Abstract
Psoriasis is a chronic autoimmune disease with complex genetic architecture. Previous genomewide association studies (GWAS) and a recent meta-analysis using Immunochip data have uncovered 36 susceptibility loci. Here, we extend our previous meta-analysis of European ancestry by refined genotype calling and imputation and by the addition of 5,033 cases and 5,707 controls. The combined analysis, consisting of over 15,000 cases and 27,000 controls, identifies five new psoriasis susceptibility loci at genomewide significance (p < 5 × 10−8). The newly identified signals include two that reside in intergenic regions (1q31.1 and 5p13.1) and three residing near PLCL2 (3p24.3), NFKBIZ (3q12.3), and CAMK2G (10q22.2). We further demonstrate that NFKBIZ is a TRAF3IP2–dependent target of IL-17 signaling in human skin keratinocytes, thereby functionally linking two strong candidate genes. These results further integrate the genetics and immunology of psoriasis, suggesting new avenues for functional analysis and improved therapies.
Introduction
Psoriasis is a chronic immune-mediated skin disease with complex genetic architecture that affects approximately 2% of the world population. It presents a significant economic burden for affected individuals and for society1, and can severely impact the patient’s quality of life2. Aided by advances in high throughput genotyping technologies, genomewide association studies have identified multiple genetic loci associated with psoriasis3–6. These studies have been accompanied by replication of the most promising signals to confirm the psoriasis-associated signals at genomewide significance levels. More recently, large consortia have utilized meta-analyses to identify additional susceptibility loci with modest effect sizes7,8.
Previously, we performed a meta-analysis combining three existing GWAS [the Collaborative Association Study of Psoriasis (CASP)3, Kiel5, the Wellcome Trust Case Control Consortium 2 (WTCCC2)6, and two Immunochip datasets (the Genetic Analysis of Psoriasis Consortium (GAPC) and the Psoriasis Association Genetics Extension (PAGE)] and identified 15 novel psoriasis susceptibility loci8, increasing the number of confirmed loci to 36 for populations of European descent.
In this study, we enhanced our previous meta-analysis8 and added replication datasets to follow up the most promising signals. First, we applied the optiCall9 genotype calling algorithm to the GAPC and PAGE datasets, and performed genomewide imputation to 1000 Genomes haplotypes. In this way, we were able to examine association at over six times the number of variants examined in our previous meta-analysis8. Using these enhancements, we perform a discovery meta-analysis of the combined GAPC/PAGE Immunochip dataset and the three psoriasis GWAS3,5–7, comprising 10,262 psoriasis cases and 21,871 controls. We then conduct replication analysis in 5,033 cases and 5,707 controls from four independent datasets, resulting in a combined analysis involving over 15,000 cases and 27,000 controls. We report five novel psoriasis susceptibility loci reaching genomewide significance (p < 5 × 10−8), which add to the collective understanding of the genetic basis of this common cutaneous disorder.
Results
Discovery meta-analysis
The discovery meta-analysis included the combined GAPC/PAGE Immunochip dataset and three existing GWAS datasets (i.e. CASP, Kiel, and WTCCC2). We first performed genomewide imputation of each dataset using 1000 Genomes haplotypes as a reference panel10. After imputation, we tested association for 696,365 markers (52,444 of them are short insertion/ deletion variations, INDELs) that had minor allele frequencies (MAF) ≥ 1% and imputation quality (r2) greater than 0.7 in all four datasets. This study analyzed more than six times the 111,236 markers examined in the original meta-analysis 8.
We have shown that linear mixed modeling could be used to correct population stratification for association analysis11 in the PAGE and GAPC datasets separately 8. This is also the case in the current combined Immunochip dataset consisting of 6,268 psoriasis cases and 14,172 controls, with the genomic inflation factor estimated to be 0.96 (Supplementary Fig. 1). The genomic inflation factors for the CASP, Kiel and WTCCC2 GWAS data were estimated to be 1.01, 1.04, and 1.04, respectively.
Among the 36 known psoriasis susceptibility loci, 35 yielded strong evidence of association (Effective sample size-weighted z-score: Pdiscovery-meta ≤ 5 × 10−7 ), of which 30 achieved genomewide significance (Pdiscovery-meta ≤ 5 × 10−8 ) in the discovery meta-analysis (Supplementary Information and Supplementary Fig. 2) involving 10,262 cases and 21,871 controls. The only previously described locus that did not yield association in this study maps near IL28RA, in which the strongest previously identified signals6 failed our quality filters in one of the GWAS datasets. However, the Immunochip data alone does show suggestive association in the IL28RA region (EMMAX: PImmunochip = 3.5 × 10−7).
