Abstract
Most men diagnosed with prostate cancer will experience indolent disease; hence discovering genetic variants that distinguish aggressive from non-aggressive prostate cancer is of critical clinical importance for disease prevention and treatment. In a multistage, case-only genome-wide association study of 12,518 prostate cancer cases, we identify two loci associated with Gleason score, a pathological measure of disease aggressiveness: rs35148638 at 5q14.3 (RASA1, P=6.49×10-9) and rs78943174 at 3q26.31 (NAALADL2, P=4.18×10-8). In a stratified case-control analysis, the SNP at 5q14.3 appears specific for aggressive prostate cancer (P=8.85×10-5) with no association for non-aggressive prostate cancer compared to controls (P=0.57). The proximity of these loci to genes involved in vascular disease suggests potential biological mechanisms worthy of further investigation.
INTRODUCTION
Prostate cancer is the most common cancer diagnosed in men and second leading cause of cancer death among men in the U.S.;1 however, little is known about why the disease progresses in some men but not others. Determining which cancers are likely to progress and cause death is of critical clinical importance. Prostate cancer aggressiveness is thought to be partially determined by genetic factors, as studies have shown an increased risk of death from prostate cancer among offspring with a family history of fatal disease.2, 3 The definitions of aggressive prostate cancer differ between studies, but one important and widely used descriptor is tumor grade at diagnosis, as measured by Gleason score, which ranks pathological changes, namely, tumor differentiation, and has been associated with disease progression and survival.4 Linkage studies using Gleason score as a measure of aggressiveness have implicated several chromosomal regions, including 1p, 5q, 6q, 7q, and 19q, but no specific genetic mutations have been conclusively identified.5-8 Although previous genetic association studies have identified or suggested markers for aggressive prostate cancer;9-11 these SNPs have either also been associated with non-aggressive disease, making them non-specific, or have not been convincingly replicated.12
Genome-wide association studies (GWAS) have successfully identified roughly 100 loci associated with prostate cancer risk;11-26 however, most loci have minor allele frequencies >10%, and so far, none conclusively differentiate aggressive from non-aggressive disease. To discover additional loci associated with risk and to identify loci specific for aggressive prostate cancer, here we conduct a multistage GWAS for prostate cancer among men of European ancestry using the Illumina HumanOmni2.5 Beadchip, which provides greater coverage of uncommon SNPs for individuals of European ancestry than microarrays used in previous GWAS of prostate cancer.
We identify two novel loci associated with Gleason score, a pathological measure of prostate cancer aggressiveness, that are located near genes involved vascular development and maintenance.
RESULTS
Case-control association results for prostate cancer
A total of 4,600 cases and 2,941 controls of European ancestry from the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial were genotyped using the Illumina HumanOmni2.5 Beadchip and passed rigorous quality control criteria (see Methods). Baseline characteristics of the cases and controls are shown in Supplementary Table 1. Based on a linear regression model, men with higher Gleason scores were more likely to be diagnosed at an older age (P<0.001). Of the SNPs genotyped, 1,531,807 passed quality control filters with a minimum call rate of 94%. Genotypes were analyzed using regression models, assuming a log-additive genetic model and adjusting for age and significant eigenvectors. A quantile-quantile (Q-Q) plot of the p-values for prostate cancer risk based on logistic regression models showed enrichment of small p-values compared to the null distribution, even after removing SNPs within 500kb of the previously published loci (Supplementary Fig. 1, λ = 1.007). Fifty-six of the previously published loci were nominally associated with risk (P<0.05) in stage 1 (Supplementary Table 2), and two previously published loci at chromosome 8q24 and 17q12 reached genome-wide significance in stage 1 (P<5×10-8, Supplementary Fig. 2). Rare variant analysis using SKAT27 for SNPs with minor allele frequencies <2% revealed five gene regions with P<5×10-8; however, all appeared to be artifacts driven by poorly clustered SNPs.
Sixteen promising SNPs with P < 2 × 10-5 based on the logistic regression models were taken forward for Taqman replication in 5,139 cases and 5,591 controls from seven studies (see Methods), but none replicated for prostate cancer overall (Supplementary Table 3). A more extensive replication was undertaken using a custom Illumina iSelect microarray comprised of 51,207 SNPs selected for prostate cancer, 10,458 SNPs for other phenotypes (e.g., obesity), and 1,435 candidate SNPs (see Methods). In stage 2, a total of 6,575 cases and 6,392 controls from five studies were genotyped with the custom iSelect and passed quality control criteria (Supplementary Table 1). In silico data was also available for stage 3 for 1,204 non-overlapping cases and 1,231 controls from a previous GWAS of advanced (defined as Gleason score ≥ 8 or stage C/D) prostate cancer,12 giving a total of 12,379 cases and 10,564 controls. As not all SNPs included on the iSelect were directly genotyped in stage 1 or stage 3, both scans were imputed using the 1000 Genomes Project release version 328 and IMPUTE2.29
In a combined meta-analysis of the primary scan together with the custom SNP microarray replication and in silico look-up in a previous GWAS, thirteen loci reached genome-wide significance (P < 5 × 10-8); however, each of them confirmed a previously reported locus13-20, 23 (Supplementary Table 4). Although not reaching genome-wide significance, two new suggestive loci at chromosome 16q22.2 (PKD1L3, rs12597458, P = 9.67 × 10-8) and 6p22.3 (CDKAL1, rs12198220, P = 2.13 × 10-7) were also identified (Supplementary Table 5, Supplementary Fig. 3). Further studies are needed to confirm these findings.
