Abstract
The renin-angiotensin system (RAS) is subject to sex-specific modulation by hormones and gene products. However, sex differences in the balance between the vasoconstrictor/proliferative ACE/ANG II/AT1 axis, and the vasodilator/antiproliferative ACE2/ANG-(1–7)/MAS axis are poorly known. Data in the rat have suggested the male-specific Y-chromosome gene Sry to contribute to balance between these two axes, but why the testis-determining gene has these functions remains unknown. A combination of in silico genetic/protein comparisons, functional luciferase assays for promoters of the human RAS, and RNA-Seq profiling in rat were used to address if regulation of Sry on the RAS is conserved in the homologous X-chromosome gene, Sox3. Both SRY and SOX3 upregulated the promoter of Angiotensinogen (AGT) and downregulated the promoters of ACE2, AT2, and MAS, likely through overlapping mechanisms. The regulation by both SRY and SOX3 on the MAS promoter indicates a cis regulation through multiple SOX binding sites. The Renin (REN) promoter is upregulated by SRY and downregulated by SOX3, likely through trans and cis mechanisms, respectively. Sry transcripts are found in all analyzed male rat tissues including the kidney, while Sox3 transcripts are found only in the brain and testis, suggesting that the primary tissue for renin production (kidney) can only be regulated by SRY and not SOX3. These results suggest that SRY regulation of the RAS is partially shared with its X-chromosome homolog SOX3, but SRY gained a sex-specific control in the kidney for the rate-limiting step of the RAS, potentially resulting in male-specific blood pressure regulation.
Keywords: renin angiotensin system, hypertension, Sox3, Sry, sex differences
the renin-angiotensin system (RAS) is one of the most studied hormonal systems in the physiological regulation of blood pressure through renal and nonrenal mechanisms (16). Dysregulation of the RAS is considered a major factor in the development of cardiovascular pathologies. It is generally subdivided into two major axes that act to maintain homeostasis, with each tissue/system having a unique combination of proteins and peptides (9). These axes are: 1) the vasoconstrictor/proliferative axis of the RAS consisting of the carboxypeptidase angiotensin-I-converting enzyme (ACE), angiotensin II (ANG II), the type 1 angiotensin II receptor (AT1) and 2) the vasodilator/antiproliferative axis consisting of angiotensin I-converting enzyme 2 (ACE2), angiotensin (1–7) [ANG-(1–7)], the MAS receptor. It is now generally accepted that both axes participate in the regulation of blood pressure and metabolism, and, even more importantly, in the pathogenesis of cardiovascular, renal, and metabolic diseases (58). Several reports have shown differences between components such as plasma renin activity and levels of angiotensinogen in human, mouse, and rat models between males and females as previously reviewed in the literature (55). However, few studies have observed and analyzed the effects of sex differences in regulation and balance of the two axes of the RAS (26).
Studies of Y-chromosome consomic rats first suggested a contribution of the male-specific chromosome to elevate blood pressure (21), with characteristics in crosses of the consomic rats to the testicular feminized rats suggesting an interplay between genes on the Y chromosome and androgen signaling (20). Dissection of the Y chromosome elucidated the Sry gene as a candidate to elevate blood pressure in the rat (22, 65). Part of the blood pressure elevation in the rat was due to changes in the RAS (19, 40), a regulation that is seen by human SRY on human RAS promoters (52, 53). Electroporation of Sry into the kidney of normotensive rats elevates plasma renin (REN) activity (19), an activity that is known to be elevated in human males relative to females (28). Evidence points to a combination of sex hormones and SRY regulating promoter activity of RAS genes, suggesting an elevation of the ACE/ANG II/AT1 axis and a depression of the ACE2/ANG-(1–7)/MAS axis (19, 23, 38, 40, 52, 53). As SRY is primarily known for its role in testis determination (4), the regulation of the RAS and blood pressure was not initially expected.
SRY is a member of the SOX transcription factor family that plays important roles in regulating cell specialization and differentiation (29). The SOX family is composed of 20 human transcription factors, all sharing a homologous high-mobility group (HMG) box domain first discovered in the SRY protein (6). The HMG box of SOX transcription factors binds to DNA with sequence specificity, resulting in both conserved transcription factor recruitment (50) and architectural modification by inducing a bend to DNA (49). Sry evolved from the Sox3 gene (12, 59), which in human is found on the X chromosome. Thus Sry could have either maintained the regulation of the RAS genes from Sox3 or gained this function de novo. With recent elucidation of critical genes maintained on the Y chromosome (3), understanding the impact these genes have on various disease phenotypes is of critical importance, especially with a focus on similarities and differences between the Y-chromosome genes and their homologous counterpart on the X chromosome. We hypothesize that there is preserved promoter regulation between SRY and SOX3 on the RAS genes, with some possibility of sex differences in the regulation of RAS components and ultimately blood pressure regulation. Therefore, the aim of this study was to further evaluate mechanisms of action, especially in the promoter regions of the RAS genes that were previously shown to be modulated by SRY.
MATERIALS AND METHODS
Sry and Sox3 in silico comparisons.
Nucleotide sequences for Sry (321 total mammalian sequences) and Sox3 (34 total vertebrate sequences from human to zebrafish) were aligned with ClustalW codon in MEGA5 (62). Phylogenetic analysis was performed with all Sry and Sox3 sequences to confirm none of the Sry sequences clustered within the SOX3 sequences. The test statistic dN-dS was used for detecting codons that have undergone selection, where dS is the rate of synonymous substitutions and dN is the rate of nonsynonymous substitutions. A positive value for the test statistic indicates an overabundance of nonsynonymous substitutions. The normalized dN-dS values were calculated for each codon, using the joint maximum likelihood reconstructions of ancestral states under a Muse-Gaut model (45) of codon substitution and Tamura-Nei model (61) of nucleotide substitution. Based on sequence alignments, those amino acids conserved in all SRY and SOX3 sequences were colored red, and those conserved in only SOX3 but not SRY in cyan onto the structure of SRY-DNA (pdb 1j46). Amino acids were swapped in the HMG box of SRY (using pdb 1j46) for those of SOX3. Models for both SRY and SOX3 interaction with DNA were then energy minimized with the AMBER03 force field (15) with a pKa of 7.4, water density of 0.997 g/ml, and 0.9% NaCl mass fraction in YASARA (32). Molecular dynamic simulations were performed for 3.25 nanoseconds.
Promoter, evolutionary conserved region, and transcription factor binding site analysis.
SOX binding sites in each promoter were detected using Genomatix MatInspector (8). All promoters cloned except the ACE promoter contain predicted SOX binding sites. Evolutionary conserved region (ECR) analysis was performed in the ECR browser (48) using the parameters listed in the figure legends for each analysis. Conserved regions were then analyzed for conserved transcription factor binding sites between human and rat using RVISTA (37) for all vertebrate transcription factor binding sites.
Cloning.
The pEF1 SOX effector plasmids cloning strategy was previously published (52). Details for the various human pGL3 luciferase reporter constructs were also previously published (53). Starting with the human REN(−1444/+8) pGL3 reporter construct, we sequentially removed the three SOX binding sites, resulting in the REN(−1444/+8) SOX mut reporter vector; i.e., the first site was removed followed by sequence confirmation, then the second site, and finally the third site. This was performed with phosphorylated primers flanking each SOX binding site (Table 1), followed by PCR with Phusion Hot Start II polymerase (Fermentas) and vector recircularized with T4 ligase. We used the following two strategies to assess SOX binding sites in the MAS promoter: 1) varying the size of the promoter insert or 2) mutating a specific SOX binding site. Both were performed using primer-directed mutagenesis (Table 1). All constructs were sequence confirmed using BigDye Sanger sequencing on an ABI 3130xl genetic analyzer (Applied Biosystems). The SRY-MAS electrophoresis mobility shift assay was performed as previously published (52).