Combined analysis identifies five new loci
In the discovery meta-analysis, we identified 6 novel loci showing significant association with Pdiscovery-meta ≤ 5 × 10−8 (Table 1, Figure 1, and Supplementary Table 1). We evaluated the most strongly associated markers (i.e. the “best markers”) identified in the new loci, and all of them have good imputation quality and none exhibited significant heterogeneity across datasets (all heterogeneity p-values > 0.1) (Supplementary Table 2). We then expanded our analysis utilizing genotyping data from four independent replication datasets, utilizing either the best markers or their best linkage disequilibrium (LD) proxies if ld-r2 ≥ 0.8. Notably, five of the six loci retained genomewide significance in the combined meta-analysis (Table 2; Supplementary Table 3). Because one of the best markers (rs4685408) was genotyped separately in a substantial fraction (3,030 cases and 2,859 controls) of two of our replication datasets (i.e. Exomechip 2 and PsA GWAS; Table 2), and the proxies for this marker in the two datasets were among the weakest of those listed in Table 2, we also treated this data as an additional independent dataset (termed as “Michigan Genotyping” in Supplementary Table 3; logistic regression: PMichigan = 9 × 10−5; combined meta-analysis: Pcombined-meta = 9 × 10−15). In all, the combined analysis consists of around 15,000 psoriasis cases and over 27,000 controls.
Table 1.
Loci with genomewide association signals identified in the discovery meta-analysis.
| Locus | Variant (annotation) | Chr | Pos | RA | NRA | RAF
|
p-value | Direction | ORs | Nearby Genes | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| case | control | ||||||||||
| 1q31.1 | rs10789285 (intergenic) | 1 | 69788482 | G | T | 0.28 | 0.25 | 1.43×10−8 | ++++ | 1.12 | LRRC7 |
| 3p24.3 | rs4685408 (intronic) | 3 | 16996035 | G | A | 0.5 | 0.48 | 8.58×10−9 | ++++ | 1.12 | PLCL2 |
| 3q12.3 | rs7637230 (intronic) | 3 | 101663555 | A | G | 0.83 | 0.8 | 2.07×10−9 | ++++ | 1.14 | NFKBIZ |
| 5p13.1 | rs114934997 (intergenic) | 5 | 40370724 | C | A | 0.92 | 0.91 | 1.27×10−8 | ++++ | 1.17 | CARD6 |
| 10q22.2 | rs2675662 (intronic) | 10 | 75599127 | A | G | 0.58 | 0.56 | 7.35×10−9 | ++++ | 1.12 | CAMK2G |
| 15q25.3 | rs35343117 (intronic) | 15 | 86079115 | G | C | 0.37 | 0.34 | 3.10×10−8 | ++++ | 1.1 | AKAP13 |
RA: risk allele; NRA: non-risk allele; RAF: risk allele frequency.
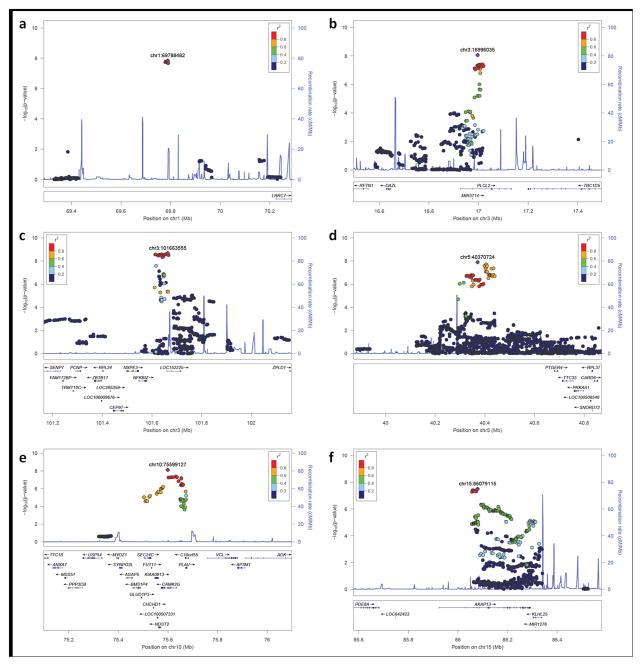
Figure 1. Regional association plots for novel psoriasis susceptibility loci.
This figure depicts LocusZoom-generated association plots for the six psoriasis susceptibility loci identified in the discovery meta-analysis (effective sample size-weighted approach). All but the 15q25.3 locus (f) showed genomewide significant results in the combined meta-analysis.
Table 2.
Results from the discovery, replication, and combined meta-analysis.
| Discovery
|
Replication
|
Combined
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CASP+Kiel+ WTCCC2+Imm | Exomechip (Utah, Sweden) | Exomechip (Michigan, Toronto, Newfoundland) | PsA GWAS | Genizon GWAS | ||||||
| 10,262 case+ 21,871controls | 913 cases+ 1,494 controls | 3,168 cases + 2,864 controls | 191 cases+ 356 controls | 761 cases+ 993 controls | ||||||
|
|
|
|
|
|
|
|||||
| Marker | P | Marker | P | Marker | P | Marker | P | Marker | P | P |
| rs10789285 | 1.4×10−8 | NA | NA | NA | NA | rs720233 | 3.0×10−1 | rs720233 | 1.4×10−1 | 2.8×10−9 |
| rs4685408 | 8.6×10−9 | rs12497667 | 3.6×10−4 | rs12497667 | 1.8×10−3 | rs1806555 | 4.4×10−1 | rs7653027 | 6.8×10−2 | 7.2×10−14 |
| rs7637230 | 2.1×10−9 | rs7637230 | 2.1×10−2 | rs7637230 | 6.7×10−1 | rs1473857 | 5.5×10−2 | rs1473857 | 1.1×10−1 | 1.7×10−10 |
| rs114934997 | 1.3×10−8 | rs56054640 | 5.1×10−1 | rs56054640 | 1.6×10−1 | rs2120854 | 9.3×10−1 | rs4957279 | 2.4×10−1 | 5.7×10−9 |
| rs2675662 | 7.4×10−9 | rs2675671 | 6.0×10−2 | rs2675671 | 2.5×10−7 | rs2664282 | 2.0×10−1 | rs2675671 | 2.3×10−1 | 1.6×10−14 |
| rs35343117 | 3.1×10−8 | rs1483578 | 9.9×10−1 | rs1483578 | 8.7×10−1 | rs2062234 | 1.3×10−3 | rs1483578 | 8.6×10−1 | 1.7×10−6 |
If the best marker identified in the discovery dataset was not genotyped in the replication dataset, the best proxy genotyped marker (r2 ≥ 0.8) was used. The p-values for replication datasets were obtained using logistic regression test; sample-size weighted approach was used in the combined meta-analysis.
