Abstract
Epigenetic mechanisms including altered DNA methylation are critical for altered gene transcription subserving synaptic plasticity and the retention of learned behavior. Here we tested the idea that one role for activity-dependent altered DNA methylation is stabilization of cognition-associated hippocampal place cell firing in response to novel place learning. We observed that a behavioral protocol (spatial exploration of a novel environment) known to induce hippocampal place cell remapping resulted in alterations of hippocampal Bdnf DNA methylation. Further studies using neurophysiological in vivo single unit recordings revealed that pharmacological manipulations of DNA methylation decreased long-term but not short-term place field stability. Together our data highlight a role for DNA methylation in regulating neurophysiological spatial representation and memory formation.
Keywords: memory, spatial memory, place cells, epigenetic, neuroepigenetics, DNA methylation, Bdnf, DNMT, hippocampus
Introduction
De novo gene expression within the hippocampus has long been recognized for its necessary role in synaptic plasticity and the long-term retention of learned behavior. Given that memory formation requires experience-driven patterns of gene expression, this has led to the search for molecular mechanisms both adequately sensitive to environmental stimuli and capable of driving and maintaining transcription-dependent cellular changes. As originally proposed by Crick nearly three decades ago (Crick 1984), studies over the past few years have implicated epigenetic mechanisms including DNA methylation as key mediators of memory formation and stabilization. For example, several studies indicate that histone acetylation, an epigenetic change that alters chromatin structure in a manner that promotes gene transcription, plays an important role in mediating hippocampal gene expression associated with contextual fear (Miller et al. 2008; Barrett et al. 2011; Oliveira et al. 2011), object recognition (Barrett et al. 2011; Oliveira et al. 2011), and spatial (Bousiges et al. 2010) memory formation. Methylation of histones in patterns permissive to gene transcription also supports the formation of contextual fear memory (Gupta et al. 2010; Gupta-Agarwal et al. 2012). Furthermore, consistent with the role of histone modifications in memory, disturbances in hippocampal histone acetylation have been associated with disruptions in hippocampal plasticity and memory capacity in aged rats and in animals with neuronal loss (Fischer et al. 2007; Peleg et al. 2010).
Similarly, studies have shown that active regulation of DNA methylation and demethylation within the hippocampus supports neural plasticity and memory formation. DNA methylation is an epigenetic mechanism typically associated with gene silencing, but studies suggest that it may also be associated with active gene transcription (Yasui et al. 2007; Chahrour et al. 2008; Uchida et al. 2011). Experience-driven changes in gene expression following contextual fear conditioning are associated with methylation of the PP1 gene (Miller and Sweatt 2007), and demethylation of the reelin (Miller and Sweatt 2007) and Bdnf (Lubin et al. 2008; Mizuno et al. 2012) genes. Changes in DNA methylation and expression of hippocampal Bdnf are also associated with object recognition memory (Munoz et al. 2010) and the memory of traumatic experiences (Roth et al. 2011). As further evidence for the role of DNA methylation in memory processes, mice with disruptions in proteins associated with DNA methylation (including MeCP2 and DNMT1), show impairments in long-term potentiation and fear memory formation (Moretti et al. 2006; Feng et al. 2010; Nelson and Monteggia 2011). Memory dysfunction in aged rats has also been linked to aberrant changes in hippocampal DNA methylation (Penner et al. 2011).
Despite this broad molecular and behavioral background, the means by which chemical modification of DNA might control cognition-associated neuronal circuit firing patterns has been left unaddressed. The general goal of the present study was to investigate the potential contribution of DNA methylation in spatial memory formation and the maintenance of neurophysiological spatial representations within the hippocampal neural circuit (Moser et al. 2008). A place cell is a hippocampal pyramidal neuron that encodes space by selectively increasing activity (firing rate) in a specific environmental location (place field). A collection of place cells with place fields in different locations may encode large environmental areas providing the animal with a neurophysiological spatial representation of the environment (O’Keefe and Dostrovsky 1971), which is commonly termed a “cognitive map” (Tolman 1948) and believed to play a key role in navigation and spatial learning and memory. To examine whether DNA methylation is involved in generating or maintaining these maps and related spatial memory processes, we performed two experiments. In Experiment 1 we examined patterns of DNA methylation in rats that were subjected to a spatial behavioral protocol (Figure 1a) similar to that used in previous place cell experiments (Roth et al. 2012), where novel environments were shown to induce place cell remapping. Specifically in our 2012 study we showed that when a rat transitioned from a familiar to novel environment some cells maintained a place field in both the familiar and novel environments located at different degrees of rotation around a track (i.e. they rotate), while other cells had fields that turned on (appear), turned off (disappear), or split into multiple fields. Together these data indicate that place fields can exhibit a variety of remapping behaviors. In Experiment 2 here we pharmacologically manipulated DNA methylation patterns as we performed single unit in vivo hippocampal place cell recordings, a procedure wherein rats performed behavioral tasks similar to Experiment 1 running laps around an elevated track in familiar and novel environments (Figures 2 and 3).
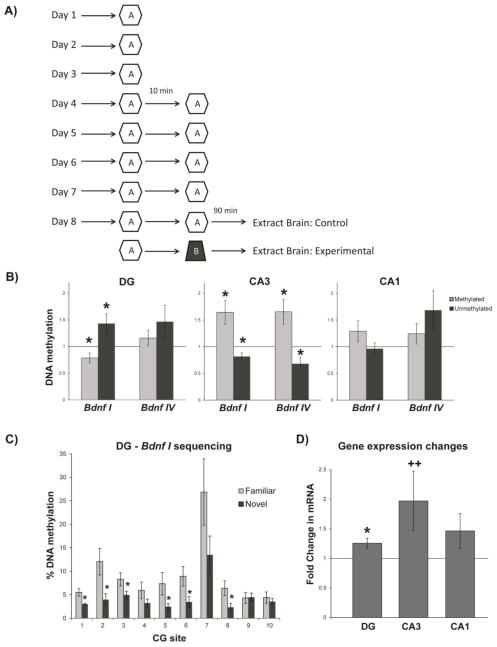
Figure 1.