Table 1.
Primers used for mutagenesis of luciferase promoters
| Construct Name | Primer Name | Primer Sequence |
|---|---|---|
| Renin SOXdel1 | R1-RenSoxdel | 5′-CAT CAG GTT TGA CTT ACA GG-3′ |
| L1-RenSoxdel | 5′-CTT GAA TGT AAT CAG ACA CAG-3′ | |
| Renin SOXdel2 | R2-RenSoxdel | 5′-TGT TAA AGA TGT ATA GGA AC-3′ |
| L2-RenSoxdel | 5′-TGT CTG TAT ATT TCC TTA TC-3′ | |
| Renin SOXdel3 | R3-RenSoxdel | 5′-TGA AAC CCC ATC TCC ACT AA-3′ |
| L3-RenSoxdel | 5′-AGG CTG GTC TGG AAC T-3′ | |
| MAS(−1123/+4) | R-MAScleavage | 5′-GGC TAG CAC GCG TAA GAG CTC G-3′ |
| L3-MAScleavage | 5′-GGA GTT CAA GAC CAG CCT CAC CAA CA-3′ | |
| MAS(−377/+4) | R-MAScleavage | 5′-GGC TAG CAC GCG TAA GAG CTC G-3′ |
| L1-MAScleavage | 5′-AGA ACT GGA AAT GGG TCC CGC ACA-3′ | |
| MutSOX-1855 | R-MASSoxdel1 | 5′-CAA CGG CCC TCA AAC TGA TGT CAG TGC GTG AA-3′ |
| L-MASSoxdel1 | 5′-CTG TTG CCG CAT TTT GCT TGA CTG CCT CCC TAC-3′ | |
| MutSOX-86 | R-MASSoxdel7 | 5′-AAA GGG AGA AAT TCA GCT ATA AAT CAT AAT CTT CAT G-3′ |
| L-MASSoxdel7 | 5′-TAT TCC AAT TCG GCG ATT TTC ATG GCT TTT TGT GTT TGT T-3′ | |
| RecSOX-86 | R-MASSoxdel7W | 5′-AAA GGG AGA AAT TCA GCT ATA AAT CAT AAT CTT CAT G-3′ |
| L-MASSoxdel7W | 5′-TAT TCC AAT TCA ACA ATT TTC ATG GCT TTT TGT GTT TGT T-3′ | |
| Mut2SOX-86 | R-MASSoxdel8 | 5′-AAA GGG AGA AAT TCA GCT ATA AAT CAT AAT CTT CAT G-3′ |
| L-MASSoxdel8 | 5′-TAT TCC AAT TCA ACT TGT TTC ATG GCT TTT TGT GTT TG-3′ |
To make each of the mutated promoter constructs the primers in this table were used. The 1st column shows the construct name as used in the study, the 2nd column the individual primer names, and the 3rd column the primer sequences. For mutations, the SOX binding site is underlined and bolded with the DNA bases to be mutated shown in italics.
Cotransfection and luciferase assays in Chinese hamster ovary cells.
Twenty-four hours prior to transfection of Chinese hamster ovary (CHO) cells, 5 × 104 cell/well were seeded on 24-well plates and incubated overnight in Ham's F12K medium (Sigma) supplemented with 10 mM HEPES and 10% fetal bovine serum (Atlanta Biologicals) in a humidified atmosphere at 37°C and 5% CO2. We selected CHO cells for the analysis for previously suggested reasons (53). Each well was transiently cotransfected with 50 ng effector plasmid (pEF1 vector), 500 ng firefly luciferase reporter construct (pGL3 vector), and 500 pg of control Renilla vector. Transfections with both SRY and SOX3 together were performed by using 50 ng effector plasmid total: for example, 25 ng of SRY with 25 ng of SOX3 for SRY+SOX or 25 ng of SRY with 25 ng of empty pEF1 vector for SRY alone. Luciferase assays were performed using the Stop & Glo luciferase kit (Promega) according to the manufacturer. Statistical analysis was performed on the SE of three or four independent experiments followed by Student's t-test to determine significant differences (P ≤ 0.05) relative to the experimental control.
Sry and Sox3 expression in the rat.
Initial work in the Fisher 344 Rat BodyMap project (71) did not annotate either Sry or Sox3 transcripts. Therefore, we used the Sry (EU984075.1) and Sox3 (XM_006227591.2) sequences from the rat to BLAST reads from the various Sequence Read Archive (SRA) datasets of the Rat BodyMap project (35). BLAST (2) was performed only on reads that were 98% homologous with an E-value better than 1e-20. The number of positive reads was converted into the RPKM, reads per kilobase per million [(# of mapped reads)/(length of transcript in kilobases)/(million mapped reads)]. For the global expression profile, all SRA files for an individual tissue were pooled together for BLAST analysis (16 total), only separated by the sex of the animal (male vs. female). Three tissues were picked (testis, brain, and kidney) for profiling the error between animals (n = 4) and at each of four ages. The error was calculated using the SE. Primate SRY (NM_003140.2, KP141769.1, NM_001032836.1, DQ977342.1, EU294189.1) and SOX3 (NM_005634.2, XM_004065361.1, NM_001193752.1, XM_001143467.3, XM_005227560.1) sequences were analyzed for SRA datasets of the kidney (SRX081994, SRX081951, SRX081930, SRX081966, SRX196347) and testis (SRX081999, SRX081954, SRX081933, SRX081969, SRX358238) for RPKM.
RESULTS
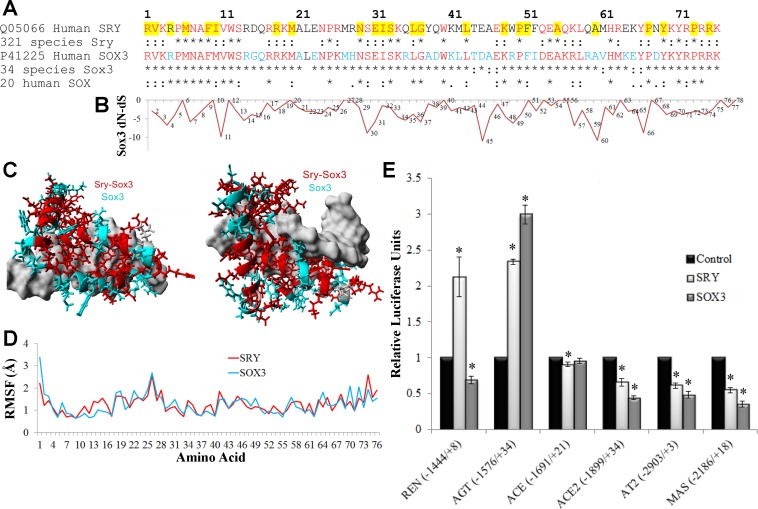
SRY has rapidly evolved from SOX3, resulting in differences in all domains outside of the DNA binding HMG box. Among 34 species sequences for the SOX3 HMG box (representing a wide range of vertebrates) only one of the 76 amino acids was not conserved (Fig. 1A). However, in the 321 sequences of SRY (found only in mammals) 26 amino acids are not conserved. The larger number of SRY sequences, compared with SOX3, represents the previous resources invested in Sanger sequencing Sry throughout mammals primarily for Y-chromosome phylogenetic work, while the few Sox3 sequences are mostly from whole genomes sequences of species. To address whether SRY and SOX3 DNA binding sites remain similar, codons of Sox3 that are under selective pressure (more negative dN-dS, Fig. 1B) were identified. Amino acids 11, 30, 45, 60, and 66 have dN-dS ∼−10 and are conserved in all or most of the SRY sequences. When conservation of amino acids for SRY and SOX3 sequences are mapped onto the protein structure, a highly conserved DNA contact is revealed (Fig. 1C), suggesting both proteins bind to DNA with similar sequence specificity. Modeling the SOX3 protein reveals similar molecular dynamics for all amino acids between SRY and SOX3 (Fig. 1D).
