Abstract
Bile acids are important physiological agents for intestinal nutrient absorption and biliary secretion of lipids, toxic metabolites, and xenobiotics. Bile acids also are signaling molecules and metabolic regulators that activate nuclear receptors and G protein-coupled receptor (GPCR) signaling to regulate hepatic lipid, glucose, and energy homeostasis and maintain metabolic homeostasis. Conversion of cholesterol to bile acids is critical for maintaining cholesterol homeostasis and preventing accumulation of cholesterol, triglycerides, and toxic metabolites, and injury in the liver and other organs. Enterohepatic circulation of bile acids from the liver to intestine and back to the liver plays a central role in nutrient absorption and distribution, and metabolic regulation and homeostasis. This physiological process is regulated by a complex membrane transport system in the liver and intestine regulated by nuclear receptors. Toxic bile acids may cause inflammation, apoptosis, and cell death. On the other hand, bile acid-activated nuclear and GPCR signaling protects against inflammation in liver, intestine, and macrophages. Disorders in bile acid metabolism cause cholestatic liver diseases, dyslipidemia, fatty liver diseases, cardiovascular diseases, and diabetes. Bile acids, bile acid derivatives, and bile acid sequestrants are therapeutic agents for treating chronic liver diseases, obesity, and diabetes in humans.
Introduction
Bile acids are the end products of cholesterol catabolism (32,34,159). Conversion of cholesterol to bile acids accounts for the daily turnover of a major fraction of cholesterol in humans. Bile acid synthesis generates bile flow and biliary secretion of bile acids, phospholipids, cholesterol, drugs, and toxic metabolites. Cholic acid (CA) and chenodeoxycholic acid (CDCA) are the major primary bile acids synthesized in human livers, and are conjugated with taurine or glycine for secretion into bile. Bile salts form mixed micelles with phospholipids and cholesterol and stored in the gallbladder, secreted into the intestinal tract to facilitate digestion and absorption of nutrients. Most bile acids are reabsorbed in the ileum and are transported back to the liver via portal blood circulation to inhibit bile acid synthesis. Enterohepatic circulation of bile acids is highly efficient in humans and is an important physiological system not only for nutrient absorption and xenobiotic disposal, but also for maintaining metabolic homeostasis. The mechanism of bile acid feedback inhibition of its own synthesis has been studied for more than 50 years, but the underlying molecular mechanism is still not clear. The recent discovery that bile acids are endogenous ligands of a nuclear receptor farnesoid X receptor (FXR) has provided some mechanistic insight into the role of bile acids in the regulation of gene transcription (28, 31, 65), but the physiological relevance of the FXR-dependent pathways in regulation of bile acid metabolism remains elusive. Many recent studies have provided strong evidence that bile acid-activate FXR plays a critical role in maintaining metabolic homeostasis (5, 33, 86, 87, 106, 189). Bile acid-activated membrane G protein-coupled receptors, TGR5 (aka Gpbar-1, G-protein-coupled bile acid receptor) appear to play a role in stimulating energy metabolism, protecting liver and intestine from inflammation and steatosis, and improving insulin sensitivity (189). Another recently identified bile acid-activated GPCR, sphingosine-1-phosphate receptor 2 (S1P2) may also play a role in lipid metabolism (183). The following sections will cover bile acid synthesis and metabolism, its regulation by nuclear receptor, the recently uncovered role of bile acids in integrated regulation of lipid, glucose, and energy metabolism. Diseases in bile acid synthesis and transport, cholestasis, and therapeutic potential of bile acids and derivatives for treating metabolic diseases are briefly reviewed. See Table 1 for a list of abbreviations.
Table 1.
Abbreviations list
| ABC | ATP binding cassette |
| ASBP | Apical sodium-dependent bile salt transporter |
| AKR | Aldo-ketose reductase |
| AMACR | α-methyl-acyl-CoA racemase |
| BACS | Bile acid-coenzyme A synthase |
| BSEP | Bile salt export pump |
| CA | Cholic acid |
| CAR | Constitutive androstane receptor A |
| CDCA | Chenodeoxycholic acid |
| CYP7A1 | Cholesterol 7α-hydroxylase |
| CYP8B1 | Sterol 12α-hydroxylase |
| CYP27A1 | Sterol 27-hydroxylase |
| CTX | Cerebrotendinous xanthomatosis |
| DCA | Deoxycholic acid |
| EGFR | Epidermal growth factor receptor |
| ERK | Extracellular signal-regulated kinase |
| FGF | Fibroblast growth factor |
| FGFR4 | FGF receptor |
| FXR | Farnesoid X receptor |
| GPCR | G protein-coupled receptor |
| G6Pase | Glucose-6-phosphatase |
| HNF4α | Hepatic nuclear factor 4α |
| HSD | Hydroxysteroid dehydrogenase |
| ICP | Intrahepatic cholestasis of pregnancy |
| Insig | Insulin induced gene |
| JNK | Jun-N-terminal kinase |
| LCA | Lithocholic acid |
| LRH-1 | Liver related homologue |
| LXR | Liver orphan receptor |
| MAPK | Mitogen-activated protein kinase |
| MCA | Muricholic acid |
| MDR | Multidrug resistant protein |
| MRP | Multidrug resistance related protein |
| NAFLD | Nonalcoholic fatty liver disease |
| NASH | Nonalcoholic steatohepatitis |
| NTCP | Na+-taurocholate cotransport peptide |
| OSTα/β | Organic solute transporter α/β |
| PBC | Primary biliary cirrhosis |
| PEPCK | Phosphoenolpyruvate carboxykinase |
| PFIC | Progressive familial intrahepatic cholestasis |
| PGC-1α | Peroxisome proliferator activated receptor γ co-activator 1α |
| PSC | Primary sclerosing cholangitis |
| PXR | Pregnane X receptor |
| RXR | Retinoid X receptor |
| SCAP | SREBP cleavage-activating protein |
| SHP | Small heterodimer partner |
| S1P | Sphingosine 1-phosphate |
| S1P2 | S1P receptor 2 |
| SREBP | Steroid response element binding protein |
| SULT | Sulfotransferas |
| UGT | UDP-glucuronosyl N-transferase |
| UDCA | Ursodeoxycholic acid |
| VDR | Vitamin D receptor |
Bile Acid Metabolism
Bile acids (or bile salts) are derived from cholesterol. In mammals, all bile acids are C24-5β-bile acids (cholanoic acid). The steroid nucleus has four fused carbon rings consisting of three 6-carbon rings and one 5-carbon ring. Conversion of cholesterol to bile acids involves hydroxylation, saturation of the double bond at C5–C6, epimerization of the 3-hydroxyl group, and oxidative cleavage of a 3-carbon unit from the side chain. The 3-hydroxyl group in all bile acids has a α-configuration. All bile acids have a 5β-hydrogen group and a cis-configuration along the plane of the fused A and B ring. In CA all three hydroxyl groups and the carboxyl group are faced to one side of the carbon skeleton forming a hydrophilic face opposing the highly hydrophobic face. Thus, bile acids are amphipathic molecules with powerful detergent properties. Most bile acids are conjugated to the amino acids glycine or taurine and form sodium salts in physiological pH to increase their solubility.
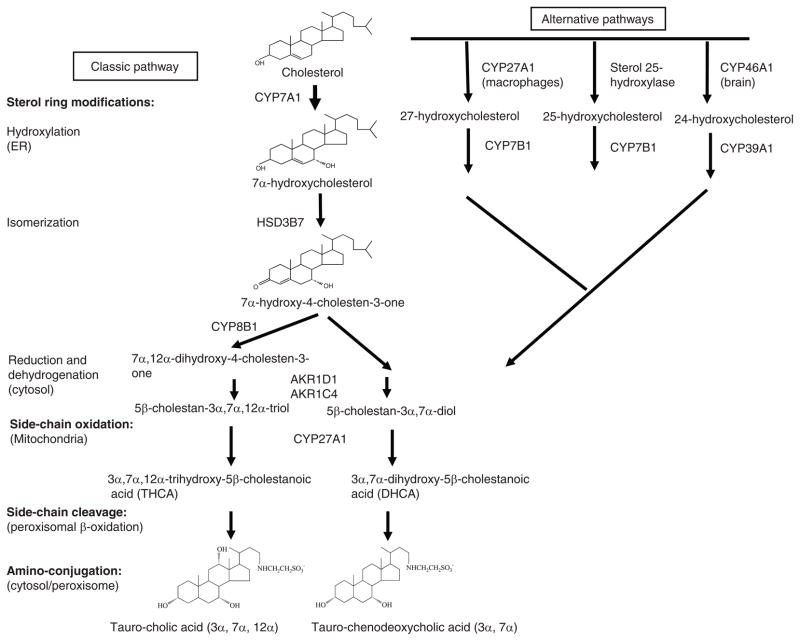
Human liver synthesizes about 200 to 600 mg bile acids per day and excretes the same amount in feces. The net daily turnover of bile acids is about 5% of a total bile acid pool of about 3 g. Conversion of cholesterol to bile acids involves 17 distinct enzymes located in the cytosol, endoplasmic reticulum, mitochondria, and peroxisomes (Fig. 1). These enzymes catalyze modifications of the steroid ring and oxidative cleavage of three carbons from the side chain of cholesterol to form C24 bile acids. There are two major bile acid biosynthetic pathways (34). In the neutral bile acid pathway (or classic pathway), steroid ring modification precedes side-chain cleavage, whereas in the acidic pathway side-chain cleavage preceded steroid ring modifications. These are five hydroxylases involved in bile acid synthesis. The classic pathway is initiated by cholesterol 7α-hydroxylase (CYP7A1), the only rate-limiting enzyme in bile acid synthesis, and synthesizes two primary bile acids, CA and CDCA in human liver. A microsomal sterol 12α-hydroxylase (CYP8B1) is required for synthesis of CA, without 12α-hydroxylase the product is CDCA. The acidic pathway (or alternative pathway) is initiated by sterol 27-hydroxylase (CYP27A1), a mitochondria cytochrome P450 enzyme, which is widely distributed in most tissues and macrophages. It has been reported that the acidic pathway may contribute about 9% of total bile acid synthesis in human hepatocytes (52). The acidic pathway may be quantitatively important in bile acid synthesis in patients with liver diseases and in the neonates.
Figure 1.
Bile acid biosynthetic pathways. Two major bile acid biosynthetic pathways are shown. The neutral (or classic) pathway is initiated by cholesterol 7α-hydroxylase (CYP7A1) located in the endoplasmic reticulum of the liver, whereas the acidic (or alternative) pathway is initiated by mitochondrial sterol 27-hydroxylase (CYP27A1). There are three sterol hydroxylases that convert cholesterol to oxysterols: CYP27A1 in macrophages and other tissues, microsomal sterol 25-hydroxylase in liver microsomes, and sterol 24-hydroxylase (CYP46A1) in the brain. Oxysterol 7α-hydroxylase (CYP7B1) is nonspecific and catalyzes hydroxylation of 27- and 25-hydroxycholesterol to 3β, 7α-dihydroxy-5-cholestenoic acid and 5-cholesten-3β, 7α, 25-triol, respectively. A brain-specific oxysterol 7α-hydroxylase (CYP39A1) catalyzes hydroxylation of 24-hydroxycholesterol to 5-cholesten-3β, 7α, 24(S)-triol. These oxysterols could be converted to CDCA if transported to the liver. In the liver, 3β-hydoxysteroid dehydrogenase (3βHSD, HSD3B7) convert 7α-hydroxycholesterol to 7α-hydroxy-4-cholesten-3-one (C4), which is converted to 7α, 12α-dihydroxy-4-cholesten-3-one by a sterol 12α-hydroxylase (CYP8B1), leading to synthesis of cholic acid (CA). Without 12α-hydroxylation, the pathway produces CDCA. Aldos-keto reductase 1D1 (AKR1D1) and AKR1C1 catalyze isomerization and saturation of the steroid ring. Then CYP27A1 catalyzes steroid side-chain oxidation to form cholestanoic acids, THCA, and DHCA. Bile acid-Co-A synthase (BACS) or very long-chain Co-A synthase (VLCS) in the endoplasmic reticulum (ER) ligates Co-A to the carboxyl groups. Bile acid thioesters are transported into peroxisomes, where an α-methylacyl-CoA racemase (AMACR) converts the methyl group from 25(R) to 25(S) conformation, and three peroxisomal β-oxidation enzymes, branched-chain acyl-CoA oxidase, D-bifunctional enzyme, and thiolase (or sterol carrier protein x) catalyze oxidative cleavage of a propionyl group from the steroid side-chain to form cholyl-CoA and chenodeoxycholyl-CoA. Cytosolic or peroxisomal bile acid: amino-acid transferase (BAAT) catalyzes conjugation of amino acids, glycine or taurine to the carboxyl group of cholyl-CoA and chenodeoxycholyl-CoA to form tauro- or glycol-conjugated CA or CDCA. ER: endoplasmic reticulum.