(a) Graphic depicting the 8 day protocol used in Experiment 1. For 8 days rats performed a foraging task in a familiar environment (Environment A). On days 4–8, rats performed the task in two 20 lap sessions separated by 10 minutes. On days 1–7, protocols were identical for the experimental and control group. On day 8 the second 20 lap session for the experimental group was conducted in a novel environment (Environment B). Brains were extracted for biochemical analyses 90 minutes after the second foraging session on day 8. (b) Experience-induced alterations of hippocampal Bdnf gene DNA methylation. Levels of methylated and unmethylated DNA associated with the Bdnf gene in dentate (Bdnf I n=10, Bdnf IV n=11), CA3 (Bdnf I n=10, Bdnf IV n=11, and CA1 (Bdnf I n=11, Bdnf IV n=11) in novel-exposed vs. control (familiar-exposed) rats. (c) Methylation analysis of individual CG dinucleotides associated with exon I in the dentate of control and novel-exposed rats (n=10/group). (d) Bdnf mRNA (exon IX) levels in novel-exposed rats relative to familiar-exposed controls (DG n=8, CA3 n=9, CA1 n=9). Error bars represent SEM; *p<0.05; ++p=0.0885.
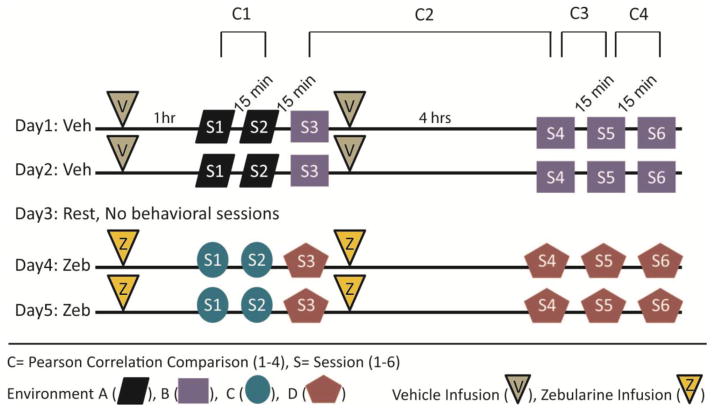
Figure 2.
Graphic depicting the timeline used for the 5 day protocol in Experiment 2. Rats performed 6 daily sessions of a foraging task in different environmental setups. A single session consisted of rats completing 13 laps around a circular track. ICV infusions were done 1 hr prior to session 1 and immediately after session 3. All intersession intervals were approximately 15 minutes (C1, C3, C4) except the interval (C2) between session 3 and session 4 which was 4 hrs.
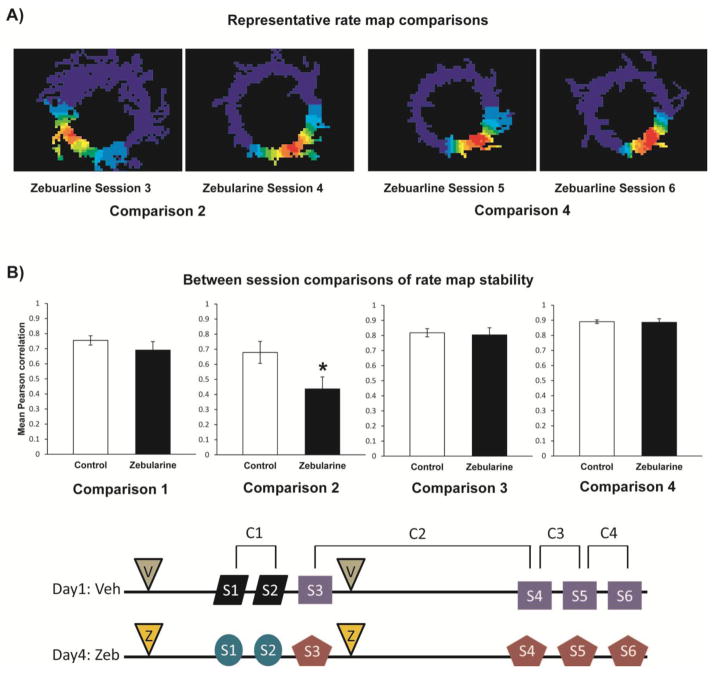
Figure 3.
Pharmacological manipulations of DNA methylation decreased place field stability. (a) Four representative rate maps used for Comparisons 2 and 4 in rats treated with zebularine. (b) Mean Pearson correlation coefficients representing place cell rate map stability across behavioral sessions are plotted. Correlations were generated comparing rate maps across sessions 1 and 2, 3 and 4, 4 and 5, and 5 and 6. The light bars represent correlations during the control treatment and the dark bars represent correlations during the zebularine treatment. Correlations between the control and zebularine treatment significantly differ (*p<0.05) in the session 3 to session 4 (3–4) comparison. Error bars represent SEM.
Results
Experiment 1: Assessment of DNA methylation following exposure to a novel environment
If a rat explores a familiar environment, place cell firing patterns remain relatively predictable and stable. However, when a rat explores a novel environment, place cell firing patterns will change (remap) as new maps and contexts are created or encoded (Hill 1978; Bostock et al. 1991). If processes generating and maintaining these spatial representations are mediated by DNA methylation, we would then expect DNA methylation patterns to change in response to spatial exposure to a novel environment. In experiment 1, we used a candidate gene approach focused on the Bdnf gene to test this prediction. Bdnf has been associated with neural plasticity (West et al. 2001), contextual fear conditioning (Hall et al. 2000), and spatial learning and memory (Mu et al. 1999; Heldt et al. 2007). Furthermore, DNA methylation has been implicated in hippocampal Bdnf activity-dependent gene regulation (Martinowich et al. 2003; Lubin et al. 2008; Roth et al. 2009; Roth et al. 2011) and consolidation of fear memory (Lubin et al. 2008).
