Abstract
Metabolic rate and lifespan are important biological parameters that are studied in a wide range of research fields. They are known to correlate with body mass, but their association with gene (protein) functions is poorly understood. In this study, we collected data on the metabolic rate and lifespan of various organisms and investigated the relationship of these parameters with their genomes. We showed that the proportion of genes in a functional category, but not genome size, was correlated with mass-specific metabolic rate and maximal lifespan. In particular, the proportion of genes in oxic reactions (which occur in the presence of oxygen) was significantly associated with these two biological parameters. Additionally, we found that temperature, taxonomy, and mode-of-life traits had little effect on the observed associations. Our findings emphasize the importance of considering the biological functions of genes when investigating the relationships between genome, metabolic rate, and lifespan. Moreover, this provides further insights into these relationships, and may be useful for estimating metabolic rate and lifespan in individuals and the ecosystem using a combination of body mass measurements and genomic data.
Metabolic processes are essential for physiological functions and maintaining homeostasis in living organisms1,2. Therefore, understanding the factors that determine metabolic processes is an important topic of scientific inquiry, not only for researchers in the field of fundamental biology but also in ecology3,4 and medical research5. In particular, metabolic rate (especially respiratory rate) is an important physiological parameter for investigators in the fields of biology and ecology because it can be used to estimate, and therefore understand, energy metabolism, lifespan6,7, and animal space use8.
Previous studies have reported that metabolic rate strongly correlates with body mass; in particular, the relationship between metabolic rate B and body mass M approximately obeys a power law4,9: B ∝ Mα In general, α = 3/4 (such a scaling law is known as Kleiber’s law4,6,9,10); however, several previous studies suggest that Kleiber’s law may be not universal. For example, White and Seymour10 reported that metabolic rate is proportional to M2/3 in mammals, as predicted from the allometric scaling between body surface and body mass, when considering body temperature, digestive state, and phylogeny. Reich et al.11 observed a linear relationship between the rate of respiratory metabolism and body mass (i.e., B ∝ M) in plants. Similarly, the mean mass-specific metabolic rates observed across life’s major domains12 also implies that B ∝ M.
According to the rate of living theory, which predicts that animals with a greater rate of metabolism should die faster6, body mass is expected to be an important factor in determining lifespan. Indeed, several previous studies report an association between lifespan, body mass, and metabolic rate6,7,9,13, although other factors (e.g., ecological and phylogenetic) may also influence lifespan14.
Genomic information may also reflect metabolic rate because metabolic networks, which are expected to determine metabolic rate, are encoded in the genome1,2. In particular, metabolic rate is considered to be the consequence of many different biological reactions15 that involve proteins encoded by genes. However, several previous studies suggest that there is only a limited association between genomic information and metabolic rate/lifespan. For example, Kozłowski et al.16 and Starostová et al.17 reported that mass-specific metabolic rate is affected by cellular mass rather than by genome size or C-value (i.e., the amount of DNA in pg) in eyelid geckos. This may be because genome size has a weak correlation with cellular mass13. Although genome size is likely to positively correlate with lifespan in birds18 and fish19, some skepticism exists regarding the importance of genome size in fish (or homeotherms)20. In short, the association of genome size with metabolic rate and lifespan is controversial and is yet to be proven conclusively.
While previous studies imply that genomic information cannot be used to accurately estimate metabolic rate and lifespan, these studies themselves have limitations. In particular, they focused only on genome size or C-value as a measure of genomic information. The genome encodes several types of biological function, and the relationship between the number of functional genes and genome size differs according to functional category (e.g., defined by the Gene Ontology21) and species domain22,23. Therefore, in this study, we hypothesized that specific functional categories determine metabolic rate and lifespan rather than genome size. For example, genes related to oxic reactions or oxic metabolism24,25 (see also Methods), which are involved in processes that occur in the presence of oxygen, (hereinafter called oxic genes) may be useful for determining metabolic rate because they are expected to be involved in oxygen consumption.
Although the above previous studies mainly focus on C-value when considering genomic information, several new technologies and high-throughput methods have generated a large amount of genomic data, and such data are collected in several databases in recent years. Thus, investigators have become to be able to evaluate the relationship between genomic information and biological features such as metabolic rate and lifespan in more detail.
In general, such a relationship remains poorly understood. Therefore, here we investigated this relationship in more detail. Specifically, we collected data on metabolic rate and lifespan from published literature, selecting data from species for which a complete sequenced genome was available (see Methods). Using these data, we evaluated the usefulness of functional categories of genes for determining metabolic rate and lifespan.
Results
Genome size does not correlate with mass-specific metabolic rate
After the data collection and integration, we had obtained the genome, metabolic rate, and body mass data for 101 organisms, including 12 mammals, 9 protozoa, and 59 prokaryotes (see Methods and Supplementary Table S1).
A previous study17, Starostová et al. reported a limited effect of genome size, defined as the C-value, on mass-specific rate in eyelid geckos. However, this conclusion is limited to a specific family. Thus, we re-evaluated the correlation between genome size, defined as genome length (i.e., number of base pairs), and mass-specific metabolic rate (i.e., metabolic rate per body mass) in a wider range of species. Note that the definitions of genome size were different in our study and that of Starostová et al.17.
We found a negative correlation between genome size and mass-specific metabolic rate (Spearman’s rank correlation coefficient rs = –0.31, p = 0.0013) (Fig. 1). However, we observed a significant positive correlation between body mass and genome size (rs = 0.83, p < 2.2 × 10–16). It remains possible that this observed relationship is a spurious correlation because it is well known that mass-specific metabolic rate shows a negative correlation with body mass9,13, despite some criticism12, In fact, in our dataset, we also found that there was a negative correlation between them (rs = –0.42, p = 1.0 × 10–5). Especially, such a relationship between mass-specific rate q and body mass M is believed to follow a power law9,13: q ∝ M−β. Although β is expected to be 1/4 according to the Kleiber’s law4,6,9,10, a regression analysis (see Method) indicates that β is not zero but smaller than 1/4: β = 0.035 ± 0.008. This result is because of the fact that mean mass-specific metabolic rates are strikingly similar across life’s major domains12.
