Abstract
Extensive evidence exists that the reaction of estrogen metabolites with DNA produces depurinating adducts that, in turn, induce mutations and cellular transformation. While it is clear that these estrogen metabolites result in a neoplastic phenotype in vitro, further evidence supporting the link between estrogen–DNA adduct formation and its role in neoplasia induction in vivo would strengthen the evidence for a genotoxic mechanism.
Diethylstilbestrol (DES), an estrogen analogue known to increase the risk of breast cancer in women exposed in utero, is hypothesized to induce neoplasia through a similar genotoxic mechanism. Cultured MCF-10F human breast epithelial cells were treated with DES at varying concentrations and for various times to determine whether the addition of DES to MCF-10F cells resulted in the formation of depurinating adducts. This is the first demonstration of the formation of DES–DNA adducts in human breast cells. A dose-dependent increase in DES–DNA adducts was observed. Demonstrating that treatment of MCF-10F cells with DES, a known human carcinogen, yields depurinating adducts provides further support for the involvement of these adducts in the induction of breast neoplasia.
Previous studies have demonstrated the ability of antioxidants such as resveratrol to prevent the formation of estrogen–DNA adducts, thus preventing a key carcinogenic event. In this study, when MCF-10F cells were treated with a combination of resveratrol and DES, a dose-dependent reduction in the level of DES–DNA adducts was also observed.
Keywords: DES–DNA adducts, Catechol estrogen quinones, Resveratrol, MCF-10F cells
1. Introduction
The important role of hormones in breast cancer pathogenesis has been accepted since the start of the 20th century [1]. Classically, it has been reported that the carcinogenic effects of estrogens originate from the downstream signaling of estrogen receptors. Another paradigm of cancer initiation by estrogens involves the genotoxicity of specific estrogen metabolites. Current research aims to understand how these specific estrogen metabolites react with DNA to form depurinating estrogen–DNA adducts [2–6] that induce mutations [7–10] and subsequent cell transformation [11–15], thereby initiating breast cancer.
The natural estrogens estrone (E1) and estradiol (E2) can be oxidized by the cytochrome P450 (CYP) system to the catechol estrogen metabolites, 4-hydroxyestrone(estradiol) [4-OHE1(E2)] and 2-OHE1(E2) [6,16]. Further oxidation of the catechol estrogen metabolites by cytochrome P450 produces genotoxic catechol estrogen quinones, E1(E2)-3,4-Q and E1(E2)-2,3-Q [6,16]. While both E1(E2)-3,4-Q and E1(E2)-2,3-Q have demonstrated genotoxic potential, it is E1(E2)-3,4-Q, derived from the more carcinogenic 4-OHE1(E2), that has the predominant reactivity with DNA [4]. The activity of enzymes such as catechol-O-methyltransferase (COMT) and NADPH quinone oxidoreductase 1 (NQO1) keeps levels of these metabolites low, but when estrogen metabolism is unbalanced, the levels of E1(E2)-3,4-Q can be elevated [16,17]. Under these conditions, E1(E2)-3,4-Q can react with DNA to form the depurinating adducts 4-OHE1(E2)-1-N3Ade and 4-OHE1(E2)-1-N7Gua [2–6]. These adducts produce apurinic sites that can generate mutations [7–10] and cellular transformation [11–15], and lead to cancer [18–21].
As an estrogen analogue, diethylstilbestrol (DES) was prescribed to pregnant women from the 1940s until the late 1970s as a form of hormone replacement with the mistaken belief that it would prevent miscarriages [22]. Therapeutic use of DES resulted in an increased risk of breast cancer both in women exposed to DES during pregnancy and in those women exposed in utero [23–26]. The mechanism responsible for the increased risk of neoplasia associated with DES has not been elucidated, and it remains of great practical and theoretical importance.
