Abstract
Kruppel-like factors (KLF) are zinc-finger DNA binding transcription factors that are critical regulators of tissue homeostasis. Emerging evidence suggests KLFs are critical regulators of muscle biology in the context of cardiovascular health and disease. The focus of this review is to provide an overview of the current state of knowledge regarding the physiologic and pathologic roles of KLFs in the three lineages of muscle: cardiac, smooth, and skeletal.
Introduction
Cardiovascular disease remains the leading cause of morbidity and mortality in the world (1). Although recent advances in clinical modalities and pharmacotheraputiecs lessen disease burden, a more detailed understanding of molecular mechanisms that drive disease initiation and progression is required for further therapeutic impact. The two predominant cell types in the heart and blood vessel are the cardiomyocyte and vascular smooth muscle cell (VSMC), respectively. The primary function of these cell types is contraction, thus enabling sufficient blood flow and oxygenation to peripheral tissues. Dysfunction of muscle leads to a broad spectrum of cardiac and vascular states that can impair their physiologic role. As such, understanding the molecular mechanisms governing cellular function in health and disease is critical for the development of novel therapies.
Kruppel-like factors: General considerations
Kruppel-like factors are members of the zinc-finger class of DNA-binding transcription factors whose name was derived from the German word kruppel (meaning “cripple”) (2). The original Kruppel gene was identified in Drosophila as a developmental gene critical in early stage body patterning and segmentation (3). The first mammalian KLF was identified in 1993, and to date, 18 family members have been identified and numbered chronologically based on their order of discovery (2). The KLFs share sequence homology in their C-terminal zinc-finger domains characterized by three Cys2/His2 zinc-finger regions connected by a conserved TGEKP(Y/F)X amino acid sequence. DNA binding and specificity are mediated through this zinc-finger region via consensus sequences including CACCC- GC- or GT- box elements located in proximal promoters and enhancers. Structural and functional divergence of the KLF family is determined by the non-DNA binding N-terminal domains that regulate protein-protein interaction and informs transcriptional activation or repression. Moreover, phylogenetic analysis of the mammalian KLF family reveals structural homologies within the N-terminal domain that correlates with functional similarities. As such, these structure / function characteristics allow for the classification of KLF family members into three distinct groups: Group 1 (KLFs 3, 8, and 12) are transcriptional repressors that interact with carboy-terminal binding protein, Group 2 (KLFs 1, 2, 4, 5, 6) are predominately transcriptional activators, and Group 3 (KLFs 9, 10, 11, 13, 14, and 16) act to repress transcriptional activity (via interaction with the co-repressor Sin3A) while KLF15 and 17 are more distantly related (2). While some KLFs are expressed ubiquitously, others display tissue restriction allowing for redundant and non-redundant roles in response to various physiological stimuli. Expressed predominately in the nucleus, KLFs are subject to various post-transcriptional modifications and responsible for recruitment of transcriptional co-activator / co-repressor complexes which modifies their DNA-binding and functional activity, respectively, to exert their cellular effects. Since their identification, these factors have been implicated as critical regulators of diverse cellular processes including metabolism, growth, proliferation, hematopoiesis, immunity, determination of pluripotency, and, important for this review, muscle remodeling and cellular differentiation / plasticity (2). This review will thus focus on the role of KLFs in the physiology and pathophysiology of muscle function.
KLFs and cardiac muscle
Despite the appreciation that transcriptional inputs guide cardiac function in health and disease, the role of KLFs are only beginning to burgeon. This topic was last reviewed seven years ago, and since this time, additional evidence has provided mechanistic insights and expanded previously known roles of KLFs in cardiac function while new biologic themes have emerged (4). As will be discussed below, seminal observations have broadly implicated KLFs as critical mediators of cardiac development, hypertrophy / remodeling, metabolism, and electrical activity.
Cardiac development
Congenital heart disease (CHD) is the leading cause of mortality in infants under the age of one (5). Inherited forms of CHD have been linked to mutations in transcription factors that are critical in heart development (6). Examples of such transcription factors include Tbx5 and Nkx2.5 that act in a coordinated fashion with GATA4 to drive cardiac development (7). Until recently, however, no known role for the KLF family in mediating cardiac development has been described. Work from the Nemer laboratory first described KLF13 as essential for cardiac development in vivo (8). Cardiac KLF13 is expressed in both atria and ventricles with expression first detected at E9.5 in the developing embryo. KLF13 expression is reduced postnatally with low levels detected in the adult valves and septum. Embryonic deletion of KLF13 in Xenopus results in septal defects with hypotrabeculation while murine deletion results in enlarged hearts suggesting a critical role for KLF13 in heart development and function. Mechanistically, KLF13 interacts with GATA4 to regulate critical cardiac promoters including BNP and ANF during development (9). In addition to KLF13, a more recent study has linked another KLF family member, KLF3, to embryonic cardiomyopathy and perinatal lethality (10). Screening for dominant mutations that affect cardiovascular function in N-ethyl-N-nitrosourea (ENU) mutagenized mice, Kelsey and colleagues identified a missense mutation in KLF3 with homozygocity resulting in embryonic lethality. Heterozygotes for the histidine to arginine mutation in the zinc finger DNA-binding region that died were characterized by biventricular cardiac hypertrophy while adult survivors exhibited reduced blood pressure along with enlarged cardiac chambers and valvular stenosis. Taken together, these data suggest KLF13 and KLF3 are critical in the developing heart and could serve as novel gene targets for therapeutic gain in congenital heart disease.
Cardiac hypertrophy / remodeling
Hypertrophy is one of the strongest predictors for the pathogenesis of heart failure, arrhythmia, and sudden cardiac death (11). Hypertrophy is an adaptive response to hemodynamic and neurohormonal stress wherein the heart enlarges to increase its primary function of contraction and maintain circulatory function while reducing myocardial wall tension. In addition to myocyte enlargement, myocyte disarray, fibrosis, and alterations in myocardial fuel utilization are linked to the pathogenesis of cardiac hypertrophy. This pathologic response occurs through the activation of molecular and genetic pathways, and over the past several years, multiple KLFs have emerged as critical regulators of cardiomyocyte remodeling.
Work largely derived from the laboratory of Dr. Ryozo Nagai, one of the first KLFs identified as critical in regulating cardiac function was KLF5 (12, 13). Early studies indicated cardiac KLF5 expression is restricted to the fibroblast with limited detection in the cardiomyocyte. KLF5 expression is upregulated in response to pro-hypertrophic stimuli such as angiotensin II while heterozygous deletion of KLF5 blunts the angiotensin II hypertrophic response. More recently, Takeda et al. showed that fibroblast KLF5 serves as a cardioprotective factor by modulating cardiomyocyte hypertrophy through a paracrine mechanism involving IGF-1 (14). These studies thus establish KLF5 as central to the interplay between cardiac myocytes and fibroblasts in response to the pathogenesis of cardiac hypertrophy and remodeling.