Figure 1. Scatter plot of mass-specific metabolic rate versus genome size.
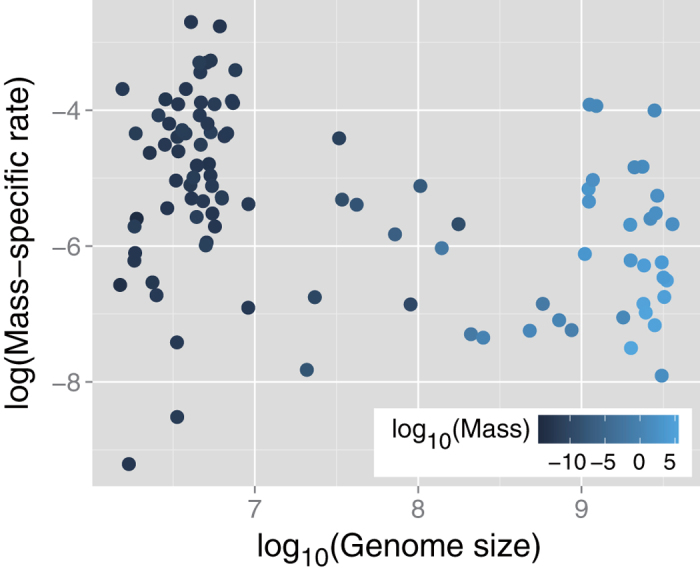
Vertical and horizontal axes are on logarithmic and base-10 logarithmic scales, respectively. Symbol color indicates base-10 logarithmic body mass. The mass-specific rate negatively correlates with genome size (Spearman’s rank correlation coefficient rs = –0.31, and p = 0.0013); however, it has no association with genome size when body mass is kept constant (partial rank correlation coefficient rsp = –0.31, p = 0.0014).
Additionally, we performed partial correlation analysis and found no correlation between genome size and mass-specific metabolic rate when body mass was kept constant (partial Spearman’s rank correlation coefficient rsp = 0.076, p = 0.45). In contrast, a negative correlation was observed between mass-specific metabolic rate and body mass when genome size was kept constant (rsp = –0.31, p = 0.0014).
This result implies a limited association between genome size and mass-specific metabolic rate, which provides evidential support for the findings of Starostová et al.17.
Temperature also affects metabolic rate4,15; therefore, we also considered temperature-corrected mass-specific metabolic rate (see Methods). In particular, we first evaluated the mass-specific metabolic rate at 25 °C (q25). Note that we only focused on approximately 80 organisms for which data on temperature was available (see Methods and Supplementary Table S1). We found no association between q25 and genome size both in a simple correlation analysis (rs = –0.051, p = 0.66) and partial correlation analysis (rsp = 0.16, p = 0.16). However, we need to consider a lower temperature because the organisms used in this study were primarily prokaryotes (see Supplementary Table S1) In particular, a lot of prokaryotes show optimal growth temperatures lower than 25 °C26. Thus, we next evaluated mass-specific metabolic rate at 10 °C (q10) (see Methods). Again, we confirmed no correlation between q10 and genome size in a simple correlation analysis (rs = –0.13, p = 0.25) and partial correlation analysis (rsp = 0.13, p = 0.23). These results suggest that a limited association between genome size and mass-specific metabolic rate can be also concluded when considering the effect of temperature on metabolic rate.
Oxic genes are associated with mass-specific metabolic rate
As a simple measure of ‘functional category’, we considered the proportion of genes in a functional category (i.e., the number genes in the category divided by the total number of genes or proteins)22,23. This measure is well used to evaluate the predominance of genes in a functional category (i.e., function) between gene sets of different sizes (organisms with different genome sizes, in this case), in the context of functional enrichment analysis27. Note that the use of the number of genes in a functional category or the total number of nucleotides of genes in the category is not suitable for comparing the predominance between organisms with different genome sizes because genome size influences these parameters.
We found a positive correlation between the proportion of oxic genes and mass-specific metabolic rate (rs = 0.52, p = 2.0 × 10–8) (Fig. 2). Because the proportion of oxic genes was also associated with body mass (rs = 0.55, p = 1.7 × 10–9), we performed a partial correlation analysis, correlating metabolic rate and the proportion of oxic genes while keeping body mass constant, and found a positive partial correlation (rsp = 0.38, p = 4.3 × 10–5). Having removed the effect of the proportion of oxic genes, a negative trend was observed between metabolic rate and body mass; however, the partial correlation was not highly significant (rsp = –0.19, p = 0.058). This result implies a limited effect of body size on mass-specific metabolic rate when considering the proportion of oxic genes in this dataset.
Figure 2. Scatter plot of the proportion of oxic genes versus mass-specific metabolic rate.
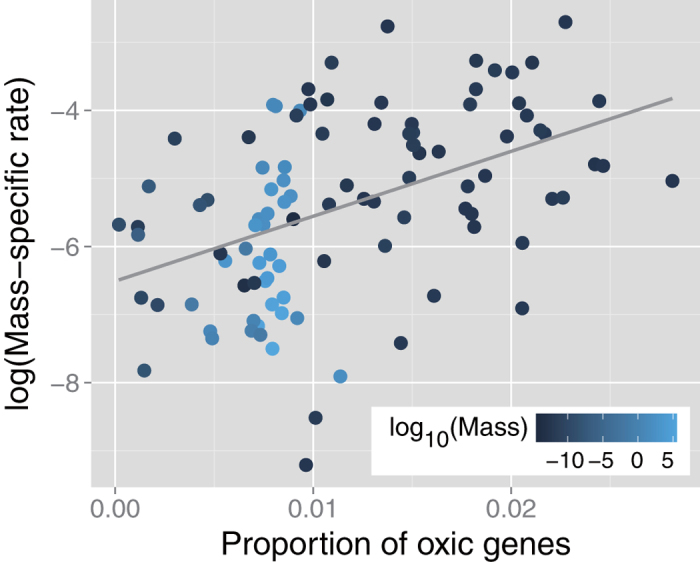
Vertical axis is on a logarithmic scale. The solid line is a linear regression fit to the data. Symbol color indicates base-10 logarithmic body mass. Both simple rank correlation analysis (rs = 0.52, p = 2.0 × 10–8) and partial rank correlation analysis (rsp = 0.38, p = 4.3 × 10–5), in which body mass is kept constant, indicate a positive association the proportion of oxic genes between the mass-specific rate.