Fig. 1.
Functional conservation in SRY and SOX3. A: sequence alignment of the high-mobility group (HMG) box of human SOX3 and SRY with conservation shown below each in multiple species. Those sites marked with an asterisk (*) are 100% conserved in all species and those with a colon (:) are functionally conserved. Amino acids in red are conserved in all SRY and SOX3, while those in cyan are conserved in SOX3 only. Amino acids highlighted in yellow are those in which mutations in SRY are associated with sex reversal (50). B: normalized values for each amino acid's (x-axis) nonsynonymous rate (dN) minus the synonymous rate (dS) for the 34 sequences of Sox3. Values more negative suggest codon selection. C: known structure of SRY bound to DNA (pdb file 1j46) with the amino acids conserved among all SRY and SOX3 sequences shown in red and those conserved in SOX3 but not SRY shown in cyan. DNA is shown in gray. D: root-mean squared fluctuation (RMSF) for each amino acid over a 3.25 nanosecond molecular dynamic simulation for the known structure of SRY (red) or the modeled SOX3 (cyan) indicate similarities of the 2 proteins. E: the pEF1 expression vectors for either human SRY (light gray) or SOX3 (darker gray) were transfected into CHO cells along with a pGL3 vector containing the human promoter (shown on the bottom) of various RAS genes driving the production of luciferase. Each sample was normalized to an empty vector control of pEF1 (black). Error bars are shown as the SE. Each sample is compared with the control for each promoter. *Significant differences (P ≤ 0.05).
Utilizing the previously designed luciferase vectors controlled by the various RAS promoters, we tested SRY and SOX3 for promoter regulations (Fig. 1E). The promoter of AGT was upregulated by both SRY and SOX3, while the promoters of ACE2, AT2, and MAS were downregulated by both SRY and SOX3. The only promoter that was differentially regulated was that of the gene REN, which was upregulated by SRY and downregulated by SOX3.
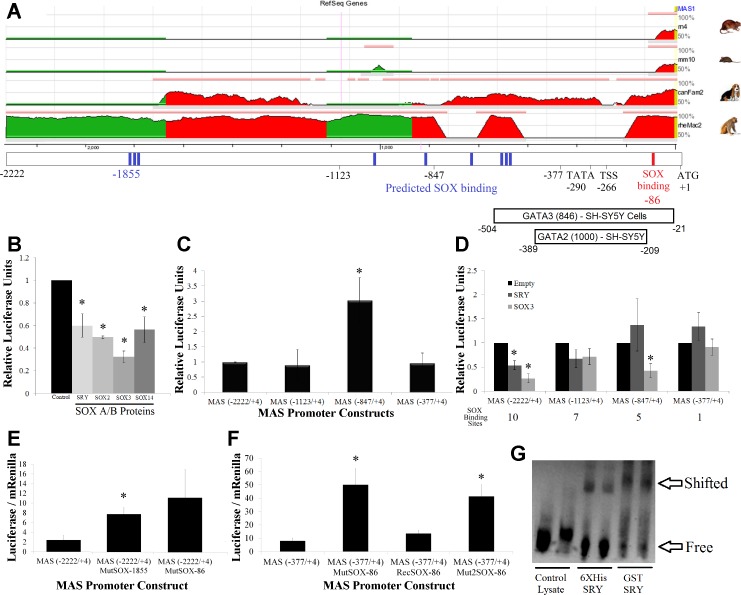
The MAS promoter was previously shown to be regulated by multiple SOX subfamily members and shown to be directly bound by SRY (52); therefore, we elected to characterize the shared binding sites between SRY and SOX3 on this promoter. The MAS promoter contains no ECRs, with sequence divergence in all species for the promoter and the 5′-untranslated region (UTR) (Fig. 2A), reducing our ability to use evolutionary genomics to decipher the molecular binding sites. Therefore, the binding sites for SOX were tested using two different approaches: 1) a cleavage series to generate MAS promoters of various sizes and 2) point mutations within two of the SOX binding sites. The full MAS promoter used [MAS(−2222/+4)] contains 10 predicted SOX binding sites. This full-length construct is regulated by all of the SOX A/B protein family members (Fig. 2B). Cleaving part of the MAS promoter such that there are seven possible SOX binding sites [MAS(−1123/+4)] or one possible binding site [MAS(−377/+4)] had little effect on relative luciferase production (Fig. 2C), with the exception being the five possible binding site MAS(−847/+4) constructs that had significant upregulation of the promoter. The addition of SRY or SOX3 to the cells treated with the various sized promoter constructs showed a complete loss of SRY/SOX3 repression only on the shortest of the constructs [MAS (−377/+4), Fig. 2D], suggesting that the cluster of SOX binding sites between −847 and −377 are important for promoter regulation.
Fig. 2.
Functional SOX binding sites in the MAS promoter. A: evolutionary conserved region (ECR) browser analysis of the MAS pGL3 promoter construct for the rat (rn4), mouse (mm10), dog (canFam2), and monkey (rh2Mac2). Regions in red and green share conservation with human based on 20 bp stretches sharing >50% homology. Below the ECR is the location of various landmarks of the promoter vector such as sites used to create cleavage series (−2222, −1123, and −377), predicted SOX binding sites (blue), in vitro confirmed SOX binding site (red), the location of the TATA box (−290), proposed transcriptional start site (TSS, −266), translational start site (ATG, +1), and ENCODE transcription factor binding with relative score and cell line of observation. B: the human MAS promoter treated with various SOX A/B proteins, all of which significantly repress the promoter. C: luciferase production of 4 MAS promoter constructs with differing lengths, all made relative to the full-length MAS (−2222/+4) construct regulation level. D: promoter analysis of 4 MAS promoter constructs transfected with control (black), SRY (light gray) or SOX3 (dark gray) vectors. Total number of SOX binding sites in each construct is shown below the graph. E: mutations to the MAS(−2222/4) promoter construct at 2 of the 10 SOX binding sites (−1855 or −86) result in elevation of promoter activity. Data are shown as the raw luciferase-Renilla ratio. F: as the MAS(−2222/+4)MutSOX-86 showed a trend in altering regulation and was previously shown to be an in vitro binding site for SRY, the site was investigated with the smaller MAS minimal promoter, MAS(−377/+4). The SOX binding site was mutated to 2 separate sequences, MAS(−377/+4)MutSOX-86 and MAS(−377/+4)Mut2SOX-86, resulting in elevation of promoter activity. One of the mutations was repaired to recover the SOX binding site, MAS(−377/+4)RecSOX-86, confirming only this site was altered in the promoter mutagenesis. G: the sequence for the SOX binding site at −86 was synthesized with a biotin tag and EMSA performed using both His- and GST-tagged Sry proteins, confirming that SOX proteins can bind to this element. Error bars for all graphs are shown as the SE. Each sample is compared with the control for each promoter. *Significant differences (P ≤ 0.05) relative to the control.