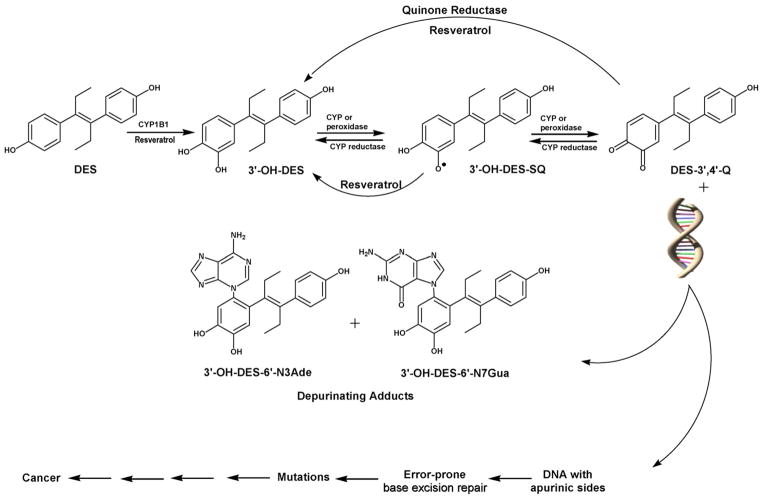
This study rests on the hypothesis that DES induces neoplasia through the genotoxic mechanism described above for the natural estrogens. Preliminary research in this area supports this hypothesis. DES is metabolized to its catechol, 3′-OH-DES (Fig. 1). Saeed et al. demonstrated that 3′-OH-DES intercalates into DNA and is then enzymatically oxidized to its quinone, which reacts with DNA to yield 3′-OH-DES-6′-N3Ade and 3′-OH-DES-6′-N7Gua adducts (Fig. 1) analogous to those formed by the natural estrogens [27]. To study the genotoxic potential of DES, we quantified the formation of depurinating DES–DNA adducts when MCF-10F cells were exposed to DES at varying concentrations using a previously developed in vitro model [16]. The MCF-10F cell line is an immortalized human breast epithelial cell line lacking estrogen receptor-α. Demonstration that the addition of DES to MCF-10F cells leads to formation of depurinating DES–DNA adducts would support the conclusion that breast cancer can be initiated by a genotoxic mechanism.
Fig. 1.
Metabolic activation of DES to form DES–DNA adducts and initiate cancer.
Under the proposed genotoxic mechanism for the induction of breast cancer, there is the possibility of preventing the crucial carcinogenic events, namely the formation of E1(E2)-3,4-Q and their reaction with DNA. The addition of natural antioxidant compounds to reaction mixtures of E2-3,4-Q or enzyme-activated 4-OHE2 and DNA was found to reduce the formation of depurinating estrogen–DNA adducts [28]. The compounds were thought to prevent oxidative and/or electrophilic damage to DNA by inhibiting the formation of the electrophilic E1(E2)-3,4-Q and/or reacting with them [28]. One of the compounds tested was resveratrol, 3,5,4′-trihydroxy-trans-stilbene, which has significant antioxidant activity [29,30]. The chemopreventive activity of resveratrol is also attributed to its ability to induce NQO1 activity in cultured cells [15,31] and to reduce E2-3,4-semiquinone to 4-OHE2, leading to a reduction in adduct formation [28].
To further characterize the antioxidant activity of resveratrol, varying concentrations were added to MCF-10F cells in culture both before and concomitantly with DES treatment. DES–DNA adducts were then quantified. This is the first study in human cell culture in which the formation of depurinating DES–DNA adducts in MCF-10F cells is reported.
2. Materials and methods
2.1. Chemicals and reagents
Standard depurinating adducts were synthesized in our laboratory, as previously described [27]. All other chemicals were purchased from Sigma (St. Louis, MO) and used without further purification.
2.2. Cell line and culture conditions
MCF-10F cells were obtained from the ATCC (Rockville, MD), and cultured in DMEM and Ham’s F12 media (Mediatech Inc.) with 20 ng/mL epidermal growth factor, 0.01 mg/mL insulin, 500 ng/mL hydrocortisone, 5% horse serum and 100 μg/mL penicillin/streptomycin mixture. Estrogen-free medium was prepared in phenol red-free DME/F12 medium with charcoal-stripped FBS (HyClone, Logan, UT).
2.3. Cytotoxicity assay
Cells were seeded at a density of 4000 cells/well in 96-well plates. After 8 h (day 0), cells in one 96-well plate were counted by the MTT [3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide] assay, while other cells were treated with different concentrations of DES (1.25–100 μM) and further incubated for 24, 48 or 72 h. DES was dissolved in DMSO (final concentration 0.001%). In the MTT assay, the media in the 96-well plate cultures were replaced with 100 μL of fresh medium containing 25 μL of MTT (5 mg/mL in PBS), and incubated for 2 h at 37 °C to allow the reduction of MTT by metabolically active cells to form a purple formazan precipitate [32]. The precipitate was then solubilized by adding 20% SDS in 1:1 DMF:H2O, pH 4.7 (100 μL) and incubating overnight at 37 °C. The purple color was read at 570 nm in a μQuant microplate spectrophotometer (Bio-Tek Instruments) and analyzed by the KCjunior (version 1.41) software. The absorbance values were converted into percentage of viable cells with respect to the blank control absorbance value.