We also observed a positive correlation between the proportion of oxic genes and temperature-corrected mass-specific metabolic rate in both cases of 25 °C (rs = 0.37, p = 7.2 × 10–4) and 10 °C (rs = 0.37, p = 5.4 × 10–4). Furthermore, a positive correlation was found when the effect of body size was removed using partial correlation analysis in both cases of 25 °C (rsp = 0.30, p = 0.0054) and 10 °C (rsp = 0.27, p = 0.011). This tendency was also observed when focusing only on prokaryotes, in both cases 25 °C (rsp = 0.28, p = 0.029) and 10 °C (rsp = 0.28, p = 0.027). In contrast, there was no significant correlation between metabolic rate and body mass when the proportion of oxic genes was kept constant, in both cases of 25 °C (rsp = –0.072, p = 0.53) and 10 °C (rsp = –0.13, p = 0.22).
However, this general finding may be biased by the organisms examined in this study, which were primarily prokaryotes (see Supplementary Table S1). For example, when focusing only on higher organisms (i.e., mammals and birds), we found a negative correlation between mass-specific metabolic rate and body mass (rsp = –0.96, p < 2.2 × 10–16) and a positive correlation between metabolic rate and the proportion of oxic genes (rsp = 0.55, p = 0.0025). This finding suggests that there are some effect of taxonomy on the observed association between the proportion of oxic genes and mass-specific metabolic rate. However, in general, our results imply that oxic genes are associated with mass-specific metabolic rate, independent of body mass and temperature.
The association between other functional categories and metabolic rate
Other functional categories are probably associated with mass-specific metabolic rate. To explore these, we evaluated Spearman’s rank correlations between the proportion of genes in functional categories, defined as a the Kyoto Encyclopedia of Genes and Genomes (KEGG) BRITE Functional Hierarchy28 (see Methods), and mass-specific metabolic rate. Additionally, we used partial correlation analysis to evaluate the association between functional categories and metabolic rate when body mass was kept constant.
Table 1 shows functional categories correlated with mass-specific metabolic rates (p < 0.05, using the partial rank correlation test). According to the effect sizes (i.e., correlation coefficients), oxic metabolism was the best estimator of mass-specific metabolic rate.
Table 1. Correlations between the proportion of genes in functional categories and mass-specific metabolic rate.
| Functional category | rs | rsp |
|---|---|---|
| Oxic metabolism | 0.52 (p = 2.0 × 10–8) | 0.38 (p = 4.3 × 10–5) |
| Cell motility | 0.33 (p = 7.3 × 10–4) | 0.35 (p = 2.7 × 10–4) |
| Xenobiotics biodegradation and metabolism | 0.42 (p = 9.9 × 10–6) | 0.27 (p = 0.0069) |
| Biosynthesis of other secondary metabolites | 0.45 (p = 2.9 × 10–6) | 0.23 (p = 0.020) |
| Energy metabolism | 0.43 (p = 8.7 × 10–6) | 0.20 (p = 0.043) |
| Membrane transport | 0.42 (p = 1.1 × 10–5) | 0.20 (p = 0.048) |
Spearman’s rank correlation coefficient (rs) and partial (i.e., body mass-controlled) rank correlation coefficients (rsp) are shown. The functional categories are displayed in descending order of rsp. Parenthetic values indicate the p-values.
Along with oxic metabolism, mass-specific metabolic rate was associated with other functional categories. In particular, the observed association with cell motility is related to metabolic rate because cell motility corresponds to energy consumption in the context of flagellar motility and actin cytoskeleton dynamics. In addition, the association with energy metabolism, in which adenosine triphosphate (ATP) is generated, is directly linked to metabolic rate because the efficient generation of ATP requires oxygen in aerobes. The observed association with membrane transport may be linked to the fact that a large part of standard metabolic costs are spent preserving ionic gradients in cell membranes16,29,30. In addition to this, this result may also support the membrane pacemaker hypothesis of metabolism31,32, which proposes fatty acid composition of membrane determines metabolic rate.
However, the associations with xenobiotic biodegradation and metabolism and biosynthesis of other secondary metabolites are more difficult to interpret. They could be explained by the concentration of oxic reactions in such peripheral metabolic pathways24,33; thus, these associations may be artifacts caused by the primary observed association between oxic metabolism and mass-specific metabolic rate.
Oxic metabolism, but not genome size, is associated with lifespan
According to the rate of living theory, metabolic rate is linked to lifespan. Consequently, we could hypothesize that gene function (specifically, the proportion of oxic genes) is also associated with lifespan. To test this hypothesis, we used data on the lifespan of 30 organisms, including 23 mammals and 7 birds, for which complete genome sequences were available (see Methods and Supplementary Table S2 for details). Based on our earlier observation of an association between the proportion of oxic genes and mass-specific metabolic rate in mammals and birds, we hypothesized that a negative correlation would exist between the proportion of oxic genes and lifespan in these animal groups.
As expected, we found a negative correlation between the proportion of oxic genes and maximum lifespan (rs = –0.42, p = 0.021) (Fig. 3). In several previous studies6,7,14, lifespan positively correlates with body mass. Here, we also observed a similar positive correlation (rs = 0.58, p = 0.00074); thus, we performed a partial correlation to remove the effect of body mass. Independent of body mass, we still found a negative correlation between the proportion of oxic genes and lifespan (rsp = –0.51, p = 0.0022). Note that this result does not directly indicate a limited effect of body mass on lifespan. We observed a positive correlation between body mass and lifespan when the proportion of oxic genes was kept constant (rsp = –0.63, p = 1.8 × 10–5).