The estimated odds ratios (ORs) for the five confirmed novel loci ranged from 1.12 to 1.17 (Table 1), similar to the 15 new loci identified in the original meta-analysis8. Among the 5 novel loci, 5q13.1 has the highest effect size (OR = 1.17). Interestingly, this signal is situated in an intergenic region (Figure 1), and was previously identified as a susceptibility locus for other autoimmune diseases including inflammatory bowel disease and multiple sclerosis (Table 3). While additional comparisons and more well-powered studies are needed, none of the five novel loci reported here have been identified as genomewide psoriasis susceptibility loci in non-European samples 12–16 (Supplementary Table 4).
Table 3.
Newly-discovered psoriasis loci that are shared with other disease susceptibility loci according to NHGRI GWAS catalog.
| Psoriasis loci
|
Susceptibility loci for other traits
|
r | |||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Position | RA | Traits | SNP | Position | RA | p-value | |
| rs4685408 | 3 | 16996035 | G | Primary biliary cirrhosis | rs1372072 | 16955259 | A | 2×10−8 | −0.54 |
| rs7637230 | 3 | 101663555 | A | Multiple sclerosis | rs771767 | 101748638 | A | 9×10−9 | −0.28 |
| rs114934997 | 5 | 40370724 | C | Self reported allergy | rs7720838 | 40486896 | G | 8×10−11 | 0.21 |
| rs114934997 | 5 | 40370724 | C | Inflammatory bowel disease | rs11742570 | 40410584 | C | 2×10−82 | 0.27 |
| rs114934997 | 5 | 40370724 | C | Multiple Sclerosis | rs4613763 | 40392728 | C | 3×10−16 | 0.10 |
| rs114934997 | 5 | 40370724 | C | Ulcerative colitis | rs6451493 | 40410935 | T | 3×10−9 | 0.27 |
| rs114934997 | 5 | 40370724 | C | Crohn’s disease | rs11742570 | 40410584 | C | 7×10−36 | 0.27 |
| rs2675662 | 10 | 75599127 | A | Inflammatory bowel disease | rs2227564 | 75673101 | C | 7×10−10 | 0.62 |
| rs2675662 | 10 | 75599127 | A | Atrial fibrillation | rs10824026 | 75421208 | A | 4×10−9 | 0.39 |
If more than one overlapping record was found for the same trait, the one with the most significant p-value is reported here. r: signed squared LD r2 between the two markers listed on the same row, with positive value indicates the two risk alleles tend to be on the same haplotype. RA = risk allele. The p-values are obtained from the reported association in the NHGRI GWAS catalog.
As assessed using ANNOVAR 17, the strongest signals from three of the confirmed loci map to intronic regions (Table 1), and the strongest signals from the other two loci map to intergenic regions. Using 1000 Genomes Project data, we did not identify any common (MAF>1%) protein-altering variants (i.e. missense/nonsense mutations) in high linkage disequilibrium (ld-r2 ≥ 0.8) with our strongest signals. We also performed conditional and interaction analyses using the five new loci identified in this study and did not identify any independent secondary signals within the five loci or evidence for epistasis effects among these loci or with previously described psoriasis loci.
Biological inferences for identified loci
Nearby genes within the three non-intergenic susceptibility loci (within ±200kb boundary of the strongest signals) include: PLCL2 on 3p24.3; NFKBIZ, ZPLD1, and CEP97 on 3q12.3; and CAMK2G, FUT11, AGAP5, PLAU, and MYOZ1 on 10q22.2 (Figure 1). Among the above genes, NFKBIZ and MYOZ1 were differentially expressed when comparing psoriatic and normal skin samples18: NFKBIZ was four-fold up-regulated (Wilcoxon rank-sum test: p = 1.1 × 10−28) and MYOZ1 was down-regulated (fold change = 0.5; p = 3.5 × 10−14) in lesional psoriatic skin vs. normal skin.
We searched the NHGRI catalog 19 for previously identified genomewide significant (p ≤ 5 × 10−8) loci within ±200kb of the best signals from the five novel loci identified in this study. Four of the five new loci are shared with other complex traits, most of which are immune-mediated inflammatory disorders (Table 3). For the most part, however, the risk variants from psoriasis and the other complex traits that map to the same loci are not in linkage disequilibrium, meaning that the psoriasis risk haplotypes do not tend to be the same as those found for the other traits (Table 3). Although intergenic in nature, the 5p13.1 locus is shared with four other complex diseases, including allergy20, multiple sclerosis (MS)21, Crohn’s disease22,23, and ulcerative colitis (UC)24.