Case-only results of disease aggressiveness
To evaluate disease aggressiveness, we modeled Gleason score as a quantitative trait among the prostate cancer cases (n=4,545) included in stage 1 in a case-only analysis using linear regression. We chose to model Gleason score as a quantitative trait as opposed to a dichotomous outcome in order to maximize our statistical power to detect variants that differentiate aggressive from non-aggressive disease. In stage 1, the Q-Q plot of the association p-values revealed a small number of SNPs with p-values less than expected under the null distribution (Supplementary Fig. 4, lambda = 0.998). We evaluated the SNPs previously reported to be associated with the risk of aggressive disease, but none were significantly associated with Gleason score among cases (Supplementary Table 6). As part of the custom SNP microarray replication, SNPs with a p-value < 0.001 from the linear regression model of Gleason score as a quantitative trait, filtered using r2>0.8, were taken forward for the custom SNP microarray replication in 5,355 cases with Gleason score from five studies (stage 2). One novel locus at chromosome 5q14.3 (rs35148638) reached genome-wide significance in the meta-analysis of stage 1 and 2. Five SNPs, including two moderately correlated SNPs at chromosome 5q14.3, with p-values < 2 × 10-6 were taken forward for replication in 2,618 cases from the Cancer of the Prostate in Sweden (CAPS) study. In the meta-analysis of the Gleason score results for all three stages including a total of 12,518 cases, three SNPs reached genome-wide significance: rs35148638 at 5q14.3 (P = 6.49 × 10-9), rs62113212 at 19q13.33 (P = 5.85 × 10-9) and rs78943174 at 3q26.31 (P = 4.18 × 10-8) (Table 1). The SNPs at chromosome 5q14.3 and 3q26.31 represent novel loci (Figure 1), whereas the chromosome 19q13.33 locus has been previously identified to be associated with prostate cancer risk overall.18
Table 1.
Loci associated with prostate cancer aggressiveness as measured by Gleason score among cases only*
| SNP | Nearest gene(s) | Chr | Position | Risk allelea | Other allele | Stage | RAFb | No. of subjects | Beta | SE | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Novel loci | |||||||||||
|
| |||||||||||
| rs35148638 | RASA1, CCNH | 5q14.3 | 86610989 | C | A | Stage 1 | 0.25 | 4,545 | 0.085 | 0.024 | 3.09×10-4 |
| Stage 2 | 0.255 | 5,353 | 0.125 | 0.022 | 1.86×10-6 | ||||||
| Stage 3 | 0.262 | 2,618 | -0.025 | 0.040 | 0.50 | ||||||
| Combined | 12,516 | 0.088 | 0.015 | 6.49×10-9 | |||||||
| rs78943174 | NAALADL2 | 3q26.31 | 175252736 | C | T | Stage 1 | 0.985 | 4,545 | 0.361 | 0.085 | 2.44×10-5 |
| Stage 2 | 0.987 | 5,336 | 0.217 | 0.085 | 0.03 | ||||||
| Stage 3 | 0.979 | 2,604 | 0.293 | 0.110 | 0.008 | ||||||
| Combined | 12,485 | 0.289 | 0.053 | 4.18×10-8 | |||||||
|
| |||||||||||
|
Previously reported loci
| |||||||||||
| rs62113212 | KLK3 | 19q13.33 | 51360840 | T | C | Stage 1 | 0.063 | 4.544 | 0.177 | 0.043 | 4.00×10-5 |
| Stage 2 | 0.061 | 5,348 | 0.124 | 0.041 | 0.009 | ||||||
| Stage 3 | 0.107 | 2,616 | 0.119 | 0.040 | 0.004 | ||||||
| Combined | 12,508 | 0.138 | 0.024 | 5.85×10-9 | |||||||
Results from case only analysis of Gleason score as a quantitative trait using linear regression.
Risk allele is the allele associated with an increased risk of aggressive disease (e.g., higher Gleason score)
Frequency of the risk allele
Figure 1.