2.4. Metabolism
The MCF-10F cells (0.75 × 105 cells) were seeded for 24 h in estrogen-containing medium. The medium was changed to estrogen-free medium and the cells were grown for another 48 h. To directly determine the relationship between the concentration of DES and formation of DNA adducts, cells were treated with increasing concentrations (1, 5, 10 and 30 μM) of DES for 24 h. Concentrations of DES were chosen on the basis of MTT assay results and previous studies by our group with MCF-10F cells [15–17]. To determine the time point at which DES induces the maximum amount of adducts, the cells were treated with 10 μM DES for the following time periods: 6, 12, 24, 48 or 72 h. To investigate the inhibiting effect of resveratrol on DES-induced DNA adduct formation, cells were first pre-incubated for 48 h with selected doses (1, 10 or 30 μM) of resveratrol, washed with PBS and, after adding fresh medium, treated with 10 μM DES for 24 h with or without fresh antioxidant. To keep the concentration of DMSO the same (0.005%) in all experiments, separate stock solutions of DES (1, 5, 10 and 30 mM) were prepared. Media from five T-150 flasks of MCF-10F cells treated with an equal amount of organic solvent were used as controls.
Once the media were removed from five flasks, each flask was incubated at 37 °C with 1 mL of trypsin. After 10 min, 10 mL of double serum media was used to combine the cells from all five flasks. Cell numbers were estimated by using a Coulter counter (Beckman Coulter Inc., USA).
2.5. Analysis of DES–DNA adducts
Following the treatments, media were harvested, 2 mM ascorbic acid (to prevent possible decomposition of the adducts) was added and processed immediately. Sample preparation and analysis by HPLC with electrochemical detection (HPLC-ECD) was carried out as follows.
2.5.1. Sample preparation
Culture media from five flasks (50 mL) were processed by passage through C8 Certify II cartridges (Varian, Harbor City, CA), which were equilibrated by sequentially passing 1 mL of methanol, distilled water, and potassium phosphate buffer (100 mM, pH 8) through them. The harvested media (50 mL) were adjusted to pH 8.0 and passed through these cartridges. The retained analytes, which included 3′-OH-DES-6′-N3Ade and 3′-OH-DES-6′-N7Gua, in the cartridges were washed with the above phosphate buffer, eluted with methanol:acetonitrile:water:trifluoroacetic acid (8:1:1:0.1), and evaporated by using a Jouan concentrator (Thermo Scientific, Waltham, MA). The residue was resuspended in 1.0 mL acetonitrile and 1.0 mg of MnO2 was added. Samples were vortexed for 1 min and then evaporated again with the Jouan concentrator. The residue was resuspended in 150 μL of methanol:water:ascorbic acid (1:1:0.001) and filtered through a 3000 MW cutoff filter (Millipore, Bedford, MA).
Identification of the DES–DNA adducts derived from oxidation of the adducts with MnO2 to form the DES-quinone adducts. Subsequent tautomerization to the dienestrol (DIES)-DNA adducts, which were identical to the standard 3′-OH-DIES-6′-N3Ade and 3′-OH-DIES-6′-N7Gua adducts [27], allowed identification and quantification of the DES–DNA adducts.
2.5.2. HPLC-ECD
Analyzes of all samples were conducted on an HPLC system equipped with dual ESA Model 580 solvent delivery modules, an ESA Model 540 autosampler, and a 12-channel CoulArray electrochemical detector (ESA, Chelmsford, MA). The two mobile phases used were A, acetonitrile:methanol:buffer:water (15:5:10:70), and B, acetonitrile:methanol:buffer:water (50:20:10:20). The buffer was a mixture of 0.25 M citric acid and 0.5 M ammonium acetate in triple-distilled water, and the pH was adjusted to 3.6 with acetic acid. The 95-μL injections were carried out on a Phenomenex Luna-2 C-18 column (250 by 4.6 mm, 5 μm; Phenomenex, Torrance, CA), initially eluted isocratically at 90% A/10% B for 15 min, followed by a linear gradient to 90% B in the next 40 min, and held there for 5 min (total 50 min gradient) at a flow rate of 1 mL/min and a temperature of 30 °C. The serial array of 12 coulometric electrodes was set at potentials of –35, 10, 70, 140, 210, 280, 350, 420, 490, 550, 620, and 690 mV. The system was controlled and the data were acquired and processed using the CoulArray software package (ESA). Three-point calibration curves were run for each standard. Triplicate samples were analyzed for each data point. The analytes were identified by their retention time and peak height ratios between the dominant peak and the peaks in the two adjacent channels. The analytes were quantified by comparison with known amounts of standards. The % recovery of each standard was used to normalize the data.
2.6. Immunoblotting analysis
The expression pattern of three (CYP1B1, COMT and NQO1) important enzymes from control and treated MCF-10F cells was analyzed by immunoblotting as described previously [17,33].
2.7. Statistical analysis
The statistical significance of the results was determined by Student’s t test and ANOVA analysis by using SAS and Graphpad Prism 4.0 software.
P < 0.05 was considered significant. All cultures and immunoblottings were repeated three times.
3. Results
To continue our study of the mechanism of carcinogenesis of DES, human breast epithelial MCF-10F cells were incubated with DES and the formation of DES–DNA adducts was quantified.