In humans, most bile acids are amino conjugated at the carboxyl group (amidation) with the ratio of glycine to taurine conjugates of about 3 to 1. In mice, most bile acids (>95%) are taurine conjugated. Conjugation of bile acids increases ionization and solubility at physiological pH, prevents Ca2+ precipitation, minimizes passive absorption, and resistant to cleavage by pancreatic carboxypeptidases. In the distal intestine, conjugated-CA and CDCA are first deconjugated, and then bacterial 7α-dehydroxylase converts CA and CDCA to DCA and lithocholic acid (LCA), respectively. DCA and LCA are the secondary bile acids (damaged bile acids). Most LCA is excreted into feces and small amounts of LCA is circulated to the liver and rapidly conjugated by sulfation and excreted into bile. Sulfation is the major pathway for detoxification of hydrophobic bile acids in humans (82). In mice, CDCA is hydroxylated to α-muricholic acid and 6β-muricholic acids, two major primary bile acids in mice. The 7α-hydroxyl groups in CDCA also can be epimerized to 7β to from ursodeoxycholic acid (UDCA). Hydroxylation at the 6α/β or 7β-position increases bile acid solubility and reduces bile acid toxicity.
Enterohepatic Circulation of Bile Acids
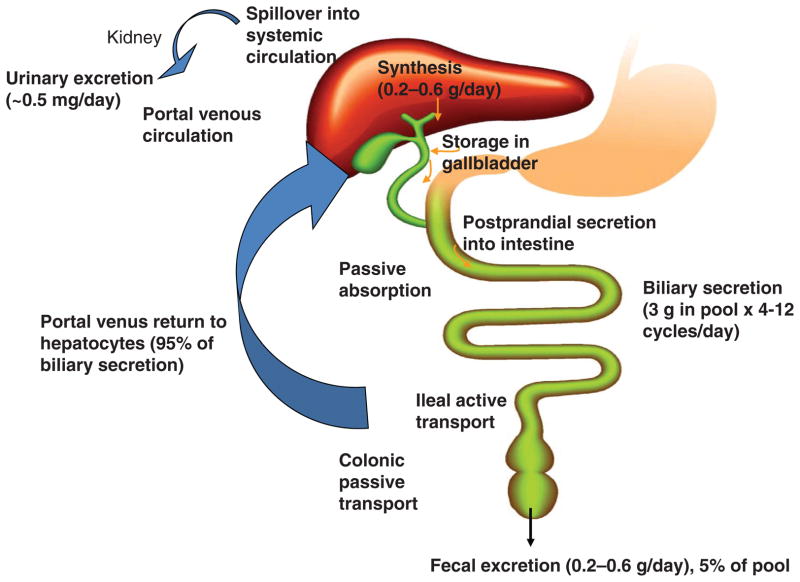
Bile acids synthesized in the liver are immediately secreted into bile, reabsorbed in the intestine and transported back to the liver. The enterohepatic circulation of bile acids (Fig. 2) is very efficient in humans. Small amounts of bile acids may spill over into the systemic circulation, reabsorbed when passing through the renal tubules in the kidney, and are then circulated back to the liver through systemic circulation. Some bile acids secreted in the bile duct are reabsorbed in the cholangiocytes (bile duct epithelial cells) and recycled back to hepatocytes (the cholangiohepatic shunt). Bile acids are stored in the gallbladder. After each meal, cholecystokinin secreted from the intestine stimulates gallbladder contraction to empty bile acids into the intestinal tract. When passing down the intestinal tract, small amounts of unconjugated bile acids are reabsorbed in the upper intestine by passive diffusion. Most bile acids (95%) are reabsorbed in the brush border membrane of the terminal ileum, transdiffused across the enterocyte to the basolateral membrane, and secreted into portal blood circulation to liver sinusoids and are taken up into hepatocytes. DCA is reabsorbed in the colon and recycled with CA and CDCA to the liver. A bile acid pool of ~3 g consisting of ~40% CA, 40% CDCA, 20% DCA, and trace amount of LCA, is recycled 4 to 12 times a day. Bile acids lost in the feces (~0.5 g/day) are replenished by de novo synthesis in the liver to maintain a constant bile acid pool.
Figure 2.
Enterohepatic circulation of bile acids. An average man produces ~0.5g bile acid per day by synthesis in the liver, and secretes ~0.5g/day. This daily turnover of bile acids accounts for about 5% of total bile acid pool. The remaining 95% of bile acids in the pool are recycled 4 to 12 times a day. Most bile acids are reabsorbed in the ileum by active transport, while a small amount is reabsorbed by passive diffusion in the upper intestine to portal blood for circulation to the liver. Small amounts of bile acids spilled over into the systemic circulation are recovered in kidney.
Regulation of Bile Acid Synthesis
Bile acid feedback regulation
Studies of bile acid metabolism in the 1960s showed that bile acids, cholesterol, thyroid hormone, glucocorticoid, insulin, and circadian rhythms regulated CYP7A1 activity and the rate of bile acid synthesis [reviewed in refs. (34,136)]. Interruption of enterohepatic circulation of bile acids by bile acid binding resins such as cholestyramine or biliary diversion by bile fistula strongly stimulates CYP7A1 enzyme activity and bile acid synthesis in rats. Feeding rats with bile acids strongly reduced CYP7A1 enzyme activity and bile acid synthesis. These results imply that bile acid synthesis is regulated by a negative feedback mechanism, and bile acids returning to the liver via enterohepatic circulation may directly or indirectly inhibit bile acid synthesis by inhibiting CYP7A1 activity. Many studies show that bile acids inhibit whereas cholesterol stimulates CYP7A1 mRNA, protein and enzyme activity and bile acid synthesis in rodents. It was concluded that CYP7A1 activity is mainly regulated by transcriptional mechanism.
It has been noted that multiple CYP7A1 transcripts exist in rat hepatocytes and the 3′-untranslated regions (3′-UTRs) of CYP7A1 mRNAs are unusually long (118). The half-life of CYP7A1 mRNA has been estimated to be very short, about 30 min (8, 139). It is possible that bile acids may reduce CYP7A1 mRNA stability via the bile acid response elements located in the 3′-UTR (2,8). However, posttranscriptional regulation of CYP7A1 has not been studied in detail. One recent study shows that apoB editing complex 1 (Apobec-1) regulates the stability of CYP7A1 by binding to the conserved AU-rich sequences in the 3′-UTR (213). The Apobec-1 locus (Lith6) encodes the RNA-specific cytidine deaminase involved in production of Apob48 in small intestine and rodent liver. In Apobec-1−/− mice, expression of Cyp7a1 mRNA and protein are significantly reduced and these mice are susceptible to lithogenic diet-induced gallstone disease. Apobec-1 mediates posttranscriptional regulation of mouse Cyp7a1 expression by stabilizing Cyp7a1 mRNAs.
Nutrient regulation of bile acid synthesis
Since liver metabolism is highly active during the postprandial period and humans undergo fasting-to-refeeding cycles several times a day, there is a physiological link between induction of bile acid synthesis and regulation of postprandial nutrient metabolism. Nutrients may play a key role in regulating bile acid synthesis, which in turn regulates nutrient absorption and metabolic homeostasis.
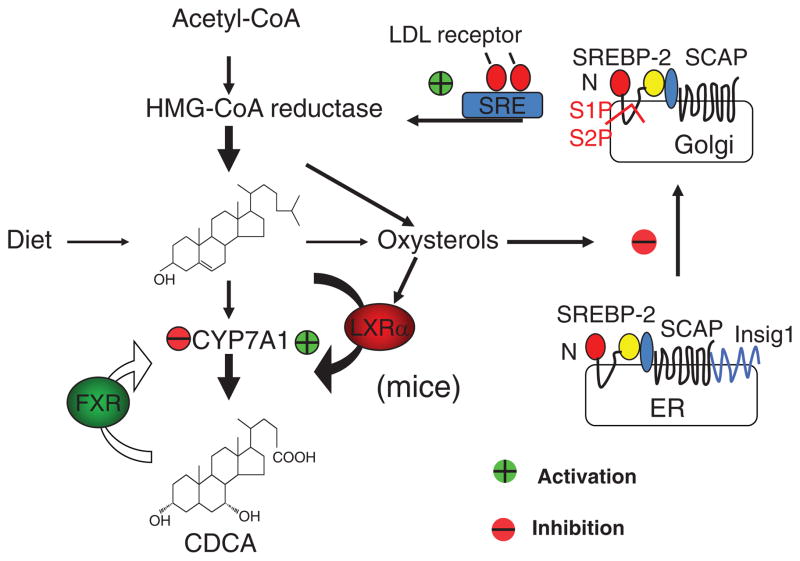
CYP7a1 is a highly specific hydroxylase that only uses cholesterol as the substrate and insert a hydroxyl group at the 7α-position. This enzyme is located in the cholesterol-poor endoplasmic reticulum. Thus the availability of cholesterol as substrate (Km effect) regulates the specific activity of CYP7A1. It has been suggested that newly synthesized cholesterol is the preferred substrate. Therefore, there is a direct link of de novo cholesterol synthesis to bile acid biosynthesis in hepatocytes. Stimulation of bile acid synthesis reduces hepatic cholesterol/oxysterol levels and results in stimulating de novo cholesterol synthesis to provide substrate for CYP7A1 (Fig. 3). When intracellular oxysterol levels decrease, a basic helix-loop-helix-leucine zipper protein called steroid response element binding protein-2 (SREBP-2) precursor (125 kDa) is dissociated from insulin induced gene-1 (Insig-1) and Insig-2 in the endoplasmic reticulum membrane and is escorted by a steroid sensitive SREBP cleavage-activating protein to the Golgi apparatus, where two oxysterol-sensitive proteases S1P and S2P cleave and release the N-terminal 68 kDa transcription factor, which enters the nucleus, binds to the SRE on the promoters of the genes encoding all enzymes in cholesterol synthesis (HMG-CoA reductase, as an example) and the LDL receptor (20, 54, 56, 85, 214) (Fig. 3). When intracellular oxysterol levels are high, SREBP-2 is retained in the endoplasmic reticulum and cholesterol synthesis is inhibited. In mice, feeding a high cholesterol diet stimulates bile acid synthesis by activating an oxysterol-activated nuclear receptor LXRα, which induces Cyp7a1 gene transcription. SREBP-1a and SREBP-1c are transcribed by the SREBP-1 gene and induce all genes in lipogenesis (64). LXRα induces the SREBP-1c gene, but not SREBP-2 gene transcription (47). Insulin and LXRα induce all genes in lipogenesis by inducing SREBP-1c gene transcription (30, 168, 169).
Figure 3.
Bile acid synthesis regulates cholesterol homeostasis in hepatocytes. Cholesterol homeostasis is maintained by dietary uptake of cholesterol, de novo cholesterol synthesis from acetyl-CoA, and conversion of cholesterol to bile acids. Oxysterols are derived from cholesterol and bile acids. When intracellular cholesterol/oxysterol levels are high, steroid response element binding protein 2 (SREBP-2) precursor (125 kDa) interacts with insulin induced gene 1/2 (Insig1/2) and is retained in endoplasmic reticulum (ER) membrane. When intracellular oxysterol levels are low, SREBP cleavage and activating protein (SCAP) escorts SREBP-2 precursor to the Golgi apparatus, where sterol sensitive proteases S1P and S2P are activated to cleave a N-terminal fragment (65 kDa), which is translocated to the nucleus to bind to the steroid response elements in the gene promoters of all cholesterogenic genes and stimulates de novo cholesterol synthesis. Oxysterols activate LXRα, which induces CYP7A1 gene transcription to stimulate bile acid synthesis in mice, but not humans. Bile acids (CDCA) activate farnesoid X receptor (FXR) to inhibit CYP7A1 gene transcription and bile acid synthesis. This may lead to increased cholesterol levels and inhibited de novo cholesterol synthesis and absorption of dietary cholesterol.