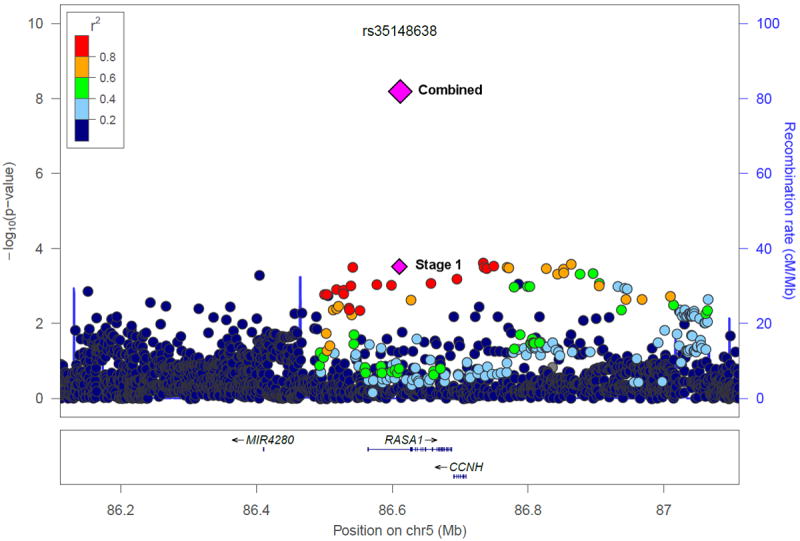
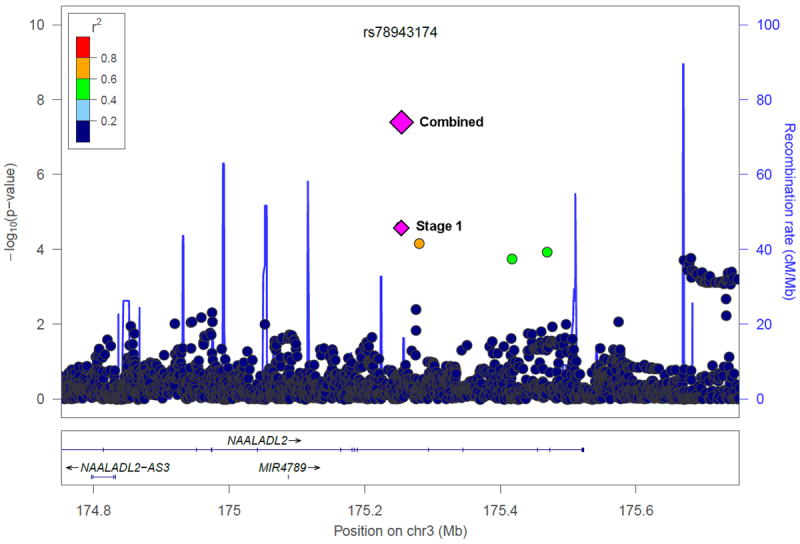
Regional association plots of the two novel loci associated with Gleason score as a quantitative trait among cases: (a) chromosome 5q14.3 (rs35148638) and (b) chromosome 3q26.31 (rs78943174). Shown are the −log10 association p-values from the linear regression model for the 4,545 cases in stage 1 (dots and lower purple diamond) and −log10 p-values from the linear regression model for the 12,518 cases in the combined stage 1-3 analysis (upper diamond).
Stratified case-control association results for novel loci
To evaluate the extent to which these three loci (identified from our case-only study of Gleason score) could be associated with aggressive prostate cancer risk, we conducted a case-control analysis stratified by overall Gleason score (e.g., 2 to 10), recognizing that our power to detect an association at genome-wide significance would be reduced. We did not have data on the individual components of Gleason 7 from most studies in order to sub-classify them as 3+4 or 4+3, and so in order to clearly differentiate between aggressive and non-aggressive disease, we stratified our cases by those with Gleason scores ≤ 6 (non-aggressive) and those with Gleason ≥ 8 (aggressive). Although it did not reach genome-wide significance, rs35148638 at 5q14.3 showed an increased risk with aggressive prostate cancer (P = 8.85 × 10-5) (Supplementary Table 7). There was no association for non-aggressive disease (P = 0.57) and the p-value for heterogeneity between the two outcomes was modestly significant (P=2.89 × 10-4). As rs78943174 at 3q26.31 was not common among controls with a minor allele frequency of 1-2%, we had limited power to detect an association with aggressive disease; however, we did observe a marginal positive association between rs78943174 and aggressive prostate cancer risk (P=0.07) and the p-value for heterogeneity between aggressive and non-aggressive prostate cancer risk was nominally significant (P=0.006). Consistent with previous studies,30 the SNP at 19q13.33 was strongly associated with non-aggressive prostate cancer (P = 3.51 × 10-13) with a weak association in the opposite direction for aggressive prostate cancer (P = 0.01) and highly significant p-value for heterogeneity (P=1.44 × 10-10).
As African Americans have an elevated risk of prostate cancer, we evaluated the extent to which these three SNPs were associated with aggressive prostate cancer risk in African Americans using data from the African American Prostate Cancer (AAPC) GWAS Consortium (see Methods). In this smaller study, none of the SNPs were significantly associated with the risk of aggressive disease (Supplementary Table 8), and only rs62113212 at chromosome 19q13.33 was nominally associated with non-aggressive disease (P=0.04). However, the direction of the effects for African Americans for the SNPs at 3q26.31 and 19q13.33 were consistent with what we observed among Europeans.
