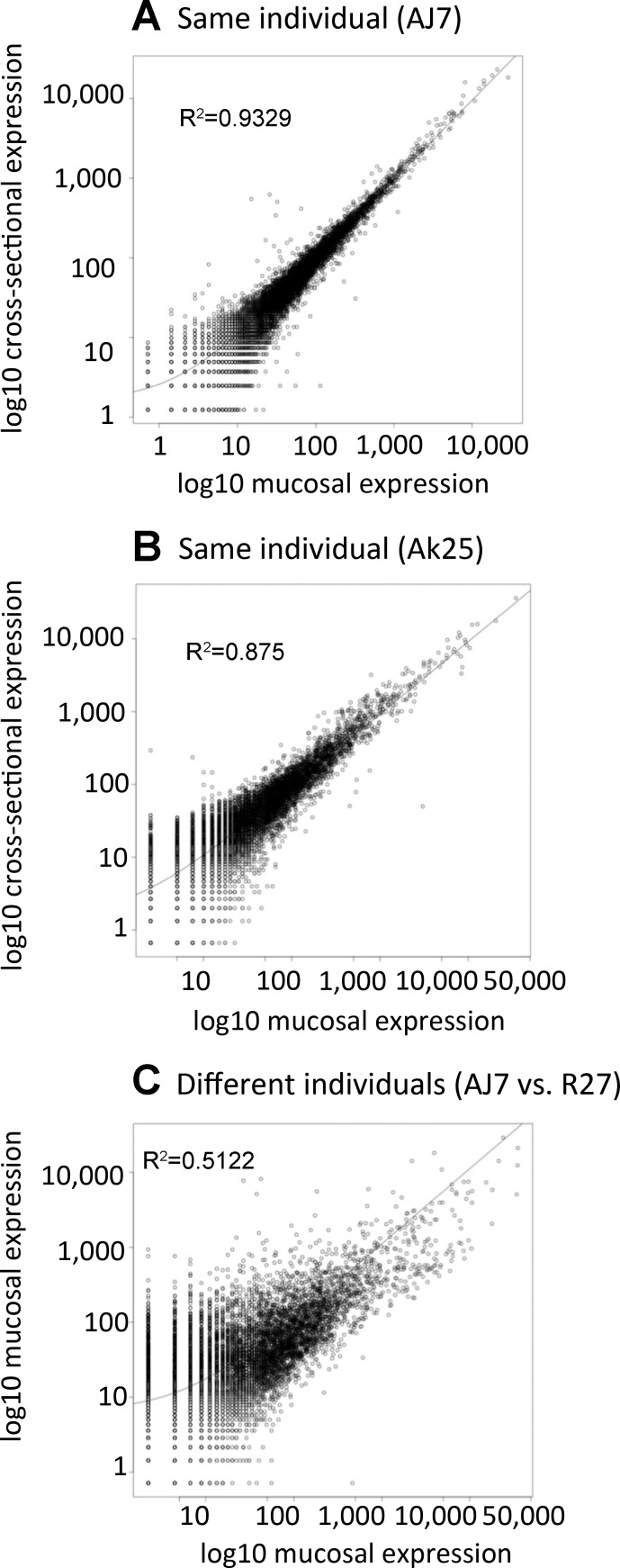
Fig. 1.
Scatterplots comparing gene expression in intestinal mucosal and intestinal cross-section samples. A: mucosal and cross-sectional samples from individual AJ7. B: mucosal and cross-sectional samples from individual Ak25. C: mucosal samples from 2 different individuals (AJ7 and R27). It is clear that the 2 different types of samples (cross-sectional and mucosal) from the same individual (A and B) are more similar than the same type of sample from different individuals (C).
The raw expression counts were normalized by trimmed mean of M-values (TMM) normalization in edgeR (50), and all statistical analyses of gene expression used these normalized data. We identified genes that were significantly differentially expressed between time-points by two different approaches. First, we estimated significant changes in gene expression between pairs of time-points with pairwise exact tests for the binomial distribution calculated in edgeR (50), which integrates both common and tagwise dispersion. Second, to accommodate the time-series nature of the experimental design of this study we also conducted step-wise regression analysis of gene expression in maSigPro (14).
To facilitate analyses of candidate genes, we annotated Burmese python genes by using reciprocal best blast and one-way tblastx (7) estimates of homology to gene sets from Anolis carolinensis, Homo sapiens, and Gallus gallus. Results of these analyses allowed a large proportion of python genes to be assigned to a homologous human gene and therefore assigned a human Ensembl ID.
We identified statistically overrepresented gene expression profile clusters with short time-series expression miner STEM (21). For STEM analyses, we input our TMM-normalized gene expression data (averaged across replicates per time-point) using default settings, further log-normalized these data as recommended in the STEM manual, and conducted STEM analyses over our entire time series dataset (fasted - 10 DPF). We set the maximum number of model profiles to 50 and limited the maximum unit change (between consecutive time-points) within model profiles to 3; together these settings allow for a large number of sampled model profiles while maintaining a conservative estimate of expression fluctuations within a profile.
RESULTS
Rapid shifts in gene expression upon feeding.
To quantify which temporal periods following feeding have the most extreme shifts in gene expression, we conducted pairwise analyses between time-points (Table 2). Our comparisons show the general trend that bursts of up- or downregulated genes early after feeding are effectively reversed around 1–4 DPF and return to near baseline by 10 DPF or earlier (Table 2). These comparisons also show that much of the observed differential expression occurs within the first 6 HPF, with 2,489 genes undergoing differential expression at this time compared with the fasted state. Nearly equal numbers of genes were up- and downregulated in the first 6 h (1,367 genes upregulated vs. 1,122 genes downregulated; Table 2). Only a few more genes were upregulated in the remaining time of the first 24 h, as seen in the 6 vs. 12 h and 12 vs. 24 h as well as the 0 vs. 24 h comparisons (Table 2). The return to near-fasting gene expression levels was not as rapid as the early postfeeding response, with only 864 significantly differentially expressed between 24 and 96 h, and 538 significantly differentially expressed genes between 96 h and 10 DPF. By 10 days, expression levels are nearly back to starting conditions, with only 31 genes significantly differentially expressed between 0 h vs. 10 days (Table 2).
Table 2.