3.1. DES cytotoxicity
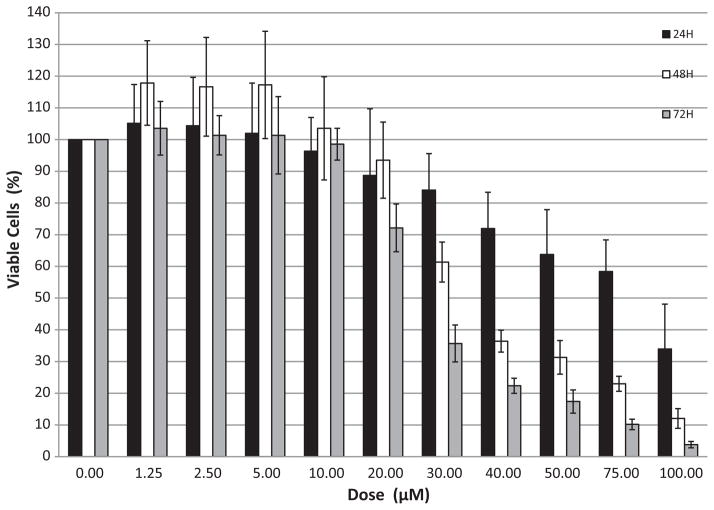
Exponentially growing MCF-10F cells were incubated with DES in doses ranging from 1.25 to 100 μM for 24, 48, or 72 h on separate 96-well plates. Plots for examining DES cytotoxicity were constructed by converting the MTT absorbance values to cell numbers at 0, 24, 48 and 72 h with respect to those in the control wells. Cell numbers were determined from optical density (OD) measurements, which were standardized by measuring the OD of cells plated on a 96-well plate in a series of decreasing concentrations starting with 40,000 cells and decreasing by half the concentration thereafter. Percent reduction in cell viability was inferred from percent reduction in cell numbers. The latter was calculated in relation to a blank control.
A dose-dependent cytotoxic response was observed between 20 and 100 μM DES. At the low and intermediate doses (1.25–20 μM), insignificant differences in cell numbers were observed compared to the blank control for all 3 days (Fig. 2). At the 30 μM dose, the highest dose used in subsequent studies, 16, 38 and 77% reductions in viable cells in relation to the blank control was observed at 24, 48 and 72 h, respectively. At the high doses (≥40 μM) cell numbers declined for all three days (Fig. 2) and there was no re-growth beyond this period (data not shown).
Fig. 2.
DES cytotoxicity in MCF-10F cells. Cells were incubated for 24, 48 or 72 h with 0–100 μM DES.
3.2. Optimal conditions for the formation of depurinating DES–DNA adducts
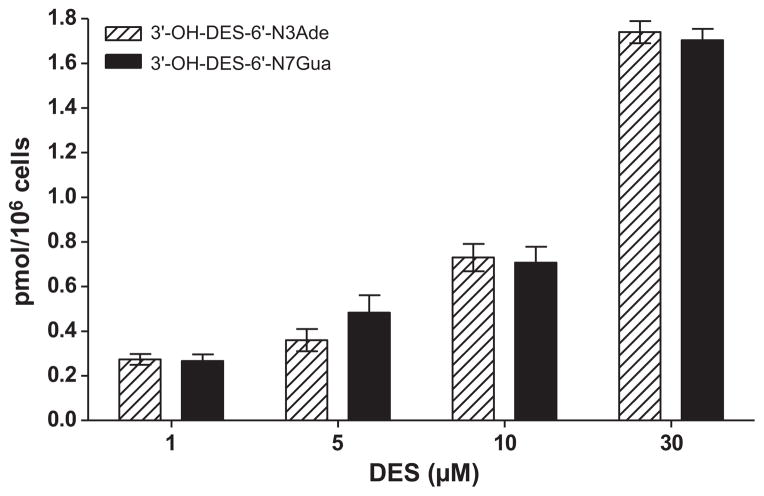
To investigate the relationship between DES concentrations and adduct formation, MCF-10F cells were incubated with increasing concentrations of DES (1–30 μM) for 24 h. Increasing concentrations of DES resulted in higher levels of the 3′-OH-DES-6′-N3Ade and 3′-OH-DES-6′-N7Gua adducts (Fig. 3). The relationship between DES and adduct concentration was linear with an R2 = 0.96 (correlation analysis). On average, there were nearly 6.5 times more adducts formed with the 30 μM dose than with the 1 μM. The two adducts observed, 3′-OH-DES-6′-N3Ade and 3′-OH-DES-6′-N7Gua, were formed in nearly equal amounts for each dose of DES (Fig. 3).
Fig. 3.
Incubation of MCF-10F cells with 1–30 μM DES for 24 h.