Further examination of novel loci
Examination of the three identified loci for Gleason score using data from ENCODE revealed significant DNase enrichment in lymphoblastoid and embryonic myoblast cells and evidence for altered motifs (Supplementary Table 9). Rs62113212 at 19q13.33 is in strong linkage disequilibrium with a missense SNP (rs17632542, r2=1). No significant expression quantitative trait locus (eQTL) associations were observed using data from the Genotype-Tissue Expression (GTEx) Project;31 however, for a proxy of rs35148638 at 5q14.3 (rs4421140, r2=0.82), we did find nominally significant eqtl associations with RASA1 and CCNH expression and meqtl associations with CpG sites in RASA1 and CCNH in adipose tissue.32, 33
DISCUSSION
Linkage studies of prostate cancer aggressiveness have reported suggestive evidence of linkage to chromosome 5q5-8 and specifically 5q14 in TMPRSS2-ERG fusion positive families.34 The 5q14.3 SNP identified in this study (rs35148638), associated with disease aggressiveness is intronic to the RAS p21 protein activator 1 (RASA1) gene, which suppresses RAS function, helps regulate cellular proliferation and differentiation,35 and controls blood vessel growth.36 Rare mutations in RASA1 lead to capillary malformation-arteriovenous malformation and Parkes-Weber syndrome37 as well as lymphatic abnormalities,38 providing an intriguing plausibility for the gene in aggressive prostate cancer. The SNP is also approximately 79kb downstream of the cyclin H (CCNH) gene, which encodes a regulatory component of a cyclin-dependent (CDK)-activating kinase (CAK), necessary for RNA polymerase II transcription, nucleotide excision repair, and p53 phosphorylation.39 CCNH has been shown to be differentially expressed between androgen-sensitive and androgen-resistant prostate cancer cell lines,40, 41 suggesting a role in prostate cancer progression.
The 3q26.31 SNP (rs78943174) is intronic to the N-acetylated alpha-linked acidic dipeptidase-like2 (NAALADL2) gene, which is part of the glutamate carboxypeptidase II family. This gene is also related to prostate-specific membrane antigen (PSMA), a well-characterized diagnostic indicator and potential drug target of prostate cancer.42 NAALADL2 has been shown to promote a pro-migratory and pro-metastatic microenvironment, and higher tumor expression of NAALAD2 is associated with higher Gleason score and poor survival following radical prostatectomy.43 Variants in NAALADL2 have also been identified to be associated with Kawasaki disease,44 a pediatric, autoimmune vascular disease. The SNP is also approximately 117kb telomeric of the microRNA, MIR4789, which is predicted to target several genes involved in the insulin resistance (e.g., IRS1, PIK3R1) among others.45, 46 A previous GWAS of prostate cancer reported a suggestive association with a SNP at 3q26.31,47 but this SNP is not in linkage disequilibrium with the SNP identified in our study (r2=0.003).
The chromosome 19q13.33 (KLK3) locus has been previously associated with prostate cancer risk overall.18 Although several studies have suggested that the risk may differ by disease aggressiveness,30, 48-50 this study shows for the first time a genome-wide significant difference between aggressive and non-aggressive disease as measured by Gleason score. KLK3 encodes the prostate-specific antigen (PSA) protein. The C allele of rs62113212 has been shown to be associated with higher PSA levels,48 suggesting the association observed with the SNP is related to early prostate cancer detection.
Although one of our goals was to identify uncommon variants for prostate cancer, we did not identify any new independent SNPs with a minor allele frequency < 10%. We did, however, identify a suggestive locus at chromosome 6p22.3 (rs12198220), which is 98 kb downstream of CDKAL1. A pooled linkage study of prostate cancer previously reported suggestive evidence of linkage to this region.51 Interestingly, SNPs at this locus have been associated with the risk of type 2 diabetes, adding to the list of susceptibility regions shared between prostate cancer and type 2 diabetes.52 We also discovered a new suggestive locus at 16q22.2, which in strong linkage disequilibrium with a missense variant (rs3213422, r2=0.74) in dihydro-orotate dehydrogenase (quinone) gene (DHODH), which encodes an enzyme necessary for the biosynthesis of pyrimidines and cell proliferation. Further studies are needed to confirm these suggestive loci.
In this study, we used Gleason score to differentiate between non-aggressive and aggressive prostate cancer. Gleason score is a powerful prognostic factor and predictor of disease behavior; however, substantial changes in Gleason scoring have changed since it was first proposed over 40 years ago, resulting in shifts toward higher scores.53 In addition, differences in scoring between pathologists remain.54 Whether these changes in Gleason scoring ultimately result in better outcome prediction and classification of disease from an etiologic standpoint remains to be seen. Unlike for breast cancer where classification by receptor status has resulted in significant advances in the etiologic understanding of the disease, clearly defining aggressive prostate cancer remains difficult. Regardless, Gleason score is an important component of prostate cancer risk assessment and is the most commonly used tool for assessing prostate cancer aggressiveness.