While KLF5 expression is restricted to the cardiac fibroblast, KLF15 expression is robust in cardiac myocytes. Initial studies by the Jain laboratory demonstrated that cardiac KLF15 expression is induced postnatally, a time at which canonical hypertrophic gene makers (e.g. ANF and BNP) are downregulated (15). Moreover, KLF15 expression is downregulated in both rodent and human biopsies of heart failure as well as in vitro in response to hypertrophic stimuli including angiotensin II, phenylephrine, and endothelin-1 (16). Germ-line deletion of KLF15 results in mice which are viable and do not display a cardiac phenotype at baseline. However, in response to pressure overload, these mice develop severe eccentric hypertrophy characterized by systolic dysfunction through a molecular mechanism involving transcriptional inhibition of MEF2 and GATA4, thus establishing KLF15 as a negative regulator of pathologic hypertrophy (15).
This molecular mechanism and the role for KLF15 in regulating cardiac hypertrophy / remodeling have been recently extended and links KLF15 to both heart failure and aortic aneurysm (discussed below) through a shared molecular mechanism involving acetylated-p53 (16). Specifically, KLF15 deficient mice develop severe heart failure in response to angiotensin II infusion resulting in p53 hyperacetylation, findings that are recapitulated in human disease. As acetylation of p53 is mediated by acetyltranferase activity of p300, pharmacologic inhibition of p300 ameliorated the cardiac (and vascular) phenotype. These results suggest KLF15 serves to not only inhibit cardiac hypertrophy through direct DNA-binding activity, but also serves as an indirect epigenetic regulator by binding p300 and thus dampening p53 hyperacetylation in response to stress. Given that p300 also acetylates additional factors critical in cardiac remodeling, including GATA4, MEF2, and histones, we speculate that KLF15 acts as a molecular brake that disrupts p300-dependent acetylation and serves as a potent repressor of multiple pathologic transcriptional circuits.
Various growth factors, including transforming growth factor-b (TGFb), have been implicated as upstream signals that convey a pro-hypertrophic phenotype in the heart. Identification in 1995 of a TGFb inducible early gene-1 (TIEG1, latter termed KLF10) by the Spelsberg laboratory revealed robust expression in the heart and has been implicated as an important factor in regulating osteoclast differentiation and bone mineralization (17, 18). KLF10 deletion in male mice (16 months of age) results in a cardiomyopathy characterized by asymmetric septal hypertrophy with increase left ventricular mass, tissue fibrosis, and myocyte disarray (18). Interestingly, this phenotype is restricted to male mice, as female mice do not display signs of hypertrophy or fibrosis at any age suggesting that KLF10 acts downstream of the estrogen pathway in the heart (18, 19). Transcriptomic profiling revealed KLF10 negatively regulates the expression of Pituitary Tumor Transforming Gene (Pttg1, also known as securin) suggesting a model wherein KLF10 acts as a hypertrophy suppressor via downregulation of Pttg1 (18). Given the heritability of hypertrophic cardiomyopathy, the Spelsberg laboratory extended their early findings by identifying six novel missense mutations within the four translated exons of the human KLF10 gene (20). Notably, 5 of the 6 KLF10 variants identified were associated with increased PTTG1 protein expression and promoter activity. Collectively, these data suggest KLF10 is a novel hypertrophic cardiomyopathy gene in humans.
While early reports identify KLF5, KLF15, and KLF10 as key transcriptional components of the cardiac hypertrophic response, KLF4 has received more recent attention with regards to cardiomyocyte physiology and pathophysiology. First identified as gut-enriched Kruppel-like factor, KLF4 expression in the heart is present from late embryonic development through adulthood (21). While our group and others have demonstrated a critical role for KLF4 in regulating endothelial, myeloid, and, as will be discussed below, smooth muscle cell biology, the role of KLF4 in cardiac muscle is only beginning to be elucidated (22). Our group provided the inaugural in vivo evidence that KLF4 serves as a negative regulator of cardiac hypertrophy (23). Importantly, this work demonstrated that mice with cardiomyocyte-specific deletion (driven the aMHC-Cre recombinase murine line) of KLF4 are characterized by high rates of mortality following pressure overload. Surviving mice display cardinal features of cardiac hypertrophy including increased cardiac mass, cavity dilation, and depressed LV systolic function with myocardial fibrosis and apoptosis. These finding were enhanced by efforts from the Owens laboratory which showed KLF4 deletion in both cardiac and vascular smooth muscle (using the SM22a-Cre recombinase murine line) gives rise to a significant postnatal mortality rate with marked growth retardation despite being born at expected Mendelian ratios (21). Surviving mice displayed attenuated cardiac output at baseline in the absence of compensatory hypertrophy along with reduced cardiac gene expression including GATA4. Moreover, using a pharmacological model of heart failure, Yoshida and colleagues showed hearts deficient in KLF4 are sensitive to chronic isoproterenol infusion via a molecular mechanism involving myocardin gene induction (24). These findings provide compelling evidence that KLF4 is requisite for cardiac development and maturation and serves as a negative regulator of cardiac hypertrophy.
Cardiac metabolism
Considered an endurance machine, the mammalian heart is one of the largest consumers of energy, thus requiring the oxidation of multiple carbon substrates to meet this energetic demand (25). In the healthy myocardium, the preferred fuel source is fat with ~70% of ATP production being derived from oxidative metabolism of fatty acids; the remainder coming from glucose (~20%) as well as lactate and ketones (26). However, the metabolic myocardium is highly plastic. During periods of heightened physiologic demand, the heart guards against energy depletion by augmenting appropriate transcriptional circuits to meet its metabolic need. Importantly, loss of this plasticity has been linked to cardiomyopathy with dysregulated lipid flux serving a causal role in heart failure progression (27, 28). Coordinated lipid flux involves appropriate sarcolemmal lipid uptake, activation of lipids to acyl-CoA moieties and import to the mitochondrial matrix for subsequent -oxidation, TCA cycle, and oxidative phosphorylation (26). This myocardial lipid flux is under robust allosteric and transcriptional control. With respect to the latter, much of this work has been ascribed the nuclear receptor family of transcription factors (i.e. peroxisome proliferator-activated receptor; PPARs) (29). In particular, PPAR and PPAR / are essential regulators of cardiac fuel utilization, and through genetic gain- and loss-of-function studies, have been linked to inappropriate lipid utilization and cardiac dysfunction (29). However, current pharmacotherapies that target PPARs are associated with unfavorable side effects (30). As such, despite being one of the largest energy consumers, there exists a paucity of evidence outside the nuclear receptor field with regards to transcriptional control of cardiac energetics in health and disease, and elucidation of such transcriptional pathways could provide novel therapeutic approaches.