The optimum time period for incubation of DES with MCF-10F cells to maximize the yield of DES–DNA adducts was evaluated by incubating MCF-10F cells with 10 μM DES. The highest amount of adducts was found at 24 h (data not shown).
3.3. Effect of DES on the expression of COMT, NQO1 and CYP1B1 in MCF-10F cells
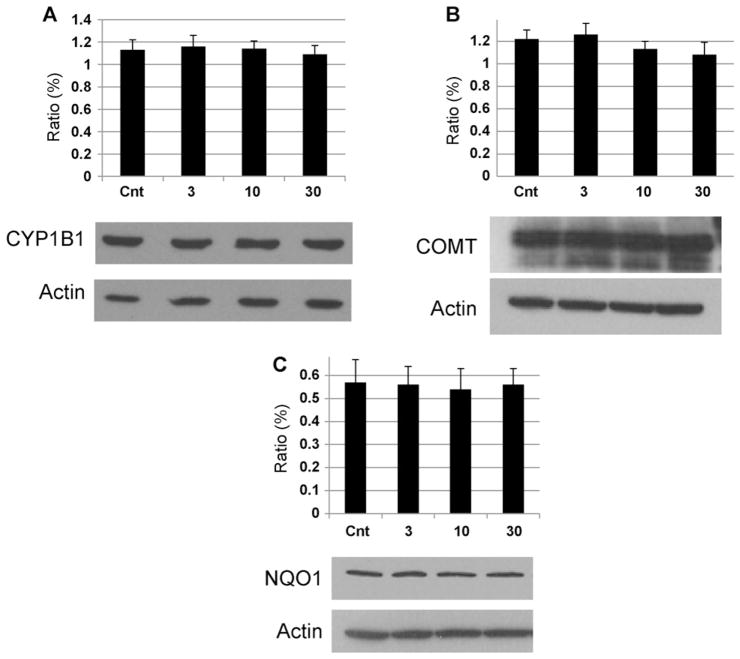
To determine the possible effect of DES on activating enzymes (CYP1B1) or protective enzymes (COMT and NQO1), MCF-10F cells were incubated with various concentrations of DES (1, 10 or 30 μM) for 24 h. These enzymes are of particular interest because breast tissue from women with breast cancer showed higher levels of activating enzymes and lower levels of protective enzymes than breast tissue from control women [34]. Immunoblotting of the samples showed that the presence of DES did not affect expression of the COMT, NQO1 or CYP1B1 proteins (Fig. 4), and, thus, did not appear to affect the balance of estrogen metabolism.
Fig. 4.
Western blot analysis of CYP1B1, COMT and NQO1 protein in MCF-10F cells after incubation with DES.
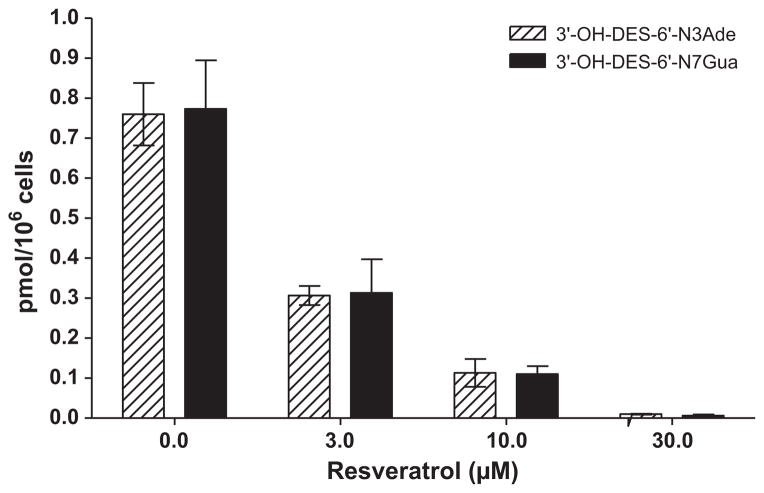
3.4. Prevention of DES–DNA adduct formation by resveratrol
To determine the effects of the antioxidant resveratrol on the formation of depurinating DES–DNA adducts, MCF-10F cells were incubated with resveratrol at increasing concentrations (3–30 μM) for 48 h before and concomitantly with 10 μM DES for 24 h. Increasing levels of resveratrol resulted in a decrease in adduct formation (Fig. 5). The relationship between the concentration of resveratrol and the level of adducts formed was linear with an R2 = 0.94 (by correlation analysis). Adduct formation was almost completely inhibited in cells treated with the 30 μM dose of resveratrol, an overall decrease of 99% from the control value. While not as extensive, resveratrol doses of 3 and 10 μM significantly reduced adduct formation by 59% and 86%, respectively.
Fig. 5.
Effect of resveratrol on the formation of depurinating DES–DNA adducts in MCF-10F cells incubated with various doses of resveratrol and 10 μM DES for 24 h.