Numbers of significantly differentially expressed genes between pre- and postfeeding time-points
| Time-point Comparisons |
||||||||
|---|---|---|---|---|---|---|---|---|
| 0 h–6 h | 6 h–12 h | 12 h–24 h | 24 h–96 h | 96 h–10 d | 0 h–24 h | 0 h–96 h | 0 h–10 d | |
| Up | 1,367 | 2 | 11 | 498 | 202 | 1,468 | 730 | 25 |
| Down | 1,122 | 22 | 5 | 366 | 336 | 1,225 | 469 | 6 |
For each pairwise time-point comparison, the number of up- and downregulated genes, as inferred by pairwise analysis (Benjamini-Hochberg corrected P value <0.05), is indicated.
Our analysis of significantly overrepresented patterns of gene expression in STEM yielded 11 significant gene profile clusters, and many of the genes identified in these clusters were significantly downregulated upon feeding, while others were significantly upregulated within the first 6–12 HPF (Fig. 2). Overall, we observed a wide variety of expression patterns for differentially expressed genes in the small intestine after feeding, with genes experiencing peak low or high expression values at various times, and many clusters either broadly corresponded with patterns of morphological and physiological changes or directly opposed them. As expected based on the comparisons in Table 1, the profiles all return to near baseline (fasted) levels of expression levels by 10 DPF or earlier (Fig. 2).
Fig. 2.
General trends in gene expression across postfed time-points in the Burmese python intestine analyzed with STEM time-series analyses. Generalized trends in gene expression that are significantly overrepresented in the python small intestine based upon cluster analysis of gene expression profiles and identification of statistically overpopulated profiles. The numbers of genes clustered and P value of these clusters is shown along with linear trends in expression.
Analyses of shifts in gene expression across all time-points via pairwise comparisons and by regression analyses both identified a large number of significantly differentially expressed genes across the entire time series before and after feeding (4,432 identified via pairwise analysis and 1,772 identified via regression analysis). Regression analysis was, however, substantially more conservative than pairwise analysis in its estimation of the number of genes significantly differentially expressed, and a large majority of genes (1,711) identified by regression analyses overlapped with those identified by pairwise comparisons. Given the large numbers of genes estimated by both methods, we focus primarily on the more conservative estimates of significance derived from regression analyses hereafter to dissect the biologically relevant features of this transcriptional response. Regression analyses identified a total of 1,772 genes that were significantly differentially expressed across the entire time course from fasted to 10 DPF (Fig. 3A). As discussed in greater detail below, it is notable that rapid shifts in gene expression correspond well to physiological and morphological changes in the intestine, returning to near baseline (fasted) levels as early as 4 DPF (Fig. 3A).
Fig. 3.
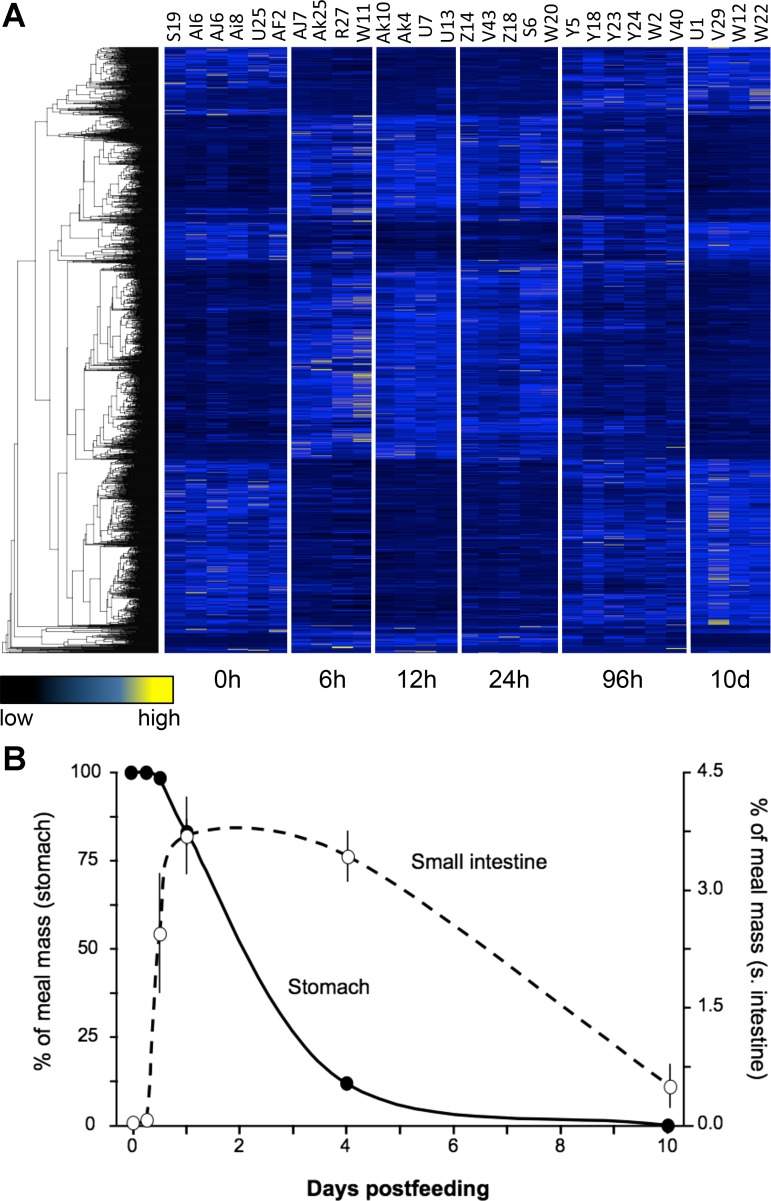
General trends of gene expression and digestion across postfed time-points. A: heat-map of gene expression for 1,772 genes found to be significantly differentially expressed across time-points based on regression analysis. Each column represents a replicate, with time-points clearly delimited, and each row represents a gene, which are clustered by similarity using average linkage hierarchical clustering. B: percent of meal mass in stomach (filled circles and solid line) and small intestine (open circles and dashed line) across postfeeding time-points.