Early studies in rats showed that the bile acid pool size increased in diabetic rats and insulin treatment reduced bile acid pool size, inhibited CYP7A1 and CYP8B1 activities, and altered bile acid composition (184). In streptozocin-treated diabetic rats CYP7A1 activity is increased suggesting that insulin represses CYP7A1 while lack of insulin induces CYP7A1 (184). It has been reported that glucagon/cAMP and fasting induce Cyp7a1 expression, which parallels the induction of Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) and phosphoenolpyruvate carboxykinase (PEPCK) in gluconeogenesis (46, 170), suggesting that Cyp7a1 expression and bile acid synthesis are up regulated during fasting as a feed forward signal for intestinal nutrient absorption. In contrast, in human patients serum 7α-hydroxy-4-cholesten-3-one (C4)-levels, reflecting the rate of bile acid synthesis, are reduced during the fasting and increased during the postprandial state (123), suggesting that bile acid synthesis is induced during the postprandial period and is inhibited during fasting. It was thought that species differences might explain the differential regulation of bile acid synthesis in the fasting-to-refeeding cycle. More recent studies show that insulin and glucose induce, while glucagon represses CYP7A1 gene expression in primary human hepatocytes (109,115,174). In vivo studies also show that glucose and insulin rapidly induce CYP7A1 gene expression and bile acid synthesis leading to an enlarged bile acid pool and elevated circulating bile acids (112). Insulin may have dual functions, stimulating CYP7A1 at physiological concentrations but inhibiting at high concentrations found in an insulin resistant state (109). Insulin signaling activates AKT/protein kinase B (PKB), and possibly also the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase 1/2 (ERK1/2) pathway to inhibit CYP7A1 gene transcription. Insulin is known to induce SREBP-1c, which may inhibit CYP7A1 expression by interacting with hepatocyte nuclear factor 4α (HNF4α). Glucose induces CYP7A1 via an epigenetic mechanism by increasing histone acetylation status of the CYP7A1 gene promoter (112). On the other hand, glucagon and cAMP strongly inhibit CYP7A1 expression via activation of PKA, which phosphorylates HNF4α to abolish its DNA-binding activity, and result in inhibition of CYP7A1 expression in human hepatocytes (174). These recent studies clearly established that nutrients play a key role in regulation of bile acid synthesis during fasting to refeeding cycles.
Conjugated bile acids activate both AKT and the MAPK/ERK1/2 pathway. Taurocholic acid activation of tyrosine phosphorylation of epidermal growth factor receptors (EGFRs) is sensitive to pertussis toxin and Gαi in rodent hepatocytes (50, 87). Unconjugated bile acids (DCA) can activate ERK1/2 and AKT pathways by stimulating the synthesis of mitochondria reactive oxidizing species, which activates EGFRs (57). AKT phosphorylates FoxO1 and inhibits PEPCK and glucose-6-phosphatase (G-6-Pase) in gluconeogenesis. AKT phosphorylates and inhibits glycogen synthase kinase 3β (GSK3β) activity to activate glycogen synthesis in rat primary hepatocytes (58). This implies that bile acids may mimic the insulin action in regulating glucose metabolism by stimulating glycogen synthesis and inhibiting gluconeogenesis. Hydrophobic bile acids are known to induce apoptosis in hepatocytes, and hydrophilic bile acids increase intracellular cAMP and activate MAPK and PI3K pathways to protect hepatocytes from apoptosis (5, 153, 157).
In diabetic patients, bile acid pool and fecal bile acids are elevated, and are decreased upon insulin treatment (12). In both mouse models of type I and type II diabetes, bile acid pool sizes increase, but the fasting-to-refeeding regulation of the cyp7a1 gene is lost (112). In diabetic mice, the CYP7A1 gene promoter is hyperacetylated, thus, these mice have higher basal CYP7A1 activity, rate of bile acid synthesis, and larger bile acid pool. The implication of bile acids in obesity and diabetes is further supported by a recent clinical study demonstrated that serum bile acids were higher in patients with prior gastric bypass than in overweight and severely obese patients without gastric bypass, and serum bile acids were positively correlated with serum glucagon-like peptide-1 (GLP-1) (141). It is likely that in gastric bypass patients, bile acid synthesis may be increased due to reduced bile acid feedback, resulting in increased bile acid synthesis and improved glucose tolerance in obese patients. This is consistent with a recent study that lowering circulating bile acids worsened diet-induced obesity and diabetes, while increasing bile acid pool size improved glucose homeostasis (206). Interestingly, a recent metabolomics study identified bile acids as the most elevated metabolites in human sera after an oral glucose challenge in patients with normal glucose tolerance, but this response was blunted in patients with impaired glucose tolerance (166). It should be clarified that increased bile acid pool is not the cause of diabetes but the consequence of dysregulation of bile acid metabolism and altered metabolic homeostasis. Serum bile acid levels have become biomarkers for diagnosis of liver diseases, diabetes, and obesity.
Bile Acid-Activated Nuclear Receptors
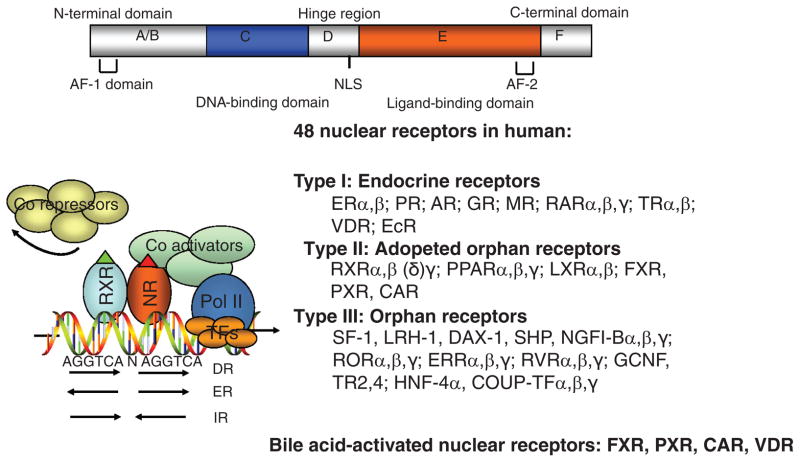
Nuclear receptors are ligand-activated transcription factors that play important roles in embryogenesis, development, and metabolism (31). Figure 4 shows a general structure of nuclear receptors. The N-terminal A and B domains contain the activation function-1 domain, which is the most variable region in nuclear receptors. The C domain is a highly conserved region containing two Zn2+ fingers responsible for binding to a hormone response element (HRE) with two tandem AGGTCA repeating sequences spaced by 1–5 nucleotides arranged in direct repeat, everted repeat, or inverted repeat. Most nuclear receptors bind to a HRE as homodimers, or heterodimers with retinoid X receptor. In general, in the absence of a ligand, nuclear receptors bind corepressors and are inactive. Upon ligand binding, coactivators are recruited to displace corepressors. Nuclear receptor/coactivators interact with other mediators to stimulate RNA polymerase II and induce gene transcription. The D or hinge domain has a nuclear localization sequence and is also involved in DNA binding. The E domain is the ligand-binding domain (LBD) conserved in nuclear receptors within the same subfamily. The LBD contains coactivator interaction motifs LXXLs and also is important in transactivation (AF-2). There are 48 nuclear receptor genes in the human genome. Nuclear receptors are classified as type 1 endocrine receptors, type II adopted receptors, and type III orphan receptors (the ligands of which have not been firmly established).
Figure 4.
Nuclear receptors. The general structure of nuclear receptors is shown on the top. The NR1 family of genes involved in metabolic regulation, and their respective endogenous ligands are shown. The putative nuclear receptor response element binding sequence, arranged in direct repeat (DR), everted repeat (ER), and inverted repeat (IR), is shown. Ligand-activated receptors recruit coactivators to replace corepressors and results in transactivation of target gene expression. AF-1-2, activation function-1 and -2; CAR, constitutive androstane receptor; FXR, farnesoid X receptor; LXR, liver orphan receptor; PPAR, peroxisome proliferator-activated receptor; PXR, pregnane X receptor; NLS, nuclear localization sequence.
Bile acid-activated nuclear receptors
Bile acids directly activate three nuclear receptors, FXR (127, 140, 199), pregnane X receptor (PXR) (212), and vitamin D receptor (VDR) (126). FXR is activated by free and conjugated-bile acids; the hydrophobic bile acid CDCA is the most efficacious bile acid ligand of FXR (EC50 = ~10 μmol/L), followed by LCA, DCA, and CA, while hydrophilic bile acids UDCA and MCA do not activate FXR. LCA and its metabolite 3-keto-LCA are the most efficacious bile acid ligands for both VDR and PXR (EC50 = ~100 nmol/L). PXR is highly expressed in the liver and intestine, and plays more important roles in detoxification of bile acids, drugs, and toxic compounds by activating phase I drug metabolizing P450 enzymes, phase II drug conjugation enzymes, and phase III drug transporters (178,194,212). PXR is known to inhibit CYP7A1 gene transcription by inhibiting HNF4α and PGC-1α transactivation of the CYP7A1 gene (15, 110). Interestingly, PXR null mice are more sensitive to lithogenic diet-induced gallstone disease and have reduced bile acid pool (81). This is because the lithogenic diet (containing 1% cholesterol and 0.5% CA) reduces Cyp7a1 expression through activation of FXR and induction of fibroblast growth factor 15 (FGF15). VDR also inhibits CYP7A1 gene transcription by interacting with HNF4α, competing for coactivators and recruiting corepressors to inhibit CYP7A1 gene transcription (78). FXR, PXR, and VDR may coordinately regulate bile acids, lipoproteins, drugs, glucose, and energy metabolism (65, 189). Bile acids also have been shown to activate several cell signaling pathways involved in regulation of bile acid metabolism (87). Thus, bile acid activation of nuclear receptors and cell signaling pathways converge to regulate a complex cellular metabolism network [see recent reviews in Refs. (38, 86, 106, 189)].
Roles of FXR in regulation of bile acid synthesis and transport
Many studies have implicated FXR in the regulation of bile acid synthesis, biliary bile acid secretion, intestinal bile acid absorption, and hepatic uptake of bile acids (24, 55, 106, 133, 172, 221). FXR activates target gene transcription mainly by binding to an inverted repeat with one-base spacing (IR1) in the gene promoter. Fxr knockout mice have increased bile acid synthesis and Cyp7a1 expression suggesting FXR-mediated bile acid inhibition of Cyp7a1 (172). The negative effect of FXR may be through an indirect mechanism by induction of a negative nuclear receptor small heterodimer partner (SHP) that inhibits transactivators of CYP7A1 and CYP8B1 by HNF4α and liver-related homolog-1 (LRH-1). FXR also regulates bile acid conjugation by inducing BACS and BAAT gene transcription. FXR induces a bile salt export pump (BSEP, a ATP binding cassette transporter, ABCB11) located in the canalicular membrane of hepatocytes (Fig. 5). BSEP is the principal bile acid efflux pump, which utilizes ATP hydrolysis to secrete conjugated-bile acids against a biliary bile acid concentration about 1000-fold higher than in hepatocytes. FXR induces a multidrug resistant protein 2/3 (MDR2/3, ABCB4), which effluxes phosphatidylcholine (Fig. 5). FXR also induces multidrug resistance-related protein 2 (MRP2, ABCC2), which effluxes glucuronidated and sulfated bile acids, organic anions and drugs (224). ABC-half transporters, ABCG5 and ABCG8 heterodimer are responsible for biliary excretion of cholesterol. FXR induces ABCG5 expression. In the gallbladder, bile acids form the mixed micelles with phosphatidylcholine and cholesterol. This allows for storage of bile acids in high concentrations (mmol/L) and prevents cholesterol from precipitating in the gallbladder. In the terminal ileum, conjugated bile acids are reabsorbed by apical sodium-dependent bile salt transporter (ASBT) located in the brush border membrane of the enterocytes (Fig. 5). Once inside the enterocytes, bile acids bind to the ileum bile acid binding protein, which is highly induced by FXR (193). Bile acids are excreted into portal circulation by the organic solute transporter α and β dimer (OSTα/β) located in the basolateral membrane of enterocytes (9,44). OSTα/β appears to be the major bile acid efflux transporter in the intestine (10). OSTα/β also acts as the secondary bile acid efflux transporter in sinusoidal membrane. FXR induces OSTα/β gene transcription (66). Bile acids are circulated via portal blood to hepatocytes, where a sinusoidal Na+-dependent taurocholate cotransport peptide (NTCP) uptakes bile acids into hepatocytes. FXR inhibits NTCP gene transcription (49). Thus, FXR plays a critical role in enterohepatic circulation of bile acids by regulating bile acid synthesis, biliary bile acid secretion, intestinal bile acid reabsorption and secretion, and bile acid uptake into hepatocytes. Defective regulation of these FXR target genes impairs enterohepatic circulation of bile acids, and contributes to cholestatic liver diseases (93, 103, 192, 223). FXR, PXR and constitutive androstane receptor (CAR) may play a complementary role in detoxification of bile acids and protection against cholestasis (76).