To assess if the SOX binding sites outside of the −847 and −377 region are also critical to MAS promoter regulation but were missed because of the nature of promoter cleavage series, we performed mutations at the distal (−1855) triple SOX binding site and the proximal SOX binding site (−86). Both mutations result in upregulation of the MAS promoter in CHO cells, with significance seen with the mutation at −1855 and a trend seen at −86 (Fig. 2E). To further validate the in vitro confirmed SOX binding site in MAS at −86, we used the short MAS promoter construct [MAS(−377/+4)], again mutating the SOX binding element, recovering the mutations, and mutating it to a different sequence than the first mutation (Fig. 2F). Mutation at the SOX binding site for both changes (MutSOX-86 and Mut2SOX-86) resulted in around fivefold upregulation of the MAS promoter, while recovery of the mutation (RecSOX-86) yielded similar values as the wild-type MAS promoter. DNA probes for the MAS promoter containing this SOX binding site can be bound by both 6X His-tagged and GST-tagged SRY protein (Fig. 2G). The data on the MAS promoter indicate that SRY and SOX3 regulate the MAS promoter in cis through a combination of multiple SOX binding sites. The ENCODE data that the MAS promoter is regulated through the GATA 2/3 transcription factors reaffirm our interests of the SOX gene regulation, as many promoters have been shown to contain competitive SOX and GATA mechanisms (34, 42, 43).
Moving from MAS to the other promoters of renin angiotensin system, for two of the promoters that are similarly regulated by SRY and SOX3, the next step was to determine if both proteins operate by the same mechanism or if they result in synergistic or additive regulation. The upregulated AGT promoter had a concentration-dependent activation by SRY and SOX3, with SRY&SOX3 simultaneous treatment resulting in a similar activation as SRY or SOX3 alone (Fig. 3A). The downregulated promoter of ACE2 was also regulated in a concentration-dependent manner with SRY&SOX3 again resulting in a similar activation as SRY or SOX3 alone (Fig. 3B). Both results suggest that SRY and SOX3 regulation have an overlapping mechanism in promoter control.
Fig. 3.
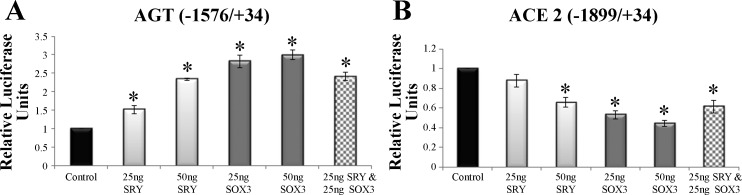
Similar mechanisms of regulation by SRY and SOX3 on promoters of AGT and ACE2. A: activation of the AGT promoter with 2 concentrations of transfected SRY (light gray) or SOX3 (dark gray). In addition, a double transfection of SRY and SOX3 (checkered) shows nonadditive activation suggesting shared mechanisms of promoter control. B: repression of the ACE2 promoter with 2 concentrations of transfected SRY (light gray) or SOX3 (dark gray). In addition a double transfection of SRY and SOX3 (checkered) shows nonadditive repression, suggesting shared mechanisms of promoter control. Error bars are shown as the SE. Each sample is compared with the control for each promoter. *Significant differences (P ≤ 0.05).
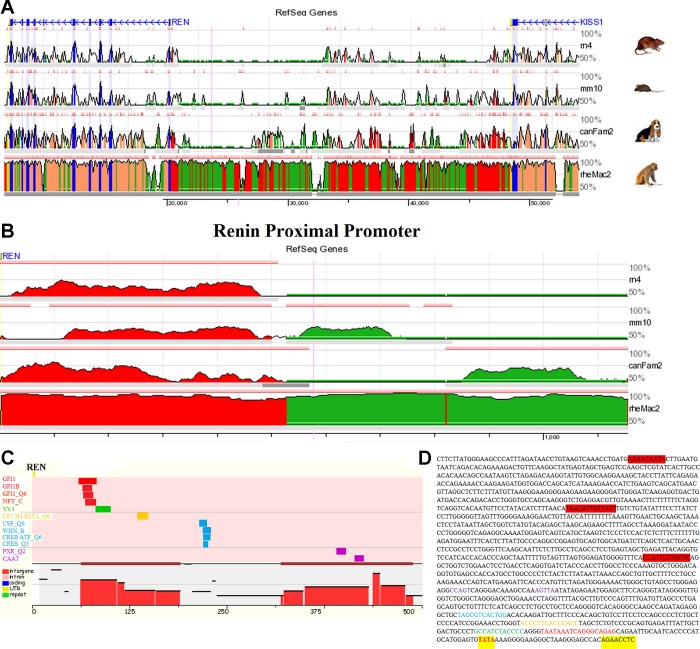
To address possible mechanistic insight into the different regulation of the Ren promoter by SRY and SOX3, an analysis of conserved transcription factor binding sites was performed on the Ren promoter to determine if there were any Sox consensus binding sequences (Fig. 4). The 5′-region of the Ren gene is located ∼30 kB from its close syntenic gene KISS1, with very few conserved ECRs located in the intergenic space (Fig. 4A). Addressing the proximal promoter of Ren, we found an ∼500 bp ECR (Fig. 4B) to contain 12 conserved transcription factor binding sites (Fig. 4C), none of which are SOX binding elements. The REN(−1444/+8) promoter driving our luciferase production contains a total of three SOX binding elements located >1 kB from the transcriptional start site (TSS), outside the ECR (Fig. 4D). A TATA box is located 29 bps away from the TSS, suggesting proper distance for transcriptional initiation (Fig. 4D).
Fig. 4.
Promoter analysis of the gene renin (Ren). A: ECR browser analysis of the intergenic space (roughly 30 kB) between Renin (REN) and the syntenic KiSS-1 metastasis-suppressor gene (KISS1) for the rat (rn4), mouse (mm10), dog (canFam2), and monkey (rh2Mac2). Regions in blue are protein coding exons, yellow are untranslated regions (UTRs), and those in red and green share conservation with human based on 20 bp stretches sharing ≥ 90% homology. B: ECR browser analysis for the proximal promoter of REN with regions in red and green showing conservation of 50 bp stretches with 50% homology. C: the large ECR in the proximal promoter of REN was analyzed for shared transcription factor (TF) binding sites between human and rat using the RVISTA software selecting for all possible TFs. D: the sequence of REN(−1444/+8) pGL3 promoter construct identifying the TATA box (red, highlighted yellow) located within the proper distance of the 5′-UTR (highlighted yellow). Additionally the conserved TF sites are colored relative to that in C. The 3 potential SOX binding sites (highlighted in red) fall outside of the ECR.