4. Discussion
A substantial amount of evidence generated by us and others has clearly demonstrated the significance of mutation and cellular transformation induced by the formation of depurinating estrogen–DNA adducts, which play a critical role in the initiation of breast cancer [4–15]. DES, a synthetic estrogen analogue and established human carcinogen, shares many physicochemical similarities with the natural estrogens E1 and E2. Therefore, it is rational to hypothesize that initiation of cancer by DES may occur by the formation of depurinating DES–DNA adducts. This mechanism has great importance for our understanding of the involvement of estrogens in breast cancer. Toward that end, we previously reported the formation and indirect detection of depurinating DES–DNA adducts following incubation of calf thymus DNA with DES at 37 °C in the presence of different activating enzymes [27]. The present study demonstrates for the first time that DES also forms depurinating adducts in cultured human breast epithelial cells. In these studies, incubation of MCF-10F cells with 30 μM DES for 24 h resulted in the highest yield of DES–DNA adducts. As a known carcinogenic estrogen analogue in humans, the similar physicochemical properties of the DES catechol quinone and E1(E2)-3,4-Q support the hypothesis that DES initiates neoplasia in vivo by a genotoxic mechanism similar to that of the natural estrogens.
In this genotoxic mechanism for the initiation of breast cancer, it is possible to prevent key carcinogenic events, namely the formation of E1(E2)-3,4-Q and their reaction with DNA. Previous studies have shown a reduction in the formation of depurinating adducts by resveratrol when E1(E2)-3,4-Q and DNA are incubated in vitro [28] and when MCF-10F cells are incubated with E2 plus resveratrol [15,31,35]. Resveratrol prevents oxidative and/or electrophilic damage to DNA by inhibiting the formation of the electrophilic E1(E2)-3,4-Q and their reaction with DNA. The effects of resveratrol include reducing estrogen semiquinones to catechols, inducing the expression of NQO1, which reduces quinones to catechols, and modulating CYP1B1 activity in MCF-10F cells (Fig. 1) [15,28,31,35]. The present study is consistent with the results obtained with the natural estrogens and demonstrates that increasing doses of resveratrol are associated with a dose-dependent reduction in DES–DNA adduct formation in MCF-10F cells.
Acknowledgments
This work was supported by Prevention LLC. Core support at the Eppley Institute was provided by grant P30 CA36727 from the National Cancer Institute.
Abbreviations
- COMT
catechol-O-methyltransferase
- CYP
cytochrome P450
- DES
diethylstilbestrol
- HPLC-ECD
high performance liquid chromatography with electrochemical detection
- MTT
3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltrazolium bromide
- NQO1
NAD(P)H quinone oxidoreductase 1
References
- 1.Abramson PD. The relationship between human breast carcinoma and sex hormones. New Orleans Med Surg J. 1950;102:637–642. [PubMed] [Google Scholar]
- 2.Cavalieri EL, Stack DE, Devanesan PD, Todorovic R, Dwivedy I, Higginbotham S, Johansson SL, Patil KD, Gross ML, Gooden JK, Ramanathan R, Cerny RL, et al. Molecular origin of cancer: catechol estrogen-3,4-quinones as endogenous tumor initiators. Proc Natl Acad Sci USA. 1997;94:10937–10942. doi: 10.1073/pnas.94.20.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li KM, Todorovic R, Devanesan P, Higginbotham S, Kofeler H, Ramanathan R, Gross ML, Rogan EG, Cavalieri EL. Metabolism and DNA binding studies of 4-hydroxyestradiol and estradiol-3,4-quinone in vitro and in female ACI rat mammary gland in vivo. Carcinogenesis. 2004;25:289–297. doi: 10.1093/carcin/bgg191. [DOI] [PubMed] [Google Scholar]
- 4.Zahid M, Kohli E, Saeed M, Rogan E, Cavalieri E. The greater reactivity of estradiol-3,4-quinone vs estradiol-2,3-quinone with DNA in the formation of depurinating adducts: implications for tumor-initiating activity. Chem Res Toxicol. 2006;19:164–172. doi: 10.1021/tx050229y. [DOI] [PubMed] [Google Scholar]