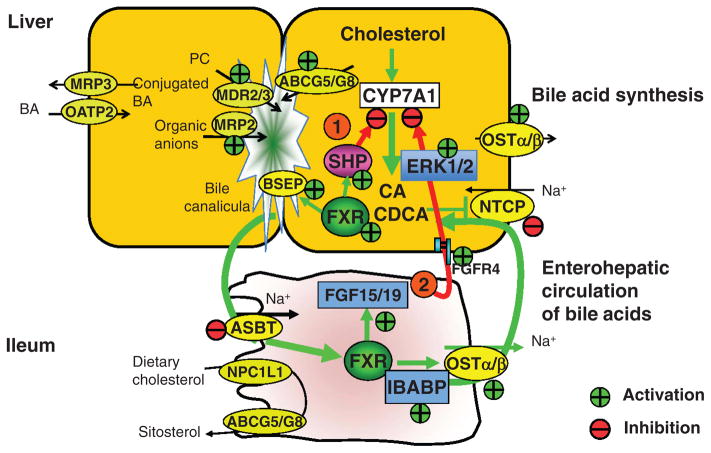
Figure 5.
Farnesoid X receptor (FXR) regulates enterohepatic circulation of bile acids. Major bile acid transporters in human hepatocytes and enterocytes are shown. Enzymes and transporters regulated by FXR are indicated. In hepatocytes, bile acids activate FXR to inhibit CYP7A1 gene transcription by two pathways: (i) FXR induces small heterodimer partner (SHP), which inhibits CYP7A1 by inhibiting nuclear receptors liver related homologue-1 (LRH-1) or hepatocyte nuclear factor (HNF4), which bind to the CYP7A1 promoter. (ii) In enterocytes, FXR induces intestinal hormone fibroblast growth factor 19 (FGF19), which is circulated to hepatocytes to activate FGF receptor 4 (FGFR4) signaling to inhibit CYP7A1 via activation of the extracellular stress-activated receptor kinase 1/2 (ERK1/2) pathway. FXR induces bile salt expert pump (BSEP) to efflux bile acids into bile; multidrug resistance protein 2/3 (MDR2/3) to efflux phosphatidylcholine (PC) to bile; and MDR related protein 2 (MRP2) to efflux organic anions including glucuronidated- and sulfated-bile acids, organic anions, and drugs into bile. Bile acids also facilitate efflux of cholesterol to bile by ATP binding casette G5/G8(ABCG5/G8). In the bile, bile acids, PC, and cholesterol form mixed micelles, which are stored in the gallbladder. In the brush border membrane of the ileum, bile acids are reabsorbed by the apical sodium bile salt transporter (ASBT). In enterocytes, bile acids activate FXR, which induces ileum bile acid binding protein (IBABP) to bind bile salts and may facilitate intracellular transport of bile acids to organic solute transporter α/β (OSTα/β) located in the basolateral membrane for efflux of bile acids into portal circulation. Bile acids in portal blood are reabsorbed into hepatocytes by Na+-dependent taurocholate cotransport peptide (NTCP). FXR inhibits NTCP transcription as a feedback inhibition of bile acid uptake to prevent liver injury. In the sinusoidal membrane of enterocytes and hepatocytes, FXR also induces MRP3/4 to efflux bile acids as an adaptive response to cholestasis. In hepatocytes, FXR also induces OSTα/β to efflux bile acids into sinusoidal blood to prevent bile acid accumulation in hepatocytes. MRP3 may be induced by FXR as an adaptive response to cholestasis. Bile acids returned to hepatic sinusoid are also taken up by Na+-independent organic anion transport proteins (OATP2). Many of these membrane transporters (ASBT, OSTα/β, and MRP2/3) also are present in cholangiocytes for reabsorption of bile acids, and in renal proximal tubule cells for reabsorption of bile acids from blood circulation and excretion of hydrophilic bile acids.
The FXR/SHP/LRH-1 pathway
FXR inhibits CYP7A1 gene transcription by indirect mechanisms. It is thought that bile acid-activated FXR induces a negative nuclear receptor SHP to inhibit CYP7A1 and CYP8B1 gene transcription (Fig. 5) (74,121). SHP is an atypical receptor without a DNA-binding domain. SHP inhibits the trans-activating activity of liver related homologue-1 (LRH-1), and results in inhibiting CYP7A1 gene transcription (74,121). SHP also interacts with HNF4α to block HNF4α interaction with peroxisome PGC-1α and results in inhibition of CYP7A1 and CYP8B1 transcription (48, 219). The FXR/SHP mechanism is supported by the finding that SHP and CYP7A1 mRNA expression levels show an inversed corelation, and CYP7A1 expression and bile acid synthesis is induced in Shp knockout mice. Paradoxically, bile acid feeding to Shp knockout mice inhibits CYP7A1 expression and bile acid synthesis suggesting that redundant pathway may exist for bile acid inhibition of CYP7A1 (101, 201). The physiological relevance of the FXR/SHP pathway in bile acid feedback inhibition remains unclear.
The FXR/FGF19/FGFR4 pathway
Another FXR-dependent mechanism is based on the observation that GW4064 induces an intestine hormone, FGF15, which activates a hepatic FGF receptor 4 (FGFR4) signaling to inhibit CYP7A1 mRNA expression (Fig. 5) (83). A subsequent study shows that FXR induces FGF15, a mouse orthologous of human FGF19 in mouse intestine, and the expression of FGF15 mRNA is inversely correlated to the CYP7A1 mRNA expression levels in mouse liver (88). Both Fgfr4−/− and Fgf15−/− mice have increased bile acid pool, fecal bile acid secretion, and CYP7A1 expression (218). Overexpression of a constitutively active FGFR4 represses CYP7A1 expression and decreases bile acid pool size in wild type mice (217). These results are consistent with the proposed role of FGF15/FGFR4 signaling in mediating bile acid inhibition of bile acid synthesis. It has been suggested that intestine-derived FGF15 functions as an enterohepatic signal to activate FGFR4 signaling, which inhibits CYP7A1 in hepatocytes (88). Furthermore, GW4064 represses CYP7A1 expression in liver Fxr knockout mice but not in intestinal Fxr knockout mice (102). Interestingly, FXR-mediated repression of CYP8B1 expression is more dependent on liver FXR than intestinal FXR, and FGF15 represses CYP7A1 but not CYP8B1 expression. These results provide strong evidences that the intestinal FXR but not liver FXR is required for bile acid inhibition of CYP7A1 gene transcription in mice. FGF19 activation of FGFR4 signaling requires a membrane-bound glycosidase β-Klotho (120). In β-Klotho knockout mice, bile acid synthesis, and secretion and CYP7A1 expression was increased, but CYP8B1 expression was not altered (90). FGF15 has not been identified in mouse sera and liver, and bile acids do not induce FGF15 expression in mouse liver. Thus, the physiological relevance of the FGF15/FGFR4 pathway in bile acid feedback regulation of bile acid synthesis in mice remains unclear.
In human patients serum FGF19 levels exhibit a diurnal rhythm with two major peaks at 3 and 9 pm, which are ~90 to 120 min following the peaks of serum bile acids and C4 (122). These observations are consistent with the concept that FGF19 is secreted from the intestine to blood circulation in response to postprandial efflux of bile acids to inhibit bile acid synthesis in the liver. In contrast to mice, GW4064 and bile acids are able to induce FGF19 synthesis and secretion in human hepatocytes (176). This study shows that CDCA and GW4064 rapidly induce FGF19 mRNA expression, FGF19 protein secretion, and tyrosine phosphorylation of FGFR4, but inhibit CYP7A1 mRNA expression in primary human hepatocytes suggesting that liver-produced FGF19 is secreted from hepatocytes to activate FGFR4 signaling in hepatocytes by an autocrine or paracrine mechanism. Furthermore, knockdown of SHP expression by siRNA does not affect FGF19 inhibition of CYP7A1 mRNA expression in primary human hepatocytes, suggesting that SHP may not be required in FGF19 signaling (176). It is also noted that induction of FGF19 is sustained for at least 24 h but induction of SHP mRNA by CDCA and GW4064 is transient, reaching the maximum in 1 to 3 h and reducing to the basal levels after 6 h of treatment (176). All these data show a lack of correlation between SHP and CYP7A1 expression levels in FGF19 signaling. The study by Song et al. (176) demonstrates that FGF19/FGFR4 signaling activates and phosphorylates mainly the MAPK/ERK1/2 signaling pathway in human primary hepatocytes (Fig. 5). However, the down stream factor(s) of the FGF19 pathway that inhibits CYP7A1 gene transcription remain unknown.
Expression of FGF19 in human sera and hepatocytes has been reported recently in a patient patients with extrahepatic cholestasis (160) Serum FGF19 levels were higher in cholestatic patients than in noncholestatic patients and postcholestatic patients who received a biliary stent to drain bile acids. FGF19 mRNA could be detected in the majority of liver specimens with a wide range of expression levels, which were much higher in the cholestatic group than in the drained and noncholestatic group. CYP7A1 mRNA expression levels were lower in the cholestatic group than in the control and drained groups. These data suggest that bile acids accumulated in cholestatic liver could induce FGF19 expression as an adaptive response to cholestatic liver injury.
Bile Acid-Activated G protein-Coupled Receptors
G protein bile acid receptor-1 (Gpbar-1, aka TGR5)
Recently, a Gαs protein-coupled receptor TGR5 has been identified as a bile acid-activated membrane receptor (96, 128). TGR5 is expressed in many tissues including gall-bladder, spleen, liver, intestine, kidney, skeletal muscle, pancreas, adipocytes, and macrophages (Fig. 6). TGR5 is not expressed in hepatocytes, but has been detected in Kupffer cells (hepatic resident macrophages), liver sinusoidal endothelial cells, and gallbladder epithelial cells (97, 98, 100). Conjugated and free bile acids bind and activate TGR5 in the order of TLCA>LCA>DCA>CDCA>CA. TGR5 signaling stimulates cAMP, which activates PKA and target gene expression. In brown adipose tissue TGR5 signaling stimulates the conversion of ATP to cAMP, which activates a type 2 iodothyronine deiodinase (DIO2). DIO2 converts thyroxine T4 to the biologically activate hormone T3, which is known to stimulate mitochondrial oxygen consumption and energy metabolism (207). TGR5 knockout mice have reduced bile acid pool and accumulate fats when fed a high fat diet (129). Interestingly, Tgr5−/− mice are protected from lithogenic diet-induced gallstone disease (196). In liver and intestine, TGR5/cAMP signaling has anti-inflammatory function by inhibiting nuclear factor NF-κB-mediated inflammatory cytokine production (204), and protects intestinal barrier integrity and prevents colitis (36). TGR5 stimulates gallbladder refilling (113). A study of human gallstone patients reports that TGR5 mRNA and protein are expressed in all patients, and TGR5 mRNA, but not protein expression levels, are increased in gallstone patients (97). This study shows localization of TGR5 in the apical membrane and recycling of endosomes in gallbladder epithelial cells. TGR5 is colocalized with cystic fibrosis transducer regulator and ASBT, suggesting the coupling of TGR5 in bile acid uptake and chloride secretion.