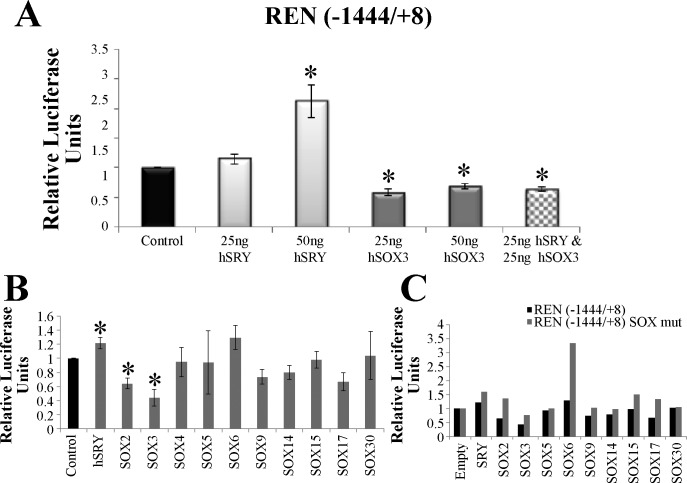
Delivery of SRY only at 50 ng, and not 25 ng, significantly upregulated the Ren promoter, while both concentrations of SOX3 result in downregulation of the promoter (Fig. 5A). Surprisingly, delivery of SRY&SOX3 results in a repression similar to SOX3 alone (Fig. 5A), suggesting that regulation mechanisms differ between SRY and SOX3 on the Ren promoter. To further understand this difference, we tested the REN(−1444/+8) promoter with members of each SOX subfamily (Fig. 5B). The promoter was significantly regulated by SRY (upregulated), SOX2 (downregulated), and SOX3 (downregulated). It should be noted that SOX2 and SOX3 are both members of the SOXB1 subfamily, with highly conserved sequence and function (30). Removal of all three SOX binding elements in this promoter construct resulted in an increase in promoter activity for all SOX constructs (except SOX30) with complete loss of promoter repression seen in the SOX2 and SOX3 treated cells (Fig. 5C). This suggests that Ren promoter repression seen by SOX3 and SOX2 is a cis-acting function, while the activation by SRY is likely a trans-acting function.
Fig. 5.
Regulation of the REN promoter by various SOX proteins. A: activation of the REN promoter with 2 concentrations of transfected SRY (light gray) or repression by 2 concentrations of SOX3 (dark gray). In addition a double transfection of SRY and SOX3 (checkered) showed downregulation of the promoter similar to that of SOX3 transfected cells. B: transfection of CHO cells with either the control pEF1 vector (black) or 1 of the SOX pEF1 constructs (gray) along with the pGL3 REN(−1444/+8) luciferase vector. Error bars are shown as the SE. Each sample is compared with the control for each promoter. *Significant differences (P ≤ 0.05). C: the REN(−1444/+8) promoter construct was mutated to remove each of the 3 SOX binding sites; this construct was named REN(−1444/+8) SOX mut. The mutant pGL3 construct (gray) was transfected with each of the SOX pEF1 expression vectors resulting in an increase in promoter activity relative to the wt pGL3 promoter construct (black) for all but the SOX30 vector (n = 1).
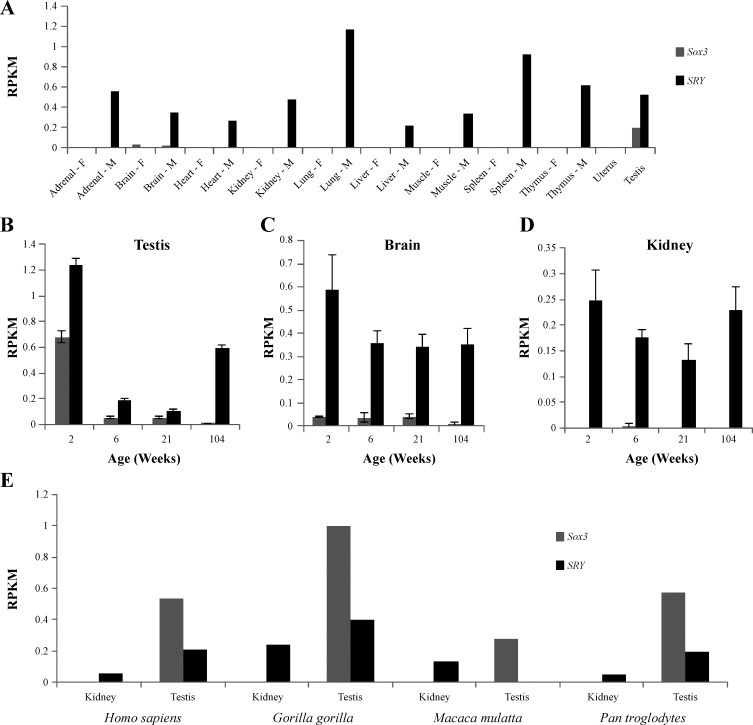
To understand the context for SRY and SOX3 regulation of the RAS, we performed an analysis of tissue expression in the rat, an organism with extensive work done to characterize the protein products of the RAS (51). Using unannotated (for Sry and Sox3) RNA-Seq datasets from the rat BodyMap project (71), we determined Sry and Sox3 expression profiles using 320 RNA-Seq experiments for 10 tissues of male and female Fisher 344 rats (Fig. 6). Global expression showed identifiable transcripts for Sry in all male tissues; however, Sox3 transcripts were detected only in male and female brain and the male testis (Fig. 6A). The testis (Fig. 6B) and brain (Fig. 6C) both had expression of Sry and Sox3 in the four age groups of male rats, while in the kidney only Sry transcripts were detected (Fig. 6D). The kidney is the primary source for Ren found in the blood and is thus the top tissue of interest for Ren regulation control in blood pressure. The rat expression profile for Sox3 closely matches that of the human protein atlas for human SOX3 (http://www.proteinatlas.org/ENSG00000134595-SOX3/tissue). In some of the human protein atlas RNA and protein expression samples, SRY was detectable in the kidney (http://www.proteinatlas.org/ENSG00000184895-SRY/tissue/kidney#img). We therefore tested primate expression of SRY and SOX3 using publically available SRA datasets for RNA-Seq of kidney and testis. While SRY and SOX3 were both detected in the testis of primates, only SRY could be detected in the kidney (Fig. 6E).
Fig. 6.
Expression profile for Sry and Sox3 in the rat using data from the Rat BodyMap project. Expression profile for Sox3 (gray) and Sry (black) in female (F) and male (M) RNA-Seq data for 10 tissues in the Fisher 344 rat (A). Each tissue is a pool of 16 individual RNA-Seq experiments (4 animals at 4 ages). Data are shown as reads per kilobase per million (RPKM). While Sry is ubiquitously expressed in male only, Sox3 transcripts are found at all 4 ages (2, 6 21, and 104 wk old rats) in the male testis (B) and brain (C). The kidney showed expression of Sry at all 4 ages with no detectable Sox3 reads (D). Error bars for the individual tissues (B–D) represent the SE of 4 independent RNA-Seq datasets at each age. E: analysis of primate (Homo sapiens, Gorilla gorilla, Macaca mulatta, Pan troglodytes) kidney and testis SRA datasets for expression of SOX3 (gray) or SRY (black). Expression of both SRY and SOX3 are seen in testis, while only SRY expression is found in kidney.
DISCUSSION
There are sex differences in the prevalence of many conditions including cardiovascular diseases, asthma, autoimmune disorders, birth defects, neurological disorders, and cancers (47). Of these diseases, the RAS has been associated with the cardiovascular, respiratory, neurological, reproductive, and immune systems along with cancer progression (24, 27, 41, 54, 68). Sry transcripts/proteins have been identified in all of these tissue types in the rat and in the human (64, 67). Part of the sex disparities for blood pressure is due to hormonal contributions (57), but additional evidence points to sex chromosome contributions (10, 11, 60). Previous research from our group has established animal models for Y chromosome-specific elevation of blood pressure (21) and behavior alteration (14, 63) in the rat, narrowing down the entire Y chromosome to the Sry gene (19). This has opened up the question of whether SRY function in blood pressure control might be conserved from the divergence of the mammalian X-chromosome gene SOX3 or whether the regulation is novel to SRY.