To illustrate this correlation of the timing of meal breakdown and passage with shifts in intestinal gene expression, we have plotted stomach and small intestinal contents (relative to meal mass) as a function of time postfeeding (Fig. 3B). Meals (adult rats) were consumed intact. After 6 h they had not experienced any discernable loss in material within the snakes' stomachs. By 12 h, a small amount of material from the rat's head had passed into the small intestine. At 24 h, on average, 17% of the rat meal had exited the stomach and was moving through the small intestine. Over the next 3 days, the majority (∼70%) of the rat meal was converted to chyme, had exited the stomach, and filled the small intestine. After 4 days, the stomach and the small intestine gradually emptied. Only remnants of the meal were found within the distal small intestine at day 10. This physiological response correlates well with the patterns of differential gene expression described above, as the most pronounced shifts in gene expression between 6 HPF and 1 DPF (as the meal is exiting the stomach and moving through the small intestine), and begin to revert to fasted expression levels by 4 DPF (when both the intestine and stomach are emptying their contents).
Intestinal structure and function.
To further dissect cellular processes regulating intestinal physiological remodeling, we identified genes known to play important roles in both cell cycle progression and apoptosis that were significantly differentially expressed across time-points. Mitogen-activating kinases as well as various cyclins and kinesin family members important to cell cycle progression experience significant differential expression during digestion of a meal, which highlights the role of both cell growth and cell division in the process of intestinal remodeling (Fig. 4A). Genes regulating apoptosis (i.e., tumor necrosis factors and caspases, among others) also experience significant differential expression across time-points, consistent with apoptosis playing a role in the atrophy of the small intestine following the completion of digestion (Fig. 4A).
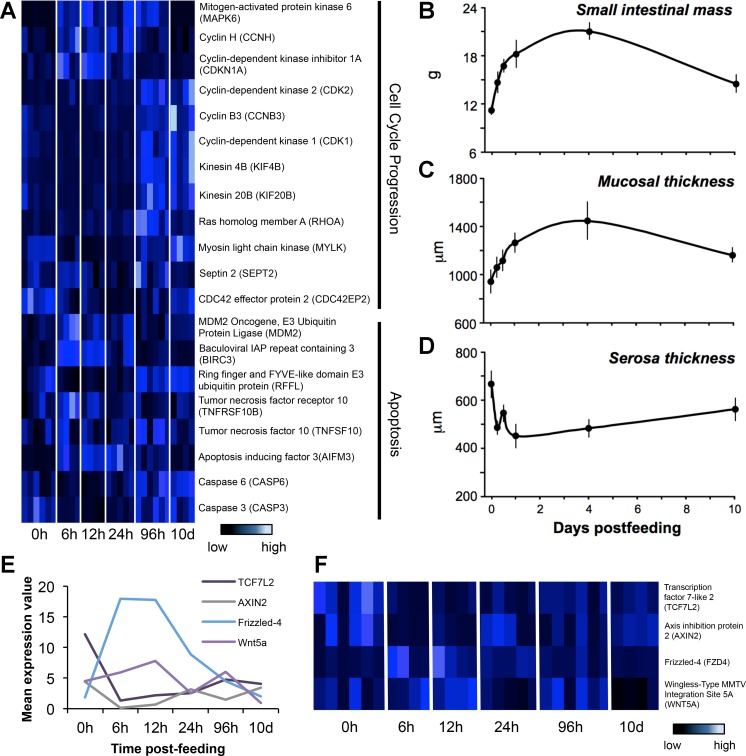
Fig. 4.
Patterns of expression for genes involved in cell cycling, apoptosis, and WNT signaling along with corresponding physiological changes in the small intestine. A: heat-map of genes involved in cell cycle progression and apoptosis that were shown to be significantly differentially expressed across time-points, identified from pairwise and regression analysis. B: change in small intestinal mass across time. C: change in mucosal thickness across time. D: change in serosa thickness across time. E: average expression values for Wnt signaling genes plotted across postfed time-points. F: heat-map of expression values for all replicates across postfed time-points, with each row representing a gene and each column representing an individual, which are manually clustered by time-points. This pathway is known to be important in development and processes such as asymmetric cell division.
At a phenotypic level, feeding generated significant trophic responses of the small intestine at multiple levels of organization. Small intestinal wet mass increased by 50% within 12 h after feeding and peaked at day 4 following an 87% increase in mass (Fig. 4B). Six days later, the small intestine had decreased in mass by 30%. Both mucosa/submucosa and muscularis/serosa thickness varied significantly (Ps < 0.034) across sampling time-points. Mucosa/submucosa thickness increased postfeeding (by 53%), whereas the thickness of the muscularis/serosa layer decreased by 32% (Fig. 4, C and D).
The WNT signaling pathway likely plays a role in growth and maintenance of the small intestine upon feeding, with multiple genes within this cascade experiencing significant differential expression throughout the digestion process. Genes such as frizzled class 4 receptor (FZD4) and wingless-type MMTV integration site 5A (WNT5A) show significant differential expression across time-points postfed, along with axis inhibition protein 2 (AXIN2) and transcription factor 7-like 2 (TCF7L2; Fig. 4, E and F).
Trafficking and cytoskeletal genes as well as genes important to microvilli structure were significantly differentially expressed upon feeding, indicative of tightly regulated patterns of expression underlying the phenotypic changes of intestinal structures across time (Fig. 5A). Significantly differentially expressed genes include those central to intestinal function, consistent with the changes in intestinal form and digestive function. Metabolism increases substantially following a large meal, and our gene expression data show corresponding significant upregulation of genes involved in oxidative phosphorylation as well as glucose uptake. Nutrient transporters and hydrolases essential to digestion and nutrient absorption also experience sharp increases in expression upon feeding (Fig. 5A).
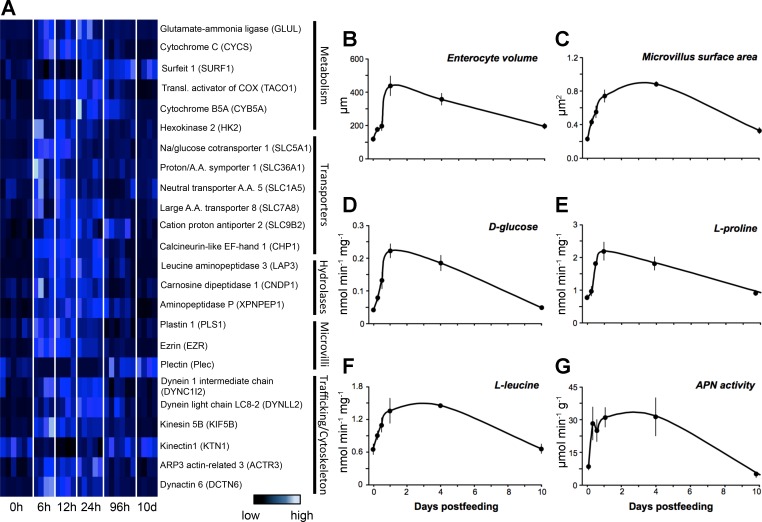
Fig. 5.