Our recent report implicates KLF15 as a novel and independent regulator of cardiac lipid metabolism (31). As postnatal maturation of the mammalian heart is characterized by increase reliance on lipids, we first observed increased KLF15 expression following birth in both rodent and human hearts. Moreover, as cardiomyopathy has been linked to diminution of lipid utilization, KLF15 levels are reduced in diseased human hearts; an effect that is reversed with mechanical unloading of the myocardium. Using the isolated working heart to model substrate flux, KLF15 knock-out hearts are characterized by a significant reduction in fatty acid oxidation with a parallel increase in glucose oxidation. Importantly, these alterations in myocardial substrate energetics occur in the presence of preserved contractile function with no difference in hemodynamic indices. Unbiased transcriptomic analysis reveals a strong KLF15-dependent signature for myocardial substrate metabolism, most notably genes involving lipid flux. These functional and genetic alterations observed in the KLF15-KO hearts occurred, however, in the absence of significant alterations in the expression of known metabolic transcriptional regulators or co-activators (e.g. PPARs and PGC1, respectively). Moreover, these observations phenocopy, at both the functional and genetic level, previous known roles ascribed to PPARs in regulating cardiac metabolism (32, 33). This allows one to speculate whether nuclear receptors and KLFs operate in a coordinated fashion. With previous reports giving rise to this possibility in other tissues and biological contexts, the interplay of KLFs and nuclear receptors in coordinating complex genetic networks to physiological responses is an emerging theme, and such investigations will provide novel avenues for therapeutic exploration (34).
Arrhythmogenesis
Worldwide, the leading cause of mortality from cardiovascular disease is sudden cardiac death secondary to ventricular arrhythmias (35). A sobering reality, however, is the current pharmacotherapeutic landscape to combat this dreadful disease is woefully inadequate with the only effective modality remaining ventricular defibrillators. Ventricular arrhythmias are driven, at least in part, by alterations in either the duration (short- and long-QT syndromes) or pattern (Brugada) during the repolarization phase of the ventricular action potential and are classified as either acquired (heart failure; short- and long-QT) or inherited (Brugada) syndromes (36–38). Although structure / function studies have revealed the importance of these repolarizing potassium currents in arrhymogenesis, little is known regarding the molecular control of cardiac electrical stability.
Interestingly, clinical observations reveal ventricular arrhythmias exhibit a diurnal variation suggesting a time of day dependence with peak abnormalities occurring in the early morning hours (39). A physiologic process that oscillates with a periodicity of ~24 hours is said to be circadian and is controlled at the molecular level by the biologic clock which consists of both positive and negative transcriptional feedback loops centered around the DNA-binding transcription factors Bmal and Clock (40, 41). However, a role for KLFs in myocardial circadian biology in general, and cardiac electrophysiology in particular, had not been elucidated.
Given the reduced expression of KLF15 in human and rodent models of heart failure, along with KLF15 serving as a negative regulator of hypertrophic remodeling, we reasoned that KLF15 might also regulate cardiac electrophysiology (42). This work led to the observation that cardiac KLF15 expression is rhythmic with peak expression observed during the transition from the inactive to active phase. Moreover, the time of day oscillation of cardiac ion-channel expression and the QT-interval duration (an index of myocardial repolarization) are KLF15-dependent. At the molecular level, this is mechanistically linked to the KLF15-dependet rhythmic expression of a critical subunit required for optimal transient outward potassium current termed the Kv channel-interactin protein 2 (KChIP2). Finally, cardiac restricted KLF15 gain- and loss-of-function studies provide a model wherein KLF15 excess or deficiency results in perturbation of rhythmic QT intervals, abnormal repolarization, and increased susceptibility to ventricular arrhythmias.
While these studies firmly link KLF15 as a clock-dependent driver of the circadian transcription of ion channels and a critical regulator of cardiac electrophysiology / arrhythmogenesis, important considerations remain. The above studies were performed in rodent hearts, and, in contrast to murine repolarization that is largely driven by Ito, human repolarization is driven by multiple repolarizing current, most notably Herg (43). Therefore, additional studies are required to establish the importance of the clock-KLF15-arrhymogenesis axis in human electrophysiology, an area of ongoing research by our group. Finally, given the recent advances implicating KLF15 as a regulator of cardiac metabolism and arrhythmogenesis, it is tempting to speculate whether KLF15 serves as a molecular bridge for electro-metabolic coupling in the heart. Electro-metabolic coupling is an emerging theme in cardiac biology supported by experimental and clinical observations including the appreciation that metabolic disease is associated with increased risk of ventricular arrhythmias (44). In addition, inborn errors of fatty acid oxidation predispose children to arrhythmogenesis and sudden death (45, 46). These inborn errors of metabolism are associated with deficiencies in carnitine-acylcarnitine translocase activity resulting in increased accumulation of intermediate metabolites such as long-chain acylcarnitines. Indeed, the long-chain acylcarnitines have been shown to affect not only potassium currents, but also sodium and calcium transients (47–49). As such, the role of KLFs in general, and KLF15 in particular, in cardiac electro-metabolic coupling is a fruitful and burgeoning area of future investigation.
KLFs and vascular smooth muscle
The principal function of the differentiated vascular smooth muscle cells (VSMC) is contraction to maintain vascular tone for blood flow distribution (50). This function is coordinated at the molecular level by the expression of multiple contractile proteins including smooth muscle actin and smooth muscle-myosin heavy chain (50). However, unlike the cardiomyocyte, VSMC exhibit a high degree of phenotype plasticity, and, in response to acute or chronic injury, are characterized by increased proliferation, migration, and loss of contractile properties that drive both vessel repair and vascular disease pathologies including stent restenosis or atherosclerosis (50). This phenotype plasticity is controlled, at least in part, at the molecular level with DNA-binding transcription factors, including serum response factor (SRF), and their coactivators (e.g. myocardin) driving the differentiated phenotypic state (50). However, with transcriptional networks and epigenetic regulation of cellular differentiation in response to injury as emerging themes, a more complete understanding of the fundamental molecular mechanisms which control VSMC biology are therefore needed. As such, KLFs offer a novel avenue for investigation, and, to date, have been implicated as critical molecular regulators of VSMC development / differentiation along with VSMC injury and repair response.
VSMC development and differentiation
Vasculogenesis, angiogenesis, and maturation are distinct biologic features required for proper blood vessel development (51). During development, the VSMC is highly migratory and undergoes rapid cellular proliferation with elaboration of extracellular matrix components. In contrast, the VSMC in adult vessels have a low rate of turnover with minimal extracellular matrix production. This differentiated state is characterized by the expression of marker genes controlled by both environmental and genetic factors. With respect to the latter, while many KLFs have been linked to cellular development and differentiation in the context of erythropoiesis (KLF1), epithelial cell mucosa (KLF4), and adipogenesis (KLF2-7, 11, 15), only a few KLFs have been linked to these processes in VSMC (2).