Since the novel signals do not tend to be in LD with any amino-acid altering variants, we then investigated their potential regulatory roles. Of the five novel loci, four of them overlapped with histone marks or transcription factor binding sites in lymphoblastoid cells or keratinocytes according to data from ENCODE25 (Supplementary Table 5). The transcription factors (e.g. NF-κB, IRF4, BATF, JUN-D) for the binding sites overlapped by these loci tend to be involved in immune responses: NF-κB and IRF4 are important transcription factors that regulate both innate and adaptive immune response; and the BATF-JUN family protein complexes are essential for IRF4-mediated transcription in T cells26. We then asked whether our best markers from the novel loci are eQTLs and could alter the expression of nearby genes. Interestingly, we found that rs7637230 (p = 2.22 × 10−8) and rs2675662 (p = 5.78 × 10−10) are both cis-eQTLs in lymphoblastoid cells27, and affect the expression levels of NFKBIZ and FUT11, respectively.
The IL-17 pathway has been shown to play an important role in psoriasis28. TRAF3IP2, a susceptibility locus for both psoriasis and psoriatic arthritis5,8,29, encodes adaptor protein Act1, one of the key components in the IL-17 signaling pathway30. NFKBIZ, a gene in one of the newly discovered loci of this study, has been shown to be an Act1-dependent IL-17 target gene in mice31,32. Because psoriasis is a uniquely human disease, and because keratinocytes appear to be a key target of IL-17 action in psoriasis 28, we set out to determine whether NFKBIZ is also an Act1-dependent target of IL-17 signaling in human keratinocytes. To do this, we investigated the time course of NFKBIZ expression in human keratinocytes engineered to silence TRAF3IP2 expression under the control of tetracycline. As shown in Figure 2, expression of NFKBIZ mRNA and protein could be significantly induced by IL-17 (p < 0.01), and these inductions were significantly (p < 0.01) blocked by TRAF3IP2 silencing. As illustrated above, variant rs7637230 is in strong LD (r2 ≥ 0.8) with markers that overlap with chromatin marks in lymphoblastoid cells and keratinocytes (Supplementary Table 5) and it is an eQTL for the expression of NFKBIZ in lymphoblastoid cells27. Together with the results from our NFKBIZ experiments, these findings nominate NFKBIZ as a strong candidate gene in the 3q12 locus and suggest a potential disease mechanism of the IL-17 pathway in psoriasis.
Figure 2. NFKBIZ mRNA (A) and IκB-zeta protein (B) are induced by IL-17 in an Act1-dependent fashion in human keratinocytes.
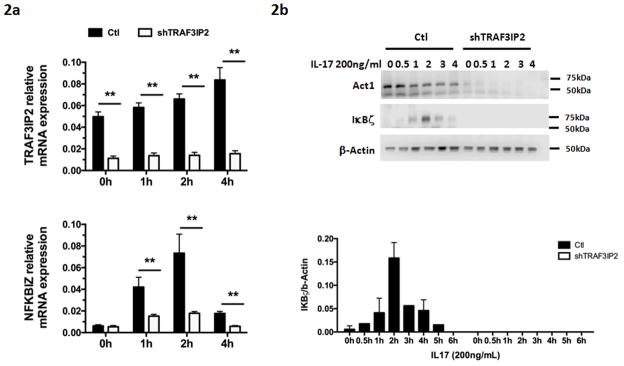
A. Time courses of TRAF3IP2 (top) and NFKBIZ (bottom) mRNA expression following IL-17 induction. Black bars represent mRNA relative expression levels measured by qPCR in the absence of tetracyline (Tet) while white bars represent levels after silencing TRAF3IP2 expression via Tet-inducible expression of shTRAF3IP2. Bars are mean +/−SEM of 3 independent experiments and ** = p<0.01; Student’s t-test.
B, Western blot analysis of time courses of IκB-zeta and Act1 expression (upper panel) and quantitations of IκB-zeta band intensity relative to β-Actin. Bars are mean + SD of 2 independent experiments (lower panel).
Discussion
In this study, we identified five novel psoriasis susceptibility loci reaching genomewide significance, increasing the number of known psoriasis susceptibility loci to 41 in Caucasians and to 49 worldwide (Supplementary Table 4).
We performed three main procedures to ensure the validity of the identified novel loci in this study: First, to ensure the imputed dosage of each marker is accurate and to avoid batch effects, we required a stringent imputation quality threshold (r2 ≥ 0.7). Furthermore, we only considered markers that passed the quality threshold in all 4 datasets when performing the associations (Supplementary Table 2). Second, we calculated the heterogeneity p-value from the meta-analysis to be sure the associations were not heterogeneous (p > 0.05) among the datasets (Supplementary Table 2). Finally, we performed a replication analysis to further validate that the 5 loci still achieve genomewide significance in the combined meta-analysis (Table 2).
While estimates of allele frequencies based on imputed dosages will have added uncertainty compared to those based on experimentally determined genotypes, the allele frequencies reported in our study do show consistent differences between cases and controls for each of the associated markers across different datasets (Supplementary Table 2). We also compared risk allele frequencies for 41 psoriasis susceptibility loci across the four discovery datasets, and we observed very high concordance (r2 ≥ 0.99) of the estimated frequencies in controls for all six possible pairs of datasets (Supplementary Fig. 3).