Figure 3. Scatter plot of the proportion of oxic genes versus maximal lifespan.
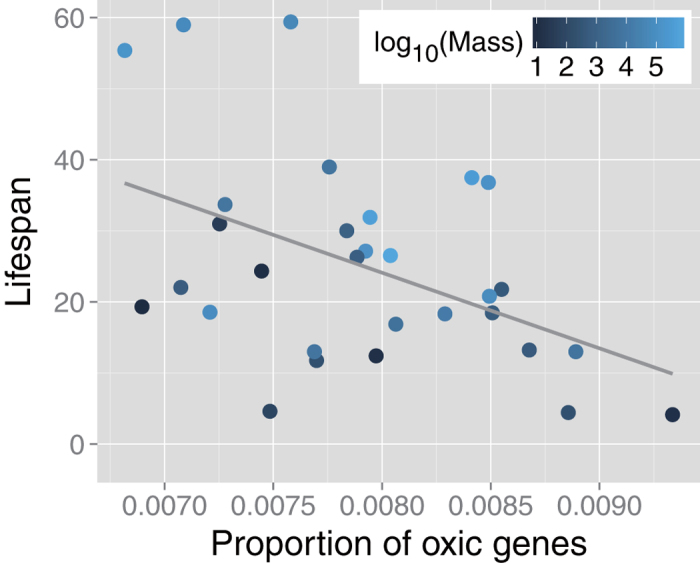
Symbol color indicates base-10 logarithmic body mass. The solid line is a linear regression fit to the data. Both simple rank correlation analysis (rs = –0.42, p = 0.021) and partial rank correlation analysis (rsp = –0.63, p = 1.8 × 10–5), in which body mass is kept constant, indicate a positive association the proportion of oxic genes between the mass-specific rate.
Ecological factors or mode-of-life traits can also affect lifespan. For example, Healy et al.14 reported that flight capability is the most important factor for longer lifespan in addition to body mass because volant species can more easily evade predators and unfavorable conditions. To test the effect of flight capability on the observed association between the proportion of oxic genes and lifespan, we separately evaluated a body mass-corrected partial correlation according to flight capability (volant/non-volant) (see Supplementary Table S2). We found negative correlations between the proportion of oxic genes and lifespan of 10 volant organisms (rsp = –0.69, p = 0.012) and 20 non-volant organisms (rsp = –0.48, p = 0.023). This result implies that the observed association between oxic metabolism and lifespan is independent of flight capability as a mode-of-life trait.
Using similar body mass-controlled correlation analysis, we found an association between lifespan and the sensory system including phototransduction and olfactory transduction (rsp = –0.41, p = 0.020) and metabolism of cofactors and vitamins (rsp = –0.38, p = 0.032). Several previous studies have shown that the sensory system is related to lifespan (reviewed in Refs.34,35). For example, Alcedo and Kenyon36 showed that gustatory neurons inhibit longevity. Our result is consistent with these previous studies. The latter association could be explained by the fact that oxic reactions or enzymes are known to frequently require iron, heme, and vitamins such as ascorbic acid as cofactors33. Similar to mass-specific metabolic rate, according to the effect size, the proportion of oxic genes was the best estimator of lifespan.
An association between genome size and lifespan is debatable because both positive18,19 and negative results20,37 have been reported. Thus, we re-evaluated this association using our dataset. Body mass-controlled partial rank correlation analysis indicated that there was no association between genome size and lifespan (rsp = 0.014, p = 0.94) (Fig. 4).
Figure 4. Limited correlation between genome size and maximal lifespan.
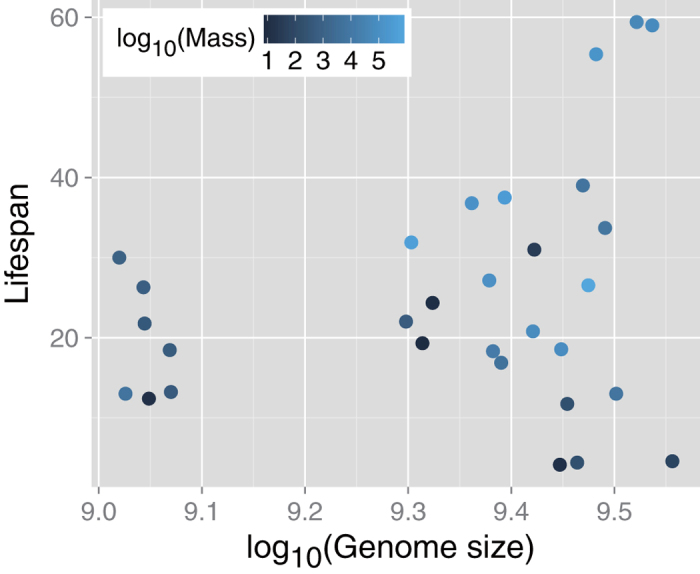
Horizontal axis is on a base-10 logarithmic scale. Symbol color indicates base-10 logarithmic body mass. A body mass-controlled (i.e., partial) rank correlation analysis indicated no association between genome size and lifespan (rsp = 0.014, p = 0.94).
Discussion
Although metabolic rate and lifespan are well known to correlate with body mass4,6,7,9,10, their association with genomic information is unclear. Several previous studies16,17,19,20,37 have suggested that genome size or C-value correlates with metabolic rate and lifespan; however, such a correlation is controversial and has received criticism (e.g., Refs.13,20). Therefore, in this study, we performed a more detailed genomic analysis using several organisms with sequenced genomes, and demonstrated that the proportion of genes in functional categories was associated with mass-specific metabolic rate and lifespan. Conversely, we found that genome size was less informative for explaining metabolic rate and lifespan.