One of the earliest described KLFs in blood vessel development and maturation was KLF2. Originally termed lung-KLF, KLF2 deficient mice die during embryogenesis (E12.5–14.5) from severe hemorrhaging characterized by endothelial cell necrosis despite normal angiogenesis and vasculogenesis (52). With regards to the VSMC, this lethality is associated with a thin tunica media along with aneurysmal dilatation (52). In the absence of KLF2, VSMC failed to form a compact tunica media and displayed a significant reduction in the number of differentiated VSMC (52). While these early studies suggest endothelial cell dysfunction as a major cause for embryonic lethality in the KLF2 deficient mice, more recent studies by the Lingrel laboratory suggest a more causal role for VSMC KLF2. While KLF2 is unlikely necessary for VSMC specification, KLF2−/− embryos display a poorly developed VSMC layer (53). In particular, VSMC were absent from the dorsal aortic surface of KLF2−/− mice suggesting a defect in the migration or proliferation of VSMC (53). Taken together, these studies suggest an important interplay between endothelial and smooth muscle KLF2 in blood vessel stabilization.
Whereas a definitive role of KLF2 in regulating differentiation in the VSMC per se has yet to be determined, a more specific role for KLF4 has been established. First identified as a critical regulator of epithelial cell differentiation, KLF4 expression levels are relatively low in VSMC under basal conditions (4). However, KLF4 expression is significantly increased with VSMC dedifferentiation (50). In keeping with this pattern of gene expression, work from the Owens laboratory has established KLF4 as a negative regulator of VSMC differentiation. Specifically, a yeast one-hybrid identified KLF4 as binding to the TGF-b control element of SM22a (54). Moreover, KLF4 expression is attenuated with TGFb stimulation in VSMC while the TGF-b induction of the SM22a promoter is KLF4 dependent (54). Additionally, as TGF-b promotes a differentiated VSMC phenotype, platelet-derived growth factor BB (PDGF-BB) is a potent and efficacious inhibitor of VSMC differentiation, thus promoting a synthetic phenotype. Importantly, KLF4 expression is induced in VSMC treated with PDGF-BB, and acute knockdown of KLF4 blunts the PDGF-BB dependent repression of SMC differentiation markers (55). Finally, KLF4 attenuates the myocardin-induced activation SMC genes by a mechanism involving inhibition of serum response factor binding to CArG box elements (55). Taken together, these data suggest KLF4 is a negative regulator of VSMC differentiation and promotes a synthetic VSMC phenotype, and, as will be discussed below, may serve as a key mediator of VSMC phenotype switching in response to injury.
VSMC injury and repair response
In response to acute (mechanical) and chronic (inflammatory) vascular injury, VSMC dedifferentiate and are characterized as a synthetic phenotype with increased proliferative and migratory properties that contribute to occlusive vascular diseases including stent restenosis an atherosclerosis (56). As stated previously, this phenotype plasticity is determined, at least in part, at the gene regulatory level whereby transcriptional inputs drive VSMC differentiation marker gene repression. As described below, recent reports demonstrate a causal role for KLFs in mediating VSMC response to injury.
Although KLF4 levels are low in the differentiated VSMC, KLF4 expression is rapidly induced following vascular injury in vivo as well as in response to PDGF-BB and oxidized phospholipids in vitro (55, 57). However, conditional deletion of KLF4 gives rise to accelerated neointimal formation suggesting the KLF4 post-injury induction is likely a negative feedback response (58). Taken together, these results suggest KLF4 is a critical regulator of VSMC proliferation following acute vessel injury. Moreover, given VSMC dedifferentiation is an early event in aneurysm formation, Salmon and colleagues recently set out to test whether KLF4 is involved in aneurysm pathogenesis. Importantly, this study demonstrated KLF4 levels are progressively increased in aortas following elastase perfusion, and VSMC-specific loss of KLF4 conferred aneurysm protection (59). Finally, whereas KLF4 expression is regulated by pathologic stimuli (i.e. 1-palmitoyl-2-(5-oxovaleroyl)-sn-glycero-3-phosphocholine and other oxidized phospholipids) that accumulate in atherosclerotic lesions, a definitive in vivo role for KLF4 in regulating chronic inflammatory vasculitis remains undetermined.
Like KLF4, KLF5 is a potent negative regulator of VSMC differentiation and drives a synthetic phenotype (2). KLF5 expression profiling reveals robust detection in fetal aortas of both humans and rodents localized to the tunica media with limited detection in adult tissue (60). However, KLF5 expression is induced following vascular injury in neointimal VSMC. A definitive role of KLF5 in vascular injury is derived from loss-of-function studies. KLF5+/- mice display a marked reduction in neointima formation and VSMC proliferative following both vascular injury and angiotensin II infusion (12). Furthermore, mechanistic studies suggest KLF5 interacts with RARa and the acetyltransferase p300 thus mediating the vascular remodeling phenotype through direct DNA-binding and chromatin remodeling, respectively (12). Taken together, these studies suggest KLF5 serves as a nodal regulator of smooth muscle proliferation and vascular injury in vivo.
In contrast to that of KLF4 and KLF5, KLF15 is minimally expressed in the developing vessel but significantly increased during gestation and into adulthood (56). In both human and rodent vessels, KLF15 expression is primarily restricted to the VSMC and is decreased following vascular injury and pathologic stimuli (16, 61). As stated previously, our initial report demonstrates KLF15−/− mice develop a severe aortopathy characterized by aortic aneurysm formation following angiotensin II stimulation in a p53-dependent and p300-dependent fashion (16). To assess the role of KLF15 in response to vascular injury, KLF15 deficient mice were subjected to femoral wire injury (61). Importantly, loss of KLF15 results in enhanced neointimal formation with increased VSMC proliferation and migration thus identifying KLF15 as a negative regulator of VSMC response to injury.
Collectively, the above studies highlight an important role for KLFs in mediating acute vascular injury; however, a role for any KLF in chronic vascular disease has only recently been determined. Inflammatory vascular disease such as atherosclerosis is characterized by activation of various cell types that either reside in or migrate to the vessel wall, most notably the VSMC (56). Given the expression profile and response to injury noted above, we reasoned that VSMC KLF15 regulated proinflammatory activation and vascular disease. Indeed, KLF15 expression is markedly reduced in human atherosclerotic tissues while smooth muscle restricted deletion of KLF15 results in aggressive inflammatory vasculopathy in response to carotid artery transplantation and diet-induced atherosclerosis (62). These data suggest the KLF gene family (KLF4, KLF5, and KLF15 reported to date) have both overlapping yet non-redundant roles in mediating VSMC biology and their response to both acute and chronic pathologic stimuli.