Patterns of expression for genes involved with intestinal form and function alongside corresponding morphological or physiological changes in the small intestine. A: heat-map of genes involved in various intestinal functional processes that were shown to be significantly differentially expressed across time-points via pairwise or regression analysis. B: change in enterocyte volume through time. C: change in microvillus surface area across time postfeeding. D: change in absorption of d-glucose across postfed time-points. E: change in absorption of l-proline across postfed time-points. F: change in absorption of l-leucine across postfed time-points. G: change in activity of aminopeptidase N (APN) across postfed time-points.
These changes in expression of genes underlying intestinal form and function correlate well with the upregulation of intestinal physiology. Feeding triggers the significant upregulation of the python's intestinal function, including increased enterocyte height and width (and thus volume) during meal digestion (Ps < 0.00001, Fig. 5B). Within 24 h of feeding, enterocyte volume had peaked, representing a 2.6-fold increase compared with the fasted state. Enterocyte volume remained elevated through day 4, before declining toward fasted values by day 10 (Fig. 5B). At the ultrastructural level, feeding generated a fourfold increase in the length of the intestinal microvilli (Fig. 5C), which regressed to prefeeding levels by between 4 and 10 DPF (Fig. 5C).
Mass-specific rates of d-glucose, l-proline, and l-leucine uptake varied significantly among sampling times (P values < 0.0009), as each nutrient significantly increased uptake within 12–24 h of feeding (Fig. 5, D–F, respectively). For all three, uptake rates peaked at 1 DPF, increasing from fasting levels 5.3-, 2.8-, and 2.1-fold, respectively. Nutrient uptake rates remained elevated through day 4, before becoming significantly downregulated by day 10 (Fig. 5, D–F). Intestinal APN exhibited a similar up-regulatory response, peaking at 1–4 DPF following a 3.6-fold increase. By day 10, APN activity had returned to fasted levels (Fig. 5G).
DISCUSSION
The Burmese python is an exceptional model for the study of extreme physiological remodeling, as few organisms exhibit this large and discretely staged response to feeding. This study, as well as previous studies (10, 15, 55, 59), has shown the process of organ remodeling in the python to be highly regulated, exhibiting controlled waves of gene expression that coincide with large changes in intestinal form and function. Despite substantial efforts, however, the cellular mechanisms that facilitate rapid organ growth and tissue remodeling in snakes remain poorly understood.
Our analysis demonstrates that extensive and rapid shifts in gene expression accompany rapid and massive changes in intestinal form and function upon feeding in the Burmese python. Perhaps the most surprising finding of the current study is that ∼2,500 genes are significantly differentially expressed within the first 6 h after feeding (Table 2). This extremely rapid response has been previously overlooked due to a lack of fine-scale sampling of postfeeding time-points following feeding (10). We find that patterns of differential expression largely mirror (and often precede) physiological changes in the intestine, with peaks of differential gene expression occurring around 1 DPF when the organ is experiencing the greatest increases in mass and functional performance (15, 51, 55). Equally remarkable, patterns of gene expression as well as physiological form and function rapidly return to baseline fasted levels within 10 DPF, indicating a tight association between differential gene expression and the rapid and cyclic physiological remodeling of the intestine. From STEM analysis, we see a number of distinct coexpressed clusters of genes that shift in expression with shifts in this remodeling process, indicating that various waves of expression are responsible for the tight regulation of intestinal remodeling (Fig. 2). A complete understanding of the roles of these discrete waves of coexpressed loci may elucidate novel and discrete mechanisms controlling organ growth and functional change. We have taken the first steps toward dissecting these roles by linking genes involved in cellular processes and intestinal function with shifts in intestinal phenotype and function.
We found that genes involved in modulating the cell cycle and cellular development are differentially expressed during the feeding response and thus direct substantial increases in organ mass and form. Previous studies of the Burmese python have shown that intestinal remodeling is accomplished by both hypertrophy (increase in cell size) and hyperplasia (cell division) (2, 25, 34, 49, 61, 66). Our gene expression results indicated tight regulation of the cell cycle through time. From observed expression patterns, it appears that the intestinal cells experience promitotic pressure 6 HPF and are actively cycling by 4 DPF, which, interestingly, is when the organ form and gene expression patterns begin to revert to fasted levels (Fig. 4A). Results of previous studies have also implicated cellular replication in intestinal remodeling in the python (25); based on these previous data, the number of replicating cells/mm increases over time and levels out at around 3 DPF (25). These clear signals of cell cycle progression indicate that hyperplastic organ growth begins relatively early in the process of intestinal remodeling.
Potent cell cycle promoters, mitogen activated protein kinase 6 (MAPK6) and cyclin H (CCNH), are upregulated 6–12 HPF. MAPK6 is a serine/threonine protein kinase that activates cyclin D3 (CCND3), which promotes cell growth and S-phase entry and is implicated in several cancers (26, 67). CCNH acts with cyclin-dependent kinase 7 (CDK7) as a broad activator of cyclin activity and is essential for cell cycle progression (30, 36). Upregulation of the cell cycle antagonist cyclin-dependent kinase inhibitor 1A (CDKN1A) during this same period suggests that intestinal cells may be preparing for, but not freely entering, the cell cycle (22). Later, at 4 DPF, we see evidence of several stages of cell division. Cyclin-dependent kinase 2 (CDK2), which drives cells into S-phase and G2 (4), is upregulated, as are cyclin B3 (CCNB3) and cyclin-dependent kinase 1 (CDK1) genes, which together are responsible for progression through mitosis (40, 42, 43) (Fig. 4A). Clearly, with increased activity of cell cycle regulators at various early and late time-points postfeeding, we can identify cell division as a key aspect to the remodeling of the intestine upon feeding.