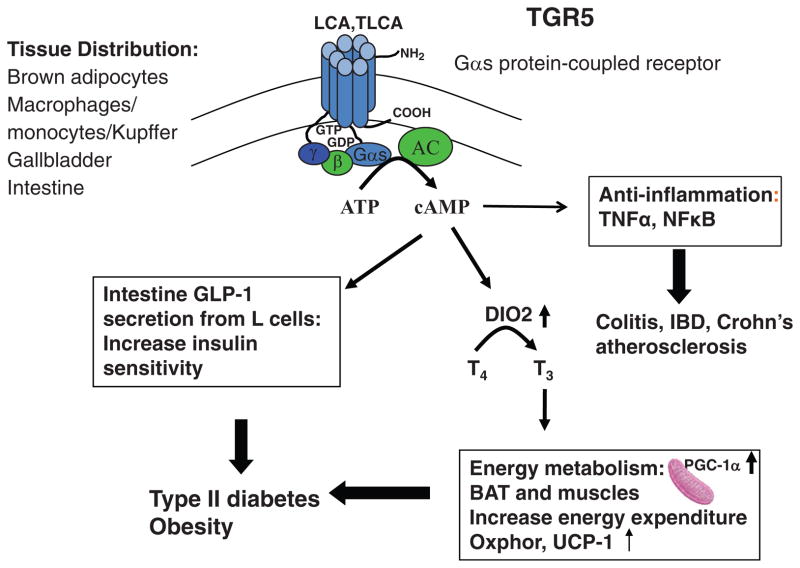
Figure 6.
Bile acid-activated TGR5 signaling. TGR5 is expressed in brown adipocytes, macrophages/monocytes and hepatic Kupffer cells, gallbladder epithelium, and intestine, with especially high levels in the colon. TGR5 is the first G protein-coupled receptor (GPCR) identified as a bile acid-activated membrane receptor. TGR5 is a Gαs GPCR activated by secondary bile acids, lithocholic acid (LCA) and TLCA to induce cAMP signaling through activation of adenylyl cyclase (AC). TGR5 signaling may increase insulin sensitivity through two mechanisms. (i) In brown adipose tissue, cAMP induces type 2 deiodinase (DIO2), which converts and activates thyroid hormone T4 to T3 to stimulate energy metabolism in mitochondria by activating oxidative phosphorylation (OXphor) and uncoupling protein-1 (UCP-1). (ii) In the intestine, cAMP stimulates glucagon like peptide-1 (GLP-1) in L cells, which stimulates insulin secretion in the pancreas. TGR5 also has anti-inflammatory functions by antagonizing of TNFα and NF-κB-dependent induction of proinflammatory cytokines in intestine and macrophages, thus, protecting against colitis, inflammatory bowel disease and Crohn’s disease, and also atherosclerosis.
Activation of TGR5 inhibits proinflammatory cytokine production by macrophages and inhibits atherosclerosis (146). Bile acid-activated TGR5 signaling may play a critical role in protection against inflammatory diseases including fatty liver disease, inflammatory bowel diseases, atherosclerosis, and diabetes. It has been reported that bile acids and TGR5 stimulate GLP-1 secretion in an enteroendocrine cell line STC-1. Knockdown of TGR5 mRNA expression by siRNA reduced GLP-1 secretion suggesting that bile acids induce GLP-1 secretion by TGR5-dependent cAMP production (95). GLP-1 plays a critical role in regulating glucose homeostasis, appetite, insulin and glucagon secretion in pancreas, and diabetes. Activation of TGR5 by a specific TGR5 agonist 6α-ethyl-23(S)-methyl-CA (6-EMCA, INT-777) stimulates GLP-1 secretion from enteroendocrine L cells, increases intracellular calcium mobilization, and improves insulin sensitivity (188). Interestingly, Tgr5 null mice did not develop gallstones when fed a lithogenic diet (196). These mice have impaired bile acid feedback, and upregulation of CYP7A1 may prevent gallstone formation in Tgr5 null mice.
Sphingosin-1-phosphate receptor 2
A recent study identified S1P2 as a bile acid-activated GPCR (183). S1P2 is expressed in hepatocytes. Conjugated bile acids, TCA, TDCA, TUDCA, GCA, and GDCA are able to activate S1P2 signaling, which has been shown to activate ERK1/2 and AKT signaling in rat hepatocytes (50) (Fig. 7). S1P receptor 2(S1P2) is a Gαi class of GPCR. Molecular modeling shows TCA is able to dock into the S1P binding site in S1P2 (183). TCA appears to have a low affinity to S1P2. Sphingosine 1-phosphate is a phosphorylated product of a membrane lipid sphingosine by sphingosine kinase 1 (SphK1) and sphingosine kinase 2 (SphK2). Extracellular signal activates SphK1, which is translocated from cytosol to the plasma membrane to convert membrane-derived sphingosine to S1P, which activates S1P receptors by an autocrine/paracrine manner. SphK2 is located in the nucleus and shuttle between the cytosol and nucleus. Phosphorylation of SphK2 by p-ERK1/2 activates SphK2. S1P is a potent pleiotropic lipid mediator that has been shown to activate at least five different GPCR. Bile acids only activate S1P2 in hepatocytes. S1P2 activates the insulin receptor/AKT pathway through activation of a tyrosine kinase, Src, and EGFRs. It has been suggested that the S1P2/ERK1/2/AKT pathway may phosphorylate and stabilize SHP, which is known to be unstable and rapidly degraded via the ubiquitin-proteasome degradation pathway (130). If this is the case then S1P may inhibit CYP7A1 and bile acid synthesis by stabilizing and activating SHP to inhibit CYP7A1 gene transcription. Similarly, the S1P/SHP pathway may inhibit peroxisome proliferator-activated receptor α (PPARα)-mediated activation of fatty acid oxidation, and SREBP-1c-mediated fatty acid synthesis. However, the mechanism of the S1P/SHP pathway in inhibition of fatty acid synthesis and PPARα-regulated fatty acid oxidation remain unclear. Bile acid-activated S1P2 signaling may activate and phosphorylate ERK1/2, which is known to inhibit Cyp7a1 resulting in inhibiting bile acid synthesis (177) (Fig. 7). S1P has been shown to bind to and inhibit histone deacetylase 1 (HDAC1) and histone deacetylase 2 (HDAC2) involved in epigenetic regulation of gene transcription, thus stimulates histone acetylation and the rate of gene transcription. It has been suggested that the S1P2/AKT pathway may regulate hepatic glucose and lipid metabolism via inhibition of GSK3β and resulting in activation of glycogen synthase. Thus S1P2 signaling may stimulate glycogenesis and reduce serum glucose. AKT is known to phosphorylate FoxO1, which is excluded from the nucleus resulting in inhibiting gluconeogenesis (151). Further in vivo study is needed to confirm that bile acids are endogenous ligands, which bind to S1P2 with high affinity and specificity, and to unveil the physiological function of this bile acid-activated S1P2/ERK1/2/AKT signaling pathway.
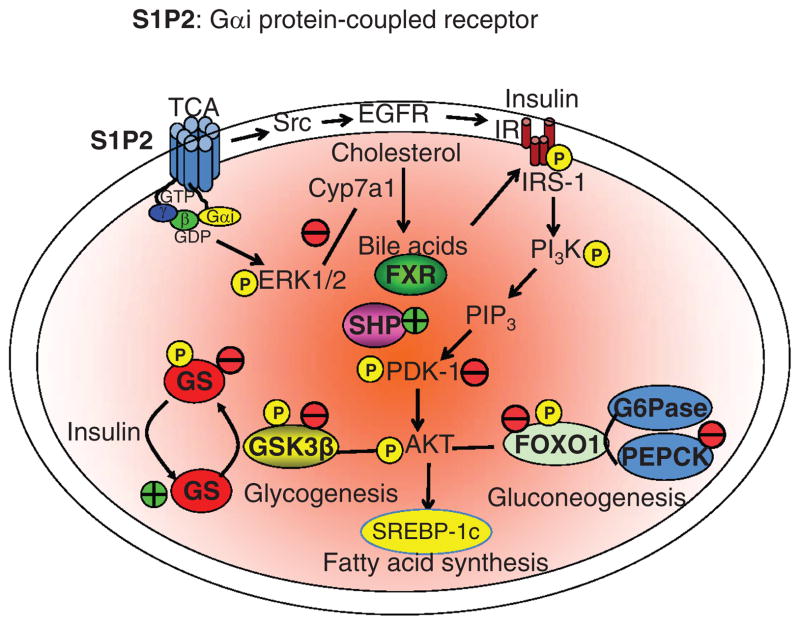
Figure 7.
Bile acid-activated S1P2 signaling. Tauro-conjugated bile acids stimulates sphingosine-1-phosphate receptor 2 (S1P2), a Gαi protein-coupled receptor, which activates the extracellular signal-regulated kinase 1/2 (ERK1/2) and AKT pathways. S1P2 signaling activates the insulin receptor/AKT pathway through activation of Src and epidermal growth factor receptors (EGFRs) leading to activation of the insulin receptor (IR), which phosphorylates and activates insulin receptor substrate-1 (IRS-1). IRS-1 phosphorylates phosphatidylinositol 3-kinase (PI3K), which phosphorylates phosphatidylinositol 4′, 5′ bis-phosphate to phosphatidylinositol 3′, 4′, 5′-trisphosphate (PIP3). PIP3 then ohosphorylates pyruvate dehydrogenase kinase-1 (PDK-1) to phosphorylate and activate AKT (also know as PKB). Phosphorylated AKT phosphorylates and inactivates glycogen synthase kinase 3β (GSK3β) and resulting in dephosphorylation and activation of glycogen synthase (GS), a key enzyme in glycogenesis. AKT also phosphorylates and inactivates a transcription factor, FoxO1 and resulted in inhibiting phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase) in gluconeogenesis. S1P2 signaling also may activate FXR/SHP pathway to inhibit peroxisome proliferator-activated receptor αγ (PPARα/γ)/PGC-1α-mediated fatty acid oxidation and steroid response element binding protein 1c (SREBP-1c)-mediated fatty acid synthesis. Therefore, SiP2 signaling may reduce serum glucose and triglycerides, and improve insulin sensitivity by stimulating glycogensis and inhibiting gluconeogenesis and lipogenesis.
Bile Acids in Inflammation
Bile acid synthesis is under tight control to prevent excessive intracellular accumulation of bile acids. Hydrophobic bile acids are potent inflammatory agents that cause injury to liver, intestine, and other tissues, whereas hydrophilic bile acids are anti-inflammatory. Bile acid-activated FXR and TGR5 signaling suppress inflammation in macrophages, intestine, and hepatocytes by inhibiting NF-κB nuclear translocation and antagonizing NF-κB-dependent induction of proinflammatory cytokines (36, 63, 146, 197). Conversely, proinflammatory cytokines could ameliorate FXR signaling and inhibit FXR target gene expression (68, 69). This inhibition may be through activation of TNFα and IL-1β signaling. The NF-κB subunits p65 and p50 may physically interact with FXR and block FXR activity. The role of FXR and TGR5 in atherosclerosis and metabolic syndrome has recently been suggested (77, 146, 147).
Bile acids are known to activate proinflammatory cytokine (TNFα and IL-1β) release from Kupffer cells (132). These cytokines may cross from sinusoid to hepatocytes to activate cytokine receptors Toll-like receptors signaling and activate the Ras/MEK4/7/JNK1/2 signal cascade, which phosphorylates HNF4α and reduces its binding to the BAREs and results in inhibiting CYP7A1 and CYP8B1 gene transcription (92, 114). On the other hand, physiological concentrations of bile acids activate FXR and TGR5 signaling to reduce NF-κB-mediated proinflammatory cytokine production. TGFβ1 secreted from Kupffer cells activates the Toll-like receptor 4 in hepatic stellate cells (HSCs), which also secrete TGFβ1 to activate the TGFβ1 receptor and the Smad signaling pathway in hepatocytes. Bile acids stimulate TGFβ1 expression in hepatocytes and activate the latent TGFβ1 in HSC and activate HSC. Smad3 recruits HDAC and a repressor mSin3A to inhibit HNF4α activation of CYP7A1 gene transcription (111). TGFβ1 and bile acids activate the Ras/MAPK/JNK pathway to phosphorylate a tumor suppressor p53 (42, 210), which interacts with HNF4α (125) and inhibits HNF4α transactivation of CYP7A1. Another cytokine, hepatocyte growth factor (HGF) has been shown to inhibit bile acid synthesis in primary human hepatocytes (175). HGF secreted from HSCs activates a tyrosine receptor kinase cMet and rapidly inhibits CYP7A1 expression. HGF induces cJun and SHP, and the ERK/1/2 and JNK pathways. This leads to recruitment of cJun and SHP, but inhibits coactivators PGC-1α and CBP binding to CYP7A1 chromatin and results in inhibited CYP7A1 gene transcription.