Patterns of DNA methylation in rats that performed the final behavioral session in a novel environment differed significantly from those that performed their final session in a familiar environment (Figure 1). DNA methylation also varied by subregion and exon of the Bdnf gene (Figure 1b). Data generated using PCR-based methylation-specific primers (MSP, see methods) indicated that for CA3, the novel group had higher levels of methylated DNA associated with Bdnf exons I (t=2.89, p=0.0180) and IV (t=2.78, p=0.0195) relative to familiar controls (Figure 1b). These results were confirmed with the unmethylated primer set, which indicated lower levels of unmethylated DNA associated with exons I (t=2.65, p=0.0267) and IV (t=2.61, p=0.0263). No significant changes were found in either methylated or unmethylated DNA levels within CA1. In the dentate (Figure 1b), the novel group had lower levels of methylated DNA associated with exon I (t=2.29, p=0.0477). This is consistent with the observation of higher levels of unmethylated DNA associated with exon I (t=2.34, p=0.0443) and our independent replication of this result using direct bisulfite sequencing (BSP) data (Figure 1c: F1,180=22.16, p<0.0001). For all subregions, no significant changes were found in either methylated or unmethylated DNA associated with exon VI (data not shown).
Thus, our MSP and BSP data indicate that DNA methylation and demethylation are evoked by a behavioral paradigm known to induce hippocampal place cell remapping. Since DNA methylation is a known regulator of Bdnf gene expression, we examined whether there were corresponding changes in mRNA levels (Figure 1d). Exon-IX containing transcripts were elevated in the novel group relative to controls in the dentate (t=3.05, p=0.0186), and there was also a trend for an increase in Bdnf expression in CA3 (t=1.94, p=0.0885). These are both regions where we detected changes in DNA methylation. Together, these data indicate that our spatial learning behavioral paradigm does induce changes in Bdnf gene expression, and that changes in DNA methylation might be one mechanism by which this occurs.
Pharmacological manipulation of DNA methylation alters place field stability
While many place cell studies have focused on defining patterns of plasticity and stability associated with these spatial representations, relatively little is known about the underlying molecular mechanisms. Some of the receptors and transcription factors known to be involved in LTP and memory consolidation have been linked to place cell encoding and stability, which include hyperpolarization-activated cyclic nucleotide-gated channels (Nolan et al. 2004; Hussaini et al. 2011), NMDA receptors (Kentros et al. 1998; Ekstrom et al. 2001; Nakazawa et al. 2003), and zif268 (Renaudineau et al. 2009). Together, these studies highlight the importance of cell-signaling cascades and gene activity to place cell function, but epigenetic factors underlying transcriptional regulation and associated gene expression patterns in spatial cognition remain unclear. If place cell properties and learning and memory processes associated with spatial representations are mediated through DNA methylation mechanisms, we would predict that manipulations of DNA methylation patterns would alter place cell firing patterns.
To assess this in Experiment 2 we pharmacologically manipulated DNA methylation patterns as we performed single unit in vivo hippocampal place cell recordings (see Figure 2 for the basic experimental design). For this experiment we utilized a procedure wherein rats performed behavioral tasks similar to Experiment 1, running laps around an elevated track in familiar and novel environments. We then measured the stability of hippocampal place cell firing rate patterns (rate maps) by making four pair-wise comparisons across six different time points of an extended spatial learning and memory episode. For each of the 4 rate map comparisons (Session 1 to 2, 3 to 4, 4 to 5, 5 to 6, see Figures 2 and 3) for Experiment 2, place field stability over time was examined using Pearson correlations for both the drug and control treatments. Each correlation comparison was calculated from an average total of 55.5 ± 5.48 (SE) place cells from the 5 rats. Although we recorded from both CA1 and CA3 cells, most recording tetrodes were placed in CA1 resulting in 78% of correlation values. Correlations from both CA1 and CA3 cells were statistically similar in session comparisons. In all ANOVA analyses the main effect and all interaction effects associated with the factor hippocampal subregion (CA1, CA3) were highly nonsignificant. Thus, CA1 and CA3 cells were collapsed into one group and subregion was removed from the statistical model. Results (Figure 3b) are reported for a two way ANOVA examining the effects of drug treatment (zebularine, vehicle) and session comparison (1–2, 3–4, 4–5, 5–6) on rate map stability (Pearson correlation values). We observed significant main effects for both drug treatment (F1,436 =7.39, p=0.0068) and session comparison (F3,436 =19.18, p<0.0001), as well as a marginally significant drug treatment by session comparison interaction effect (F3,436 =2.57, p=0.0538). Across all 4 session comparisons, mean rate map correlations for the zebularine treatment were less than controls (Figure 3b), but within a session comparison the differences between drug treatments were not statistically significant except in the 3–4 (long term novel to familiar) session comparison where correlations in the zebularine treatment were significantly lower (F1,64 =4.81, p=0.032). Post hoc Fisher’s protected least significant difference pairwise comparisons revealed that all session comparisons were significantly different (all p’s<0.05) from each other. As would be expected rate map correlations were generally lower in both zebularine and control treatments in the novel to familiar session comparisons (1–2, 3–4), and then generally increased as familiarity with the environment increased with multiple exposures (sessions 4–5, 5–6). As this familiarity with the environment increased from session 3 to session 6, the mean differences between the drug and control groups decreased. Overall these data indicate a significant destabilization of hippocampal place cell firing patterns, in a within-subject design +/- drug, upon administration of a DNMT inhibitor: an observation consistent with the hypothesis that DNA methylation contributes to regulating the stability of hippocampal place cell firing patterns.