In particular, we found that the proportion of oxic genes was the best estimator of metabolic rate and lifespan because it is associated with both metabolic rate and lifespan. Because oxic genes are related to reactions that occur in the presence of oxygen, their association may reflect the importance of oxygen consumption in metabolic rate and lifespan. In addition, other functional categories such as cell mortality, membrane transport, and sensory systems were also associated with metabolic rate and lifespan, and the observed associations are is consistent with previous studies, as mentioned in Results.
However, more careful examinations are required to conclude what functional categories dominantly determine mass-specific metabolic rate and lifespan because we only performed correlation analyses. In particular, the result that multiple functional categories are linked to metabolic rate and lifespan may be because genes are overlapped among functional categories (i.e., multicollinearity). Ideally, we may need to perform a higher-level statistical analysis such as stepwise regression analyses. However, assumptions, such an analysis requires, (e.g., linearity and normality) are not satisfied in this study. In particular, several studies reported no universal scaling relationship in genome22,23. In addition to this, some outliers are observed. We were afraid that linear model-based analyses result in misleading conclusions. Thus, we could not considered linear model-based analyses. Although our results are consistent with biological understanding, they are still debatable. To avoid this problem, we need to consider a higher-level analysis and collection of biological data (details will be described below).
Our results do not entirely discount the possibility that genome size is associated to lifespan. Rather, they highlight the need for more detailed examination of relationships between genomic information and lifespan. In particular, we emphasize the importance of considering the biological functions encoded by the genome when exploring these relationships.
Although metabolic rate may be the result of many different biological reactions15, the specific reactions are not entirely clear. Our study suggests that the contribution of biological reactions to metabolic rate differs according to the reaction types. This finding is similar to a previous study, which reported that the body mass–metabolic rate relationship results from the sum of the influences of multiple contributors to metabolism and control38. Based on our results, oxic reactions are particularly useful for investigating metabolic rate and its association with environmental factors.
Metabolic rate is a primary focus in ecology because it is useful for estimating values such as animal space use8, lifespan14, and feeding rate4. In particular, the body mass–metabolic rate relationship has been actively investigated4,6,7,9,10. Our study suggests a possibility that genomic data are also used to estimate these ecological parameters. For example, machine learning methods (e.g., support vector machine and neural networks) may be useful. Support vector machine and neural networks are supervised learning models, and they are well used when predicting parameters from multidimensional data. Especially, these models are also applicable to nonlinear regression. For these reasons, machine learning methods have already been widely applied to predict biological features using genomic data in bioinformatics39. These methods require variables explaining (e.g., well correlated with) an objective variable (metabolic rate or lifespan, in this case). Thus, it is important to explore such explanatory variables. Indeed, we found that the proportions of genes in the several functional categories, including oxic metabolism, are associated with metabolic rate and lifespan. Using the patterns of functional gene contents (i.e., vectors of the proportions of these functional genes), machine learning methods may estimate metabolic rate and lifespan. Sequencing analyses are now beginning to be applied in ecology (e.g., in population ecology40 and for identification of species–species interactions41). This approach, in which genomic data are used to investigate the ecosystem, is known as reverse ecology42,43. We suggest that the findings in our study may be usefully applied in such research fields. However, more careful examinations are required to complete the prediction of ecological parameters using genomic data. This study merely reported an association between genomic data and ecological parameters, and it does not show the cause-effect relationship between them. In such a case, a prediction of ecological parameters from genomic data may be ineffective in real-world cases because of overfitting problem. To avoid this problem, we need to consider better methods of data analysis and data collection (details will be described below).
We acknowledge that the analysis we present here has some limitations. For example, the definition of functional categories is controvertible. Our conclusions are limited to the context of functional categories defined by the KEGG BRITE Functional Hierarchy28, and these definitions may be somewhat arbitrary (i.e., they depend on the database administrators). For a deeper understanding of the association between biological functions and metabolic rate and lifespan, better definition of functional categories will be required. For example, several studies focus on detecting functional modules (i.e., categories) using biological networks. This challenge is related to graph clustering or community detection of networks44,45,46. In addition, it is important to consider biological information such as reaction mechanisms, direction of reaction (i.e., reversible vs. irreversible), chemical structure of metabolites, and gene clusters. In this context, methods for finding biologically meaningful modules of biological networks based on gene clusters and chemical transformation patterns47,48 may be useful.
In contrast to some other functional categories, oxic genes or reactions24,25 have been defined by considering biological information using Scope49, a computational framework used to characterize the biosynthetic capability of a network when it is provided with certain external resources. Therefore, these genes/reactions may be more useful for explaining metabolic rate and lifespan. An extension of the method for detecting metabolic scope will also be important for exploring the association between genomic information and metabolic rate/lifespan.
The definition of the predominance of functional genes is controversial. In this study, we considered a simple measure: the proportion of genes in a functional category. It is still datable that this measure really reflects the predominance of functions. Ideally, we may need to consider the expression and activity of functional genes and metabolic enzymes using microarray and mass spectrometry50 because several previous studies51,52 have reported that activities of specific enzymes are linked to metabolic rate (i.e., oxygen consumption rate).
Here, we investigated only organisms for which genomes were complete and available; thus, our study was somewhat biased toward lower organisms such as prokaryotes. The observed correlation between genomic information and metabolic rate may, therefore, be more applicable to prokaryotes. However, we also observed a positive correlation between these factors in higher organisms such as mammals and birds. Additionally, we took steps in our methodology to reduce phylogenetic signals. Thus, we believe that the effect of taxonomy is unlikely to change our conclusion; however, we acknowledge that further careful examination will be required. The importance of phylogeny for evaluating associations between biological features is well known in terms of comparative phylogenetic analysis53,54,55. For example, several previous studies10,12 have reported that Kleiber’s law may be not observed when considering phylogenetic information. However, comparative phylogenetic analysis generally assumes a simple evolutionary model, which deems random Brownian-motion-like traits to be change on a phylogenetic tree with accurate branch lengths, and may, therefore, result in misleading conclusions. For instance, Griffith et al.19 pointed out the loss of statistical power that occurs when a dataset reduces in size because of phylogenetic corrections. Because our dataset contained only a few samples for higher organisms and, thus, falls into condition described by Griffith et al., we did not consider comparative phylogenetic analysis.