At 4 DPF, we also see increased activity of genes responsible for the regulation of the cytoskeleton related to mitotic processes. Kinesin 4B (KIF4B) and kinesin 20B (KIF20B) are responsible intracellular trafficking during metaphase to anaphase, and anaphase to telophase/cytokinesis transitions respectively (16). Several indicators of cells undergoing cytokinesis are also observed at 4 DPF, notably components of the contractile ring apparatus Ras homolog gene family, member A (RhoA), myosin light chain kinase (MYLK), and Septin 2 (SEPT2), which are essential for cell shape reconfigurations (27, 47). For the Burmese python, as for other infrequently feeding snakes, the surface area of the intestine is massively expanded primarily due to an increase in microvillus length of up to sixfold following feeding (34, 51, 52, 62), entailing a substantial increase in the ability of the small intestine to absorb nutrients (15, 55). In contrast, snakes that feed more frequently in nature (along with fishes, amphibians, other reptiles, birds, and mammals) maintain consistent microvillus length with feeding and fasting and hence experience only modest regulatory changes in intestinal function (9, 18, 20, 39, 52, 54, 62). It is also notable that the pulse of contractile machinery at 4 DPF likely additionally plays a role in initiating the shortening of microvilli that begins at this time (Fig. 4A).
A particularly intriguing result from our analysis of gene expression underlying intestinal remodeling is evidence that WNT signaling may play a central role in coordinating rapid shifts in cellular processes in response to feeding. The WNT signaling pathway is a key signal transduction pathway crucial to appropriate embryonic development and includes signaling that leads to correct axis formation as well as proliferation, cell fate specification, cell migration, and planar cell polarity (12, 46). Dysregulation of WNT signaling also contributes to several cancer pathologies, including breast and colorectal cancer (48). We found that increased expression of both WNT and its extracellular receptor, Frizzled, occurs 6–12 HPF. During this same period, AXIN2, an intracellular inhibitor of WNT signaling, is substantially downregulated within 6 h. We also find that TCF7L2 is downregulated within the first 6 h and remains low through 10 DPF. TCF7L2 acts in concert with β-catenin in response to WNT signaling but, in the absence of WNT pathway activity, inhibits expression of WNT target genes (6). Interestingly, the TCF7L2 gene is known to be necessary for the maintenance of stem cell compartments in the epithelium of the small intestine (41). These and earlier observations implicate WNT signaling in the proliferative and physiological response to feeding (Fig. 4, E and F). Collectively, these results indicate that the python intestinal remodeling response may provide a unique and novel system for investigating the role of WNT signaling in trophic physiological contexts such as feeding.
Once digestion is complete, we see a complete reversal in intestinal remodeling as the organ atrophies and metabolism and nutrient uptake are downregulated. This and previous studies have shown that in the Burmese python, the postfeeding increase in enterocyte proliferation and sharp increase in intestinal activity are followed by an increase in apoptosis upon completion of digestion and a sharp decline in absorption and digestion (25, 55). Our results here indicate that this apoptotic response appears to occur as a two-stage process, primed first by the expression of apoptosis regulators and completed by the expression of potent downstream factors that execute apoptosis.
We observed the differential expression of genes regulating apoptosis by 6 HPF, indicating that regulation of apoptosis begins early in digestion. At 6 HPF there is increased expression of TNF-related apoptosis-inducing ligand (TRAIL), which activates extracellular death receptors and promotes of apoptosis (71). Apoptosis inducing factor 3 (AIFM3) is also expressed at this point. AIF proteins are mitochondrial flavoproteins that normally act as NADH oxidases but can trigger caspase-independent cell death in cells under proapoptotic pressure (8, 17). In contrast, two negative regulators of apoptosis, MDM2 proto-oncogene, known for its role in inhibiting p53, and Baculoviral IAP repeat containing 3 (BIRC3), a caspase inhibitor, showed increased expression at 6 HPF. Together these observations indicate that by 6 HPF some intestinal cells are exposed to extracellular and intracellular signaling promoting cell death, but many cells exhibit intracellular resistance to this process by expressing inhibitors of apoptosis. Competing factors that promote or antagonize apoptosis are also expressed later. Around 96 HPF there is a significant increase in expression of both tumor necrosis factor receptor 10 (TNFSF10), an apoptosis inducing death receptor (35, 71), and ring finger and FYVE-like domain E3 ubiquitin protein (RFFL), a negative regulator of apoptosis (38) (Fig. 4A).
While expression of upstream activators and inhibitors of apoptosis are highly variable, we see a clear pattern of caspase gene expression: both caspase 6 and caspase 3 are activated at 4 DPF, which, as clarified above, is the time when the organ begins to atrophy and revert to fasted levels of gene expression and nutrient absorption. Many types of caspases are known to play key roles in the execution of apoptosis upon activation by various intra- and extracellular signals. The caspases identified here are known as executioner caspases, activated by initiator caspases, which are capable of irreversibly triggering cell death (32). This controlled activation and execution of apoptosis has not been documented by previous studies on the Burmese python small intestine (Fig. 4A).
Genes involved in intestinal function that are significantly differentially expressed.
In addition to gene expression programs related to regulation of cellular processes (e.g., cell division and apoptosis), we observed major shifts in expression in diverse classes of genes associated with the upregulation of digestive and absorptive functionality of the intestine upon feeding (15, 55). This included genes related to metabolism, transport, and hydrolase activity, which allow for increased absorption and processing of large meals. Physiological data also indicates rapid increases in absorption of d-glucose, l-proline, and l-leucine within the same time frame that genes for transporters and hydrolases for these types of nutrients are being rapidly upregulated (Fig. 5, A and D–F). Genes related to trafficking and microvillus regulation are also upregulated rapidly upon feeding and act to expand (and later reduce) the surface area of the small intestine following feeding by facilitating the sixfold increase in microvillus length (Figs. 6A and 5C). A likely method for this lengthening of intestinal microvilli upon feeding is the triggering of existing pools of cytoskeletal and linking proteins to migrate to the microvilli and insert at the tips; this is likely followed by de novo construction of new cytoskeletal proteins, membranes, regulatory, and trafficking molecules (51). The expansion of enterocyte volume and increase in microvillus length observed in physiological experiments (Fig. 5, B and C) coincides well with this pattern of significant differential expression of candidate genes related to microvillus regulation and transport. Here, and in previous studies, it has been demonstrated that extreme increases in nutrient absorption occur within the first 24–48 h of feeding, along with large increases in metabolic rates and the mass of the organ (55, 57). In this study we have identified a number of associated candidate genes related to these shifts in phenotype and show coincident shifts in gene expression such that their expression levels mirror the physiology of the organ throughout digestion (Fig. 5A).