Discussion
In Experiment 1, we predicted that in behavioral paradigms known to induce hippocampal place cell remapping, we would also observe alterations to DNA methylation patterns as novel environments are encoded or learned. Indeed, the rats that performed the final behavioral session in a novel environment showed changes in patterns of hippocampal DNA methylation compared to controls. Specifically, rats with the novel spatial experience exhibited decreased Bdnf DNA methylation in the dentate gyrus and increased Bdnf DNA methylation in CA3. A rat’s spatial experience likely involves a complex interaction of contextual learning elements, exercise, and stress, factors on their own capable of inducing epigenetic modifications and altering gene expression (Bilang-Bleuel et al. 2005; Chandramohan et al. 2007; Lubin et al. 2008; Penner et al. 2011; Roth et al. 2011). Rats were over-trained for an extended period of time in the same context to try to minimize the influence of contextual learning and other factors associated with the behavioral task. To control for exercise, the number of laps during the behavioral paradigm were held constant between the two groups of animals. Although our animals did not appear stressed, it is possible that our changes in DNA methylation patterns also reflect a response to a stressor (novel environment) rather than a spatial learning and memory mechanism per se, or perhaps both as spatial learning and stress are likely not mutually exclusive processes. Nonetheless, these results provide the first demonstration that a novel spatial experience can alter DNA methylation patterns in the hippocampus in a subregion and exon-specific manner for a given gene.
In Experiment 2 we directly recorded the neurobiological hippocampal circuit spatial representation while pharmacologically altering DNA methylation. Prior to experimentation we had very little detailed knowledge about what, where, and when spatial learning and memory machinery and associated gene expression might be affected by DNA methylation. We did know from our previous in vivo work with zebularine that it is capable of altering hippocampal DNA methylation and gene expression patterns 40 min post-infusion (Lubin et al., 2008). To increase our chances of detecting changes in spatial representation in response to manipulation of methylation, experiments were designed to examine many different contexts and time points. For session comparison 1–2, session 1 represents the rat’s first exposure to a novel environment. After a brief 15 minute rest, session 2 represents the rat’s second exposure to that same environment. Here we tested if an ICV infusion of a demethylating agent 1 hour prior to session 1 alters the encoding or acquisition of a spatial representation (map) and/or the re-expression of this map in session 2. Under these conditions we did not see any significant effects of the drug on place field stability compared to control trials (Figure 3). These data indicate that the zebularine infusion does not non-specifically disrupt the ability of the hippocampal circuit to form a space field firing pattern. For session comparison 3–4, session 3 also represents the rat’s first exposure to another novel environment and session 4 is the first re-exposure to this environment. This is similar to the 1–2 comparison except there is a 4 hour delay between sessions. Under these long term delay conditions, we did see significant decreases in place field stability in DNMT inhibitor-treated rats compared to control trials. The session 4–5 and 5–6 comparisons were re-exposures to the 3–4 environment, but the delay between sessions was reduced to 15 minutes. Under these conditions place field stability returned to levels similar to control trials. In summary, we only detected a drug effect on place field stability when a long delay period was used between sessions. These data suggest that DNA methylation controls place field stabilization over time, but is not necessary for initial map formation.
These short-term vs. long-term results coincide well with previous reports. Injections of anisomycin, a protein synthesis blocker, revealed that protein synthesis was necessary for long term (6 hours between sessions) place field stability but not necessary for short term stability (1 hr between sessions) (Agnihotri et al. 2004). Additionally, injections of CPP, a competitive NMDA receptor antagonist, did not significantly disrupt short term place field stability (1.5 hrs) while abolishing long term stability (16–24 hrs) (Kentros et al. 1998). Mice lacking the transcription factor zif268 exhibited normal short term (1 hr) place field stability, but long term (24 hrs) stability was disrupted (Renaudineau et al. 2009). Under our current protocol, the ability to form stable fields in the short term comparisons (1–2, 4–5, 5–6) before and after the long term comparison (3–4) suggests that the drug effect in session 3–4 comparison likely represents a disruption of a memory consolidation mechanism rather than acquisition. Similarly, our study suggests that long term place field stability (4 hrs) may require molecular memory processes that are influenced by epigenetic mechanisms that include DNA methylation.
It is important to note in our within-subjects design, the drug treatment always came after the control treatment because we were concerned that effects from the drug may otherwise carry over into the control treatment. Thus we cannot experimentally rule out sequence effects. However, the significant loss of place field stability in the 3–4 comparison in the drug treatment is likely not a sequence effect. Nothing in the place field literature would predict this instability from a sequence effect. Actually, if any sequence effects were present, we would expect the reverse to happen as place field stability is more likely to increase as the animal becomes more experienced with the task. Finally, for future studies, another important factor to consider relative to timing and consolidation processes is that the second daily ICV infusion occurred immediately after the 3rd session. It is unclear if this infusion was necessary to disrupt consolidation processes between sessions 3 and 4 or if the first daily infusion was sufficient to produce this effect.
In summary, our results support the hypothesis that DNA methylation plays a role in regulating neurophysiological correlates of space and spatial memory formation (Miller and Sweatt 2007; Lubin et al. 2008; Feng et al. 2010). To our knowledge, this is the first study to demonstrate that a novel spatial experience alters DNA methylation patterns that vary by subregion and exon of the Bdnf gene in the hippocampus. Likewise, we provide novel evidence regarding a potential role of DNA methylation in place field stability and spatial memory formation. Although many questions remain and additional research is needed to provide detailed mechanistic explanations, this study provides valuable insight into the potential molecular mechanisms influencing long term stability of neurobiological spatial representations and memory. Additionally, given that DNA methylation controls memory capacity in aging (Oliveira et al. 2012) this work provides support for the speculative notion that the decline in spatial memory associated with aging and neurological disease could reflect dysregulation of hippocampal circuit function due to alterations in DNA methylation.