Three of the five newly identified loci contained protein-coding genes; however, we found no evidence suggesting that our signals are missense or nonsense mutations, nor are they in LD with such variations. Our results are in agreement with data for other known psoriasis susceptibility loci, in that fewer than 25% of the psoriasis-associated signals are in linkage disequilibrium with codon-changing variations8. While no deleterious functional variants were identified in the three protein-coding loci (PLCL2 at 3p24.3; NFKBIZ, ZPLD1, and CEP97 at 3q12.3; and CAMK2G, FUT11, AGAP5, PLAU, and MYOZ1 at 10q22.2), variation in PLCL2 has previously been shown to be associated at genomewide significance (p = 2.3 × 10−8) with primary biliary cirrhosis 33 and nominally (p = 1.7 × 10−3) in a cohort of 982 Caucasian cases of psoriatic arthritis and 8,676 Caucasian controls 34. In mice, NFKBIZ deletion has been functionally associated with inflammatory skin eruptions35 and CAMK2G has been functionally implicated in thymic development36. Additionally, PLAU, encoding urokinase-type plasminogen activator, has been reported to be overexpressed in psoriatic skin37 and was up-regulated 1.49-fold (p = 3.7 × 10−13) in our psoriasis RNA-seq transcriptome data, albeit short of the 2-fold change threshold we used to declare significance18. Of the remaining protein-coding genes in these three loci, MYOZ1 encodes myozenin, a muscle protein of no obvious relevance to psoriasis, and very little information is available for ZPLD1, CEP97, and AGAP5 in Online Mendelian Inheritance in Man38. The identification of the disease-associated SNPs rs7637230 (p = 2.22 × 10−8) and rs2675662 (p = 5.78 × 10−10) as cis-eQTLs in lymphoblastoid cells27 prioritizes NFKBIZ and FUT11 as strong candidate genes in their respective loci.
Several recent clinical studies have shown benefit of blockade of IL-17 and its receptors in psoriasis39. In this context, the connection between NFKBIZ and TRAF3IP2 is of biological and therapeutic interest. TRAF3IP2 encodes Act1 (also known as CIKS), an ubiquitin ligase that binds to IL-17 receptors 40,41. NFKBIZ encodes IκB-zeta, a transcriptional regulator that binds to the p50 subunit of NF-kB 42. Act1 and IκB-zeta are required for IL-17-dependent signaling associated with autoimmune and inflammatory diseases 43,44. Nfkbiz-deficient mice manifest defective Th17 development, and IκB-zeta regulates this process by cooperating with ROR nuclear receptors 44. While Act1 has previously been shown to regulate Nfkbiz expression in mouse embryonic fibroblasts 31 and mouse skin keratinocytes 32, this is the first demonstration of Act1-dependent NFKBIZ / IκB-zeta expression in human keratinocytes. Interestingly, TNFAIP3 encoding A20 is also a strong candidate gene in psoriasis and several other inflammatory diseases, which appears to act as a brake on cytokine-medicated signaling45. Interestingly TNFAIP3 is also an Act1-dependent target of IL-17 signaling, which in turn functions as a negative regulator of IL-17 receptor function30. These results highlight NFKBIZ as an IL17 target and identify an immunoregulatory network downstream of the IL-17 receptor involving at least three psoriasis candidate genes: TRAF3IP2, NFKBIZ, and TNFAIP3.
Evidence is accumulating to indicate that genetic variants in regulatory regions may play important biological roles in complex genetic disorders46,47, and large scale projects such as ENCODE25 have been using sequencing technologies and integrative approaches to illuminate the functional elements of the human genome. Our results illustrate the potential regulatory roles of the psoriasis susceptibility loci, and will facilitate the design of future functional analyses.
Similar to other complex traits and diseases48,49, large inter-continental consortia have been forming to gather hundreds of thousands of samples to identify susceptibility loci with very mild effect sizes for psoriasis. Moreover, the study of psoriasis genetics is entering a new era, with more efforts on investigating the missing heritability explained by secondary independent signals (fine-mapping analysis50,51) and rare variants (by using exome-array or target-/exome-sequencing16) in known loci. Not only will these studies provide a higher explained variance by the genetic components, more importantly, they will also shed light on the pathogenesis and disease mechanisms, and ultimately provide new approaches and targets for effective drug discovery.
