Abstract
Objectives: AmpC β-lactamases are classified as Amber Class C and Bush Group 1. AmpC β-lactamases can hydrolyze broad and extended-spectrum cephalosporins, and are not inhibited by β-lactamase inhibitors such as clavulanic acid. This study was conducted to identify DHA-23, a novel plasmid-mediated and inducible AmpC β-lactamase obtained from Enterobacteriaceae.
Methods: A total of 210 carbapenem-resistant Enterobacteriaceae isolates were collected from a medical center (comprising two branches) in Northern Taiwan during 2009–2012. AmpC β-lactamase genes were analyzed through a polymerase chain reaction using plasmid DNA templates and gene sequencing. The genetic relationships of the isolates were typed using pulsed-field gel electrophoresis following the digestion of intact genomic DNA by using XbaI.
Results: Three enterobacterial isolates (one Escherichia coli and two Klebsiella pneumoniae) were obtained from three hospitalized patients. All three isolates were resistant or intermediately susceptible to all β-lactams, and exhibited reduced susceptibility to carbapenems. These three isolates expressed a novel AmpC β-lactamase, designated DHA-23, approved by the curators of the Lahey website. DHA-23 differs from DHA-1 and DHA-6 by one amino acid substitution (Ser245Ala), exhibiting three amino acid changes compared with DHA-7 and DHA-Morganella morganii; three amino acid changes compared with DHA-3; four amino acid changes compared with DHA-5; and eight amino acid changes compared with DHA-2 (>97% identity). This AmpC β-lactamase is inducible using a system involving ampR.
Conclusion: This is the first report to address DHA-23, a novel AmpC β-lactamase. DHA-type β-lactamases are continuous threat in Taiwan.
Keywords: AmpC beta-lactamase, Enterobacteriaceae, antimicrobial resistance epidemiology
Introduction
AmpC enzymes are located in the bacterial periplasm, with the exception of the AmpC β-lactamase of Psychrobacter immobilis, which is secreted mainly into the external medium (Feller et al., 1997). They are active on cephalosporins, cephamycins (such as cefoxitin), oxyiminocephalosporins (such as ceftazidime and cefotaxime), and monobactams (such as aztreonam). AmpC β-lactamases are classified according to their Amber molecular structure as belonging to Class C, whereas according to function, they are classified into Group 1 (Bush and Jacoby, 2010). The sequence of the ampC gene differed from the sequence of penicillinase-type β-lactamases such as TEM-1 but similarly, had serine at its active site (Knott-Hunziker et al., 1982). AmpC β-lactamases, in contrast to extended-spectrum β-lactamases (ESBLs), can hydrolyze broad and extended-spectrum cephalosporins, and are not inhibited by β-lactamase inhibitors such as clavulanic acid. AmpC β-lactamases are assumed to be chromosomally mediated; however, Klebsiella pneumoniae, K. oxytoca, Proteus mirabilis, and Salmonella sp. lack a chromosomal blaAmpC gene (Bergstrom et al., 1983; Bauernfeind et al., 1989). Described plasmid-mediated AmpC genes in a K. pneumoniae isolate from South Korea. There are various types of plasmid-mediated AmpC β-lactamases: CMY, MIR, MOX, LAT, FOX, DHA, ACT, ACC, and CFE (Jacoby, 2009). AmpR is a member of the LysR transcriptional regulator family. During normal growth, in the absence of β-lactam as an inducer, the AmpR regulator binds with a peptidoglycan precursor uridine pyrophosphoryl-N-acetyl muramyl-L-alanyl-D-glutamylmeso-diaminopimelic acid-D-alanyl-D-alanine (UDP-N-acetylmuramic acid peptide). AmpR- UDP-N-acetylmuramic acid peptide complex binds to the operator site between the ampC and ampR structural genes, leading to the repression of ampC expression. Displacement of the UDP-N-acetylmuramic acid peptides signals a conformational change in AmpR, which activates the transcription of ampC. AmpR mutations are less common but can also result in high-constitutive or hyperinducible phenotypes (Kaneko et al., 2005). Resistance caused by plasmid-mediated AmpC β-lactamase is less common than the production of ESBLs, but may be more difficult to detect. The purpose of this study was to identify DHA-23, a novel plasmid-mediated and inducible AmpC β-lactamase identified in clinical enterobacterial isolates that were obtained from a hospital in Northern Taiwan.