In conclusion, we identified two new loci associated with prostate cancer aggressiveness as measured by Gleason score in a case-only study of prostate cancer. Although additional studies are needed to confirm these findings and reveal the underlying biological mechanism, the proximity of these SNPs to genes involved in vascular disease, cell migration, and metastasis makes them intriguing loci for further study.
Methods
Stage 1. Discovery population and genotyping
A new genome-wide association study was conducted in prostate cancer cases and controls of European ancestry from the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. PLCO is a randomized trial for the early detection of prostate, lung, colorectal and ovarian cancers.55 In brief, 76,693 men were enrolled in the trial from 10 centers in the United States from 1993 to 2001 and randomized to receive annual screening with prostate-specific antigen (PSA) for six years and digital rectal exam for the four years or referred to their physician for routine care. Men with positive screening results were referred to their primary physician for further evaluation. All prostate cancer cases detected during screening or reported during the trial were pathologically confirmed, and information on stage and grade was abstracted from medical records. Blood or buccal cells were collected from participants in the trial.56 The study was approved by the institutional review board at each center and NCI; all study participants provided written consent.
A total of 4,838 prostate cancer cases and 3,053 controls of European ancestry, matched on age and year of randomization, were selected for stage 1. The sample size was chosen based on statistical power estimates for detecting a modest association in a multistage genome-wide association study. Including quality control duplicates, 8,222 samples were genotyped on the Illumina HumanOmni2.5 Beadchip. Extensive quality control metrics were employed to ensure that only high quality genotype data was analyzed using the GLU software package. Samples with a missing rate > 6% (n=323) or heterozygosity < 16% or > 21% (n=7) were excluded, and 221 samples were removed due to technical issues. Gender discordance based on chromosome X heterozygosity was evaluated, but no subjects were removed. One unexpected duplicate (>99.9% concordance) and 28 full sibling pairs based on an identity-by-descent threshold of 0.70 were detected and one subject from each pair was removed (n=29). Ancestry was estimated using a set of population informative markers57 and the GLU struct.admix module, which is similar to the method proposed by Pritchard et al.58 Five subjects (3 cases and 2 controls) were determined to have <80% European ancestry and were removed from analysis (Supplementary Fig. 5). Principal components analysis was performed to evaluate population substructure in greater detail (Supplementary Fig. 6) and two significant eigenvectors (P<0.05) were include in the analytic model. Expected duplicates yielded 99.9% concordance. SNPs without genotype calls, a completion rate <94%, Hardy-Weinberg proportion test p-value < 1 × 10-8, or minor allele frequency < 1% were excluded, leaving 1,531,807 SNPs for analysis. After quality control exclusions, 4,600 cases and 2,840 controls remained. An additional 101 male controls from PLCO Cancer Screening Trial, genotyped previously on the HumanOmni2.5,59 were also included, resulting in 4,600 cases and 2,941 controls for the primary prostate cancer analysis (Supplementary Table 1 and Supplementary Fig. 7). Of those cases, 4,545 men had information on Gleason score available. Regression models were fit adjusting for significant principal components and age. Sixteen different models were fitted for prostate cancer related outcomes, including overall prostate cancer risk and Gleason score.
Stage 2. Follow-up studies and genotyping
Replication was conducted using a set of independent prostate cancer cases and controls from five studies: Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study60 (ATBC, n=1092 cases, 1099 controls), Cancer Prevention Study II61 (CPSII, n=2,770 cases/2,669 controls), Health Professional Follow-up Study62 (HPFS, n=963 cases/1,047 controls), French Prostate Cancer Case-Control Study (CeRePP, n=1,494 cases/1,546 controls), and PLCO55 (n=990 cases/922 controls). Including quality control duplicates, 14,592 samples were genotyped using a custom Illumina iSelect microarray comprised of 51,207 SNPs selected for prostate cancer, 10,458 SNPs for other phenotypes (e.g., smoking, obesity), and 1,435 candidate SNPs. The SNPs for prostate cancer were filtered using r2<0.7 and selected based on the most significant results from sixteen prostate cancer models with the primary models being: an overall prostate cancer risk model assuming a log-additive effect for each SNP and case-only Gleason score model, where Gleason was modeled as a quantitative linear trait among cases and each SNP was assumed to have an additive effect. SNPs with p-values <0.05 and <0.001 from each model, respectively, were advanced for possible replication.