Genes on the Y chromosome evolve much faster than their X-chromosome homologs or other autosomal genes due to a loss of crossover, the founder effect, and increased mutagenesis in the testis (25). In our data we confirm this, with SOX3 protein sequences from a broad range of vertebrates highly conserved for the HMG box; however, the HMG box of SRY found in a small subset of vertebrates (mammals) has a higher rate of divergence (Fig. 1). Still, structural and functional assessment of these sequences confirms that DNA binding through the HMG box is similar between SRY and SOX3, maintaining the critical amino acids for proper DNA contact, with sequence divergence between SRY and SOX3 found at amino acids that do not contribute to the folding of the domain or DNA contact.
The solid number of studies pointing to sex-specific differences in RAS modulation (26) and our previous data showing SRY regulation of the RAS genes both in vitro (40, 53) and in vivo (19), in combination with the conserved DNA binding between SRY and SOX3, lead us to hypothesize that the promoter regions of the RAS genes may be regulated by both SRY and SOX3 transcription factors, which may potentially contribute to sex differences in blood pressure regulation. The promoter of AGT was upregulated by both SRY and SOX3, while the promoters of ACE2, AT2, and MAS were downregulated by both SRY and SOX3. The only promoter that was differentially regulated was that of REN, which was upregulated by SRY and downregulated by SOX3. This suggests that SRY has gained a novel protein function from its homolog, SOX3, in trans-regulating Ren (or loss of cis-regulation seen by SOX3) while maintaining the regulation of genes such as AGT, ACE2, AT2, and MAS. As many of the experiments shown in this study are based on luciferase promoter constructs, the size of these constructs may influence the outcomes of the SRY/SOX3 regulation with potential regulation left out of the construct design. However, the constructs as designed show promoter regulation that is supported by the in vivo Sry electroporation studies in the rat (19) and thus likely contain the minimal promoter for Sry regulation in vivo. It is also possible that regions outside of the HMG box of SRY and SOX3 control the differences seen in regulation of Ren. Ren, primarily produced in the kidney, is the rate-limiting step of the RAS. Thus the activation by SRY, which is expressed in the kidney, might contribute to elevation of blood pressure through increased ANG II peptide; however, SOX3 does not appear to regulate this step. A decrease in the ANG-(1–7) arm of the RAS maintained in SRY- and SOX3-treated cells further potentiates blood pressure elevation control by SRY. The androgen receptor is known to directly interact with SRY and regulate gene promoters (70), suggesting potential for synergistic effects between Sry and androgen signaling to contribution to sex differences in blood pressure.
The rat is a well-established model organism for understanding the contributions of the Y chromosome on phenotype. Almost 25 years ago, crosses of the Y chromosome from the spontaneously hypertensive rat (SHR) (21) and the SHR stroke prone (13) with the normotensive WKY rat resulted in blood pressure elevation, while the reciprocal crosses reduces blood pressure in the SHR rat. Crosses of the Brown Norway (BN) Y chromosome onto the SHR were also able to reduce blood pressure (31). Recently, crosses for the Y chromosome from the salt-sensitive rat, with salt-dependent blood pressure increases, to the normotensive BN altered kidney function (39). These rat consomic animals establish needed model organisms to translate the literature of human Y-chromosome contributions to blood pressure (11, 18, 60). Details in this article provide the first molecular mechanisms for differences in homologous transcription factors found on the X or Y chromosome to contribute to sex differences in blood pressure regulation.
In the rat and human, Sry is expressed in numerous tissues including testis, brain, heart, and kidney (64); however, Sox3 expression appears to be limited to the brain and testis. In addition to testis development (69), Sox3 has been proposed to be critical for brain development, particularly for the hypothalamic-pituitary-adrenal axis (HPA) (46, 56). In the brain, proteins of the RAS have been shown to contribute to the HPA (36), an axis with known sex differences (33, 66). In the classical RAS, ANG II acts on the pituitary to release vasopressin and ACTH. These pituitary cell lineages are developmentally regulated by SOX3 (1), such that they are likely activated by the ACE/ANG II/AT1 axis of the RAS. In concert, the ACE 2/ANG-(1–7)/MAS axis of the RAS is repressed in the pituitary (16). It is of interest to note that in the rat models of Y-chromosome crosses, behavioral differences have also been seen in males (14). With the brain RAS shown to be involved in neurological development (5, 44) and RAS inhibitors given during pregnancy associated with mental delays in the offspring (7), our data also suggest a potential impact on sex differences in brain development through the RAS regulation.
In summary, the fact that SOX3 functions in the brain combined with the documented literature above on RAS in brain development leads us to suggest that regulation of multiple RAS proteins in the brain was inherited and selected on following the X/Y split in mammals, with SRY gaining additional expression in tissues such as the kidney and trans regulation of genes such as Renin resulting in a male-specific modulation of blood pressure.
GRANTS
This work was supported by American Heart Association Grant 11PRE7380033 and National Institutes of Health Office of the Director Grant 1K01ES-025435 to J. W. Prokop, Brazilian National Institute of Science and Technology in Hormones and Women's Health to R. A. S. Santos, and the Federal University of Minas Gerais.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: F.C.A., A.M., H.L.D.P., R.A.S.S., F.M.R., and J.W.P. conception and design of research; F.C.A., I.K.M.W., and J.W.P. performed experiments; F.C.A., I.K.M.W., and J.W.P. analyzed data; F.C.A., A.M., H.L.D.P., R.A.S.S., J.L., F.M.R., and J.W.P. interpreted results of experiments; F.C.A. and J.W.P. prepared figures; F.C.A. and J.W.P. drafted manuscript; F.C.A., A.M., I.K.M.W., H.L.D.P., R.A.S.S., J.L., F.M.R., and J.W.P. edited and revised manuscript; F.C.A., A.M., I.K.M.W., H.L.D.P., R.A.S.S., J.L., F.M.R., and J.W.P. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Leming Shi of Fudan University for approval and help in utilizing the Rat BodyMap datasets.