Materials and Methods
Isolates and Data Collection
A total of 210 carbapenem-resistant Enterobacteriaceae isolates were collected from a medical center (comprising two branches) in Northern Taiwan from 2009 to 2012, including 100 K. pneumoniae, 53 Escherichia coli, 41 Enterobacter cloacae, and 16 other isolates (one K. oxytoca, one Citrobacter freundii, two Providencia rettgeri, eight Serratia sp., and four E. aerogenes). The isolates were identified using the VITEK2 system (bioMérieux Vitek Systems Inc., Hazelwood, MO, USA).
Polymrase Chain Reaction Detection of Carbapenemase Genes and Insertion Sequences
The carbapenemase-encoding genes were detected using polymerase chain reaction (PCR) methods as previously suggested by Woodford et al. (2006) and Ellington et al. (2007). All primers used in this study, such as blaTEM-type, blaSHV-type, blaCTX-M-type, blaCMY-type, and blaDHA-type primers, were as described previously (Tenover et al., 1995; Chung et al., 2011; Qi et al., 2011; Huang et al., 2012). Sequence similarity searches were conducted using the BLAST program1.
Pulsed-Field Gel Electrophoresis
The isolates were compared using pulsed-field gel electrophoresis (PFGE; Seifert et al., 2005) following the digestion of intact genomic DNA by using XbaI (Biolabs, UK). The XbaI restriction profiles were initially compared using visual inspection according to the criteria of Tenover et al. (1995). A computer-assisted analysis was performed using BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium) software.
Conjugation and Electrotransformation Experiments
Plasmid DNAs were extracted using the Qiagen Plasmid Purification Midi Kits (Qiagen, Courtaboeuf, France). Plasmid conjugation experiments were performed using E. coli DH5α as the recipient (Poirel et al., 1999b). Transconjugants were selected on Luria–Bertani agar plates supplemented with sodium azide (100 mg/L) and cefotaxime (2 mg/L). In addition, cefoxitin (8 mg/L) was added to prevent the selection of ESBL-producing transconjugants. Electrotransformants were selected on agar containing cefotaxime (2 mg/L) or cefoxitin (8 mg/L).
Minimum Inhibitory Concentrations
Minimum inhibitory concentrations (MICs) were determined using the agar dilution method, which was repeated twice for each sample. MICs of tested antibiotics were interpreted according to Clinical and Laboratory Standards Institute guidelines (M7–A9, CLSI, 2012). Antibiotics were purchased from Sigma–Aldrich (St. Louis, MO, USA). Quality control was assured by testing Staphylococcus aureus ATCC 29213, E. coli ATCC25922, and Pseudomonas aeruginosa ATCC 27853.
Results
Novel AmpC β-Lactamase Discovered in Three Enterobacterial Isolates
One E. coli (EC56) and two K. pneumoniae (KP11 and KP19) isolates that exhibited resistance to cefoxitin, cefotaxime, and ceftazidime were isolated from 3 adult patients hospitalized in Northern Taiwan (Table 1). We had the designated DHA-23 approved by the curators of the Lahey website 2. Using specific primers for blaDHA-1, we obtained PCR fragments from plasmid DNA preparations of E. coli EC56 and K. pneumoniae KP11 and KP19. The deduced amino-acid sequence (Figure 1) indicated that DHA-23 exhibited only one amino-acid change compared with DHA-1 and DHA-6 (Ser245Ala), two amino acid changes compared with DHA-7 and DHA-Morganella morganii (DHA-MM; Barnaud et al., 1998), three amino acid changes compared with DHA-3 (Wu et al., 2005), four amino acid changes compared with DHA-5, and eight amino acid changes compared with DHA-2 (>97% identity; Fortineau et al., 2001).
Table 1.