KLFs and skeletal muscle
Despite the early identification of Kruppel by Ruiz-Gomez and colleagues nearly two decades ago through myogenic fate mapping studies in the gestating fly, the role of KLFs in skeletal muscle has, until recently, been under-developed compared to cardiac and smooth muscle (63). Due to space limitations, we refer the reader to a recent review on the role of KLFs in skeletal muscle biology and will only highlight key insights recently described as it pertains to myogenesis / muscle fusion and skeletal muscle metabolism (64).
Myogenesis and muscle fusion
Skeletal muscle myogenesis is a multistep process in which multinucleated myotubes are formed from mononucleate myoblast cell cycle withdrawal followed by fusion. This differentiation process is regulated at the molecular level by myogenic regulatory factors, including MyoD, Myogenin, and MEF, yet recent findings have demonstrated a critical role of KLFs in muscle maturation (65). As such, Himeda and colleagues identified KLF3 as capable of binding the Muscle Creatine Kinase (MCK) promoter thereby driving muscle differentiation in an SRF-independent manner (66). In contrast, KLF6 has been recently identified as a downstream effector of TGFb and molecular regulator of skeletal myoblast proliferation (67). Acute knockdown of KLF6 drives myogenic differentiation phenotype suggesting KLF6 is required for cell proliferation. Finally, as muscle fusion is critical for myogenesis, Nishida and colleagues recently identified ERK5 inducible KLFs 2/4 as critical for muscle fusion in a MyoD and MEF2 independent manner (68). These studies collectively link the KLF gene family to myogenesis and muscle maturation.
Skeletal muscle metabolism
Skeletal muscle comprises roughly 40% of body mass and accounts for ~30% of energy expenditure (69). Importantly, skeletal muscle structural and metabolic abnormalities are often associated with cardiovascular disease (70). Like cardiac muscle, skeletal muscle metabolism is efficiently coordinated at the gene regulatory level with PPARs, along with their associated co-regulators, serving a central role (71). However, a role for the KLF gene family in regulating skeletal muscle metabolism has only recently been elucidated. Early work by the Nagai laboratory provided inaugural evidence that KLF5 is a nodal regulator of skeletal muscle lipid utilization. In particular, KLF5+/− mice are resistant to diet induced obesity, despite increased food intake, due to increased energy expenditure through a molecular mechanism involving the nuclear receptor PPARd (72). As in cardiac muscle, these data add to the growing appreciation that KLFs and nuclear receptors coordinate their effects on metabolic gene targets for appropriate tissue homeostasis.
Our group established KLF15 as a fasting responsive gene that coordinates nutrient flux across multiple tissue beds (73, 74). These early studies provided the framework for the recent identification that KLF15 is a critical determinant of skeletal muscle lipid flux and exercise capacity (75). This work implicates KLF15 as an important regulator of nutrient catabolism and suggests upstream catabolic signals could regulate KLF15 expression. Indeed, recent reports demonstrate glucocorticoids induce KLF15 expression (76, 77). The in vivo significance of this axis was recently examined by Shimizu et al. who suggest that the glucocorticoid induced muscle wasting is the result of enhanced KLF15-dependent BCAA catabolism and diminished mTOR activity (78). However, in the absence of KLF15 loss-of-function studies in vivo, a definitive role for KLF15 in mediating this glucocorticoid-induced atrophy response remains tenuous and should be the focus of future investigation.
Concluding remarks
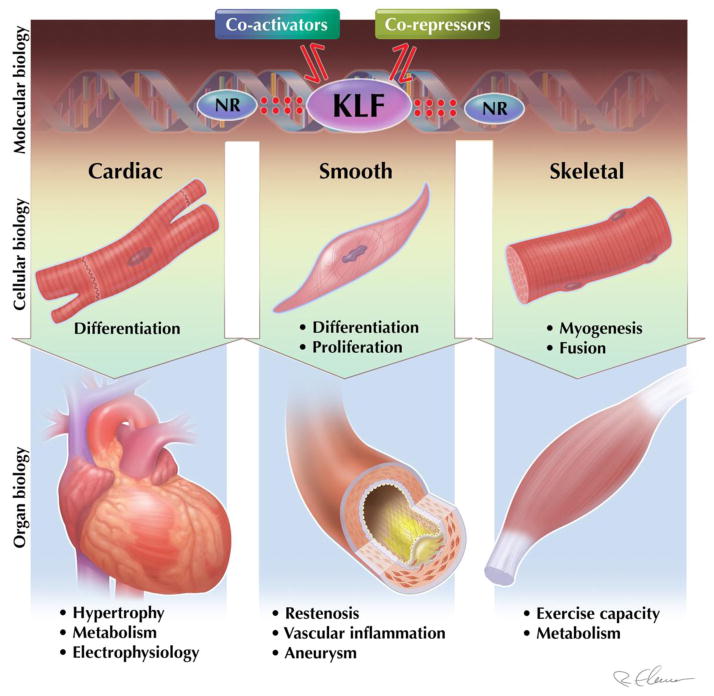
Cardiovascular function is directed by the dynamic interplay between cardiac, smooth, and skeletal muscle. The KLFs have emerged as important regulators of cellular differentiation / proliferation along with various aspects of physiology / pathophysiology in muscle (Figure 1). Given that KLFs have been shown to regulate multiple biological processes, it is tempting to speculate that a unifying mechanism may be operative that explains diverse cellular effects. For example, cardiac KLF15 is known to regulate cardiac and vascular physiology and disease. The same factor is also increasingly viewed as a master regulator of metabolism, thereby raising the possibility that KLF15 regulation of cellular metabolism may underlie the physiologic role of KFL15 in muscle tissues. In addition, this review illuminates the importance of KLF interactions with co-activators/co-repressors (e.g. p300 and HDACs) as well as other families of transcription factors (e.g. nuclear receptors) to control the genetic and epigenetic landscape as a means to regulate cellular function; an emerging theme of transcriptional control of cardiovascular disease in general, and KLF biology in particular. Moreover, as multiple KLFs are expressed in each muscle bed, future exploration should focus on the redundant vs. non-redundant roles of KLFs in health and disease. Finally, a deeper understanding of upstream factors that regulate KLF expression in the various biological processes mentioned above will help inform pharmacologic targeting of KLFs for therapeutic gain. Critical to this endeavor, high-throughput screening mediated elucidation of novel small-molecules that regulate, directly or indirectly, the expression of KLFs will provide novel therapeutic insights. Importantly, using a cell-based ultrahigh-throughput screening approach, the Yang laboratory identified two potent compounds that attenuated KLF5 expression and reduced viability of several colorectal cancer cell lines (79). Although there exists a paucity of data using this high-throughput screening approach as it pertains to KLFs and cardiovascular disease, these reports suggest this screening approach is feasible and likely to be fruitful.
Figure 1. Kruppel-like factors (KLF) in muscle biology.
KLFs are emerging as critical regulators of a transcriptional circuitry, along with nuclear receptors (NR) and co-activator / repressor complexes, which governs muscle biology at the cellular and organ level.