Similar to stage 1, samples genotyped in stage 2 underwent rigorous quality control procedures. Samples with missing rate > 10% (n=1,158) or mean heterozygosity < 20% or > 26% (n=4) were excluded. In addition, 21 subjects without phenotype information were removed. Fifteen unexpected duplicates with concordance rates >99.9% were observed, and twenty-five first degree relative pairs were detected assuming an identity-by-descent threshold of 0.7. For each unexpected duplicate and relative pair, one subject was removed. Using the GLU struct.admix module, ancestry was estimated based on genotyped SNPs with a MAF>10% and HapMap data as the fixed reference population. Sixty-six subjects with <80% European ancestry were removed from analysis. Principal components analysis was conducted using a set of SNPs selected for traits unrelated to prostate cancer (e.g., smoking, alcohol intake). After quality control exclusions, a total of 6,575 cases and 6,392 controls remained for the primary analysis (Supplementary Table 1), including 5,355 cases with Gleason score. SNPs with a minor allele frequency <1% or completion rate <90% were excluded from analysis, leaving 55,497 SNPs for analysis (Supplementary Fig. 7). Regression models were fit adjusting for age, significant principal components, and study.
In addition to the custom SNP microarray replication, 16 promising SNPs (P<2×10-5) were taken forward for fast-track replication in the five studies listed above (n=2,495 cases/2,532 controls) as well as three additional studies: Agricultural Health Study63 (AHS, n=579 cases/1,172 controls), Fred Hutchinson Cancer Research Center (FHCRC, n=1,315 cases/1,152 controls), and the Multiethnic Cohort64 (MEC, n=750 cases/735 controls). In total, 5,139 cases and 5,591 controls, all of European ancestry, were genotyped (Supplementary Fig. 7). The SNPs were genotyped using individual TaqMan assays (Applied Biosystems, Inc) and quality control duplicates yielded >99.9% concordance.
Stage 3a. In silico replication of prostate cancer findings
For replication of the overall prostate cancer results, non-overlapping in silico GWAS data was available from 1,204 cases and 1,231 controls of European ancestry from four studies from a previous GWAS of advanced prostate cancer12: European Prospective Investigation into Cancer and Nutrition (EPIC; 431 cases / 426 controls),65 Multiethnic Cohort (MEC; 244 cases/ 259 controls),64 Physicians Health Study (PHS; 298 cases / 255 controls), and American Cancer Society Cancer Prevention Study II (CPSII; 231 cases / 291 controls not included in stage 2)61 (Supplementary Fig. 7). Subjects were genotyped using the Illumina HumanHap610K and extensive quality control filters applied as described previously. All data was imputed using IMPUTE229 and 1000 Genomes Project release version 328 as the reference panel, and data analyzed using SNPTEST assuming a log-additive genetic model and adjusting for age, study, and significant principal components. Only SNPs with an information score >0.3 were included in the meta-analysis.
Stage 3b. Additional replication for Gleason score findings
For further replication of the results for Gleason score, we genotyped five of the most significant SNPs (P<2 × 10-6) in the Cancer of the Prostate in Sweden (CAPS), a population-based case-control study of 2,618 cases and 1,728 controls using Sequenom (Supplementary Fig. 7). Regression models were fit adjusting for age.
Meta-analysis
Data from all three stages were meta-analyzed using the fixed effects inverse variance method based on the beta estimates and standard errors from each stage.
Further follow-up analyses
To evaluate the associations observed in our study of men of European ancestry with those observed in African Americans, we obtained association results for three genome-wide significant hits from the African American Prostate Cancer (AAPC) GWAS Consortium.22 Although it was not possible to evaluate Gleason score as a quantitative trait among cases in this consortium, we were able to obtain stratified association results for cases with Gleason ≤ 6 versus controls and cases with Gleason ≥ 8 versus controls.
Using 1000 Genomes Project data, we identified SNPs with r2>0.8 with the lead SNPs identified to be associated with Gleason score and evaluated whether they were non-synonymous coding variants. We utilized HaploReg66 to assess non-coding functional markers in the regions containing our lead SNPs and related proxy SNPs (r2>0.8) (Supplementary Table 9). We explored possible cis expression quantitative trait loci (eQTL) associations with our lead SNPs and related proxy SNPs (r2>0.8) in adipose tissue, lymphoblastoid cell lines, and skin using data from the MuTHER resource32 and all available tissues, including whole blood, in the Genotype-Tissue Expression Project (GTEx).31 We also examined possible methylation quantitative trait loci (meQTL) associations in adipose tissue using the MuTHER resource.33
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH (Z01-CP010119, ZIA-CP01015211) and in part by the National Institute of Environmental Health Sciences (Z01-ES049030). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organization indicate endorsement by the U.S. Government. The authors thank Drs. Christine Berg and Philip Prorok, Division of Cancer Prevention, NCI, the screening center investigators and staff of the PLCO Cancer Screening Trial, Mr. Thomas Riley and staff at Information Management Services, Inc., and Ms. Barbara O’Brien and staff at Westat, Inc. for their contributions to the PLCO. Finally, we are grateful to the study participants for donating their time and making this study possible.