Materials and Methods
Subjects
All three experiments used adult male Long-Evans rats (400–500g). Rats were housed individually in plastic cages with a 12 hour day and night cycle, with food and water provided ad libitum. In Experiments 1 and 2, approximately 2 weeks prior to beginning behavioral experiments, food was restricted to maintain rats at 85–90% of their ad libitum weights (but ad libitum access to water remained). All procedures were approved by University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Apparatus and training
In Experiments 1 and 2, rats performed a foraging task. To perform this task rats were trained to run laps clockwise around an elevated circular track as they foraged for chocolate sprinkles randomly placed on the track by an experimenter. Although tracks were painted different colors and had different textures in different environmental setups, all tracks were the same size (76 cm outer diameter, 55 cm inner diameter) and mounted 13 cm above a platform that was 75 cm above the floor.
Histology
To confirm tetrode and/or cannula locations, transcardial perfusions (10% formalin) were performed after Experiments 2 and 3. Brains were removed and placed in 30% sucrose formalin solution. Brains were sectioned at 40 μm on a microtome, mounted, and stained with cresyl violet. To aid in confirming tetrode locations after Experiment 2, approximately 24 h before perfusions electrolytic lesions were generated (10 μA for 15 sec) on a subset of tetrodes.
Drug infusions
Experiment 2 included intracerebroventricular drug infusions of zebularine to manipulate DNA methylation. Zebularine is a demethylating agent known to reverse DNA methylation (Cheng et al. 2003; Marquez et al. 2005) and has been used to alter experience-dependent DNA methylation changes in other paradigms (Lubin et al. 2008; Roth et al. 2009). For all infusions, a guide cannula was placed in the left lateral ventricle and 3 μL (infusion rate of 1 μL/min) of either vehicle (control treatment: 10% dimethyl sulfoxide (DMSO)) or zebularine (drug treatment: 600 ng/μL in 10% DMSO) were delivered via a microinfusion pump.
Experiment 1: Assessment of DNA methylation following exposure to a novel environment
Behavioral training
Speakers were placed under the platform to provide background noise. Two behavioral rooms that differed in environmental cues (differences in visual cues, floor and ceiling patterns, platform texture, and track type) were used. One room served as the training environment (thus labeled familiar), while the other room was used as the novel environment. Rooms were randomly assigned as the familiar or novel environment prior to training. During the 3 day training period, 22 male Long-Evans rats were trained (30 minutes per day) on the foraging task on the circular track in the familiar environment. The rats were also familiarized with a staging location where they were permitted to rest in a small circular dish between sessions.
Behavioral testing
After training, 11 rats were assigned to the control group and 11 were assigned to the experimental group. On any given experimental day, the same general experimental procedure was followed (Figure 1a). Each rat in the control group was first transported in an enclosed opaque plastic box (disorientation chamber) from the home cage to the staging location or resting dish and permitted to habituate in the dish for 5 minutes. The rat was then placed at a random start location on the track in the familiar training environment. After completing 20 laps of the foraging task, the rat was returned to the disorientation box, and walked around the laboratory. Tracks were cleaned with 70% ethanol between all behavioral sessions. After a 10 minute rest period, the rat was again placed on the track for a second 20 lap session in the familiar environment. These two daily behavioral sessions in the familiar environment were repeated for 5 days. For the rats in the novel group, the protocol was identical except the 2nd behavioral session on the 5th day was conducted in the novel behavioral room. 90 minutes after the 2nd behavioral session on the 5th day, each rat was retrieved from its home cage and its brain was removed for biochemical analyses.
Biochemical analyses
90 minutes following completion of the spatial protocol, brains were removed, sectioned (1mm) and brushed onto glass slides, and the tissue was stored at −80 °C. DNA was extracted from the hippocampal subregions (dentate gyrus, CA3, CA1) using an AllPrep DNA/RNA kit (Qiagen). Following bisulfite-treatment (Qiagen EpiTect kit), methylation status was assessed via methyl-specific real-time PCR (MSP) in a Bio-Rad iQ5 system. PCR primer sequences designed to detect methylated vs. unmethylated DNA within target Bdnf exons (I, IV, VI) were used as previously described (Lubin et al. 2008; Roth et al. 2009). Product specificity was confirmed via a melting curve analysis performed in increasing 1 °C increments (real-time PCR) or gel electrophoresis. Samples were normalized to tubulin and the comparative Ct method and two-tailed one-sample t-tests were used to calculate differences in methylation in animals from the novel vs. familiar group.
For a subset of samples, methylation status of exon I was also independently assessed via direct bisulfite sequencing (BSP) using a similar protocol as previously described (Lubin et al. 2008; Roth et al. 2009; Roth et al. 2011; Parrish et al. 2012). Bisulfite-treated samples were amplified by the following primer sequences: 5′-TTT ATT TTT TGG AGT TTG TGG TAT G-3′ (forward) and 5′-ACT TCT CAA ATA AAA ATT AAC AAC CTC TAT-3′ (reverse). The PCR products were purified using a gel extraction kit (Qiagen), and sequenced using the reverse primer. The percent methylation of each CG site within the amplified region was determined by the ratio between peaks values of G and A (G/[G+A]) on the electropherograms (determined using Chromas software). Universally unmethylated and methylated standards (EpigenDx) were run in parallel with samples, which indicated that the ratio of cytosine methylation increased proportionately with expected methylation rates (r2=0.9, p<0.001). Differences in BSP data were analyzed by analysis of variance tests with Bonferroni’s post hoc tests when appropriate.