Minimum inhibitory concentrations (MICs) of three DHA-23 carrying enterobacterial isolates to antimicrobial agents.
| Antimicrobial agent | MIC (mg/L) |
||
|---|---|---|---|
| KP11* | KP19* | EC56* | |
| Ampicillin | ≥32 | ≥32 | ≥32 |
| Cefazolin | ≥64 | ≥64 | ≥64 |
| Cefoxitin | ≥64 | ≥64 | ≥64 |
| Cefuroxime | ≥64 | ≥64 | ≥64 |
| Cefotaxime | 128 | 128 | 128 |
| Ceftazidime | 64 | 32 | 128 |
| Flomoxef | ≥64 | ≥64 | ≥64 |
| Cefpirome | 16 | 16 | ≥64 |
| Ciprofloxacin | ≥4 | ≥4 | ≥4 |
| Moxifloxacin | ≥8 | ≥8 | ≥8 |
| Piperacillin-tazobactam | ≥128 | ≥128 | ≥128 |
| Trimethoprim-sulfamethoxazole | ≥320 | ≥320 | ≥320 |
| Amikacin | ≥64 | ≥64 | ≥64 |
| Gentamicin | ≥16 | ≥16 | ≥16 |
| Tigecycline | 4 | 2 | ≤0.5 |
| Colistin | ≤0.5 | ≥16 | ≤0.5 |
| Ertapenem | 2 | 2 | 4 |
| Meropenem | 0.12 | 0.12 | 0.12 |
| Imipenem | 2 | 2 | 1 |
| Doripenem | 0.5 | 0.25 | 0.06 |
*KP11 and KP19 expressed TEM-1, SHV-11, CTX-M-14, and DHA-23, EC56 expressed TEM-1, CTX-M-14, CMY-2, and DHA-23.
FIGURE 1.
Alignment of the deduced amino-acid sequences of DHA-23 with those of other DHA type AmpC enzymes. Identical amino-acids are marked with dashes. The underlined amino-acids are those that may be involved in the catalytic site of these AmpC enzymes, including the β-lactamase active site S–V–S–K and the conserved triad K–T–G. DHA-MM, DHA-Morganella morganii.
ampR Gene Found Immediately Upstream from the ampC Gene
The 110-bp intercistronic region of ampC and ampR contained the promoter sequences for ampC and ampR expression. This region of blaDHA-23 was identical to the corresponding region of blaDHA-1 (Barnaud et al., 1998) and was 97.3% identical to that in the chromosome of M. morganii (Poirel et al., 1999a). The ampR gene had an overlapping and divergently oriented promoter, as described for other ampC–ampR regulatory systems (Bennett and Chopra, 1993). Comparing the deduced amino-acid sequences of the AmpR of DHA-23 with those of DHA-1 and M. morganii indicated >99% identity, with only one amino-acid change in each case (Figure 2). A comparison of the AmpR of DHA-2 or the AmpR of DHA-3 revealed that they were at least >98% similar, exhibiting two and three amino acid changes, respectively. Similar to DHA-1, DHA-2, and DHA-3 (Barnaud et al., 1998; Fortineau et al., 2001; Wu et al., 2005), the sequences surrounding blaDHA showed that hybF was found upstream from the ampC gene in DHA-1, DHA-2, and DHA-3, whereas orf-1 was found downstream from the ampC gene only in DHA-1 (Barnaud et al., 1998). Subsequently, hybF and orf-1 were investigated in the sequences surrounding blaDHA-23. The hybF was identified downstream of ampR, but orf-1 was not detected (Figure 3). The hybF was coded for HybF and shared sequence homologies with hydrogenase subunits of E. coli (Menon et al., 1994). Although ampR and blaDHA were mobilized from the chromosome of M. morganii into a complex of In6–In7–sulI-type class-1 integrons in a strain of Salmonella enterica serovar Enteritidis (Verdet et al., 2000), an integron carrying blaDHA-23 was not detected in E. coli EC56 through PCR experiments employing 5′-CS and 3′-CS specific primers of class one integrons.
FIGURE 2.

Alignment of the deduced amino-acid sequences of the AmpR protein of DHA-23 with those of other DHA type AmpC enzymes.
FIGURE 3.
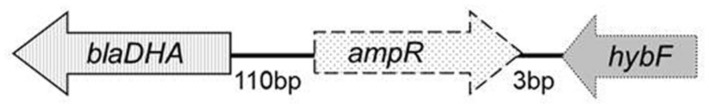
Genetic environment of blaDHA-23.