Methods
Discovery meta-analysis datasets
Samples were collected at the participating institutions after obtaining informed consent, and enrollment of human subjects for this study was approved by the following ethics boards: Estonian Biobank (EGCUT) – Research Ethics Committee of the University of Tartu; Michigan samples - Institutional Review Board of the University of Michigan Medical School (IRBMED); German samples - Ethical review board of the Medical Faculty of the CAU (Christian-Albrechts-University of Kiel, Germany); Toronto samples - the ethics board is the University Health Network; Swedish samples - the ethics board is the Local Ethical Review Board at Linköping University; Newfoundland samples- the ethics board is the Health Research Ethics Authority; Utah samples - the ethics board is the Institutional Review Board of the University of Utah. We first performed the discovery meta-analysis using four datasets: i) combined GAPC/PAGE Immunochip dataset; ii) CASP GWAS; iii) Kiel GWAS; and iv) WTCCC2 GWAS. Since the algorithm implemented in optiCall, which uses both within and across sample signal intensities, can outperform standard methods (e.g. GenCall or GenoSNP)9 when applied to Immunochip, we used optiCall9 to re-call the PAGE and GAPC datasets. We removed samples with outlying intensity values based on optiCall default settings by using the “–meanintfilter” flag. Samples with more than 2% missing genotypes and markers with more than 5% missing values were rejected. We merged the PAGE and GAPC datasets to form a unified Immunochip dataset, re-applying the above call rate criteria to the combined data. These and subsequent genotype-dependent quality control procedures resulted in the removal of 1,222 (5.6%) samples from this analysis, compared to the previous study8. The quality-controlled Immunochip dataset consisted of 2,858 cases and 8,636 controls in GAPC, and 3,410 cases and 5,536 controls in PAGE. For each of the three GWAS datasets, we first obtained the quality controlled genotyped data used by the previous studies3,5,6. We then applied additional filters (sample/marker call rate ≥ 0.95; HWE p >= 1 × 10−6) to the genotyped data before phasing/imputation (see below). Supplementary Table 6 shows the quality measurements for each dataset.
Imputation and association
For each of the GWAS/Immunochip datasets, we performed haplotype phasing of the genotypes using ShapeIT52, we then performed imputation using minimac53 using haplotypes from the EUR subset of version 3 of the 1000 Genomes Project Phase I release as the reference panel10. After imputation, we analyzed markers with minor allele frequency (MAF) ≥ 1% and with imputation quality r2 ≥ 0.7 in all four datasets.
For the Immunochip data, a linear mixed model was used to perform association tests as implemented in EMMAX11. The kinship matrix was computed using common (MAF > 1%) independent markers located outside the 36 known psoriasis loci, having pGWA ≥ 0.5 (where pGWA represents the association p-values obtained from the meta-analysis using only the three independent GWAS datasets; this choice is common for analyses using ImmunoChip markers which are enriched for variants associated with psoriasis in our original GWAS). Using the same procedures described in the previous study for quality control8 (i.e. SNPs with call rate below 95% or with a hardy-Weinberg equilibrium p-value < 1 × 10−6 were excluded; samples with less than 95% call rate were excluded), we performed logistic regression on the three GWAS datasets (i.e. CASP, Kiel, and WTCCC2), using principal components as covariates to correct for population stratification 3,5,6.
Discovery meta-analysis
We used METAL to perform meta-analysis, using the sample size weighted approach54, adjusting the genomic control inflation factor separately for each dataset. To estimate the odds ratios (ORs) for each marker, we first computed the ORs from the Immunochip data using logistic regression, and then used a sample-size weighted approach to compute the ORs from the full datasets8. The regional association plots were created by using the software LocusZoom55. We used ANNOVAR to perform variant annotations17 using Gencode v1956 as gene references.
Replication datasets and combined meta-analysis
For each of the most strongly associated genomewide significant (p < 5 × 10−8) markers (i.e. the “best markers”) from the novel signals identified in the discovery meta-analysis, we used genotyping data obtained from four different collaborative datasets (here referred to as Exomechip 1, Exomechip 2, PsA GWAS, and Genizon GWAS) for replication. If the best associated markers were not genotyped in the replication datasets, we identified the best proxies based on the linkage disequilibrium (ld-r2) in the European ancestry subset of version 3 of the 1000 Genomes Project Phase I release10. Only proxies with ld-r2 ≥ 0.8 against our best signals were considered. Using only genetically-independent samples, the 5,033 additional cases and 5,707 controls were combined with the discovery meta-analysis using the sample size weighted approach54.
NFKBIZ expression
N/TERT-TR keratinocytes (KCs) were engineered to express a tetracycline (Tet)-inducible shRNA as described 57. These cells (N/TERT-TR-shTRAF3IP2) were seeded at 8,000 cells cm−2 in the presence or absence of 1 μgxml−1 Tet. When confluent (after 7 days), cells were stimulated for 1 to 4 hours with 200ngxml−1 recombinant human (rh) IL-17A (R&D Systems). RNA was isolated using RNAeasy columns (Qiagen) and reverse-transcribed using the High Capacity cDNA Reverse Transcription Kit (Life Technologies). Quantitative real-time polymerase chain reaction (qPCR) was performed using Taqman primers (Life Technologies) specific for nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta (NFKBIZ, Hs00230071-m1), TNF receptor associated factor 3 interacting protein 2 (TRAF3IP2, Hs00974570-m1) and the control gene RPLP0 (Hs99999902_m1). From the same experiments, NFKBIZ and TRAF3IP2/Act1 proteins were assessed by western blotting, utilizing rabbit polyclonal IgG (respectively from Cell Signal Transduction #9244 and Santa Cruz Biotechnology sc-11444, both diluted 1:1000) to detect protein, with β-actin (Cell Signal Transduction #4967 dilution 1:5000) as a loading control. Statistical analysis was performed using GraphPad software (Prizm).The uncropped scans of the blots are shown in Supplementary Fig. 4.
Overlap with ENCODE genomic features
We investigated whether the best signals identified from the novel loci are in LD with markers within chromatin marks or transcription factor binding sites. For each best associated marker, we first identified a set of high LD-markers based on the European population of the 1000 Genomes. We then examined human keratinocytes and lymphoblastoid cell lines (relevant cell types for psoriasis) and identified any chromatin marks or transcription factor binding sites overlapping with the LD-marks.