REFERENCES
- 1.Alatzoglou KS, Kelberman D, Dattani MT. The role of SOX proteins in normal pituitary development. J Endocrinol 200: 245–258, 2009. [DOI] [PubMed] [Google Scholar]
- 2.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 215: 403–410, 1990. [DOI] [PubMed] [Google Scholar]
- 3.Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, Koutseva N, Zaghlul S, Graves T, Rock S, Kremitzki C, Fulton RS, Dugan S, Ding Y, Morton D, Khan Z, Lewis L, Buhay C, Wang Q, Watt J, Holder M, Lee S, Nazareth L, Rozen S, Muzny DM, Warren WC, Gibbs RA, Wilson RK, Page DC. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508: 494–499, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berta P, Hawkins JB, Sinclair AH, Taylor A, Griffiths BL, Goodfellow PN, Fellous M. Genetic evidence equating SRY and the testis-determining factor. Nature 348: 448–450, 1990. [DOI] [PubMed] [Google Scholar]
- 5.Bodiga VL, Bodiga S. Renin angiotensin system in cognitive function and dementia. Asian J Neurosci 2013: e102602, 2013. [Google Scholar]
- 6.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol 227: 239–255, 2000. [DOI] [PubMed] [Google Scholar]
- 7.Bullo M, Tschumi S, Bucher BS, Bianchetti MG, Simonetti GD. Pregnancy outcome following exposure to angiotensin-converting enzyme inhibitors or angiotensin receptor antagonists: a systematic review. Hypertension 60: 444–450, 2012. [DOI] [PubMed] [Google Scholar]
- 8.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942, 2005. [DOI] [PubMed] [Google Scholar]
- 9.Chappell MC. Nonclassical renin-angiotensin system and renal function. Compr Physiol 2: 2733–2752, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charchar FJ, Bloomer LD, Barnes TA, Cowley MJ, Nelson CP, Wang Y, Denniff M, Debiec R, Christofidou P, Nankervis S, Dominiczak AF, Bani-Mustafa A, Balmforth AJ, Hall AS, Erdmann J, Cambien F, Deloukas P, Hengstenberg C, Packard C, Schunkert H, Ouwehand WH, Ford I, Goodall AH, Jobling MA, Samani NJ, Tomaszewski M. Inheritance of coronary artery disease in men: an analysis of the role of the Y chromosome. Lancet 379: 915–922, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charchar FJ, Tomaszewski M, Padmanabhan S, Lacka B, Upton MN, Inglis GC, Anderson NH, McConnachie A, Zukowska-Szczechowska E, Grzeszczak W, Connell JMC, Watt GCM, Dominiczak AF. The Y chromosome effect on blood pressure in two European populations. Hypertension 39: 353–356, 2002. [DOI] [PubMed] [Google Scholar]
- 12.Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, Norris D, Rastan S, Stevanovic M, Goodfellow PN, Lovell-Badge R. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development 122: 509–520, 1996. [DOI] [PubMed] [Google Scholar]
- 13.Davidson AO, Schork N, Jaques BC, Kelman AW, Sutcliffe RG, Reid JL, Dominiczak AF. Blood pressure in genetically hypertensive rats. Influence of the Y chromosome. Hypertension 26: 452–459, 1995. [DOI] [PubMed] [Google Scholar]
- 14.Dickey C, Toot J, Terwilliger M, Payne R, Turner M, Ely D. The SHR Y chromosome increases cardiovascular, endocrine, and behavioral responses to stress compared to the WKY Y chromosome. Physiol Behav 106: 101–108, 2012. [DOI] [PubMed] [Google Scholar]
- 15.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24: 1999–2012, 2003. [DOI] [PubMed] [Google Scholar]
- 16.Dzau VJ, Ellison KE, Brody T, Ingelfinger J, Pratt RE. A comparative study of the distributions of renin and angiotensinogen messenger ribonucleic acids in rat and mouse tissues. Endocrinology 120: 2334–2338, 1987. [DOI] [PubMed] [Google Scholar]
- 17.Elased KM, Cunha TS, Marcondes FK, Morris M. Brain angiotensin-converting enzymes: role of angiotensin-converting enzyme 2 in processing angiotensin II in mice. Exp Physiol 93: 665–675, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellis JA, Stebbing M, Harrap SB. Association of the human Y chromosome with high blood pressure in the general population. Hypertension 36: 731–733, 2000. [DOI] [PubMed] [Google Scholar]
- 19.Ely D, Boehme S, Dunphy G, Hart M, Chiarappa F, Miller B, Martins AS, Turner M, Milsted A. The Sry3 Y chromosome locus elevates blood pressure and renin-angiotensin system indexes. Gend Med 8: 126–138, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ely DL, Daneshvar H, Turner ME, Johnson ML, Salisbury RL. The hypertensive Y chromosome elevates blood pressure in F11 normotensive rats. Hypertension 21: 1071–1075, 1993. [DOI] [PubMed] [Google Scholar]
- 21.Ely DL, Turner ME. Hypertension in the spontaneously hypertensive rat is linked to the Y chromosome. Hypertension 16: 277–281, 1990. [DOI] [PubMed] [Google Scholar]
- 22.Ely D, Milsted A, Bertram J, Ciotti M, Dunphy G, Turner M. Sry delivery to the adrenal medulla increases blood pressure and adrenal medullary tyrosine hydroxylase of normotensive WKY rats. BMC Cardiovasc Disord 7: 6, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer M, Baessler A, Schunkert H. Renin angiotensin system and gender differences in the cardiovascular system. Cardiovasc Res 53: 672–677, 2002. [DOI] [PubMed] [Google Scholar]
- 24.Gard PR. Implications of the angiotensin converting enzyme gene insertion/deletion polymorphism in health and disease: a snapshot review. Int J Mol Epidemiol Genet 1: 145–157, 2010. [PMC free article] [PubMed] [Google Scholar]
- 25.Graves JAM. Sex chromosome specialization and degeneration in mammals. Cell 124: 901–914, 2006. [DOI] [PubMed] [Google Scholar]
- 26.Hilliard LM, Sampson AK, Brown RD, Denton KM. The “his and hers” of the renin-angiotensin system. Curr Hypertens Rep 15: 71–79, 2013. [DOI] [PubMed] [Google Scholar]
- 27.Irani RA, Xia Y. The functional role of the renin-angiotensin system in pregnancy and preeclampsia. Placenta 29: 763–771, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James GD, Sealey JE, Müller F, Alderman M, Madhavan S, Laragh JH. Renin relationship to sex, race and age in a normotensive population. J Hypertens Suppl4: S387–S389, 1986. [PubMed] [Google Scholar]
- 29.Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development 140: 4129–4144, 2013. [DOI] [PubMed] [Google Scholar]
- 30.Kiefer JC. Back to basics: Sox genes. Dev Dyn 236: 2356–2366, 2007. [DOI] [PubMed] [Google Scholar]
- 31.Kren V, Qi N, Krenova D, Zidek V, Sladká M, Jáchymová M, Míková B, Horky K, Bonne A, Van Lith HA, Van Zutphen BF, Lau YF, Pravenec M, St Lezin E. Y-chromosome transfer induces changes in blood pressure and blood lipids in SHR. Hypertension 37: 1147–1152, 2001. [DOI] [PubMed] [Google Scholar]
- 32.Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77, Suppl 9: 114–122, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudielka BM, Kirschbaum C. Sex differences in HPA axis responses to stress: a review. Biol Psychol 69: 113–132, 2005. [DOI] [PubMed] [Google Scholar]
- 34.Kuwahara M, Yamashita M, Shinoda K, Tofukuji S, Onodera A, Shinnakasu R, Motohashi S, Hosokawa H, Tumes D, Iwamura C, Lefebvre V, Nakayama T. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-β and suppresses T(H)2 differentiation. Nat Immunol 13: 778–786, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leinonen R, Sugawara H, Shumway M. The Sequence Read Archive. Nucleic Acids Res 39: D19–D21, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liebl C, Panhuysen M, Pütz B, Trümbach D, Wurst W, Deussing JM, Müller MB, Schmidt MV. Gene expression profiling following maternal deprivation: involvement of the brain Renin-Angiotensin system. Front Mol Neurosci 2: 1, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res 32: W217–W221, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maric-Bilkan C, Manigrasso MB. Sex differences in hypertension: contribution of the renin-angiotensin system. Gend Med 9: 287–291, 2012. [DOI] [PubMed] [Google Scholar]