Integration of Bile Acid Signaling and Liver Metabolism
Bile acid synthesis rate is correlated to serum triglyceride levels in hyperlipidemia patients (6). Bile acid sequestrants increase bile acid and triglyceride synthesis, while CDCA treatment reduces serum triglycerides in hyperlipoproteinemic patients (7). There early studies in human patients suggest an integrated regulation of bile acid synthesis and serum triglycerides. More recent studies show that bile acid signaling through FXR and TGR5 not only regulates triglycerides, but also glucose and energy metabolisms (106,147,148,187,189). Figure 8 illustrates the role of FXR and TGR5 in regulation of lipid and energy metabolism in the liver, adipocytes, and intestine. Figure 9 shows the role of FXR in regulation of glucose metabolism in the liver.
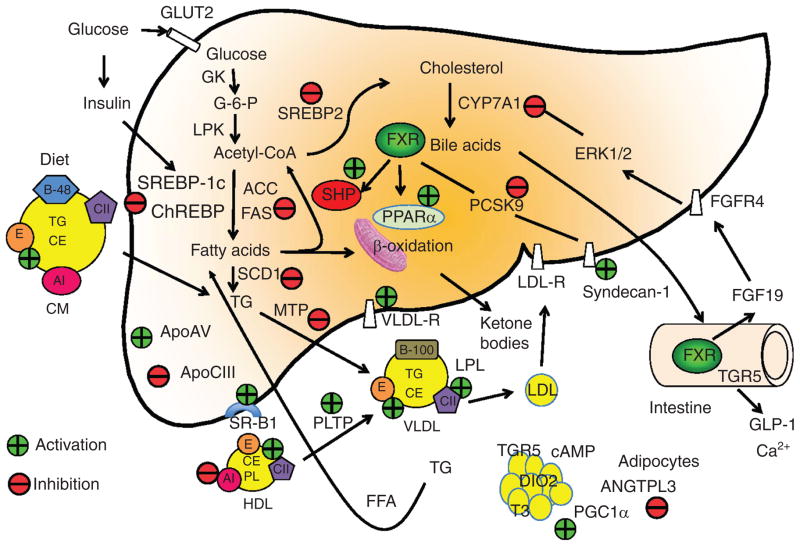
Figure 8.
Farnesoid X receptor (FXR) regulation of hepatic glucose and lipid metabolism in liver, adipocytes, and intestine. Glucose and insulin stimulate glycolysis to form acetyl-CoA, which is a precursor of cholesterol and fatty acids. The FXR/small heterodimer partner (SHP) pathway may inhibit steroid response element binding protein 1c (SREBP-1c), which induces all genes involved in lipogenesis, acetyl CoA carboxylase (ACC), fatty acid synthase (FAS), and stearoyl CoA desaturase (SCD). The FXR/SHP pathway also inhibits SREBP-2, which induces all genes in de novo cholesterol synthesis. FXR activates mitochondria fatty acid β-oxidation by inducing peroxisome proliferator-activated receptor α (PPARα). FXR inhibits PCSK9, which is an inhibitor of LDL receptor. Thus FXR induces LDL-R and also syndecan-1 involved in cholesterol uptake. FXR inhibits mitochondria triglyceride transport protein (MTP), which is required for assembly of VLDL particles with ApoB100. FXR also induces ApoE, which is a high affinity ligand of LDL receptor and scavenger receptor B1 (SR-B1), and a component of VLDL and chylomicron remnants, which are taken up by ApoE receptors. FXR also induces phospholipid transport protein (PLTP) involved in reverse cholesterol transport of cholesterol from peripheral tissues to liver by HDL/SR-B1 receptor-mediated mechanisms. On the other hand, FXR inhibits ApoA1, a component of HDL, and ANGPTL3, which is involved in hydrolysis of triglycerides in liver and adipocytes. FXR induces FGF19 synthesis in the intestine, which activates FGFR4 receptor in hepatocytes to activate ERK1/2 signaling to inhibit CYP7A1 and bile acid synthesis. In colon, bile acids activate TGR5 signaling to stimulate GLP-1 release. GLP-1 increases insulin sensitivity. TGR5 in brown adipocytes stimulates cAMP production, which induce deiodinase 2 (DIO2) to convert T4 to T3, which stimulates mitochondrial energy metabolism via activation of PGC-1α.
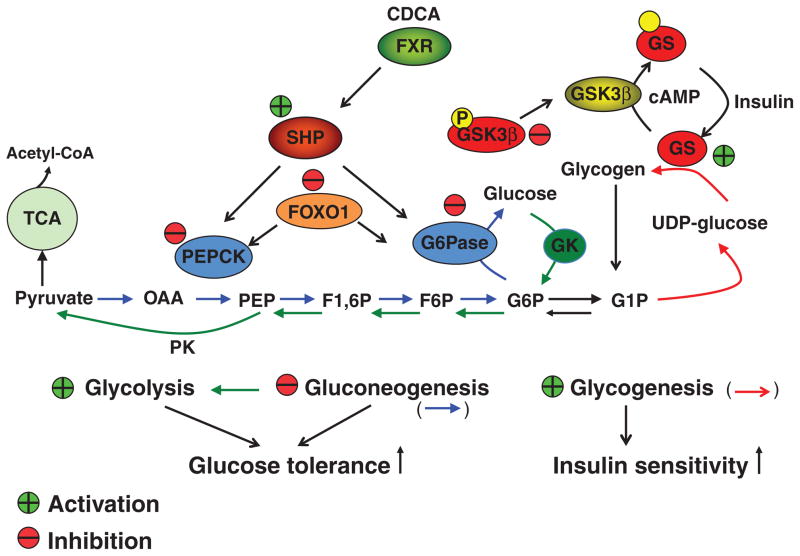
Figure 9.
FXR regulation of hepatic glucose metabolism. FXR signaling phosphorylates and inhibits glycogen synthase kinase 3β (GSK3β), which is an inhibitor of glycogen synthase activity. This results in stimulating glycogenesis. The FXR/SHP pathway inhibits PEPCK and glucose 6-phosphatase (G6Pase) to inhibit gluconeogenesis. This results in increasing glucose tolerance and insulin sensitivity.
Lipid metabolisms
The finding that FXR null mice have increased hepatic bile acids, cholesterol, triglycerides, and proatherogenic serum lipoprotein profiles provides the first experimental evidence that bile acid-activated FXR plays a role in regulation of lipid metabolism (172). It has been shown that FXR induces expression of human apolipoprotein C-II (94), which is an activator of lipoprotein lipase involved in lipolysis of triglycerides carried by triglyceride-rich lipoproteins, VLDL and chylomicron (Fig. 8). FXR also regulates several other genes involved in lipoprotein and triglyceride metabolism, including ApoA-V, ApoC-III, apoE, PPARα, and syndecan-1 (38,86). The role of FXR in triglyceride metabolism is also supported by the finding that overexpression of FXR by adenovirus-mediated transduction or treatment with GW4064 reduces serum triglyceride levels in wild-type and diabetic mice (222), and the FXR/SHP pathway inhibits SREBP-1c and other lipogenic genes and results in reducing serum triglycerides and VLDL production in mice (208).
The liver and biliary system are an integral part of the reverse cholesterol transport system (21). The liver contributes to more than 50% of reverse cholesterol transport system that transports cholesterol from liver and peripheral tissues to the liver for conversion to bile acids. Reverse cholesterol transport plays a critical role in preventing accumulation of cholesterol and oxysterols in macrophages and protecting against atherosclerosis (75, 84, 108, 186, 209, 220). Transgenic over-expression of CYP7A1 in mice prevents high fat diet-induced atherosclerosis in wild-type and LDL receptor-deficient mice (131, 154), and prevents high fat diet-induced obesity and insulin resistance (117). Increasing bile acid synthesis stimulates de novo cholesterol synthesis, but increases biliary cholesterol secretion, without increasing intestinal cholesterol absorption, thus maintains whole body cholesterol homeostasis (116).
Glucose metabolism
Many recent studies have implicated FXR in regulation of hepatic glucose metabolism (Fig. 9). An early study reports that bile acids inhibit PEPCK suggesting that bile acid synthesis and gluconeogenesis are coordinately regulated and linked to the fasting-to-refeeding cycle in mice (46). It has been reported that activation of FXR inhibits PEPCK by SHP-dependent inhibition of HNF4α and FoxO1 (215). In contrast, another report shows that activation of FXR by a FXR agonist GW4064 stimulates PEPCK in hepatocytes (181). Furthermore, bile acid activation of FXR stimulates the insulin/AKT pathway, which phosphorylates and inactivates GSK3β, and results in stimulating glycogen synthesis (117). Activation of FXR by GW4064 or a specific FXR agonist 6-ethyl-CDCA in mouse models of diabetes inhibits gluconeogenesis (37, 124, 222). Thus, FXR may inhibit gluconeogenesis but stimulates glycolysis and glycogenesis to improve glucose tolerance and insulin sensitivity. It has been reported that high concentration of insulin inhibits while glucose increases FXR expression (53). In pancreatic β cell line, FXR stimulates glucose-induced insulin transcription and secretion (158). At high glucose concentrations, FXR stimulates insulin secretion via AKT phosphorylation and translocation of glucose transporter-2 (GLUT2) at the plasma membrane of pancreatic β cells and GLUT4 in hepatocytes.
Several animal studies report that FXR plays a role in regulation of serum glucose and insulin resistance in mouse models (124,222). FXR null mice developed severe fatty liver, increased circulating free fatty acids associated with elevated serum glucose and impaired glucose and insulin tolerance (124). FXR-deficiency also reduces adipose tissue mass, lowers serum leptin concentrations and impairs glucose tolerance and insulin resistance, and treatment with GW4064 improved insulin resistance in obese ob/ob mice (26). In diabetic db/db mice, activation of FXR by GW4064 or overexpressing FXR lowers serum glucose (222). In contrast, a recent study reports that loss of FXR in obese and diabetic mice reduces body weight and improves peripheral insulin sensitivity (150). Another report shows that feeding GW4064 to high fat diet-fed mice caused increased susceptibility to diet-induced obesity, fatty liver and hypertriglyceridemia (206). In principal, activation of FXR should reduce bile acid synthesis, bile acid pool size, and energy expenditure, whereas administration of bile acids restores bile acid pool and should reverse these abnormalities. This apparent paradox of FXR and bile acid actions in glucose metabolism may be explained by TGR5 signaling. Activation of TGR5 signaling has been shown to stimulate intestinal GLP-1 secretion and improve liver and pancreatic function and enhance glucose tolerance in obese mice (Fig. 6) (188). Activation of TGR5 signaling by a TGR5 specific agonist 6α-ethyl-23(S)-methyl-CA (INT-777) stimulates GLP-1 release in enteroendocrine cells. This may be due to an increase of intracellular ATP/ADP ration and calcium mobilization.
Energy metabolism
Fatty acids and glucose are two major fuel molecules in the body. By controlling lipid and glucose metabolism, bile acids play a central role in energy metabolism. The role of FXR in energy metabolism is implicated by the finding that FGF19 increases metabolic rate, reduces adiposity and reverses diet-induce and leptin-deficient diabetic mice (67, 191). FGF19 induces hepatic leptin receptor, reduces acetyl-CoA carboxylase 2 expression, and increases fatty acid oxidation. FXR deficiency in mice increases susceptibility to torpor, supporting the role FXR plays in energy metabolism (25). It has been reported recently that FGF19 suppresses insulin-induced fatty acid synthesis in hepatocytes by inhibiting lipogenic gene expression and PGC-1α by inducing SHP (16). Shp null mice show increased energy expenditure, PGC-1α expression, and diet-induced obesity, suggesting that SHP may be involved in energy production in brown adipocye tissue by inhibiting PGC-1α expression (202).