Clavulanic Acid Acts as An Inducer of DHA-23
E. coli EC56 contained 4 β-lactamases that corresponded to TEM-1, CTX-M-14, CMY-2, and DHA-23. K. pneumoniae KP11 and KP19 both contained β-lactamases that corresponded to TEM-1, SHV-11, CTX-M-14, and DHA-23. The genotyping of isolates was conducted using PFGE by using XbaI digestion. The results indicated that these two K. pneumoniae isolates had similar pulsotypes. Conjugation experiments were subsequently conducted, regardless of whether cefoxitin was used as a selecting agent, and the transfer of a plasmid coding the DHA-23 cephalosporinase into E. coli DH5α was not observed. The data suggested that the blaDHA-23 plasmid is not self-transferable. Electroporation experiments were then performed using plasmid DNA preparation of E. coli EC56 and K. pneumoniae KP11 and KP19. Amplification and gene sequencing were used to analyze the plasmid DNA, and the results revealed that the E. coli DH5α (pEC56) transformants contained two β-lactamases, TEM-1 and DHA-23. The E. coli DH5α (pKP11) and E. coli DH5α (pKP19) transformants contained only one β-lactamase, TEM-1. However, repeated electrotransformation experiments failed to obtain transformants carrying DHA-23 from KP11 and KP19. The MIC of ceftazidime (128 mg/L) for E. coli EC56 was not reduced by clavulanic acid. However, the MICs of cefotaxime (0.12 mg/L) and ceftazidime (1.0 mg/L) for E. coli DH5α (pEC56) increased in the presence of clavulanic acid, suggesting that clavulanic acid may act as an inducer of DHA-23 (Table 2).
Table 2.
Minimum inhibitory concentrations of β-lactams for clinical isolate Klebsiella pneumoniae KP19, Escherichia coli EC56, electroporant E. coli DH5α (pPK19), E. coli DH5α (pEC56), and reference strain E. coli DH5α.
| Antimicrobial agent | MIC (mg/L) |
||||
|---|---|---|---|---|---|
| KP19 | EC56 | E. coli DH5α (pKP19)# | E. coli DH5α (pEC56)† | E. coli DH5α | |
| Ampicillin | ≥32 | ≥32 | ≥32 | ≥32 | ≤2 |
| Cefoxitin | ≥64 | ≥64 | ≤4 | 32 | ≤4 |
| Cefotaxime | 128 | 128 | 0.06 | 0.12 | 0.03 |
| Ceftazidime | 32 | 128 | 0.25 | 1 | 0.25 |
| Ceftazidime-clavulanate* | 32 | ≥128 | 0.25 | 2 | 0.25 |
| Cefpirome | 16 | ≥64 | ≤1 | ≤1 | ≤1 |
| Ciprofloxacin | ≥4 | ≥4 | ≤0.25 | ≤0.25 | ≤0.25 |
| Ertapenem | 2 | 4 | ≤0.5 | ≤0.5 | ≤0.5 |
| Imipenem | 2 | 1 | ≤1 | ≤1 | ≤1 |
*Clavulanate was tested at a fixed concentration of 4 mg/L.
#E. coli DH5α (pKP19) expressed the plasmid-mediated TEM-1from K. pneumoniae 19.
†E. coli DH5α (pEC56) expressed the plasmid-mediated TEM-1 and DHA-23 from E. coli 56.
Discussion
This study presented a novel plasmid-mediated and inducible AmpC β-lactamase obtained from Enterobacteriaceae, designated DHA-23 and approved by the curators of the Lahey website. There are currently 22 DHA variants listed on the site, most of which are assigned with no reference to the sequence.