In addition to a high-throughput screening approach, studies focused on identifying KLF single nucleotide polymorphisms (SNP) or mutations within their gene regulatory regions will provide novel insights into the signaling pathways involved in the pathogenesis of human cardiovascular disease. This is highlighted by the aforementioned missense mutations identified in KLF10 from patients with hypertrophic cardiomyopathy that led to the elucidation of Pttg1 as a potential biomarker in maladaptive cardiac remodeling (20). In addition, recent work from the Nagai laboratory identified a novel angiotensin-II-MEF2A signaling axis via a SNP in the proximal promoter of KLF5 that is associated with an increase risk of hypertension (80). Using this combinatorial approach, future studies should focus on KLF SNP / mutations to aid in the elucidation of novel cardiovascular disease associated gene regulatory programs in humans.
Footnotes
There are no conflicts of interest to disclose
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Turk-Adawi K, Sarrafzadegan N, Grace SL. Global availability of cardiac rehabilitation. Nature reviews Cardiology. 2014;11(10):586–96. doi: 10.1038/nrcardio.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiological reviews. 2010;90(4):1337–81. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 4.Haldar SM, Ibrahim OA, Jain MK. Kruppel-like Factors (KLFs) in muscle biology. Journal of molecular and cellular cardiology. 2007;43(1):1–10. doi: 10.1016/j.yjmcc.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Bom T, Zomer AC, Zwinderman AH, Meijboom FJ, Bouma BJ, Mulder BJ. The changing epidemiology of congenital heart disease. Nature reviews Cardiology. 2011;8(1):50–60. doi: 10.1038/nrcardio.2010.166. [DOI] [PubMed] [Google Scholar]
- 6.Andersen TA, de Troelsen KL, Larsen LA. Of mice and men: molecular genetics of congenital heart disease. Cellular and molecular life sciences : CMLS. 2014;71(8):1327–52. doi: 10.1007/s00018-013-1430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCulley DJ, Black BL. Transcription factor pathways and congenital heart disease. Current topics in developmental biology. 2012;100:253–77. doi: 10.1016/B978-0-12-387786-4.00008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lavallee G, Andelfinger G, Nadeau M, Lefebvre C, Nemer G, Horb ME, et al. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. The EMBO journal. 2006;25(21):5201–13. doi: 10.1038/sj.emboj.7601379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grepin C, Dagnino L, Robitaille L, Haberstroh L, Antakly T, Nemer M. A hormone-encoding gene identifies a pathway for cardiac but not skeletal muscle gene transcription. Molecular and cellular biology. 1994;14(5):3115–29. doi: 10.1128/mcb.14.5.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelsey L, Flenniken AM, Qu D, Funnell AP, Pearson R, Zhou YQ, et al. ENU-induced mutation in the DNA-binding domain of KLF3 reveals important roles for KLF3 in cardiovascular development and function in mice. PLoS genetics. 2013;9(7):e1003612. doi: 10.1371/journal.pgen.1003612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Berlo JH, Maillet M, Molkentin JD. Signaling effectors underlying pathologic growth and remodeling of the heart. The Journal of clinical investigation. 2013;123(1):37–45. doi: 10.1172/JCI62839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, et al. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nature medicine. 2002;8(8):856–63. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- 13.Nagai R, Shindo T, Manabe I, Suzuki T, Kurabayashi M. KLF5/BTEB2, a Kruppel-like zinc-finger type transcription factor, mediates both smooth muscle cell activation and cardiac hypertrophy. Advances in experimental medicine and biology. 2003;538:57–65. doi: 10.1007/978-1-4419-9029-7_5. discussion 6. [DOI] [PubMed] [Google Scholar]
- 14.Takeda N, Manabe I, Uchino Y, Eguchi K, Matsumoto S, Nishimura S, et al. Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. The Journal of clinical investigation. 2010;120(1):254–65. doi: 10.1172/JCI40295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisch S, Gray S, Heymans S, Haldar SM, Wang B, Pfister O, et al. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(17):7074–9. doi: 10.1073/pnas.0701981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haldar SM, Lu Y, Jeyaraj D, Kawanami D, Cui Y, Eapen SJ, et al. Klf15 deficiency is a molecular link between heart failure and aortic aneurysm formation. Science translational medicine. 2010;2(26):26ra. doi: 10.1126/scitranslmed.3000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic acids research. 1995;23(23):4907–12. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajamannan NM, Subramaniam M, Abraham TP, Vasile VC, Ackerman MJ, Monroe DG, et al. TGFbeta inducible early gene-1 (TIEG1) and cardiac hypertrophy: Discovery and characterization of a novel signaling pathway. Journal of cellular biochemistry. 2007;100(2):315–25. doi: 10.1002/jcb.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leenders JJ, Pinto YM, Creemers EE. Tapping the brake on cardiac growth-endogenous repressors of hypertrophic signaling. Journal of molecular and cellular cardiology. 2011;51(2):156–67. doi: 10.1016/j.yjmcc.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 20.Bos JM, Subramaniam M, Hawse JR, Christiaans I, Rajamannan NM, Maleszewski JJ, et al. TGFbeta-inducible early gene-1 (TIEG1) mutations in hypertrophic cardiomyopathy. Journal of cellular biochemistry. 2012;113(6):1896–903. doi: 10.1002/jcb.24058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida T, Gan Q, Franke AS, Ho R, Zhang J, Chen YE, et al. Smooth and cardiac muscle-selective knock-out of Kruppel-like factor 4 causes postnatal death and growth retardation. The Journal of biological chemistry. 2010;285(27):21175–84. doi: 10.1074/jbc.M110.112482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain MK, Sangwung P, Hamik A. Regulation of an inflammatory disease: Kruppel-like factors and atherosclerosis. Arteriosclerosis, thrombosis, and vascular biology. 2014;34(3):499–508. doi: 10.1161/ATVBAHA.113.301925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao X, Haldar SM, Lu Y, Jeyaraj D, Paruchuri K, Nahori M, et al. Kruppel-like factor 4 regulates pressure-induced cardiac hypertrophy. Journal of molecular and cellular cardiology. 2010;49(2):334–8. doi: 10.1016/j.yjmcc.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida T, Yamashita M, Horimai C, Hayashi M. Kruppel-like Factor 4 Protein Regulates Isoproterenol-induced Cardiac Hypertrophy by Modulating Myocardin Expression and Activity. The Journal of biological chemistry. 2014;289(38):26107–18. doi: 10.1074/jbc.M114.582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldberg IJ, Trent CM, Schulze PC. Lipid metabolism and toxicity in the heart. Cell metabolism. 2012;15(6):805–12. doi: 10.1016/j.cmet.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC. Myocardial fatty acid metabolism in health and disease. Physiological reviews. 2010;90(1):207–58. doi: 10.1152/physrev.00015.2009. [DOI] [PubMed] [Google Scholar]