To determine whether there were changes in Bdnf gene expression consistent with methylation changes, RNA was extracted from the same tissue using an AllPrep DNA/RNA kit (Qiagen). Reverse transcription was performed using a cDNA synthesis kit (Qiagen). cDNA was amplified by real-time PCR (Bio-Rad iQ5 system) using Taqman probes targeted at exon IX (Applied Biosystems), with tubulin serving as a reference gene. The comparative Ct method was used to calculate differences in gene expression between samples, and group differences in mRNA levels were analyzed by two-tailed one-sample t-tests.
Experiment 2: Hippocampal single unit recordings
Surgical Procedures
Five male rats were anesthetized by isoflurane inhalation and a microdrive array was implanted above the right dorsal hippocampus (3.9 mm posterior to bregma, 3.6 mm lateral to midline). The microdrive array was made up of 20 recording probes or tetrodes. A stainless steel guide cannula (23 gauge) was placed in the left lateral ventricle (1.4 mm posterior to bregma, 2.1 mm lateral from midline, and 3.0 mm ventral to brain surface) for drug infusions. The guide cannula and microdrive were secured with stainless steel screws and dental cement. Rats were allowed one week to recover before further behavioral training.
Single-unit Neuronal Recordings
Tetrodes were constructed with 4 fine (0.0005 inches) insulated nichrome electrode wires twisted together. Each electrode was goldplated to obtain an impedance between 200–300 kΩ measured at 1 kHz. Neuronal signals were passed through a headstage (HS 36, Neuralynx) to a data acquisition system (Digital Lynx, Neuralynx) and filtered between 600 Hz and 6 kHz. Light-emitting diodes mounted on the headstage were used to track the animal’s location at 60 Hz.
Behavioral training
Similar to Experiment 1, rats were trained on the foraging task. Generally 9–16 training sessions were required before the experiment to assure rats could meet behavioral criteria (i.e. attentively foraging for sprinkles with limited distractions, completing at least 15 laps within 10 minutes). Rats were also familiarized with another room (quiet room) where rats were permitted to rest in a small circular dish. During the post-surgical training period (10–18 days), electrodes were gradually advanced to pyramidal cell layers in CA1 and CA3.
Behavioral apparatus and environment
The behavioral recording environment was cylindrical (2.3 m diameter) with the outer perimeter defined by curtains extending from floor to ceiling. The platform with a circular track was located in the center of the room. Light was provided by a single 25 W bulb mounted in the ceiling in the center of the room. A commutator with recording tethers and video camera were ceiling mounted slightly offset from center (≤12 cm) surrounding the central light. Speakers were mounted behind the ring of curtains in the four corners of the room to provide background noise. By altering floor, ceiling, platform, and wall coverings, as well as exchanging circular tracks and directional audio cues, 4 distinct environments were created for behavioral testing. Environmental pairings were randomly determined for each rat. The first environment that the rats experienced was always labeled as Environment A (Env.A), and the second environment was labeled as Environment B (Env.B).
Behavioral Testing
The within subject experimental design contained a control treatment (infusion of DMSO) and a drug treatment (infusion of Zebularine). The drug and control treatments consisted of 2 days of single-unit recordings during multiple sessions of the foraging task. Each foraging task session consisted of the rat completing 13 laps around the track. Each of the 5 rats sequentially received 2 days of the control treatment followed by a day of rest (with no behavioral tasks) followed by 2 days of drug treatment.
On any given experimental day, the same general experimental procedure was followed (Figure 2). Each day was split into 2 sets (morning and afternoon) of 3 behavioral (13 lap) recording sessions (i.e., a total of 6 behavioral recording sessions per day). The two sets were separated by 4 h while the 3 within-set behavioral recording sessions were separated by approximately 15 minutes. The first 2 sessions were in the same environment (e.g., Env.A). The 3rd through 6th sessions were in another environment (e.g., Env.B). Rats were infused 1 hour prior to the first session and immediately after the third session. After the rats received the first infusion, they were permitted to rest quietly in a dish in a quiet room as we performed single-unit recordings to collect baseline sleep data for approximately 20 minutes. These data were used to assess recording stability. One hour after infusion, the rat was placed in a covered box and briefly walked around the laboratory for disorientation before entering the behavior room. Once in the behavior room (e.g., Env.A) the rat was removed from the disorientation box and placed on a pedestal in the center of the track and left undisturbed for 2 minutes. We then connected the rat’s microdrive to the Neuralynx headstage and tethers that transmit signals to the acquisition system. The rat was then placed at a random start location on the track to begin session 1 as we recorded neuronal signals and tracked the rat’s location. After completing 13 laps, the rat was returned to the disorientation box, and transported to the dish in the quiet room. The rat rested in the dish for 15 minutes and was then placed back in the disorientation box.
This process was repeated for sessions 2 and 3. However, between sessions 2 and 3 the behavioral room was converted to the other environmental setup (e.g., from Env.A to Env.B.). After all 3 behavioral sessions were completed in the first (morning) set, another (5 min) baseline sleep data session was recorded and the rat was infused for the second time. In the afternoon set (4 hours later), this protocol was repeated. However, all 3 behavioral sessions were in the same environment as the third session (e.g., Env.B), no infusions were administered, and the final baseline sleep data session was 20 min. Drug and control treatments used the same protocols except each treatment had a unique set of behavioral environments (e.g., treatment 1 control: Env.A/Env.B, treatment 2 drug: Env.C/Env.D).