The results of our study also depend on the quality of genome annotation. Furthermore, it is possible that our results are influenced by the percentage of functionally-unknown proteins in the study organisms. For metabolic networks, we already confirmed the difference in the fraction of functionally-unknown proteins between species categories in our previous study25. Thus, we believe that the quality of genome annotation would affect our conclusions; however, further research is required in this area. For example, metabolic networks are not fully understood. In particular, the existence of enzyme promiscuity56, which implies that enzymes can catalyze multiple reactions, act on more than one substrate, or exert a range of suppressions57, suggests the possibility of many hidden metabolic reactions, which may be related to metabolic robustness against changing environments58. Consideration of these hidden metabolic reactions will be important for designing metabolic pathways and for developing our understanding of metabolic evolution.
It will also be necessary to test the association between gene (protein) functions and metabolic rate and lifespan using additional organisms. Therefore, the continued sequencing of genomes from a wide range of organisms (including microorganisms such as extremophiles, mammals, fish, and insects) is obviously important. The development of high-throughput sequencing techniques will enable the collection of such data. For example, metagenomic techniques can now help to complete the sequencing of an organism’s genome.
Despite the limitations of our data analysis, our findings enhance the current understanding of the relationship between genomic information and the parameters metabolic rate and lifespan. Furthermore, they may be usefully applied in future research for estimating metabolic rate and lifespan using genomic data.
Methods
Data on metabolic rate, lifespan, and genome
We obtained data on metabolic rate and mass-specific metabolic rate from previously published literature12,14,59, which comprehensively reported the relationship between metabolic rate and body mass. We also extracted data on body mass and temperature from this literature.
The data on maximal lifespan were obtained from Healy et al.14. Additionally, data on body mass and species’ mode-of-life traits (i.e., volancy, fossoriality, foraging environment, and daily activity) were collected.
To prevent redundancy and reduce phylogenetic signals, we averaged the biological parameters (i.e., metabolic rate, body mass, and temperature) according to genus after unit conversion. The units of metabolic rate, mass-specific metabolic rate, body mass, and maximal lifespan were watts [W], watts per gram [W/g], grams [g], and years, respectively. We selected organisms for which genomes were available in the KEGG database28. We selected one species as a representative of a genus according to the year in which the species genome was first completely determined, and we used this genome for investigating the association between the biological parameters and the genomic information.
For investigating the association between metabolic rate and genomic information, we obtained data on 101 organisms including 12 mammals, 6 birds, 4 ectothermic vertebrates, 5 insects, 9 protozoa, and 59 prokaryotes (see Supplementary Table S1 for full details). In this dataset, the data on temperature were available for 84 organisms, including 12 mammals, 4 ectothermic vertebrate, 9 protozoa, and 59 prokaryotes.
For investigation of the association between lifespan and genomic information, we obtained data on 30 organisms, including 23 mammals and 7 birds (see Supplementary Table S2).
Temperature-corrected mass-specific metabolic rate
To remove the effect of temperature15, mass-specific metabolic rate q obtained at different temperatures T [°C] was transformed to 25 °C: q25 = q × 10–3 × 2(25 – T)/10, according to a previous study12. In our study, we did not consider ectothermic vertebrates because these organisms do not live at a body temperatures of 25 °C12. Our dataset include only 4 (5% of the total) ectothermic vertebrates; thus, the exclusion of these organisms does not affect the conclusion. We can confirm that the similar conclusions can be obtained even if these ectothermic vertebrates using the dataset (Supplementary Table S1).
In addition to this, according to a previous study15, we also considered to transform q at observed at different temperature T [°C] to a given temperature X [°C]: qX = q × exp[–E/k {1/(X + 273.15) −1/(T + 273.15)}], where E and k indicate an average activation energy [eV] for enzyme-catalyzed biochemical reactions and Boltzmann’ constant (i.e., k = 8.6173 × 10–5 [eV/K]). According to a previous study15, we considered E = 0.65 although E ranges between 0.4 and 0.8; but, we confirmed that similar conclusions can be obtained when E = 0.4 and 0.8.
Identification of the oxic genes of each species
According to a database24 (prelude.bu.edu/O2/networks.html), we obtained an oxic reactions list based on Enzyme Commission (EC) numbers and metabolic reaction notations (i.e., reaction or ‘R’ numbers such as R00010) in the KEGG database28. Via the KEGG FTP site (ftp.bioinformatics.jp/kegg/xml/kgml/metabolic/organisms), on March 17, 2014 we downloaded XML files (version 0.7.1) containing the data on gene–reaction (i.e., gene identifier–R number) relationships of 111 organisms from the KEGG database. Based on this data, we defined oxic genes as genes associated with at least one oxic reaction.
Functional categories of genes
In this study, we used the second level of KEGG BRITE Functional Hierarchy of the KEGG metabolic map (www.genome.jp/kegg-bin/get_htext?br08901.keg) for identifying functional categories of genes. We downloaded the data on functional category–gene identifier relationships of species S from the KEGG FTP site (ftp.bioinformatics.jp/kegg/brite/organisms/S/S00001.keg) on 17 March 2014, where S indicates the KEGG organism identifier (see Supplementary Tables S1 and S2).
In this study, we did not consider Gene Ontology21 as a definition of functional category because of fewer organisms whose GO annotations were completed, compared to KEGG BRITE Functional Hierarchy.
Functional genome size
We computed the number of genes in a functional category and functional genome size (i.e., total number of nucleotides of genes in the functional category) (Supplementary Tables S1 and S2). For 111 organisms, we downloaded the nucleotide sequence data of species S from the KEGG FTP site (ftp.bioinformatics.jp/kegg/genes/organisms/S/) on 17 March 2014. On the basis of the functional category–gene identifier relationships, obtained as above, we calculated these two parameters.