In Table 1, MICs of the DHA-23 carrying enterobacterial isolates E. coli EC56 and K. pneumoniae KP11 and KP19 to antimicrobial agents show that cephamycins (such as cefoxitin) are not hydrolyzed by ESBLs, but are hydrolyzed by associated AmpC β-lactamase. Class-C enzymes hydrolyze cephamycins but do not hydrolyze extended-spectrum cephalosporins effectively. DHA-23 showed that the MICs did not substantially increase for cefotaxime and ceftazidime when DHA-23 was present in the transformants, whereas the MICs of cefoxitin increased to 32 mg/L (Table 2). Resistance to the carbapenems was variable. All strains were fully susceptible to meropenem and doripenem (Table 2). In general, carbapenems are regarded as the preferred agent for treatment. However, the production of AmpC β-lactamase significantly increased the MICs of carbapenems was reported by Bradford et al. with the ACT-1 β-lactamase (Bradford et al., 1997), and by Lee et al. (2010) with the DHA-1 β-lactamase. Based on the recombinant experiments, Martinez-Martinez et al. (1999) demonstrated that MICs of carbapenems increased significantly in the recombinant K. pneumoniae strain harboring over expressing AmpC β-lactamase and loss of porins. They proposed that the spread of strains that express the plasmid-mediated AmpC β-lactamases and lack porins may create serious therapeutic problems in the future. Furthermore, proteomic investigation of the inner-membrane fraction of carbapenem-resistant strain of Acinetobacter baumannii supported a model for the importance of upregulated AmpC β-lactamases and down-regulated OmpW production in the mediation of carbapenem resistance in A. baumannii (Tiwari et al., 2012).
Enterobacteriaceae isolates producing a DHA-1-like enzyme have been identified previously in Taiwan (Yan et al., 2002; Yu et al., 2004; Wu et al., 2005). We report that DHA-type β-lactamases remain a threat in this country. Further nationwide surveillance should be conducted, antibiotic stewardship should be advocated, and strict infection control measures should be enforced.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The present study was supported by a research grant (TMU99-AE1-B02) from Taipei Medical University, Taiwan, and (MMH-10144) from Mackay Memorial Hospital, Taiwan.
Footnotes
References
- Barnaud G., Arlet G., Verdet C., Gaillot O., Lagrange P. H., Philippon A. (1998). Salmonella enteritidis: AmpC plasmid-mediated inducible beta-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob. Agents Chemother. 42 2352–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind A., Chong Y., Schweighart S. (1989). Extended broad spectrum beta-lactamase in Klebsiella pneumoniae including resistance to cephamycins. Infection 17 316–321 10.1007/BF01650718 [DOI] [PubMed] [Google Scholar]
- Bennett P. M., Chopra I. (1993). Molecular basis of beta-lactamase induction in bacteria. Antimicrob. Agents Chemother. 37 153–158 10.1128/AAC.37.2.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom S., Lindberg F. P., Olsson O., Normark S. (1983). Comparison of the overlapping frd and ampC operons of Escherichia coli with the corresponding DNA sequences in other gram-negative bacteria. J. Bacteriol. 155 1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford P. A., Urban C., Mariano N., Projan S. J., Rahal J. J., Bush K. (1997). Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1 a plasmid-mediated AmpC beta-lactamase, and the foss of an outer membrane protein. Antimicrob. Agents Chemother. 41563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K., Jacoby G. A. (2010). Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 54 969–976 10.1128/AAC.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K. P., Tseng S. P., Huang Y. T., Tsai T. H., Teng L. J., Hsueh P. R. (2011). Arrival of Klebsiella pneumoniae carbapenemase (KPC)-2 in Taiwan. J. Antimicrob. Chemother. 66 1182–1184 10.1093/jac/dkr025 [DOI] [PubMed] [Google Scholar]
- CLSI. (2012). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard-Ninth Edition. CLSI Document M7-A9. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Ellington M. J., Kistler J., Livermore D. M., Woodford N. (2007). Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J. Antimicrob. Chemother. 59 321–322 10.1093/jac/dkl481 [DOI] [PubMed] [Google Scholar]
- Feller G., Zekhnini Z., Lamotte-Brasseur J., Gerday C. (1997). Enzymes from cold-adapted microorganisms. The class C beta-lactamase from the antarctic psychrophile Psychrobacter immobilis A5. Eur. J. Biochem. 244 186–191 10.1111/j.1432-1033.1997.00186.x [DOI] [PubMed] [Google Scholar]