Data Analyses
Custom software was used to isolate single units by examining relative signal amplitudes and other waveform parameters on each tetrode wire. Spatial information scores were calculated (Skaggs et al. 1993). A place cell was defined as any cell with a statistically significant (p ≤ 0.05) information score of ≥ 0.7 with ≥ 100 spikes. Rate maps were generated for each cell in each session by dividing the number of spikes in a pixel by the amount of time spent in each pixel. Rate maps were smoothed as 640–480 pixel arrays were collapsed into 64–48 bins (bin size, 2.2 cm × 2.2 cm). Spatial correlations (Pearson) were calculated between rate map distributions in sessions 1 and 2, 3 and 4, 4 and 5, as well as 5 and 6. Correlation coefficients were only generated if a cell met the criteria above defining a place cell in at least one of the sessions being compared and generated at least 50 spikes in the other session. These Pearson correlations were analyzed by ANOVA with session comparison (session 1–2, 3–4, 4–5, 5–6), drug (vehicle, zebularine), and sub-region (CA1, CA3) as factors. To avoid potential confounding interactions across days, analyses were limited to comparisons of day 1 of the drug and control treatment.
Acknowledgments
University of Delaware undergraduates Kenneth Chen and Samantha Jones provided help with sequencing data analysis. University of Alabama-Birmingham (UAB) undergraduates Kendra Crews, Emily Ellis, Jessica Hembree, Jamie Holt, Whitney Hudman, Galen Jeong, Matthew Ostrander, and Russell Warr assisted in behavioral training of animals and other elements of experimental support. UAB graduate students Frankie Heyward and Luke Coddington aided in data collection for Experiment 2. Special thanks to Felecia Hester and Dr. Cristin Gavin (UAB) for countless aspects of laboratory management, and Dr. Farah Lubin (UAB), Dr. Jim Knierim (Johns Hopkins University), and Geeta Rao (Johns Hopkins University) for valuable advice and support on many aspects of the experiments. Research was supported by grants from the NIMH (MH091122), NINDS, NIA, NIH Blueprint, the Ellison Medical Foundation, Brain & Behavior Research Foundation, and the Evelyn F. McKnight Brain Research Foundation.
Footnotes
Author contributors
Authors EDR, TLR, and JDS designed the study and wrote the paper. For Experiment 1, EDR performed the behavioral training and TLR performed the biochemical analyses. For Experiment 2, EDR, KMM, SS, and DEE performed the surgeries, behavioral training, and neuronal recordings. All authors took part in the analysis of data and interpretation of the results.
Conflict of Interest Statement
The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agnihotri NT, Hawkins RD, Kandel ER, Kentros C. The long-term stability of new hippocampal place fields requires new protein synthesis. Proc Natl Acad Sci U S A. 2004;101:3656–3661. doi: 10.1073/pnas.0400385101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA. Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology. 2011;36:1545–1556. doi: 10.1038/npp.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilang-Bleuel A, Ulbricht S, Chandramohan Y, De Carli S, Droste SK, Reul JM. Psychological stress increases histone H3 phosphorylation in adult dentate gyrus granule neurons: involvement in a glucocorticoid receptor-dependent behavioural response. European Journal of Neuroscience. 2005;22:1691–1700. doi: 10.1111/j.1460-9568.2005.04358.x. [DOI] [PubMed] [Google Scholar]
- Bostock E, Muller RU, Kubie JL. Experience-dependent modifications of hippocampal place cell firing. Hippocampus. 1991;1:193–205. doi: 10.1002/hipo.450010207. [DOI] [PubMed] [Google Scholar]
- Bousiges O, de Vasconcelos AP, Neidl R, Cosquer B, Herbeaux K, Panteleeva I, Loeffler JP, Cassel JC, Boutillier AL. Spatial Memory Consolidation is Associated with Induction of Several Lysine-Acetyltransferase (Histone Acetyltransferase) Expression Levels and H2B/H4 Acetylation-Dependent Transcriptional Events in the Rat Hippocampus. Neuropsychopharmacology. 2010;35:2521–2537. doi: 10.1038/npp.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramohan Y, Droste SK, Reul JM. Novelty stress induces phospho-acetylation of histone H3 in rat dentate gyrus granule neurons through coincident signalling via the N-methyl-D-aspartate receptor and the glucocorticoid receptor: relevance for c-fos induction. Journal of Neurochemistry. 2007;101:815–828. doi: 10.1111/j.1471-4159.2006.04396.x. [DOI] [PubMed] [Google Scholar]
- Cheng JC, Matsen CB, Gonzales FA, Ye W, Greer S, Marquez VE, Jones PA, Selker EU. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst. 2003;95:399–409. doi: 10.1093/jnci/95.5.399. [DOI] [PubMed] [Google Scholar]
- Crick F. Memory and molecular turnover. Nature. 1984;312:101. doi: 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- Ekstrom AD, Meltzer J, McNaughton BL, Barnes CA. NMDA receptor antagonism blocks experience-dependent expansion of hippocampal “place fields”. Neuron. 2001;31:631–638. doi: 10.1016/s0896-6273(01)00401-9. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nature Neuroscience. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Gupta-Agarwal S, Franklin AV, Deramus T, Wheelock M, Davis RL, McMahon LL, Lubin FD. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. Journal of Neuroscience. 2012;32:5440–5453. doi: 10.1523/JNEUROSCI.0147-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. Journal of Neuroscience. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J, Thomas KL, Everitt BJ. Rapid and selective induction of BDNF expression in the hippocampus during contextual learning. Nature Neuroscience. 2000;3:533–535. doi: 10.1038/75698. [DOI] [PubMed] [Google Scholar]