- 27.Kolwicz SC, Jr, Tian R. Metabolic therapy at the crossroad: how to optimize myocardial substrate utilization? Trends in cardiovascular medicine. 2009;19(6):201–7. doi: 10.1016/j.tcm.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertrand L, Horman S, Beauloye C, Vanoverschelde JL. Insulin signalling in the heart. Cardiovascular research. 2008;79(2):238–48. doi: 10.1093/cvr/cvn093. [DOI] [PubMed] [Google Scholar]
- 29.Madrazo JA, Kelly DP. The PPAR trio: regulators of myocardial energy metabolism in health and disease. Journal of molecular and cellular cardiology. 2008;44(6):968–75. doi: 10.1016/j.yjmcc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 30.Wright MB, Bortolini M, Tadayyon M, Bopst M. Minireview: Challenges and opportunities in development of PPAR agonists. Molecular endocrinology. 2014:me20131427. doi: 10.1210/me.2013-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prosdocimo DA, Anand P, Liao X, Zhu H, Shelkay S, Artero-Calderon P, et al. Kruppel-like factor 15 is a critical regulator of cardiac lipid metabolism. The Journal of biological chemistry. 2014;289(9):5914–24. doi: 10.1074/jbc.M113.531384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell FM, Kozak R, Wagner A, Altarejos JY, Dyck JR, Belke DD, et al. A role for peroxisome proliferator-activated receptor alpha (PPARalpha ) in the control of cardiac malonyl-CoA levels: reduced fatty acid oxidation rates and increased glucose oxidation rates in the hearts of mice lacking PPARalpha are associated with higher concentrations of malonyl-CoA and reduced expression of malonyl-CoA decarboxylase. The Journal of biological chemistry. 2002;277(6):4098–103. doi: 10.1074/jbc.M106054200. [DOI] [PubMed] [Google Scholar]
- 33.Cheng L, Ding G, Qin Q, Huang Y, Lewis W, He N, et al. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-delta deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nature medicine. 2004;10(11):1245–50. doi: 10.1038/nm1116. [DOI] [PubMed] [Google Scholar]
- 34.Knoedler JR, Denver RJ. Kruppel-like factors are effectors of nuclear receptor signaling. General and comparative endocrinology. 2014;203:49–59. doi: 10.1016/j.ygcen.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair GM, Nery PB, Redpath CJ, Birnie DH. Ventricular arrhythmias in patients with heart failure secondary to reduced ejection fraction: a current perspective. Current opinion in cardiology. 2014;29(2):152–9. doi: 10.1097/HCO.0000000000000035. [DOI] [PubMed] [Google Scholar]
- 36.Goldenberg I, Moss AJ. Long QT syndrome. Journal of the American College of Cardiology. 2008;51(24):2291–300. doi: 10.1016/j.jacc.2008.02.068. [DOI] [PubMed] [Google Scholar]
- 37.Patel U, Pavri BB. Short QT syndrome: a review. Cardiology in review. 2009;17(6):300–3. doi: 10.1097/CRD.0b013e3181c07592. [DOI] [PubMed] [Google Scholar]
- 38.Antzelevitch C. Role of spatial dispersion of repolarization in inherited and acquired sudden cardiac death syndromes. American journal of physiology Heart and circulatory physiology. 2007;293(4):H2024–38. doi: 10.1152/ajpheart.00355.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller JE, Ludmer PL, Willich SN, Tofler GH, Aylmer G, Klangos I, et al. Circadian variation in the frequency of sudden cardiac death. Circulation. 1987;75(1):131–8. doi: 10.1161/01.cir.75.1.131. [DOI] [PubMed] [Google Scholar]
- 40.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330(6009):1349–54. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 42.Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, Hu K, et al. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature. 2012;483(7387):96–9. doi: 10.1038/nature10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimizu W, Horie M. Phenotypic manifestations of mutations in genes encoding subunits of cardiac potassium channels. Circulation research. 2011;109(1):97–109. doi: 10.1161/CIRCRESAHA.110.224600. [DOI] [PubMed] [Google Scholar]
- 44.Bonnet D, Martin D, Pascale De L, Villain E, Jouvet P, Rabier D, et al. Arrhythmias and conduction defects as presenting symptoms of fatty acid oxidation disorders in children. Circulation. 1999;100(22):2248–53. doi: 10.1161/01.cir.100.22.2248. [DOI] [PubMed] [Google Scholar]
- 45.Saudubray JM, Martin D, de Lonlay P, Touati G, Poggi-Travert F, Bonnet D, et al. Recognition and management of fatty acid oxidation defects: a series of 107 patients. Journal of inherited metabolic disease. 1999;22(4):488–502. doi: 10.1023/a:1005556207210. [DOI] [PubMed] [Google Scholar]
- 46.Wilcken B. Fatty acid oxidation disorders: outcome and long-term prognosis. Journal of inherited metabolic disease. 2010;33(5):501–6. doi: 10.1007/s10545-009-9001-1. [DOI] [PubMed] [Google Scholar]
- 47.Yamada KA, Kanter EM, Newatia A. Long-chain acylcarnitine induces Ca2+ efflux from the sarcoplasmic reticulum. Journal of cardiovascular pharmacology. 2000;36(1):14–21. doi: 10.1097/00005344-200007000-00002. [DOI] [PubMed] [Google Scholar]
- 48.Xu Z, Rozanski GJ. K+ current inhibition by amphiphilic fatty acid metabolites in rat ventricular myocytes. The American journal of physiology. 1998;275(6 Pt 1):C1660–7. doi: 10.1152/ajpcell.1998.275.6.C1660. [DOI] [PubMed] [Google Scholar]
- 49.Wu J, Corr PB. Palmitoylcarnitine increases [Na+]i and initiates transient inward current in adult ventricular myocytes. The American journal of physiology. 1995;268(6 Pt 2):H2405–17. doi: 10.1152/ajpheart.1995.268.6.H2405. [DOI] [PubMed] [Google Scholar]
- 50.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiological reviews. 2004;84(3):767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 51.Jain RK. Molecular regulation of vessel maturation. Nature medicine. 2003;9(6):685–93. doi: 10.1038/nm0603-685. [DOI] [PubMed] [Google Scholar]
- 52.Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes & development. 1997;11(22):2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu J, Bohanan CS, Neumann JC, Lingrel JB. KLF2 transcription factor modulates blood vessel maturation through smooth muscle cell migration. The Journal of biological chemistry. 2008;283(7):3942–50. doi: 10.1074/jbc.M707882200. [DOI] [PubMed] [Google Scholar]