Bile acids increase energy expenditure in brown adipose tissue and TGR5-dependent cAMP activation of a DIO2 is required for the bile acid effect on energy metabolism (Fig. 6) (207). In DIO2−/− mice, bile acids are unable to increase energy production. Knockout of the Tgr5 gene reduces bile acid pool size by 25%, and female Tgr5 null mice show significant weight gain and fat accumulation when fed a high fat diet (129). These phenotypes are consistent with the role of TGR5 in energy metabolism. However, adult humans have very little brown adipose tissue and the role of TGR5 in energy metabolism in man is not clear. An in vitro study shows that TGR5 regulates energy metabolism in human muscle cells. However, TGR5 levels in human skeletal muscle, adipose tissue and intestine are very low (96). The role of TGR5 in muscle energy metabolism is not known.
Bile Acids in Human Diseases
Bile acid metabolism defects
Bile acids increase cell proliferation and apoptosis in the liver and intestine (5, 173). Accumulation of toxic endobiotics and xenobiotics causes damage to cells and organs in the digestive tract. Inborn errors in bile acid metabolism have been identified by analysis of abnormal bile acid metabolites in human patients (163) and identification of mutations or enzyme deficiencies in bile acid metabolism. Decrease in bile acid synthesis can be caused by a primary defect in the enzymes involved in the bile acid biosynthetic pathways. Inborn error of bile acid synthesis can produce abnormal bile acid metabolites with altered steroid side chains or steroid nucleus structures. These metabolites are toxic and can cause cholestatic liver disease early in infancy and progressive neurological disease later in childhood and into adulthood. The diseases are characterized by various phenotypes depending on specific deficiency of enzymes in the pathways, such as jaundice, hyperbilirubinemia, giant cell hepatitis, neurologic dysfunctions, cholesterol gallstones, premature heart disease, cholestatic liver diseases, malabsorption of fat and fat-soluble vitamins, etc. These diseases can be treated effectively with bile acid replacement therapy if diagnosed early. Nine inborn errors of bile acid synthesis have been reported, including CYP7A1 deficiency (152), CYP7B1 deficiency (39, 165), 3β-Δ5-C27 steroid oxidoreductase (HSD3B7) mutations (40,161,185), Δ4-3-oxysteroid 5β-reductase deficiency (51, 73, 107, 164, 171), CYP27A1 mutations (cerebrotendinous xanthomatosis (17,18), bile acid conjugation deficiency (27, 35), and two or three defect in peroxisomal enzymes or biogenesis (19, 60, 61, 72, 80, 162).
Cholestatic liver diseases
Cholestasis is caused by a disruption of bile flow, which results in a lack of bile in the intestine, accumulation of toxic bile acids and other metabolites in the liver, and increased bile acids in the systemic circulation (192). Obstruction of the bile ducts by tumors or stones, genetic mutations of bile acid transporter genes, and acquired dysregulation of bile transport system by drugs, pregnancy, and pathophysiological conditions causes intra- and extrahepatic cholestasis (198).
Transporter defects
Congenital or acquired defects in canalicular membrane transporters can cause accumulation of bile acids in hepatocytes and in systemic circulation. Congenital cholestasis occurs early in life and patients have jaundice, pruritus, low absorption of fat-soluble vitamins leading to slow growth, and progressive liver damage by increased hepatic and serum levels of bile acids. Familial progressive intrahepatic cholestasis (FPIC) and benign recurrent intrahepatic cholestasis are autosomal recessive diseases linked to mutations in ATP8B1 (Type 1, PFIC1), BSEP (Type 2, PFIC2), and MDR3 (Type 3, PFIC3). PFIC1 (also known as Byler disease) is linked to mutations in the ATP8B1 gene, which codes a P-type ATPase, functioning as an aminophospholipid flippase that maintains membrane asymmetry by inward flipping of phosphatidylserine from the outer leaflet of the plasma membrane. The mechanism of ATP8B1 mutations in pathogenesis of PFIC1 is not clear. More recent data suggest that ATP8B1 counterbalances the effect of ABCB4 (PFIC3), a phospholipid floppase, and that ATP8B1 may be an anchor of the actin cytoskeleton for microvilli formation in the brush border membrane. Several studies have shown reduced hepatic FXR expression levels in PFIC1 patients (4, 29). A recent study reports that hepatic FXR mRNA expression is not altered and ATP8B1 deficiency may disrupt the bile canalicular membrane structure and cause cholestasis (23).
PFIC2 is linked to BSEP mutations (182). Bile acids accumulated in the liver and in systemic circulation cause hepatitis and liver damage requiring liver transplant in pediatric patients. BSEP mutations and polymorphisms have been linked to intrahepatic cholestasis of pregnancy (ICP) (105,137,142) and drug-induced liver injury (104). ICP is a reversible form of intrahepatic cholestasis associated with adverse pregnancy outcomes (55).
PFIC3 is linked to mutation of MDR3 (ABCB4), a phospholipid floppase that flops phosphatidylcholine to the outer leaflet of the canalicular membrane. PFIC3 patients have high levels of γ-glutamyl transpeptidase activity, progressive cholestasis, bile duct damage, and may require liver transplant. Patients have low phospholipids in bile, which are required for mixed micelle formation with bile acids and cholesterol (91, 142). Without forming mixed micelles, bile acids damage the canalicular membrane and cholangiocytes. MDR3 mutations may cause cholesterol gallstone diseases and has been linked to ICP (91, 205).
Genetic polymorphisms and heterozygote mutations of the PFIC1, PFIC2, and PFIC3 genes may increase susceptibility to acquired cholestasis in adults including ICP, drug-induced liver injury, primary biliary cirrhosis (PBC), and primary sclerosing cholangitis (PSC). FXR variants have been identified in ICP patients (195), and affect FXR target genes ABCB11, and ABCB4, and ATP8B1 expression in ICP (135, 138).
Mutations in the MRP2 gene have been linked to Dubin-Johnson syndrome, a disease characterized by chronic hyperbilirubinemia (99). MRP2 excretes conjugated bile acids, bilirubin, and other organic anions. Patients have elevated bile acids and cholestasis and MRP2 mutations have been linked to ICP.
Flow defects
Congenital or acquired defects in bile flow can cause obstructive cholestasis. Blockage of the bile duct by gallstones or tumors causes intrahepatic cholestasis due to accumulation of bile acids in the liver and absent in the intestine. The FXR gene has been identified as a candidate gene for the cholesterol gallstone susceptibility locus Lith7 in mice (211). A study of obstructive cholestasis in human patients shows that bile acid synthesis is suppressed but CYP7A1 expression is not altered (13). In contrast, another recent study reports that CYP7A1 mRNA expression is repressed in human patients with obstructive extrahepatic cholestasis, likely due to increased FGF19 expression in hepatocytes (160).
Bile Acids as Therapeutic Agents
Bile acids are therapeutic agents for cholestatic liver diseases. In principal, activation of FXR inhibits CYP7A1 and reduces bile acid synthesis, and inhibits NTCP and OATPs to reduce sinusoidal uptake of bile acids. FXR also may upregulate MRP3, MRP4, and OSTα/β in the sinusoidal membrane as an adaptive response to efflux bile acids into systemic circulation in obstructive cholestasis. Bile acids also play a protective role in controlling bacterial overgrowth in the intestine (89). Thus obstruction of bile flow or knockout of the Fxr gene in mice increases bacterial growth and mucosal injury in the intestine, and bile acid administration reduces bacterial growth in obstructive cholestasis.
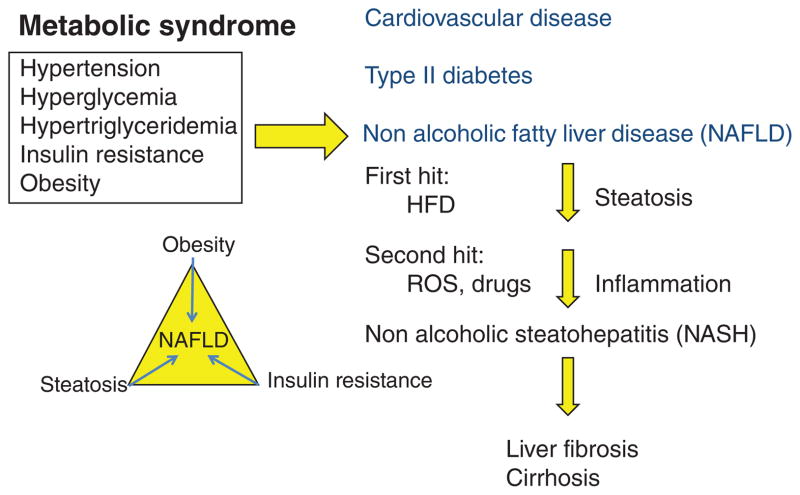
The therapeutic potential of bile acids and derivatives for treating hepatic and biliary diseases, nonalcoholic fatty liver diseases (NAFLDs), and metabolic syndrome are now well recognized (62, 189, 224). NAFLD is the most prevalent chronic liver disease affecting ~30% of the population in developed countries. Metabolic syndrome is a collection of five clinical symptoms including hypertension, hyperglycemia, hypertriglyceridemia, insulin resistance, and central obesity (155, 156) (Fig. 10). Metabolic syndrome contributes to chronic heart disease atherosclerosis, type II diabetes, and NAFLD. Dyslipidemia causes insulin resistance and inflammation, and pathogenesis of NAFLD (41,70). NAFLD is a spectrum of chronic liver abnormalities from simple steatosis to nonalcoholic steatohepatitis (NASH) to liver cirrhosis (216). In hepatic steatosis, more than 5% of hepatocytes accumulate lipid droplets. Hepatic steatosis is caused by alcohol, drugs, hepatitis, insulin resistance, high fat diet, and other factors, and is reversible. About 30% of steatotic patients develop to NASH, which involves hepatic ballooning, inflammatory infiltrates, and cell death. About 10% to 29% of NASH patients will progress to cirrhosis and hepatocellular carcinoma, the end stage of liver disease. NAFLD is closely associated with obesity, insulin resistance, and hepatic steatosis (59, 190). The progression of NAFLD to NASH requires at least “two-hits,” hepatic steatosis is the first hit, followed by inflammation as the second hit (45). Other factors such as insulin resistance and oxidative stress accelerate the progression from NAFLD to NASH.
Figure 10.
Mechanism of nonalcoholic fatty liver disease (NAFLD). Metabolic syndrome is a constellation of five clinical symptoms, hypertension, hyperglycemia, hypertriglyceridemia, insulin resistance, and obesity. Metabolic syndrome is linked to cardiovascular disease, type II diabetes, and NAFLD. NAFLD is progressed to nonalcoholic steatohepatitis (NASH) by many factors. The first hit is high fat diet (HFD)-induced hepatic steatosis, followed by inflammation involving reactive oxidizing species, drugs, and endoplasmid reticulum stress to NASH. NASH patients have prevalence for liver fibrosis and cirrhosis, while NAFLD is linked to obesity, steatosis, and insulin resistance. HFD, high fat diet; ROS, reactive oxidizing species.
Based on the results from CYP7A1 overexpressing mouse model (116, 117), increasing bile acid synthesis with reduced CA in the pool may be a strategy for preventing high fat diet-induced NAFLD and improving insulin resistance through activating FXR and TGR5 signaling to stimulate energy metabolism and increasing glucose tolerance and insulin sensitivity.
Many studies in animals and human diabetic patients report that bile acids may improve glycemic control (38). Disruption of enterohepatic circulation of bile acids by bile fistula and feeding bile acid sequestrants may reduce bile acid pool, which is enlarged in diabetes. Increasing hepatic bile acid synthesis may inhibit gluconeogenesis and lipogenesis, and stimulate glycolysis, fatty acid oxidation, and glycogenesis. Gastric bypass is currently being used as a treatment for reducing weight and type 2 diabetes mellitus (T2DM). However, the mechanism for improving insulin resistance and glycemic control is not known. Increases of serum bile acids after Roux-en-Y gastric bypass surgery may be linked to improved glucose and lipid metabolism (141). It is likely that increased serum bile acids after gastric bypass may play a role in improving glycemic control. Another recent study shows that Roux-en-Y gastric bypass significantly increases bile flow and fasting serum bile acids and FGF19 in patients (149).