- Fortineau N., Poirel L., Nordmann P. (2001). Plasmid-mediated and inducible cephalosporinase DHA-2 from Klebsiella pneumoniae. J. Antimicrob. Chemother. 47 207–210 10.1093/jac/47.2.207 [DOI] [PubMed] [Google Scholar]
- Huang S. R., Liu M. F., Lin C. F., Shi Z. Y. (2012). Molecular surveillance and clinical outcomes of carbapenem-resistant Escherichia coli and Klebsiella pneumoniae infections. J. Microbiol. Immunol. Infect. 47 187–196 10.1016/j.jmii.2012.08.029 [DOI] [PubMed] [Google Scholar]
- Jacoby G. A. (2009). AmpC beta-lactamases. Clin. Microbiol. Rev. 22 161–182 10.1128/CMR.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K., Okamoto R., Nakano R., Kawakami S., Inoue M. (2005). Gene mutations responsible for overexpression of AmpC beta-lactamase in some clinical isolates of Enterobacter cloacae. J. Clin. Microbiol. 43 2955–2958 10.1128/JCM.43.6.2955-2958.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott-Hunziker V., Petursson S., Jayatilake G. S., Waley S. G., Jaurin B., Grundstrom T. (1982). Active sites of beta-lactamases. The chromosomal beta-lactamases of Pseudomonas aeruginosa and Escherichia coli. Biochem. J. 201 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. H., Su L. H., Li C. C., Chien C. C., Tang Y. F., Liu J. W. (2010). Microbiologic and clinical implications of bacteremia due to extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae with or without plasmid-mediated AmpC beta-lactamase DHA-1. Antimicrob. Agents Chemother. 54 5395–5398 10.1128/AAC.00083-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Martinez L., Pascual A., Hernandez-Alles S., Alvarez-Diaz D., Suarez A. I., Tran J., et al. (1999). Roles of beta-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 43 1669–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon N. K., Chatelus C. Y., Dervartanian M., Wendt J. C., Shanmugam K. T., Peck H. D., Jr., et al. (1994). Cloning, sequencing, and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase 2. J. Bacteriol. 176 4416–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Guibert M., Girlich D., Naas T., Nordmann P. (1999a). Cloning, sequence analyses, expression, and distribution of ampC-ampR from Morganella morganii clinical isolates. Antimicrob. Agents Chemother. 43 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Naas T., Guibert M., Chaibi E. B., Labia R., Nordmann P. (1999b). Molecular and biochemical characterization of VEB-1 a novel class A extended-spectrum beta-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Wei Z., Ji S., Du X., Shen P., Yu Y. (2011). ST11 the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66 307–312 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- Seifert H., Dolzani L., Bressan R., van der Reijden T., van Strijen B., Stefanik D., et al. (2005). Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43 4328–4335 10.1128/JCM.43.9.4328-4335.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V., Vashistt J., Kapil A., Moganty R. R. (2012). Comparative proteomics of inner membrane fraction from carbapenem-resistant Acinetobacter baumannii with a reference strain. PLoS ONE 7:e39451 10.1371/journal.pone.0039451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdet C., Arlet G., Barnaud G., Lagrange P. H., Philippon A. (2000). A novel integron in Salmonella enterica serovar Enteritidis, carrying the bla(DHA-1) gene and its regulator gene ampR, originated from Morganella morganii. Antimicrob. Agents Chemother. 44 222–225 10.1128/AAC.44.1.222-225.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N., Ellington M. J., Coelho J. M., Turton J. F., Ward M. E., Brown S., et al. (2006). Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27 351–353 10.1016/j.ijantimicag.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Wu L. T., Hung S. W., Chuang Y. C., Chen H. E., Jones R. N., Yu W. L. (2005). Identification of a novel cephalosporinase (DHA-3) in Klebsiella pneumoniae isolated in Taiwan. Clin. Microbiol. Infect 11 893–897 10.1111/j.1469-0691.2005.01252.x [DOI] [PubMed] [Google Scholar]
- Yan J. J., Ko W. C., Jung Y. C., Chuang C. L., Wu J. J. (2002). Emergence of Klebsiella pneumoniae isolates producing inducible DHA-1 beta-lactamase in a university hospital in Taiwan. J. Clin. Microbiol. 40 3121–3126 10.1128/JCM.40.9.3121-3126.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W. L., Chuang Y. C., Jones R. N. (2004). A pragmatic approach to identify extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in Taiwan: in vitro activity of newer and established antimicrobial agents. Diagn. Microbiol. Infect Dis. 48 277–282 10.1016/j.diagmicrobio.2003.11.001 [DOI] [PubMed] [Google Scholar]