The discrete temporal regulation and the large number of coordinated, significant changes in gene expression observed in the python intestine postfeeding make it an excellent potential model for understanding the regulation of a number of important pathways, including cell cycle regulation, apoptosis, and WNT signaling in a physiological context in differentiated tissues of an adult vertebrate organ, as opposed to a developmental context in an undifferentiated embryonic tissue, for example. In most systems, the continuous nature of metabolism and growth obscures discrete gene expression regimes underlying such shifts in form and function because of the need for persistent and moderate activity. In contrast, examining the extreme physiological responses observed in the python may provide the opportunity to clarify fundamental relationships between gene expression and basic changes in vertebrate physiology. Our exploration of gene expression and analysis of candidate genes demonstrates that intestinal remodeling in pythons involves the regulation of gene expression programs associated with cell division, cytoskeletal remodeling, and apoptosis during growth and regression of the small intestine to modulate intestinal form, in addition to other gene expression programs regulating intestinal function (e.g., hydrolases and transporters).
Studies such as this one, which investigate extreme physiology and dramatic shifts in structural, functional, and transcriptional dynamics, provide a new avenue for gaining novel insights into basic mechanisms controlling key aspects of vertebrate biology. This may offer new information about the links between genotypes, phenotypes, and adaptation in nature and may also provide new information relevant to understanding and treating human disease. Damage of the intestinal tract can occur for various reasons in humans, including damage from cancer treatments using radiation or chemotherapeutics (33, 65), which can have chronic or even lethal effects, or complications due to inflammatory bowel disease, such as Crohn's Disease or ulcerative colitis (3). Using the Burmese python as a model system for understanding how the vertebrate intestine may be regulated to induce shifts in form and function is therefore potentially relevant for understanding diseases and conditions that affect the intestine and for understanding how intestinal regulation may be modulated to treat various diseases. Additionally, colorectal and intestinal cancers (5, 24, 48) and many other types of cancers (48, 68) involve dysregulation of WNT signaling. Evidence that python intestinal remodeling involves cell division and apoptosis, modulated at least in part by WNT signaling, suggests the python intestine may represent a valuable model for studying the interactions of metabolism with the regulation of cell division/death and WNT signaling relevant to cancer.
GRANTS
Support was provided from an National Institutes of Health - Library of Medicine postdoctoral training fellowship to R. P. Ruggiero (NIH 5R01 LM008111-08), National Science Foundation Grant IOB-0466139 to S. M. Secor, setup funds from of the University of Colorado School of Medicine to D. D. Pollock, and from startup funds from the University of Texas at Arlington to T. A. Castoe.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: A.L.A., D.C.C., S.M.S., and T.A.C. performed experiments; A.L.A., S.M.S., and T.A.C. analyzed data; A.L.A., R.P.R., S.M.S., and T.A.C. interpreted results of experiments; A.L.A., D.R.S., and S.M.S. prepared Figs.; A.L.A. and T.A.C. drafted manuscript; A.L.A., D.C.C., R.P.R., D.R.S., R.H.A., D.D.P., S.M.S., and T.A.C. edited and revised manuscript; A.L.A., D.C.C., R.P.R., D.D.P., S.M.S., and T.A.C. approved final version of manuscript; S.M.S. and T.A.C. conception and design of research.
REFERENCES
- 1.Adade CM, Cons BL, Melo PA, Souto-Padron T. Effect of Crotalus viridis viridis snake venom on the ultrastructure and intracellular survival of Trypanosoma cruzi. Parasitology 138: 46–58, 2011. [DOI] [PubMed] [Google Scholar]
- 2.Andersen JB, Rourke BC, Caiozzo VJ, Bennett AF, Hicks JW. Physiology: postprandial cardiac hypertrophy in pythons. Nature 434: 37–38, 2005. [DOI] [PubMed] [Google Scholar]
- 3.Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet 369: 1641–1657, 2007. [DOI] [PubMed] [Google Scholar]
- 4.Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol 14: 518–528, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell 103: 311–320, 2000. [DOI] [PubMed] [Google Scholar]
- 6.Brantjes H, Barker N, van Es J, Clevers H. TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling. Biol Chem 383: 255–261, 2002. [DOI] [PubMed] [Google Scholar]
- 7.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformat 10: 421, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cande C, Cecconi F, Dessen P, Kroemer G. Apoptosis-inducing factor (AIF): key to the conserved caspase-independent pathways of cell death? J Cell Sci 115: 4727–4734, 2002. [DOI] [PubMed] [Google Scholar]
- 9.Carey HV, Sills NS. Hibernation enhances D-glucose uptake by intestinal brush border membrane vesicles in ground squirrels. J Comp Physiol B 166: 254–261, 1996. [DOI] [PubMed] [Google Scholar]