- 54.Adam PJ, Regan CP, Hautmann MB, Owens GK. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. The Journal of biological chemistry. 2000;275(48):37798–806. doi: 10.1074/jbc.M006323200. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. The Journal of biological chemistry. 2005;280(10):9719–27. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 56.Alaiti MA, Orasanu G, Tugal D, Lu Y, Jain MK. Kruppel-like factors and vascular inflammation: implications for atherosclerosis. Current atherosclerosis reports. 2012;14(5):438–49. doi: 10.1007/s11883-012-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pidkovka NA, Cherepanova OA, Yoshida T, Alexander MR, Deaton RA, Thomas JA, et al. Oxidized phospholipids induce phenotypic switching of vascular smooth muscle cells in vivo and in vitro. Circulation research. 2007;101(8):792–801. doi: 10.1161/CIRCRESAHA.107.152736. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida T, Kaestner KH, Owens GK. Conditional deletion of Kruppel-like factor 4 delays downregulation of smooth muscle cell differentiation markers but accelerates neointimal formation following vascular injury. Circulation research. 2008;102(12):1548–57. doi: 10.1161/CIRCRESAHA.108.176974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salmon M, Johnston WF, Woo A, Pope NH, Su G, Upchurch GR, Jr, et al. KLF4 regulates abdominal aortic aneurysm morphology and deletion attenuates aneurysm formation. Circulation. 2013;128(11 Suppl 1):S163–74. doi: 10.1161/CIRCULATIONAHA.112.000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoshino Y, Kurabayashi M, Kanda T, Hasegawa A, Sakamoto H, Okamoto E, et al. Regulated expression of the BTEB2 transcription factor in vascular smooth muscle cells: analysis of developmental and pathological expression profiles shows implications as a predictive factor for restenosis. Circulation. 2000;102(20):2528–34. doi: 10.1161/01.cir.102.20.2528. [DOI] [PubMed] [Google Scholar]
- 61.Lu Y, Haldar S, Croce K, Wang Y, Sakuma M, Morooka T, et al. Kruppel-like factor 15 regulates smooth muscle response to vascular injury--brief report. Arteriosclerosis, thrombosis, and vascular biology. 2010;30(8):1550–2. doi: 10.1161/ATVBAHA.110.207050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y, Zhang L, Liao X, Sangwung P, Prosdocimo DA, Zhou G, et al. Kruppel-like factor 15 is critical for vascular inflammation. The Journal of clinical investigation. 2013;123(10):4232–41. doi: 10.1172/JCI68552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruiz-Gomez M, Romani S, Hartmann C, Jackle H, Bate M. Specific muscle identities are regulated by Kruppel during Drosophila embryogenesis. Development. 1997;124(17):3407–14. doi: 10.1242/dev.124.17.3407. [DOI] [PubMed] [Google Scholar]
- 64.Haldar SM. Experimental Medicine. Tokyo, Japan: Yodosha; 2013. Emerging Role of Kruppel-like Factors (KLFs) in Skeletal Muscle Biology; pp. 1–8. [Google Scholar]
- 65.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Seminars in cell & developmental biology. 2005;16(4–5):585–95. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 66.Himeda CL, Ranish JA, Pearson RC, Crossley M, Hauschka SD. KLF3 regulates muscle-specific gene expression and synergizes with serum response factor on KLF binding sites. Molecular and cellular biology. 2010;30(14):3430–43. doi: 10.1128/MCB.00302-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dionyssiou MG, Salma J, Bevzyuk M, Wales S, Zakharyan L, McDermott JC. Kruppel-like factor 6 (KLF6) promotes cell proliferation in skeletal myoblasts in response to TGFbeta/Smad3 signaling. Skeletal muscle. 2013;3(1):7. doi: 10.1186/2044-5040-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sunadome K, Yamamoto T, Ebisuya M, Kondoh K, Sehara-Fujisawa A, Nishida E. ERK5 regulates muscle cell fusion through Klf transcription factors. Developmental cell. 2011;20(2):192–205. doi: 10.1016/j.devcel.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 69.Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell metabolism. 2013;17(2):162–84. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 70.Zizola C, Schulze PC. Metabolic and structural impairment of skeletal muscle in heart failure. Heart failure reviews. 2013;18(5):623–30. doi: 10.1007/s10741-012-9353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rowe GC, Safdar A, Arany Z. Running forward: new frontiers in endurance exercise biology. Circulation. 2014;129(7):798–810. doi: 10.1161/CIRCULATIONAHA.113.001590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oishi Y, Manabe I, Tobe K, Ohsugi M, Kubota T, Fujiu K, et al. SUMOylation of Kruppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-delta. Nature medicine. 2008;14(6):656–66. doi: 10.1038/nm1756. [DOI] [PubMed] [Google Scholar]
- 73.Gray S, Wang B, Orihuela Y, Hong EG, Fisch S, Haldar S, et al. Regulation of gluconeogenesis by Kruppel-like factor 15. Cell metabolism. 2007;5(4):305–12. doi: 10.1016/j.cmet.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jeyaraj D, Scheer FA, Ripperger JA, Haldar SM, Lu Y, Prosdocimo DA, et al. Klf15 orchestrates circadian nitrogen homeostasis. Cell metabolism. 2012;15(3):311–23. doi: 10.1016/j.cmet.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haldar SM, Jeyaraj D, Anand P, Zhu H, Lu Y, Prosdocimo DA, et al. Kruppel-like factor 15 regulates skeletal muscle lipid flux and exercise adaptation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(17):6739–44. doi: 10.1073/pnas.1121060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Masuno K, Haldar SM, Jeyaraj D, Mailloux CM, Huang X, Panettieri RA, Jr, et al. Expression profiling identifies Klf15 as a glucocorticoid target that regulates airway hyperresponsiveness. American journal of respiratory cell and molecular biology. 2011;45(3):642–9. doi: 10.1165/rcmb.2010-0369OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sasse SK, Mailloux CM, Barczak AJ, Wang Q, Altonsy MO, Jain MK, et al. The Glucocorticoid Receptor and KLF15 Regulate Gene Expression Dynamics and Integrate Signals through Feed-Forward Circuitry. Molecular and cellular biology. 2013;33(11):2104–15. doi: 10.1128/MCB.01474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shimizu N, Yoshikawa N, Ito N, Maruyama T, Suzuki Y, Takeda S, et al. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell metabolism. 2011;13(2):170–82. doi: 10.1016/j.cmet.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 79.Bialkowska AB, Crisp M, Bannister T, He Y, Chowdhury S, Schurer S, et al. Identification of small-molecule inhibitors of the colorectal cancer oncogene Kruppel-like factor 5 expression by ultrahigh-throughput screening. Molecular cancer therapeutics. 2011;10(11):2043–51. doi: 10.1158/1535-7163.MCT-11-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oishi Y, Manabe I, Imai Y, Hara K, Horikoshi M, Fujiu K, et al. Regulatory polymorphism in transcription factor KLF5 at the MEF2 element alters the response to angiotensin II and is associated with human hypertension. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24(6):1780–8. doi: 10.1096/fj.09-146589. [DOI] [PubMed] [Google Scholar]