Supplementary Material
Acknowledgments
Major support for this study was provided by the National Institutes of Health (R01AR042742, R01AR050511, R01AR054966, R01AR062382, R01AR065183 to JTE), the Wellcome Trust, and the German Research Foundation. We acknowledge use of the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council (G0000934) and the Wellcome Trust (068545/Z/02), and of the UK National Blood Service controls funded by the Wellcome Trust. FC, RCT, and JNB were supported by the Medical Research Council Stratified Medicine award (MR/L011808/1). AE and EE received infrastructure support through the DFG Clusters of Excellence 306 “Inflammation at Interfaces” and is supported by the German Ministry of Education and Research (BMBF) through the e:Med sysINFLAME grant. JEG was supported by Doris Duke Foundation. We acknowledge support from the Department of Health via the NIHR comprehensive Biomedical Research Center award to GSTT NHS Foundation Trust in partnership with King’s College London and KCH NHS Foundation Trust. We acknowledge the Collaborative Association Study of Psoriasis (CASP) and the Wellcome Trust Case Control Consortium 2 (WTCCC2) for the contribution of GWAS data, as well as the provision of control DNA samples by the Cooperative Research in the Region of Augsburg (KORA) and genotyping data generated by the Dietary, Lifestyle, and Genetic determinants of Obesity and Metabolic syndrome (DILGOM) consortium. We also thank the Genetic Analysis of Psoriasis Consortium (GAPC) and Psoriasis Association Genetics Extension (PAGE) for the contribution of Immunochip data. We thank the Barbara and Neal Henschel Charitable Foundation for their support of the National Psoriasis Victor Henschel BioBank. The Heinz Nixdorf Recall (HNR) cohort was established with the support of the Heinz Nixdorf Foundation. The Estonian Psoriasis cohort was supported by institutional research funding IUT20-46 of the Estonian Ministry of Education and Research, by the Centre of Translational Genomics of University of Tartu (SP1GVARENG) and by the European Regional Development Fund (Centre of Translational Medicine, University of Tartu). The genotyping of control individuals from the HNR cohort was financed by the German Federal Ministry of Education and Research (BMBF), within the context of the NGFNplus Integrated Genome Research Network MooDS (Systematic Investigation of the Molecular Causes of Major Mood Disorders and Schizophrenia). JTE is supported by the Ann Arbor Veterans Affairs Hospital. Detailed consortium contributor lists are detailed in the Supplementary Notes.
Footnotes
Author Contributions
J.T.E., R.C.T. and G.R.A. designed and directed the study. R.P.N., M.W., J.J.V., J.T.E., F.C., J.N.B., T.T., V.C., C.F.R., M.H.A., A.R., R.C.T., A.F., S.W., S.K., K.K., T.E., G.G.K., C.E., A.M., D.D.G., P.R., and S.L.S. contributed to sample collection and phenotyping. J.K. coordinated the GAPC samples and datasets. J.T.E. coordinated the PAGE samples and datasets. J.K., P.E.S, G.R.A., and H.M.K. advised on the statistical analysis. E.E., and R.P.N. performed genotyping. S.L.S., L.C.T., and E.E. performed the genotype calling. L.C.T. and S.L.S. and S.D. performed genotype imputation. L.C.T. performed statistical analysis. S.L. and S.W.S. performed the NFKBIZ expression experiments. F.C., T.T., S.L., S.S., J.E.G., J.N.B., R.C.T., J.T.E. and L.C.T. contributed to interpretation and biological inference for the results. L.C.T. and J.T.E. drafted the manuscript. L.C.T. and S.L. prepared the figures and tables. All authors approved the final draft.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Mustonen A, Mattila K, Leino M, Koulu L, Tuominen R. The costs of psoriasis medications. Dermatol Ther (Heidelb) 2013;3:169–77. doi: 10.1007/s13555-013-0040-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hawro T, et al. Impact of psoriasis severity on family income and quality of life. J Eur Acad Dermatol Venereol. 2014 doi: 10.1111/jdv.12572. [DOI] [PubMed] [Google Scholar]
- 3.Nair RP, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun LD, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 2010;42:1005–9. doi: 10.1038/ng.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellinghaus E, et al. Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2. Nat Genet. 2010;42:991–5. doi: 10.1038/ng.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genetic Analysis of Psoriasis C et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuart PE, et al. Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat Genet. 2010;42:1000–4. doi: 10.1038/ng.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–8. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah TS, et al. optiCall: a robust genotype-calling algorithm for rare, low-frequency and common variants. Bioinformatics. 2012;28:1598–603. doi: 10.1093/bioinformatics/bts180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1, 092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–54. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun LD, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 2010;42:1005–9. doi: 10.1038/ng.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, et al. Association analyses identifying two common susceptibility loci shared by psoriasis and systemic lupus erythematosus in the Chinese Han population. J Med Genet. 2013;50:812–8. doi: 10.1136/jmedgenet-2013-101787. [DOI] [PubMed] [Google Scholar]
- 14.Zhang XJ, et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat Genet. 2009;41:205–10. doi: 10.1038/ng.310. [DOI] [PubMed] [Google Scholar]
- 15.Sheng Y, et al. Sequencing-based approach identified three new susceptibility loci for psoriasis. Nat Commun. 2014;5:4331. doi: 10.1038/ncomms5331. [DOI] [PubMed] [Google Scholar]