Note that the KEGG FTP site was available only to paid subscribers as of 1 July 2011. Because the use of our data may be desirable to ensure reproducibility, our datasets are available upon request.
Statistical tests
For measuring statistical dependence between parameters, we computed the Spearman’s rank correlation coefficient rs (a non-parametric measure, which is relatively robust to outliers and can be used to analyze nonlinear relationships) and its associated p value using R version 3.1.1 (www.r-project.org).
To ensure that the results of the Spearman’s rank correlation analysis were robust, we also performed partial Spearman’s rank correlation analysis using R software. Specifically, we used the function pcor, available in the R package ppcor version 1.0.
To estimate the exponent of a power-law relationship, we performed a linear regression analysis using logarithmic values. In particular, we used the function lm in R software.
Author Contributions
K. T. and Y. K. designed this study. K. T. and Y. K. collected the data, and K. T. performed data analysis. K. T. and Y. K. completed the manuscript and approved the final manuscript.
Additional Information
How to cite this article: Takemoto, K. and Kawakami, Y. The proportion of genes in a functional category is linked to mass-specific metabolic rate and lifespan. Sci. Rep. 5, 10008; doi: 10.1038/srep10008 (2015).
Supplementary Material
Acknowledgments
This study was supported by a Grant-in-Aid for Young Scientists (A) from the Japan Society for the Promotion of Science (no. 25700030). KT was partly supported by the Chinese Academy of Sciences Fellowships for Young International Scientists (no. 2012Y1SB0014), and the International Young Scientists Program of the National Natural Science Foundation of China (no. 11250110508).
References
- Takemoto K. Current understanding of the formation and adaptation of metabolic systems based on network theory. Metabolites 2, 429–457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K. & Oosawa C. in Stat. Mach. Learn. Approaches Netw. Anal. 77–108 (2012). doi: 10.1002/9781118346990.ch3 [DOI] [Google Scholar]
- Savage V. M. et al. The predominance of quarter-power scaling in biology. Funct. Ecol. 18, 257–282 (2004). [Google Scholar]
- Brown J. H., Gillooly J. F., Allen A. P., Savage V. M. & West G. B. Toward a metabolic theory of ecology. Ecology 85, 1771–1789 (2004). [Google Scholar]
- Milotti E., Vyshemirsky V., Sega M., Stella S. & Chignola R. Metabolic scaling in solid tumours. Sci. Rep. 3, 1938 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman J. R. Body size, energy metabolism and lifespan. J. Exp. Biol. 208, 1717–1730 (2005). [DOI] [PubMed] [Google Scholar]
- Niitepõld K. & Hanski I. A long life in the fast lane: positive association between peak metabolic rate and lifespan in a butterfly. J. Exp. Biol. 216, 1388–1397 (2013). [DOI] [PubMed] [Google Scholar]
- Jetz W., Carbone C., Fulford J. & Brown J. H. The scaling of animal space use. Science 306, 266–268 (2004). [DOI] [PubMed] [Google Scholar]
- West G. B., Woodruff W. H. & Brown J. H. Allometric scaling of metabolic rate from molecules and mitochondria to cells and mammals. Proc. Natl. Acad. Sci. U. S. A. 99, 2473–2478 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. R. & Seymour R. S. Mammalian basal metabolic rate is proportional to body mass2/3. Proc. Natl. Acad. Sci. U. S. A. 100, 4046–4049 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich P. B., Tjoelker M. G., Machado J.-L. & Oleksyn J. Universal scaling of respiratory metabolism, size and nitrogen in plants. Nature 439, 457–61 (2006). [DOI] [PubMed] [Google Scholar]
- Makarieva A. M. et al. Mean mass-specific metabolic rates are strikingly similar across life’s major domains: Evidence for life’s metabolic optimum. Proc. Natl. Acad. Sci. U. S. A. 105, 16994–16999 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- West G. B. & Brown J. H. The origin of allometric scaling laws in biology from genomes to ecosystems: towards a quantitative unifying theory of biological structure and organization. J. Exp. Biol. 208, 1575–92 (2005). [DOI] [PubMed] [Google Scholar]
- Healy K. et al. Ecology and mode-of-life explain lifespan variation in birds and mammals. Proc. R. Soc. B 281, 20140298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly J. F., Brown J. H., West G. B., Savage V. M. & Charnov E. L. Effects of size and temperature on metabolic rate. Science 293, 2248–2251 (2001). [DOI] [PubMed] [Google Scholar]
- Kozłowski J., Konarzewski M. & Gawelczyk A. T. Cell size as a link between noncoding DNA and metabolic rate scaling. Proc. Natl. Acad. Sci. U. S. A. 100, 14080–14085 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starostová Z., Kubicka L., Konarzewski M., Kozłowski J. & Kratochvíl L. Cell size but not genome size affects scaling of metabolic rate in eyelid geckos. Am. Nat. 174, E100–E105 (2009). [DOI] [PubMed] [Google Scholar]
- Monaghan P. & Metcalfe N. B. Genome size and longevity. Trends Genet. 16, 331–332 (2000). [DOI] [PubMed] [Google Scholar]
- Griffith O. L., Moodie G. E. E. & Civetta A. Genome size and longevity in fish. Exp. Gerontol. 38, 333–337 (2003). [DOI] [PubMed] [Google Scholar]
- Gregory T. R. Genome size is not correlated positively with longevity in fishes (or homeotherms). Exp. Gerontol. 39, 859–860 (2004). [DOI] [PubMed] [Google Scholar]
- Ashburner M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina N. & van Nimwegen E. Scaling laws in functional genome content across prokaryotic clades and lifestyles. Trends Genet. 25, 243–7 (2009). [DOI] [PubMed] [Google Scholar]