Footnotes
AUTHOR CONTRIBUTIONS
S.I.B., Z.W., M.Y., L.A., J.S., A.B., K.J., R.N.H., M.T., S.J.C. organized and designed the study. Z.W., A.H., M.Y., S.J.C. conducted/supervised the genotyping of the samples S.I.B., Z.W., M.Y., M.M., J.S., H.Z., W.W., K.Y., K.J., M.T., S.J.C. contributed to the design and execution of the statistical and/or bioinformatic analysis. S.I.B. and S.J.C. drafted the manuscript. S.I.B., M.C.A., D.A., G.A., L.B.F., D.C., G.C-T., F.C., J-N.C., O.C., W.R.D., S.M.G., H.G., C.A.H., B.H., D.H., T.J.K., S.K., S.K., P.K., L.L.M., S.L., E.A.O., E.R., F.S., A.S., J.S., V.L.S., R.C.T., K.T., J.V., S.W., F.W., J.X., S.L.Z., A.B. R.N.H. conducted the epidemiologic studies and contributed samples to the GWAS and/or follow-up genotyping. All authors contributed to the writing of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
There are no conflicts of interest.
ACCESSION NUMBER
The GWAS data from stage 1 is available on dbGaP under accession number phs000882.v1.p1.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Lindstrom LS, et al. Familial concordance in cancer survival: a Swedish population-based study. Lancet Oncol. 2007;8:1001–1006. doi: 10.1016/S1470-2045(07)70282-6. [DOI] [PubMed] [Google Scholar]
- 3.Hemminki K. Familial risk and familial survival in prostate cancer. World J Urol. 2012;30:143–148. doi: 10.1007/s00345-011-0801-1. [DOI] [PubMed] [Google Scholar]
- 4.Gleason DF. Histologic grading of prostate cancer: a perspective. Hum Pathol. 1992;23:273–279. doi: 10.1016/0046-8177(92)90108-f. [DOI] [PubMed] [Google Scholar]
- 5.Witte JS, et al. Genomewide scan for prostate cancer-aggressiveness loci. Am J Hum Genet. 2000;67:92–99. doi: 10.1086/302960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witte JS, et al. Genome-wide scan of brothers: replication and fine mapping of prostate cancer susceptibility and aggressiveness loci. Prostate. 2003;57:298–308. doi: 10.1002/pros.10304. [DOI] [PubMed] [Google Scholar]
- 7.Slager SL, et al. Genome-wide linkage scan for prostate cancer aggressiveness loci using families from the University of Michigan Prostate Cancer Genetics Project. Prostate. 2006;66:173–179. doi: 10.1002/pros.20332. [DOI] [PubMed] [Google Scholar]
- 8.Schaid DJ, et al. Genome-wide linkage scan of prostate cancer Gleason score and confirmation of chromosome 19q. Hum Genet. 2007;121:729–735. doi: 10.1007/s00439-007-0368-5. [DOI] [PubMed] [Google Scholar]
- 9.Sun J, et al. Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res. 2009;69:10–15. doi: 10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J, et al. Inherited genetic variant predisposes to aggressive but not indolent prostate cancer. Proc Natl Acad Sci U S A. 2010;107:2136–2140. doi: 10.1073/pnas.0914061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al Olama AA, et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet. 2013;22:408–415. doi: 10.1093/hmg/dds425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumacher FR, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20:3867–3875. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 14.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 15.Gudmundsson J, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 16.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 17.Gudmundsson J, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 19.Gudmundsson J, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eeles RA, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takata R, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42:751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 22.Haiman CA, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43:570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kote-Jarai Z, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43:785–791. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, et al. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat Genet. 2012;44:1231–1235. doi: 10.1038/ng.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eeles RA, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45:385–2. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al Olama AA, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. 2014;46:1103–1109. doi: 10.1038/ng.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu MC, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindstrom S, et al. Characterizing associations and SNP-environment interactions for GWAS-identified prostate cancer risk markers--results from BPC3. PLoS One. 2011;6:e17142. doi: 10.1371/journal.pone.0017142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grundberg E, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93:876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luedeke M, et al. Predisposition for TMPRSS2-ERG fusion in prostate cancer by variants in DNA repair genes. Cancer Epidemiol Biomarkers Prev. 2009;18:3030–3035. doi: 10.1158/1055-9965.EPI-09-0772. [DOI] [PubMed] [Google Scholar]
- 35.McCormick F. ras GTPase activating protein: signal transmitter and signal terminator. Cell. 1989;56:5–8. doi: 10.1016/0092-8674(89)90976-8. [DOI] [PubMed] [Google Scholar]
- 36.Henkemeyer M, et al. Vascular system defects and neuronal apoptosis in mice lacking ras GTPase-activating protein. Nature. 1995;377:695–701. doi: 10.1038/377695a0. [DOI] [PubMed] [Google Scholar]