- 5.Cavalieri E, Chakravarti D, Guttenplan J, Hart E, Ingle J, Jankowiak R, Muti P, Rogan E, Russo J, Santen R, Sutter T. Catechol estrogen quinones as initiators of breast and other human cancers. Implications for biomarkers of susceptibility and cancer prevention. BBA-Rev Cancer. 2006;1766:63–78. doi: 10.1016/j.bbcan.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Cavalieri EL, Rogan EG. Depurinating estrogen–DNA adducts in the etiology and prevention of breast and other human cancers. Future Oncol. 2010;6:75–91. doi: 10.2217/fon.09.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarti D, Mailander P, Li KM, Higginbotham S, Zhang H, Gross ML, Cavalieri EL, Rogan EG. Evidence that a burst of DNA depurination in SENCAR mouse skin induces error prone repair and forms mutations in the H-ras gene. Oncogene. 2001;20:7945–7953. doi: 10.1038/sj.onc.1204969. [DOI] [PubMed] [Google Scholar]
- 8.Mailander PC, Meza JL, Higginbotham S, Chakravarti D. Induction of A.T. to G.C. mutations by erroneous repair of depurinated DNA following estrogen treatment of the mammary gland of ACI rats. J Steroid Biochem Mol Biol. 2006;101:204–215. doi: 10.1016/j.jsbmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez SV, Russo IH, Russo J. Estradiol and its metabolites 4-hydroxyestradiol and 2-hydroxyestradiol induce mutations in human breast epithelial cells. Int J Cancer. 2006;118:1862–1868. doi: 10.1002/ijc.21590. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Z, Kosinska W, Khmelnitsky M, Cavalieri EL, Rogan EG, Chakravarti D, Sacks P, Guttenplan JB. Mutagenic activity of 4-hydroxyestradiol, but not 2-hydroxyestradiol, in BB2 rat embryonic cells, and the mutational spectrum of 4-hydroxyestradiol. Chem Res Toxicol. 2006;19:475–479. doi: 10.1021/tx0502645. [DOI] [PubMed] [Google Scholar]
- 11.Russo J, Lareef MH, Balogh G, Guo S, Russo IH. Estrogen and its metabolites are carcinogenic agents in human breast epithelial cells. J Steroid Biochem Mol Biol. 2003;87:1–25. doi: 10.1016/s0960-0760(03)00390-x. [DOI] [PubMed] [Google Scholar]
- 12.Lareef MH, Garber J, Russo PA, Russo IH, Heulings R, Russo J. The estrogen antagonist ICI-182-780 does not inhibit the transformation phenotypes induced by 17-Beta-estradiol and 4-OH estradiol in human breast epithelial cells. Int J Oncol. 2005;26:423–429. [PubMed] [Google Scholar]
- 13.Russo J, Fernandez SV, Russo PA, Fernbaugh R, Sheriff FS, Lareef HM, Garber J, Fusso IH. 17-Beta-estradiol induces transformation and tumorigenesis in human breast epithelial cells. Faseb J. 2006;20:1622–1634. doi: 10.1096/fj.05-5399com. [DOI] [PubMed] [Google Scholar]
- 14.Venugopal D, Zahid M, Mailander PC, Meza JL, Rogan EG, Cavalieri EL, Chakravarti D. Reduction of estrogen-induced transformation of mouse mammary epithelial cells by N-acetylcysteine. J Steroid Biochem Mol Biol. 2008;109:22–30. doi: 10.1016/j.jsbmb.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu F, Zahid M, Wang C, Saeed M, Cavalieri EL, Rogan EG. Resveratrol prevents estrogen–DNA adduct formation and neoplastic transformation in MCF-10F cells. Cancer Prev Res. 2008;1:135–145. doi: 10.1158/1940-6207.CAPR-08-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zahid M, Saeed M, Lu F, Gaikwad N, Rogan E, Cavalieri E. Inhibition of catechol-O-methyltransferase increases estrogen–DNA adduct formation. Free Radic Biol Med. 2007;43:1534–1540. doi: 10.1016/j.freeradbiomed.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu F, Zahid M, Saeed M, Cavalieri EL, Rogan EG. Estrogen metabolism and formation of estrogen–DNA adducts in estradiol-treated MCF-10F cells. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin induction and catechol-O-methyltransferase inhibition. J Steroid Biochem Mol Biol. 2007;105:150–158. doi: 10.1016/j.jsbmb.2006.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liehr JG, Fang WF, Sirbasku DA, Ari-Ulubelen A. Carcinogenicity of cat-echolestrogens in Syrian hamsters. J Steroid Biochem Mol Biol. 1986;24:353–356. doi: 10.1016/0022-4731(86)90080-4. [DOI] [PubMed] [Google Scholar]
- 19.Li JJ, Li SA. Estrogen carcinogenesis in Syrian hamster tissue: role of metabolism. Fed Proc. 1987;46:1858–1863. [PubMed] [Google Scholar]