- Koonin E. V. Are there laws of genome evolution? PLoS Comput. Biol. 7, e1002173 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J. & Segrè D. The effect of oxygen on biochemical networks and the evolution of complex life. Science 311, 1764–1767 (2006). [DOI] [PubMed] [Google Scholar]
- Takemoto K. & Yoshitake I. Limited influence of oxygen on the evolution of chemical diversity in metabolic networks. Metabolites 3, 979–992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. K., Kremer C. T., Klausmeier C. A. & Litchman E. A global pattern of thermal adaptation in marine phytoplankton. Science 338, 1085–1088 (2012). [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. & Lempicki R. A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter R. K. & Brand M. D. Body mass dependence of H+ leak in mitochondria and its relevance to metabolic rate. Nature 362, 628–630 (1993). [DOI] [PubMed] [Google Scholar]
- Szarski H. Cell size and the concept of wasteful and frugal evolutionary strategies. J. Theor. Biol. 105, 201–209 (1983). [DOI] [PubMed] [Google Scholar]
- Hulbert A. J. Life, death and membrane bilayers. J. Exp. Biol. 206, 2303–2311 (2003). [DOI] [PubMed] [Google Scholar]
- Rodriguez E. et al. Setting the pace of life: membrane composition of flight muscle varies with metabolic rate of hovering orchid bees. Proc. R. Soc. B Biol. Sci. 282, 20142232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.-Y. et al. The impact of oxygen on metabolic evolution: a chemoinformatic investigation. PLoS Comput. Biol. 8, e1002426 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. A conserved regulatory system for aging. Cell 105, 165–168 (2001). [DOI] [PubMed] [Google Scholar]
- Guarente L. & Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature 408, 255–262 (2000). [DOI] [PubMed] [Google Scholar]
- Alcedo J. & Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron 41, 45–55 (2004). [DOI] [PubMed] [Google Scholar]
- Morand S. & Ricklefs R. E. Genome size , longevity and development time in birds Genome size , longevity and development time. Trends Genet. 17, 567–568 (2001). [DOI] [PubMed] [Google Scholar]
- Darveau C.-A., Suarez R. K., Andrews R. D. & Hochachka P. W. Allometric cascade as a unifying principle of body mass effects on metabolism. Nature 417, 166–170 (2002). [DOI] [PubMed] [Google Scholar]
- Larrañaga P. et al. Machine learning in bioinformatics. Brief. Bioinform. 7, 86–112 (2006). [DOI] [PubMed] [Google Scholar]
- Takahara T., Minamoto T., Yamanaka H., Doi H. & Kawabata Z. Estimation of fish biomass using environmental DNA. PLoS One 7, e35868 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toju H., Guimarães P. R., Olesen J. M. & Thompson J. N. Assembly of complex plant–fungus networks. Nat. Commun. 5, 5273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R. & Borenstein E. Reverse ecology: from systems to environments and back. Adv. Exp. Med. Biol. 751, 329–345 (2012). [DOI] [PubMed] [Google Scholar]
- Klitgord N. & Segrè D. Ecosystems biology of microbial metabolism. Curr. Opin. Biotechnol. 22, 541–546 (2011). [DOI] [PubMed] [Google Scholar]
- Ahn Y.-Y., Bagrow J. P. & Lehmann S. Link communities reveal multiscale complexity in networks. Nature 466, 761–764 (2010). [DOI] [PubMed] [Google Scholar]
- Becker E., Robisson B., Chapple C. E., Guénoche A. & Brun C. Multifunctional proteins revealed by overlapping clustering in protein interaction network. Bioinformatics 28, 84–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato S. Community detection in graphs. Phys. Rep. 486, 75–174 (2010). [Google Scholar]
- Kanehisa M. Chemical and genomic evolution of enzyme-catalyzed reaction networks. FEBS Lett. 587, 2731–2737 (2013). [DOI] [PubMed] [Google Scholar]
- Muto A. et al. Modular architecture of metabolic pathways revealed by conserved sequences of reactions. J. Chem. Inf. Model. 53, 613–622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handorf T., Ebenhöh O. & Heinrich R. Expanding metabolic networks: scopes of compounds, robustness, and evolution. J. Mol. Evol. 61, 498–512 (2005). [DOI] [PubMed] [Google Scholar]
- De Rond T., Danielewicz M. & Northen T. High throughput screening of enzyme activity with mass spectrometry imaging. Curr. Opin. Biotechnol. 31, 1–9 (2015). [DOI] [PubMed] [Google Scholar]
- Thuesen E. V. & Childress J. J. Enzymatic activities and metabolic rates of pelagic chaetognaths: Lack of depth-related declines. Limnol. Oceanogr. 38, 935–948 (1993). [Google Scholar]
- Thuesen E. V. & Childress J. J. Metabolic rates, enzyme activities and chemical compositions of some deep-sea pelagic worms, particularly Nectonemertes mirabilis (Nemertea; Hoplonemertinea) and Poeobius meseres (Annelida; Polychaeta). Deep Sea Res. Part I 40, 937–951 (1993). [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 125, 1–15 (1985). [DOI] [PubMed] [Google Scholar]
- Garland T., Harvey P. H. & Ives A. R. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst. Biol. 41, 18–32 (1992). [Google Scholar]
- Garland T., Bennett A. F. & Rezende E. L. Phylogenetic approaches in comparative physiology. J. Exp. Biol. 208, 3015–3035 (2005). [DOI] [PubMed] [Google Scholar]
- Khersonsky O. & Tawfik D. S. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505 (2010). [DOI] [PubMed] [Google Scholar]
- Patrick W. M., Quandt E. M., Swartzlander D. B. & Matsumura I. Multicopy suppression underpins metabolic evolvability. Mol. Biol. Evol. 24, 2716–2722 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H. et al. Network context and selection in the evolution to enzyme specificity. Science 337, 1101–4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolokotrones T., Van Savage, Deeds E. J. & Fontana W. Curvature in metabolic scaling. Nature 464, 753–756 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