- 16.Tang H, et al. A large-scale screen for coding variants predisposing to psoriasis. Nat Genet. 2014;46:45–50. doi: 10.1038/ng.2827. [DOI] [PubMed] [Google Scholar]
- 17.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, et al. Transcriptome Analysis of Psoriasis in A Large Case-Control Sample: Rna-Seq Provides Insights Into Disease Mechanisms. J Invest Dermatol. doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinds DA, et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 2013;45:907–11. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Julia A, et al. A genome-wide association study on a southern European population identifies a new Crohn’s disease susceptibility locus at RBX1-EP300. Gut. 2013;62:1440–5. doi: 10.1136/gutjnl-2012-302865. [DOI] [PubMed] [Google Scholar]
- 24.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–52. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li P, et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490:543–6. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang L, et al. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res. 2013;23:716–26. doi: 10.1101/gr.142521.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowes MA, Suarez-Farinas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. 2014;32:227–55. doi: 10.1146/annurev-immunol-032713-120225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huffmeier U, et al. Common variants at TRAF3IP2 are associated with susceptibility to psoriatic arthritis and psoriasis. Nat Genet. 2010;42:996–9. doi: 10.1038/ng.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol. 2014;14:585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang SH, Park H, Dong C. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J Biol Chem. 2006;281:35603–7. doi: 10.1074/jbc.C600256200. [DOI] [PubMed] [Google Scholar]
- 32.Sonder SU, Paun A, Ha HL, Johnson PF, Siebenlist U. CIKS/Act1-mediated signaling by IL-17 cytokines in context: implications for how a CIKS gene variant may predispose to psoriasis. J Immunol. 2012;188:5906–14. doi: 10.4049/jimmunol.1103233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mells GF, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–32. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowes J, et al. Comprehensive assessment of rheumatoid arthritis susceptibility loci in a large psoriatic arthritis cohort. Ann Rheum Dis. 2012;71:1350–4. doi: 10.1136/annrheumdis-2011-200802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto M, et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. 2004;430:218–22. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- 36.Bui JD, et al. A role for CaMKII in T cell memory. Cell. 2000;100:457–67. doi: 10.1016/s0092-8674(00)80681-9. [DOI] [PubMed] [Google Scholar]
- 37.Spiers EM, Lazarus GS, Lyons-Giordano B. Expression of plasminogen activator enzymes in psoriatic epidermis. J Invest Dermatol. 1994;102:333–8. doi: 10.1111/1523-1747.ep12371792. [DOI] [PubMed] [Google Scholar]
- 38.Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM) Hum Mutat. 2000;15:57–61. doi: 10.1002/(SICI)1098-1004(200001)15:1<57::AID-HUMU12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 39.Martin DA, et al. The emerging role of IL-17 in the pathogenesis of psoriasis: preclinical and clinical findings. J Invest Dermatol. 2013;133:17–26. doi: 10.1038/jid.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu C, et al. Act1, a U-box E3 ubiquitin ligase for IL-17 signaling. Sci Signal. 2009;2:ra63. doi: 10.1126/scisignal.2000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonder SU, et al. IL-17-induced NF-kappaB activation via CIKS/Act1: physiologic significance and signaling mechanisms. J Biol Chem. 2011;286:12881–90. doi: 10.1074/jbc.M110.199547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamazaki S, Muta T, Takeshige K. A novel IkappaB protein, IkappaB-zeta, induced by proinflammatory stimuli, negatively regulates nuclear factor-kappaB in the nuclei. J Biol Chem. 2001;276:27657–62. doi: 10.1074/jbc.M103426200. [DOI] [PubMed] [Google Scholar]
- 43.Qian Y, et al. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat Immunol. 2007;8:247–56. doi: 10.1038/ni1439. [DOI] [PubMed] [Google Scholar]
- 44.Okamoto K, et al. IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature. 2010;464:1381–5. doi: 10.1038/nature08922. [DOI] [PubMed] [Google Scholar]
- 45.Martin F, Dixit VM. A20 edits ubiquitin and autoimmune paradigms. Nat Genet. 2011;43:822–3. doi: 10.1038/ng.916. [DOI] [PubMed] [Google Scholar]
- 46.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakabe NJ, Savic D, Nobrega MA. Transcriptional enhancers in development and disease. Genome Biol. 2012;13:238. doi: 10.1186/gb-2012-13-1-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willer CJ, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knight J, et al. Conditional analysis identifies three novel major histocompatibility complex loci associated with psoriasis. Hum Mol Genet. 2012;21:5185–92. doi: 10.1093/hmg/dds344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan X, et al. Fine mapping of the psoriasis susceptibility locus PSORS1 supports HLA-C as the susceptibility gene in the Han Chinese population. PLoS Genet. 2008;4:e1000038. doi: 10.1371/journal.pgen.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 53.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoll SW, Johnson JL, Li Y, Rittie L, Elder JT. Amphiregulin carboxy-terminal domain is required for autocrine keratinocyte growth. J Invest Dermatol. 2010;130:2031–40. doi: 10.1038/jid.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.