- 20.Newbold RR, Liehr JG. Induction of uterine adenocarcinoma in CD-1 mice by catechol estrogens. Cancer Res. 2000;60:235–237. [PubMed] [Google Scholar]
- 21.Yue W, Santen RJ, Wang JP, Li Y, Verderame MF, Bocchinfuso WP, Korach KS, Devanesan P, Todorovic R, Rogan EG, Cavalieri EL. Genotoxic metabolites of estradiol in breast: Potential mechanism of estradiol induced carcinogenesis. J Steroid Biochem Mol Biol. 2003;86:477–486. doi: 10.1016/s0960-0760(03)00377-7. [DOI] [PubMed] [Google Scholar]
- 22.Newbold R. Prenatal exposure to diethylstilbestrol (DES) Fertil Steril. 2008 Feb;89(Suppl 1) doi: 10.1016/j.fertnstert.2008.01.062. [DOI] [PubMed] [Google Scholar]
- 23.Greenberg ER, Barnes AB, Resseguie L, Barrett JA, Burnside S, Lanza LL, Neff RK, Stevens M, Young RH, Colton T. Breast cancer in mothers given diethylstilbestrol in pregnancy. N Engl J Med. 1984;311:1393–1398. doi: 10.1056/NEJM198411293112201. [DOI] [PubMed] [Google Scholar]
- 24.Colton T, Greenberg ER, Noller K, Resseguie L, Van Bennekom C, Heeren T, Zhang Y. Breast cancer in mothers prescribed diethylstilbestrol in pregnancy. Further follow-up. JAMA. 1993;269:2096–2100. [PubMed] [Google Scholar]
- 25.Palmer JR, Wise LA, Hatch EE, Troisi R, Titus-Ernstoff L, Strohsnitter W, Kaufman R, Herbst AL, Noller KL, Hyer M, Hoover RN. Prenatal diethylstilbestrol exposure and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1509–1514. doi: 10.1158/1055-9965.EPI-06-0109. [DOI] [PubMed] [Google Scholar]
- 26.Troisi R, Hatch EE, Titus-Ernstoff L, Hyer M, Palmer JR, Robboy SJ, Strohsnitter WC, Kaufman R, Herbst AL, Hoover RN. Cancer risk in women pre-natally exposed to diethylstilbestrol. Int J Cancer. 2007;121:356–360. doi: 10.1002/ijc.22631. [DOI] [PubMed] [Google Scholar]
- 27.Saeed M, Rogan E, Cavalieri E. Mechanism of metabolic activation and DNA adduct formation by the human carcinogen diethylstilbestrol: the defining link to natural estrogens. Int J Cancer. 2009;124:1276–1284. doi: 10.1002/ijc.24113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zahid M, Gaikwad NW, Rogan EG, Cavalieri EL. Inhibition of depurinating estrogen–DNA adduct formation by natural compounds. Chem Res Toxicol. 2007;20:1947–1953. doi: 10.1021/tx700269s. [DOI] [PubMed] [Google Scholar]
- 29.Stivala LA, Savio M, Carafoli F, Perucca P, Bianchi L, Maga G, Forti L, Pagnoni UM, Albini A, Prosperi E, Vannani V. Specific structural determinants are responsible for the antioxidant activity and the cell cycle effects of resveratrol. J Biol Chem. 2001;276:22586–22594. doi: 10.1074/jbc.M101846200. [DOI] [PubMed] [Google Scholar]
- 30.Jang YJ, Kang NJ, Lee KW, Lee HJ. Protective effects of red wine flavonols on 4-hydroynonenal-induced apoptosis in PC12 Cells. Ann N Y Acad Sci. 2009;1171:170–175. doi: 10.1111/j.1749-6632.2009.04720.x. [DOI] [PubMed] [Google Scholar]
- 31.Zahid M, Gaikwad NW, Ali MF, Lu F, Saeed M, Yang L, Rogan EG, Cavalieri EL. Prevention of estrogen–DNA adduct formation in MCF-10F cells by resveratrol. Free Radic Biol Med. 2008;45:136–145. doi: 10.1016/j.freeradbiomed.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denizot F, Lang R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J Immunol Methods. 1986;89:271–277. doi: 10.1016/0022-1759(86)90368-6. [DOI] [PubMed] [Google Scholar]
- 33.Zahid M, Saeed M, Rogan E, Cavalieri E. N-acetylcysteine blocks formation of cancer-initiating estrogen–DNA adducts in cells. Free Radic Biol Med. 2010;49:392–400. doi: 10.1016/j.freeradbiomed.2010.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh S, Chakravarti D, Edney JA, Hollins RR, Johnson PJ, West WW, Higginbotham S, Cavalieri E, Rogan E. Relative imbalance in the expression of estrogen-metabolizing enzymes in the breast tissue of women with breast carcinoma. Oncol Rep. 2005;14:1091–1096. [PubMed] [Google Scholar]
- 35.Zahid M, Saeed M, Beseler C, Rogan EG, Cavalieri EL. Resveratrol and N-acetylcysteine block the cancer-initiating step in MCF-10F cells. Free Radic Biol Med. 2011;50:78–85. doi: 10.1016/j.freeradbiomed.2010.10.662. [DOI] [PMC free article] [PubMed] [Google Scholar]