- 37.Eerola I, et al. Capillary malformation-arteriovenous malformation, a new clinical and genetic disorder caused by RASA1 mutations. Am J Hum Genet. 2003;73:1240–1249. doi: 10.1086/379793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burrows PE, et al. Lymphatic abnormalities are associated with RASA1 gene mutations in mouse and man. Proc Natl Acad Sci U S A. 2013;110:8621–8626. doi: 10.1073/pnas.1222722110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lolli G, Johnson LN. CAK-Cyclin-dependent Activating Kinase: a key kinase in cell cycle control and a target for drugs? Cell Cycle. 2005;4:572–577. [PubMed] [Google Scholar]
- 40.Karan D, Kelly DL, Rizzino A, Lin MF, Batra SK. Expression profile of differentially-regulated genes during progression of androgen-independent growth in human prostate cancer cells. Carcinogenesis. 2002;23:967–975. doi: 10.1093/carcin/23.6.967. [DOI] [PubMed] [Google Scholar]
- 41.Romanuik TL, et al. LNCaP Atlas: gene expression associated with in vivo progression to castration-recurrent prostate cancer. BMC Med Genomics. 2010;3:43. doi: 10.1186/1755-8794-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ristau BT, O’Keefe DS, Bacich DJ. The prostate-specific membrane antigen: Lessons and current clinical implications from 20 years of research. Urol Oncol. 2013 doi: 10.1016/j.urolonc.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitaker HC, et al. N-acetyl-L-aspartyl-L-glutamate peptidase-like 2 is overexpressed in cancer and promotes a pro-migratory and pro-metastatic phenotype. Oncogene. 2013 doi: 10.1038/onc.2013.464. [DOI] [PubMed] [Google Scholar]
- 44.Burgner D, et al. A genome-wide association study identifies novel and functionally related susceptibility Loci for Kawasaki disease. PLoS Genet. 2009;5:e1000319. doi: 10.1371/journal.pgen.1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 47.FitzGerald LM, et al. Genome-wide association study identifies a genetic variant associated with risk for more aggressive prostate cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:1196–1203. doi: 10.1158/1055-9965.EPI-10-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parikh H, et al. Fine mapping the KLK3 locus on chromosome 19q13.33 associated with prostate cancer susceptibility and PSA levels. Hum Genet. 2011;129:675–685. doi: 10.1007/s00439-011-0953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kote-Jarai Z, et al. Identification of a novel prostate cancer susceptibility variant in the KLK3 gene transcript. Hum Genet. 2011;129:687–694. doi: 10.1007/s00439-011-0981-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bensen JT, et al. Genetic polymorphism and prostate cancer aggressiveness: a case-only study of 1,536 GWAS and candidate SNPs in African-Americans and European-Americans. Prostate. 2013;73:11–22. doi: 10.1002/pros.22532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaid DJ, et al. Pooled genome linkage scan of aggressive prostate cancer: results from the International Consortium for Prostate Cancer Genetics. Hum Genet. 2006;120:471–485. doi: 10.1007/s00439-006-0219-9. [DOI] [PubMed] [Google Scholar]
- 52.Machiela MJ, et al. Association of type 2 diabetes susceptibility variants with advanced prostate cancer risk in the Breast and Prostate Cancer Cohort Consortium. Am J Epidemiol. 2012;176:1121–1129. doi: 10.1093/aje/kws191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilliland FD, et al. Trends in Gleason score for prostate cancer diagnosed between 1983 and 1993. J Urol. 2001;165:846–850. [PubMed] [Google Scholar]
- 54.Allsbrook WC, Jr, et al. Interobserver reproducibility of Gleason grading of prostatic carcinoma: urologic pathologists. Hum Pathol. 2001;32:74–80. doi: 10.1053/hupa.2001.21134. [DOI] [PubMed] [Google Scholar]
- 55.Andriole GL, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayes RB, et al. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin Trials. 2000;21:349S–355S. doi: 10.1016/s0197-2456(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 57.Yu K, et al. Population substructure and control selection in genome-wide association studies. PLoS One. 2008;3:e2551. doi: 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z, et al. Improved imputation of common and uncommon SNPs with a new reference set. Nat Genet. 2012;44:6–7. doi: 10.1038/ng.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.The alpha-tocopherol, beta-carotene lung cancer prevention study: design, methods, participant characteristics, and compliance. The ATBC Cancer Prevention Study Group. Ann Epidemiol. 1994;4:1–10. doi: 10.1016/1047-2797(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 61.Calle EE, et al. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer. 2002;94:2490–2501. doi: 10.1002/cncr.101970. [DOI] [PubMed] [Google Scholar]
- 62.Giovannucci E, et al. A prospective study of dietary fat and risk of prostate cancer. J Natl Cancer Inst. 1993;85:1571–1579. doi: 10.1093/jnci/85.19.1571. [DOI] [PubMed] [Google Scholar]
- 63.Alavanja MC, et al. The Agricultural Health Study. Environ Health Perspect. 1996;104:362–369. doi: 10.1289/ehp.96104362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kolonel LN, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Riboli E, et al. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5:1113–1124. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 66.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