- Heldt SA, Stanek L, Chhatwal JP, Ressler KJ. Hippocampus-specific deletion of BDNF in adult mice impairs spatial memory and extinction of aversive memories. Mol Psychiatry. 2007;12:656–670. doi: 10.1038/sj.mp.4001957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AJ. First occurrence of hippocampal spatial firing in a new environment. Exp Neurol. 1978;62:282–297. doi: 10.1016/0014-4886(78)90058-4. [DOI] [PubMed] [Google Scholar]
- Hussaini SA, Kempadoo KA, Thuault SJ, Siegelbaum SA, Kandel ER. Increased Size and Stability of CA1 and CA3 Place Fields in HCN1 Knockout Mice. Neuron. 2011;72:643–653. doi: 10.1016/j.neuron.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentros C, Hargreaves E, Hawkins RD, Kandel ER, Shapiro M, Muller RV. Abolition of long-term stability of new hippocampal place cell maps by NMDA receptor blockade. Science. 1998;280:2121–2126. doi: 10.1126/science.280.5372.2121. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. Journal of Neuroscience. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez VE, Kelley JA, Agbaria R, Ben-Kasus T, Cheng JC, Yoo CB, Jones PA. Zebularine: a unique molecule for an epigenetically based strategy in cancer chemotherapy. Ann N Y Acad Sci. 2005;1058:246–254. doi: 10.1196/annals.1359.037. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiology of Learning and Memory. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Dempster E, Mill J, Giese KP. Long-lasting regulation of hippocampal Bdnf gene transcription after contextual fear conditioning. Genes Brain and Behavior. 2012;11:651–659. doi: 10.1111/j.1601-183X.2012.00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. Journal of Neuroscience. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser EI, Kropff E, Moser MB. Place cells, grid cells, and the brain’s spatial representation system. Annu Rev Neurosci. 2008;31:69–89. doi: 10.1146/annurev.neuro.31.061307.090723. [DOI] [PubMed] [Google Scholar]
- Mu JS, Li WP, Yao ZB, Zhou XF. Deprivation of endogenous brain-derived neurotrophic factor results in impairment of spatial learning and memory in adult rats. Brain Research. 1999;835:259–265. doi: 10.1016/s0006-8993(99)01592-9. [DOI] [PubMed] [Google Scholar]
- Munoz PC, Aspe MA, Contreras LS, Palacios AG. Correlations of recognition memory performance with expression and methylation of brain-derived neurotrophic factor in rats. Biol Res. 2010;43:251–258. [PubMed] [Google Scholar]
- Nakazawa K, Sun LD, Quirk MC, Rondi-Reig L, Wilson MA, Tonegawa S. Hippocampal CA3 NMDA receptors are crucial for memory acquisition of one-time experience. Neuron. 2003;3:305–315. doi: 10.1016/s0896-6273(03)00165-x. [DOI] [PubMed] [Google Scholar]
- Nelson ED, Monteggia LM. Epigenetics in the mature mammalian brain: effects on behavior and synaptic transmission. Neurobiology of Learning and Memory. 2011;96:53–60. doi: 10.1016/j.nlm.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan MF, Malleret G, Dudman JT, Buhl DL, Santoro B, Gibbs E, Vronskaya S, Buzsaki G, Siegelbaum SA, Kandel ER, et al. A behavioral role for dendritic integration: HCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell. 2004;119:719–732. doi: 10.1016/j.cell.2004.11.020. [DOI] [PubMed] [Google Scholar]
- O’Keefe J, Dostrovsky J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Research. 1971;34:171–175. doi: 10.1016/0006-8993(71)90358-1. [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Estevez MA, Hawk JD, Grimes S, Brindle PK, Abel T. Subregion-specific p300 conditional knock-out mice exhibit long-term memory impairments. Learn Mem. 2011;18:161–169. doi: 10.1101/lm.1939811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AMM, Hemstedt TJ, Bading H. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat Neurosci. 2012;15:1111–1113. doi: 10.1038/nn.3151. [DOI] [PubMed] [Google Scholar]
- Parrish RR, Day JJ, Lubin FD. Direct bisulfite sequencing for examination of DNA methylation with gene and nucleotide resolution from brain tissues. Curr Protoc Neurosci. 2012;Chapter 7(Unit7):24. doi: 10.1002/0471142301.ns0724s60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, et al. Altered Histone Acetylation Is Associated with Age-Dependent Memory Impairment in Mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging. 2011;32:2198–2210. doi: 10.1016/j.neurobiolaging.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudineau S, Poucet B, Laroche S, Davis S, Save E. Impaired long-term stability of CA1 place cell representation in mice lacking the transcription factor zif268/egr1. Proc Natl Acad Sci U S A. 2009;106:11771–11775. doi: 10.1073/pnas.0900484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth ED, Yu X, Rao G, Knierim JJ. Functional Differences in the Backward Shifts of CA1 and CA3 Place Fields in Novel and Familiar Environments. PLoS One. 2012;7:e36035. doi: 10.1371/journal.pone.0036035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Zoladz PR, Sweatt JD, Diamond DM. Epigenetic modification of hippocampal Bdnf DNA in adult rats in an animal model of post-traumatic stress disorder. Journal of Psychiatric Research. 2011;45:919–926. doi: 10.1016/j.jpsychires.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaggs WE, McNaughton BL, Gothard KM, Markus EJ. An information-theoretic approach to deciphering the hippocampal code. In: Hanson SJ, Cowan JD, Giles CL, editors. Advances in neural information processing systems. Morgan Kaufman; San Mateo, CA: 1993. pp. 1030–1037. [Google Scholar]
- Tolman EC. Cognitive Maps in Rats and Men. Psychol Rev. 1948;55:189–208. doi: 10.1037/h0061626. [DOI] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T, Suzuki T, Miyata N, Watanabe Y. Epigenetic Status of Gdnf in the Ventral Striatum Determines Susceptibility and Adaptation to Daily Stressful Events. Neuron. 2011;69:359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, Greenberg ME. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci U S A. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, Lasalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci U S A. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]