- 10.Castoe TA, de Koning AP, Hall KT, Card DC, Schield DR, Fujita MK, Ruggiero RP, Degner JF, Daza JM, Gu W, Reyes-Velasco J, Shaney KJ, Castoe JM, Fox SE, Poole AW, Polanco D, Dobry J, Vandewege MW, Li Q, Schott RK, Kapusta A, Minx P, Feschotte C, Uetz P, Ray DA, Hoffmann FG, Bogden R, Smith EN, Chang BS, Vonk FJ, Casewell NR, Henkel CV, Richardson MK, Mackessy SP, Bronikowski AM, Yandell M, Warren WC, Secor SM, Pollock DD. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci USA 110: 20645–20650, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castoe TA, Fox SE, Jason de Koning A, Poole AW, Daza JM, Smith EN, Mockler TC, Secor SM, Pollock DD. A multi-organ transcriptome resource for the Burmese Python (Python molurus bivittatus). BMC Res Notes 4: 310, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell 127: 469–480, 2006. [DOI] [PubMed] [Google Scholar]
- 13.Cohn MJ, Tickle C. Developmental basis of limblessness and axial patterning in snakes. Nature 399: 474–479, 1999. [DOI] [PubMed] [Google Scholar]
- 14.Conesa A, Nueda MJ, Ferrer A, Talon M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics 22: 1096–1102, 2006. [DOI] [PubMed] [Google Scholar]
- 15.Cox CL, Secor SM. Matched regulation of gastrointestinal performance in the Burmese python, Python molurus. J Exp Biol 211: 1131–1140, 2008. [DOI] [PubMed] [Google Scholar]
- 16.Cross RA, McAinsh A. Prime movers: the mechanochemistry of mitotic kinesins. Nat Rev Mol Cell Biol 15: 257–271, 2014. [DOI] [PubMed] [Google Scholar]
- 17.Daugas E, Nochy D, Ravagnan L, Loeffler M, Susin SA, Zamzami N, Kroemer G. Apoptosis-inducing factor (AIF): a ubiquitous mitochondrial oxidoreductase involved in apoptosis. FEBS Lett 476: 118–123, 2000. [DOI] [PubMed] [Google Scholar]
- 18.Day R, Tibbetts IR, Secor SM. Physiological responses to short-term fasting among herbivorous, omnivorous, and carnivorous fishes. J Comp Physiol B 184: 497–512, 2013. [DOI] [PubMed] [Google Scholar]
- 19.Di-Poi N, Montoya-Burgos JI, Miller H, Pourquie O, Milinkovitch MC, Duboule D. Changes in Hox genes' structure and function during the evolution of the squamate body plan. Nature 464: 99–103, 2010. [DOI] [PubMed] [Google Scholar]
- 20.Dunel-Erb S, Chevalier C, Laurent P, Bach A, Decrock F, Le Maho Y. Restoration of the jejunal mucosa in rats refed after prolonged fasting. Comp Biochem Physiol A Mol Integr Physiol 129: 933–947, 2001. [DOI] [PubMed] [Google Scholar]
- 21.Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7: 191, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res 65: 3980–3985, 2005. [DOI] [PubMed] [Google Scholar]
- 23.Gomez C, Ozbudak EM, Wunderlich J, Baumann D, Lewis J, Pourquie O. Control of segment number in vertebrate embryos. Nature 454: 335–339, 2008. [DOI] [PubMed] [Google Scholar]
- 24.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev 19: 877–890, 2005. [DOI] [PubMed] [Google Scholar]
- 25.Helmstetter C, Reix N, T'Flachebba M, Pope RK, Secor SM, Le Maho Y, Lignot JH. Functional changes with feeding in the gastro-intestinal epithelia of the Burmese python (Python molurus). Zoolog Sci 26: 632–638, 2009. [DOI] [PubMed] [Google Scholar]
- 26.Hoeflich KP, Eby MT, Forrest WF, Gray DC, Tien JY, Stern HM, Murray LJ, Davis DP, Modrusan Z, Seshagiri S. Regulation of ERK3/MAPK6 expression by BRAF. Int J Oncol 29: 839–849, 2006. [PubMed] [Google Scholar]
- 27.Joo E, Surka MC, Trimble WS. Mammalian SEPT2 is required for scaffolding nonmuscle myosin II and its kinases. Dev Cell 13: 677–690, 2007. [DOI] [PubMed] [Google Scholar]
- 28.Karasov WH, Diamond JM. A simple method for measuring intestinal solute uptake invitro. J Comp Physiol 152: 105–116, 1983. [Google Scholar]
- 29.Karasov WH, Pond RS 3rd Solberg DH, Diamond JM. Regulation of proline and glucose transport in mouse intestine by dietary substrate levels. Proc Natl Acad Sci USA 80: 7674–7677, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim KK, Chamberlin HM, Morgan DO, Kim SH. Three-dimensional structure of human cyclin H, a positive regulator of the CDK-activating kinase. Nat Struct Biol 3: 849–855, 1996. [DOI] [PubMed] [Google Scholar]
- 31.Kohlsdorf T, Cummings MP, Lynch VJ, Stopper GF, Takahashi K, Wagner GP. A molecular footprint of limb loss: sequence variation of the autopodial identity gene Hoxa-13. J Mol Evol 67: 581–593, 2008. [DOI] [PubMed] [Google Scholar]
- 32.Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death Different 14: 44–55, 2007. [DOI] [PubMed] [Google Scholar]
- 33.Lee CS, Ryan EJ, Doherty GA. Gastro-intestinal toxicity of chemotherapeutics in colorectal cancer: the role of inflammation. World J Gastroenterol 20: 3751–3761, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lignot JH, Helmstetter C, Secor SM. Postprandial morphological response of the intestinal epithelium of the Burmese python (Python molurus). Comp Biochem Physiol A Mol Integr Physiol 141: 280–291, 2005. [DOI] [PubMed] [Google Scholar]
- 35.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104: 487–501, 2001. [DOI] [PubMed] [Google Scholar]
- 36.Lolli G, Johnson LN. CAK-cyclin-dependent activating kinase: a key kinase in cell cycle control and a target for drugs? Cell Cycle 4: 572–577, 2005. [PubMed] [Google Scholar]
- 37.Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K, Matsuda Y. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci USA 103: 18190–18195, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald ER 3rd, El-Deiry WS. Suppression of caspase-8- and -10-associated RING proteins results in sensitization to death ligands and inhibition of tumor cell growth. Proc Natl Acad Sci USA 101: 6170–6175, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misch DW, Giebel PE, Faust RG. Intestinal microvilli: responses to feeding and fasting. Eur J Cell Biol 21: 269–279, 1980. [PubMed] [Google Scholar]
- 40.Morgan DO. Principles of CDK regulation. Nature 374: 131–134, 1995. [DOI] [PubMed] [Google Scholar]