- 39.Mattson DL, Dwinell MR, Greene AS, Kwitek AE, Roman RJ, Jacob HJ, Cowley AW. Chromosome substitution reveals the genetic basis of Dahl salt-sensitive hypertension and renal disease. Am J Physiol Renal Physiol 295: F837–F842, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milsted A, Underwood AC, Dunmire J, DelPuerto HL, Martins AS, Ely DL, Turner ME. Regulation of multiple renin-angiotensin system genes by Sry. J Hypertens 28: 59–64, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitchell AL, Pearce SHS. Autoimmune Addison disease: pathophysiology and genetic complexity. Nat Rev Endocrinol 8: 306–316, 2012. [DOI] [PubMed] [Google Scholar]
- 42.Montserrat N, Nivet E, Sancho-Martinez I, Hishida T, Kumar S, Miquel L, Cortina C, Hishida Y, Xia Y, Esteban CR, Izpisua Belmonte JC. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell 13: 341–350, 2013. [DOI] [PubMed] [Google Scholar]
- 43.Murakami A, Shen H, Ishida S, Dickson C. SOX7 and GATA-4 are competitive activators of Fgf-3 transcription. J Biol Chem 279: 28564–28573, 2004. [DOI] [PubMed] [Google Scholar]
- 44.Murck H, Uhr M, Ziegenbein M, Künzel H, Held K, Antonijevic IA, Schüssler P, Steiger A. Renin-angiotensin-aldosterone system, HPA-axis and sleep-EEG changes in unmedicated patients with depression after total sleep deprivation. Pharmacopsychiatry 39: 23–29, 2006. [DOI] [PubMed] [Google Scholar]
- 45.Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol 11: 715–724, 1994. [DOI] [PubMed] [Google Scholar]
- 46.Neves J, Kamaid A, Alsina B, Giraldez F. Differential expression of Sox2 and Sox3 in neuronal and sensory progenitors of the developing inner ear of the chick. J Comp Neurol 503: 487–500, 2007. [DOI] [PubMed] [Google Scholar]
- 47.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet 9: 911–922, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res 32: W280–W286, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phillips NB, Jancso-Radek A, Ittah V, Singh R, Chan G, Haas E, Weiss MA. SRY and human sex determination: the basic tail of the HMG box functions as a kinetic clamp to augment DNA bending. J Mol Biol 358: 172–192, 2006. [DOI] [PubMed] [Google Scholar]
- 50.Prokop JW, Leeper TC, Duan ZH, Milsted A. Amino acid function and docking site prediction through combining disease variants, structure alignments, sequence alignments, and molecular dynamics: a study of the HMG domain. BMC Bioinformatics 13, Suppl 2: S3, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prokop JW, Petri V, Shimoyama ME, Watanabe IKM, Casarini DE, Leeper TC, Bilinovich SM, Jacob HJ, Santos RAS, Martins AS, Araujo FC, Reis FM, Milsted A. Structural libraries of protein models for multiple species to understand evolution of the renin-angiotensin system. Gen Comp Endocrinol [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prokop JW, Rauscher FJ 3rd, Peng H, Liu Y, Araujo FC, Watanabe I, Reis FM, Milsted A. MAS promoter regulation: a role for Sry and tyrosine nitration of the KRAB domain of ZNF274 as a feedback mechanism. Clin Sci (Lond) 126: 727–738, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prokop JW, Watanabe IKM, Turner ME, Underwood AC, Martins AS, Milsted A. From rat to human: regulation of Renin-Angiotensin system genes by sry. Int J Hypertens 2012: 724240, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramser J, Abidi FE, Burckle CA, Lenski C, Toriello H, Wen G, Lubs HA, Engert S, Stevenson RE, Meindl A, Schwartz CE, Nguyen G. A unique exonic splice enhancer mutation in a family with X-linked mental retardation and epilepsy points to a novel role of the renin receptor. Hum Mol Genet 14: 1019–1027, 2005. [DOI] [PubMed] [Google Scholar]
- 55.Reckelhoff JF. Gender differences in the regulation of blood pressure. Hypertension 37: 1199–1208, 2001. [DOI] [PubMed] [Google Scholar]
- 56.Rizzoti K, Brunelli S, Carmignac D, Thomas PQ, Robinson IC, Lovell-Badge R. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat Genet 36: 247–255, 2004. [DOI] [PubMed] [Google Scholar]
- 57.Rossouw JE. Hormones, genetic factors, and gender differences in cardiovascular disease. Cardiovasc Res 53: 550–557, 2002. [DOI] [PubMed] [Google Scholar]
- 58.Santos RAS, Ferreira AJ, Verano-Braga T, Bader M. Angiotensin-converting enzyme 2, angiotensin-(1–7) and Mas: new players of the renin-angiotensin system. J Endocrinol 216: R1–R17, 2013. [DOI] [PubMed] [Google Scholar]
- 59.Sato Y, Shinka T, Sakamoto K, Ewis A, Nakahori Y. The male-determining gene SRY is a hybrid of DGCR8 and SOX3, and is regulated by the transcription factor CP2. Mol Cell Biochem 337: 267–275, 2010. [DOI] [PubMed] [Google Scholar]
- 60.Shankar RR, Charchar FJ, Eckert GJ, Saha C, Tu W, Dominiczak AF, Pratt JH. Studies of an association in boys of blood pressure and the Y chromosome. Am J Hypertens 20: 27–31, 2007. [DOI] [PubMed] [Google Scholar]
- 61.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10: 512–526, 1993. [DOI] [PubMed] [Google Scholar]
- 62.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toot J, Dunphy G, Turner M, Ely D. The SHR Y-chromosome increases testosterone and aggression, but decreases serotonin as compared to the WKY Y-chromosome in the rat model. Behav Genet 34: 515–524, 2004. [DOI] [PubMed] [Google Scholar]
- 64.Turner ME, Ely D, Prokop J, Milsted A. Sry, more than testis determination? Am J Physiol Regul Integr Comp Physiol 301: R561–R571, 2011. [DOI] [PubMed] [Google Scholar]
- 65.Turner ME, Farkas J, Dunmire J, Ely D, Milsted A. Which Sry locus is the hypertensive Y chromosome locus? Hypertension 53: 430–435, 2008. [DOI] [PubMed] [Google Scholar]
- 66.Uhart M, Chong RY, Oswald L, Lin PI, Wand GS. Gender differences in hypothalamic-pituitary-adrenal (HPA) axis reactivity. Psychoneuroendocrinology 31: 642–652, 2006. [DOI] [PubMed] [Google Scholar]
- 67.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Björling L, Ponten F. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28: 1248–1250, 2010. [DOI] [PubMed] [Google Scholar]
- 68.Wegman-Ostrosky T, Soto-Reyes E, Vidal-Millán S, Sánchez-Corona J. The renin-angiotensin system meets the hallmarks of cancer. J Renin-Angiotensin-Aldosterone Syst (August9, 2013). doi: 10.1177/1470320313496858. [DOI] [PubMed] [Google Scholar]
- 69.Weiss J, Meeks JJ, Hurley L, Raverot G, Frassetto A, Jameson JL. Sox3 is required for gonadal function, but not sex determination, in males and females. Mol Cell Biol 23: 8084–8091, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan X, Lu ML, Li T, Balk SP. SRY interacts with and negatively regulates androgen receptor transcriptional activity. J Biol Chem 276: 46647–46654, 2001. [DOI] [PubMed] [Google Scholar]
- 71.Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, Bannon DI, Lancashire L, Bao W, Du T, Luo H, Su Z, Jones WD, Moland CL, Branham WS, Qian F, Ning B, Li Y, Hong H, Guo L, Mei N, Shi T, Wang KY, Wolfinger RD, Nikolsky Y, Walker SJ, Duerksen-Hughes P, Mason CE, Tong W, Thierry-Mieg J, Thierry-Mieg D, Shi L, Wang C. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun 5: 3230, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]