- 41.Muncan V, Faro A, Haramis AP, Hurlstone AF, Wienholds E, van Es J, Korving J, Begthel H, Zivkovic D, Clevers H. T-cell factor 4 (Tcf7l2) maintains proliferative compartments in zebrafish intestine. EMBO Rep 8: 966–973, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen TB, Manova K, Capodieci P, Lindon C, Bottega S, Wang XY, Refik-Rogers J, Pines J, Wolgemuth DJ, Koff A. Characterization and expression of mammalian cyclin b3, a prepachytene meiotic cyclin. J Biol Chem 277: 41960–41969, 2002. [DOI] [PubMed] [Google Scholar]
- 43.Norbury C, Nurse P. Animal cell cycles and their control. Ann Rev Biochem 61: 441–470, 1992. [DOI] [PubMed] [Google Scholar]
- 44.Ott BD, Secor SM. Adaptive regulation of digestive performance in the genus Python. J Exp Biol 210: 340–356, 2007. [DOI] [PubMed] [Google Scholar]
- 45.Overgaard J, Busk M, Hicks JW, Jensen FB, Wang T. Respiratory consequences of feeding in the snake Python molorus. Comp Biochem Physiol A Mol Integr Physiol 124: 359–365, 1999. [DOI] [PubMed] [Google Scholar]
- 46.Petersen CP, Reddien PW. Wnt signaling and the polarity of the primary body axis. Cell 139: 1056–1068, 2009. [DOI] [PubMed] [Google Scholar]
- 47.Piekny A, Werner M, Glotzer M. Cytokinesis: welcome to the Rho zone. Trends Cell Biol 15: 651–658, 2005. [DOI] [PubMed] [Google Scholar]
- 48.Polakis P. Wnt signaling and cancer. Genes Dev 14: 1837–1851, 2000. [PubMed] [Google Scholar]
- 49.Riquelme CA, Magida JA, Harrison BC, Wall CE, Marr TG, Secor SM, Leinwand LA. Fatty acids identified in the Burmese python promote beneficial cardiac growth. Science 334: 528–531, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Secor SM. Digestive physiology of the Burmese python: broad regulation of integrated performance. J Exp Biol 211: 3767–3774, 2008. [DOI] [PubMed] [Google Scholar]
- 52.Secor SM. Evolutionary and cellular mechanisms regulating intestinal performance of amphibians and reptiles. Integr Comp Biol 45: 282–294, 2005. [DOI] [PubMed] [Google Scholar]
- 53.Secor SM. Gastric function and its contribution to the postprandial metabolic response of the Burmese python Python molurus. J Exp Biol 206: 1621–1630, 2003. [DOI] [PubMed] [Google Scholar]
- 54.Secor SM. Physiological responses to feeding, fasting and estivation for anurans. J Exp Biol 208: 2595–2608, 2005. [DOI] [PubMed] [Google Scholar]
- 55.Secor SM, Diamond J. Adaptive responses to feeding in Burmese pythons: pay before pumping. J Exp Biol 198: 1313–1325, 1995. [DOI] [PubMed] [Google Scholar]
- 56.Secor SM, Diamond J. Determinants of the postfeeding metabolic response of Burmese pythons, Python molurus. Physiol Zool 70: 202–212, 1997. [DOI] [PubMed] [Google Scholar]
- 57.Secor SM, Diamond J. Effects of meal size on postprandial responses in juvenile Burmese pythons (Python molurus). Am J Physiol Regul Integr Comp Physiol 272: R902–R912, 1997. [DOI] [PubMed] [Google Scholar]
- 58.Secor SM, Diamond J. A vertebrate model of extreme physiological regulation. Nature 395: 659–662, 1998. [DOI] [PubMed] [Google Scholar]
- 59.Secor SM, Diamond JM. Evolution of regulatory responses to feeding in snakes. Physiol Biochem Zool 73: 123–141, 2000. [DOI] [PubMed] [Google Scholar]
- 60.Secor SM, Fehsenfeld D, Diamond J, Adrian TE. Responses of python gastrointestinal regulatory peptides to feeding. Proc Natl Acad Sci USA 98: 13637–13642, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Secor SM, Lane JS, Whang EE, Ashley SW, Diamond J. Luminal nutrient signals for intestinal adaptation in pythons. Am J Physiol Gastrointest Liver Physiol 283: G1298–G1309, 2002. [DOI] [PubMed] [Google Scholar]
- 62.Secor SM, Ott BD. Adaptive correlation between feeding habits and digestive physiology for boas and python. In: Biology of the Boas and Pythons. Eagle Mountain, UT: Eagle Mountain Publishing, 2007. [Google Scholar]
- 63.Secor SM, Stein ED, Diamond J. Rapid upregulation of snake intestine in response to feeding: a new model of intestinal adaptation. Am J Physiol Gastrointest Liver Physiol 266: G695–G705, 1994. [DOI] [PubMed] [Google Scholar]
- 64.Secor SM, White SE. Prioritizing blood flow: cardiovascular performance in response to the competing demands of locomotion and digestion for the Burmese python, Python molurus. J Exp Biol 213: 78–88, 2010. [DOI] [PubMed] [Google Scholar]
- 65.Shadad AK, Sullivan FJ, Martin JD, Egan LJ. Gastrointestinal radiation injury: symptoms, risk factors and mechanisms. World J Gastroenterol 19: 185–198, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Starck JM, Beese K. Structural flexibility of the intestine of Burmese python in response to feeding. J Exp Biol 204: 325–335, 2001. [DOI] [PubMed] [Google Scholar]
- 67.Sun MY, Wei YY, Yao LY, Xie JH, Chen XN, Wang HZ, Jiang JH, Gu JX. Identification of extracellular signal-regulated kinase 3 as a new interaction partner of cyclin D3. Biochem Biophys Res Commun 340: 209–214, 2006. [DOI] [PubMed] [Google Scholar]
- 68.Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature 411: 349–354, 2001. [DOI] [PubMed] [Google Scholar]
- 69.Tempone AG, Sartorelli P, Mady C, Fernandes F. Natural products to anti-trypanosomal drugs: an overview of new drug prototypes for American Trypanosomiasis. Cardiovasc Hematol Agents Med Chem 5: 222–235, 2007. [DOI] [PubMed] [Google Scholar]
- 70.Wall CE, Cozza S, Riquelme CA, McCombie WR, Heimiller JK, Marr TG, Leinwand LA. Whole transcriptome analysis of the fasting and fed Burmese python heart: insights into extreme physiological cardiac adaptation. Physiol Genomics 43: 69–76, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, Goodwin RG. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 3: 673–682, 1995. [DOI] [PubMed] [Google Scholar]