Bile acid displacement and replacement
CDCA, CA, and UDCA have been used for effective gallstone dissolution for many years. CDCA and CA are not toxic to humans. CDCA has been used to treat bile acid deficient patients as a replacement of bile acids in the pool. CA is converted to DCA, which is highly toxic and is a colon cancer promoter. CA is more efficient in intestinal absorption of cholesterol than other bile acids and may cause gallstones in human patients (200). UDCA has been used in traditional Chinese medicine for treating digestive disease for several centuries. UDCA (Ursodiol™) is a highly soluble and nontoxic bile acid, and has been approved by FDA for gallstone dissolution and primary biliary cirrhosis. UDCA reduces the cytotoxicity of the circulating bile acid pool, and thus, protects cholangiocytes, stimulates hepatobiliary secretion, and inhibits liver cell apoptosis (173). UDCA may also activate PXR and induce PXR target genes, CYP3A4, SULTs, UGTs, BSEP, MDR3, and MRP4 for detoxification of bile acids. Norursodeoxycholic acid (norUDCA) is a side chain-shortened C23 homolog of UDCA that cannot be conjugated and is secreted into bile, reabsorbed by cholangiocytes, and returned to liver. Cholehepatic shunt of norUDCA leads to increase in bicarbonate in bile and hypercholeresis. This bile acid derivative reverses sclerosing cholangitis in the Mdr2−/−(Abcb4) model of cholangiopathy by ameliorating of bile acid hydrophobicity, stimulating bile flow, inducting bile acid detoxification and anti-inflammation and antifibrosis (134). Clinical trials of norUDCA for PBC and PSC are underway in Europe.
Bile acid sequestrants
Bile acid sequestrants, like cholestyramine, colestipol, and colesevelam, have been used for gallstone dissolution and lipid lowering in humans. Colesevelam is the second-generation bile acid sequestrants that have been used for lipid lowering in combination with statins, and with antidiabetic drugs for increasing glycemic control and insulin resistance (3, 11, 22, 43, 71, 167). These drugs bind bile acids in the intestine and interrupt enterohepatic circulation of bile acids, and result in stimulating bile acid synthesis, increasing LDL receptors and reducing serum cholesterol levels. Cholestyramine and colesevelam also have glucose-lowering effect and improve glycemic control for T2DM in some clinical studies (11, 14, 71, 179, 180). The underlying mechanism of glucose-lowering effect of bile acid sequestrants is not clear. Recent reports show that colesevelam may mediate its action through regulation of FXR/FGF19 and TGR5/GLP-1 signaling pathways (167). Colesevelam reduces bile acids in the intestine and should reduce FGF15 synthesis and consequently induces hepatic CYP7A1 expression and bile acid synthesis. However, effect of colesevelam on improving glucose tolerance and insulin sensitivity are not conclusive in human studies. A recent study shows that colesevelam increases GLP-1 levels and improves β cell function, but has no effect on insulin sensitivity (14). Colesevelam improves glycemic control in T2DM patients, but is not correlated with changes in bile acid metabolism (22). In T2DM patients, CA synthesis increase and colesevelam treatment preferentially increases CA but decreased CDCA and DCA, thus decreasing the hydrophobicity of bile acid pool without changing total bile acid pool size. There is no correlation between bile acid kinetics and glucose metabolism in these patients. Several clinical trials of colestipol, cholestyramine and colesevelam for type II diabetes have been approved by US FDA (79).
Bile acids and derivatives for liver and metabolic diseases
A synthetic bile acid derivative, obeticholic acid (OCA, 6-ethyl-CDCA or INT-747) is a potent and selective FXR agonist that has anticholestatic effects (143). In animal studies, OCA increases insulin sensitivity, inhibits gluconeogenesis, inhibits lipogenesis, and also has anti-inflammatory and anti-fibrotic properties (1). OCA ameliorates high fat diet-induced obesity and insulin resistance in mice, and insulin resistance and fatty liver in Zucker rats (fa/fa) (37). OCA antagonizes NF-κB-stimulated inflammation in liver (203), modulates innate immunity in animal models of colitis (197), inhibits and preserves the intestinal barrier in inflammatory bowel disease (69), and inhibits vascular smooth muscle cell inflammation and migration (119). OCA has been in phase II clinical trials for type 2 diabetic patients with NAFLD. Patients have improved insulin sensitivity, liver γ-glutamlytransferase levels (a marker of NASH) and weight loss. In phase II clinical studies of PBC patients, OCA significantly lowered alkaline phosphatase after 12 weeks of treatment. However, the majority of patients have pruritus, a common symptom of cholestasis and side effect of bile acid therapies. A larger National Institute of Diabetes and Digestive and Kidney Diseases-sponsored phase IIb trial OCA for NASH network will start soon.
Another bile acid derivative 6α-ethyl-23(S)-methyl-CA (EMCA or INT-777) is a selective and potent TGR5 agonist (144, 145). In animal studies, EMCA improves glucose tolerance, stimulates GLP-1 secretion from enteroendocrine L cells and improves insulin sensitivity, and increases intracellular ATP/ADP ratio and calcium mobilization in obese mice (188). Activation of TGR5 also reduces atherosclerotic lesions by reducing macrophage inflammation and lipid loading (146). TGR5 agonists also reduce inflammation in liver and intestine to prevent liver inflammation and IBD (147, 204).
Conclusion
Basic research in bile acid metabolism and signaling in last 20 years has greatly improved our current knowledge and understanding of bile acid-mediated integration of liver metabolism and diseases. Bile acid-based therapies have great potential for treatment of liver diseases and diabetes. Current research in bile acid metabolism relies on genetically modified mouse models. Future research using proteomic, metabolomic, and lipidomic approaches will help in identifying bile acid biomarkers for diagnosis and treatment of human metabolic diseases.
Acknowledgments
This work is supported by NIH grants R37DK58379 and R01DK44442.
References
- 1.Adorini L, Pruzanski M, Shapiro D. Farnesoid X receptor targeting to treat nonalcoholic steatohepatitis. Drug Discov Today. 2012;17:988–997. doi: 10.1016/j.drudis.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Agellon LB, Cheema SK. The 3′-untranslated region of the mouse cholesterol 7α-hydroxylase mRNA contains elements responsive to post-transcriptional regulation by bile acids. Biochem J. 1997;328:393–399. doi: 10.1042/bj3280393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldridge MA, Ito MK. Colesevelam hydrochloride: A novel bile acid-binding resin. Ann Pharmacother. 2001;35:898–907. doi: 10.1345/aph.10263. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez L, Jara P, Sanchez-Sabate E, Hierro L, Larrauri J, Diaz MC, Camarena C, De La Vega A, Frauca E, Lopez-Collazo E, Lapunzina P. Reduced hepatic expression of Farnesoid X Receptor in hereditary cholestasis associated to mutation in ATP8B1. Hum Mol Genet. 2004;13:2451–2460. doi: 10.1093/hmg/ddh261. [DOI] [PubMed] [Google Scholar]
- 5.Amaral JD, Viana RJ, Ramalho RM, Steer CJ, Rodrigues CM. Bile acids: Regulation of apoptosis by ursodeoxycholic ccid. J Lipid Res. 2009;50:1721–1734. doi: 10.1194/jlr.R900011-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angelin B, Einarsson K, Hellstrom K, Leijd B. Bile acid kinetics in relation to endogenous triglyceride metabolism in various types of hyperlipoproteinemia. J Lipid Res. 1978;19:1004–1016. [PubMed] [Google Scholar]
- 7.Angelin B, Einarsson K, Hellstrom K, Leijd B. Effects of cholestyramine and chenodeoxycholic acid on the metabolism of endogenous triglyceride in hyperlipoproteinemia. J Lipid Res. 1978;19:1017–1024. [PubMed] [Google Scholar]
- 8.Baker DM, Wang SL, Bell DJ, Drevon CA, Davis RA. One or more labile proteins regulate the stability of chimeric mRNAs containing the 3′-untranslated region of cholesterol-7alpha - hydroxylase mRNA. J Biol Chem. 2000;275:19985–19991. doi: 10.1074/jbc.M002351200. [DOI] [PubMed] [Google Scholar]
- 9.Ballatori N, Christian WV, Lee JY, Dawson PA, Soroka CJ, Boyer JL, Madejczyk MS, Li N. OSTalpha-OSTbeta: A major basolateral bile acid and steroid transporter in human intestinal, renal, and biliary epithelia. Hepatology. 2005;42:1270–1279. doi: 10.1002/hep.20961. [DOI] [PubMed] [Google Scholar]
- 10.Ballatori N, Fang F, Christian WV, Li N, Hammond CL. Ostalpha-Ostbeta is required for bile acid and conjugated steroid disposition in the intestine, kidney, and liver. Am J Physiol Gastrointest Liver Physiol. 2008;295:G179–G186. doi: 10.1152/ajpgi.90319.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bays HE, Goldberg RB, Truitt KE, Jones MR. Colesevelam hydrochloride therapy in patients with type 2 diabetes mellitus treated with metformin: Glucose and lipid effects. Arch Intern Med. 2008;168:1975–1983. doi: 10.1001/archinte.168.18.1975. [DOI] [PubMed] [Google Scholar]
- 12.Bennion LJ, Grundy SM. Effects of diabetes mellitus on cholesterol metabolism in man. N Engl J Med. 1977;296:1365–1371. doi: 10.1056/NEJM197706162962401. [DOI] [PubMed] [Google Scholar]
- 13.Bertolotti M, Carulli L, Concari M, Martella P, Loria P, Tagliafico E, Ferrari S, Del Puppo M, Amati B, De Fabiani E, Crestani M, Amorotti C, Manenti A, Carubbi F, Pinetti A, Carulli N. Suppression of bile acid synthesis, but not of hepatic cholesterol 7alpha-hydroxylase expression, by obstructive cholestasis in humans. Hepatology. 2001;34:234–242. doi: 10.1053/jhep.2001.25958. [DOI] [PubMed] [Google Scholar]
- 14.Beysen C, Murphy EJ, Deines K, Chan M, Tsang E, Glass A, Turner SM, Protasio J, Riiff T, Hellerstein MK. Effect of bile acid sequestrants on glucose metabolism, hepatic de novo lipogenesis, and cholesterol and bile acid kinetics in type 2 diabetes: A randomised controlled study. Diabetologia. 2012;55:432–442. doi: 10.1007/s00125-011-2382-3. [DOI] [PubMed] [Google Scholar]
- 15.Bhalla S, Ozalp C, Fang S, Xiang L, Kemper JK. Ligand-activated PXR interferes with HNF-4 signaling by targeting a common coactivator PGC-1alpha : Functional implications in hepatic cholesterol and glucose metabolism. J Biol Chem. 2004;279:45139–45147. doi: 10.1074/jbc.M405423200. [DOI] [PubMed] [Google Scholar]
- 16.Bhatnagar S, Damron HA, Hillgartner FB. Fibroblast growth factor-19, a novel factor that inhibits hepatic fatty acid synthesis. J Biol Chem. 2009;284:10023–10033. doi: 10.1074/jbc.M808818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjorkhem I. Inborn errors of metabolism with consequences for bile acid biosynthesis. A minireview. Scand J Gastroenterol Suppl. 1994;204:68–72. doi: 10.3109/00365529409103629. [DOI] [PubMed] [Google Scholar]
- 18.Bjorkhem I, Leitersdorf I. Sterol 27-hydroxylase deficiency: A rare cause of xanthomas in normocholesterolemic humans. Trends Endocrinol Metab. 2000;11:180–183. doi: 10.1016/s1043-2760(00)00255-1. [DOI] [PubMed] [Google Scholar]
- 19.Bove KE, Daugherty CC, Tyson W, Mierau G, Heubi JE, Balistreri WF, Setchell KD. Bile acid synthetic defects and liver disease. Pediatr Dev Pathol. 2000;3:1–16. doi: 10.1007/s100240050001. [DOI] [PubMed] [Google Scholar]
- 20.Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 21.Brufau G, Groen AK, Kuipers F. Reverse cholesterol transport revisited: Contribution of biliary versus intestinal cholesterol excretion. Arterioscler Thromb Vasc Biol. 2011;31:1726–1733. doi: 10.1161/ATVBAHA.108.181206. [DOI] [PubMed] [Google Scholar]
- 22.Brufau G, Stellaard F, Prado K, Bloks VW, Jonkers E, Boverhof R, Kuipers F, Murphy EJ. Improved glycemic control with colesevelam treatment in patients with type 2 diabetes is not directly associated with changes in bile acid metabolism. Hepatology. 2010;52:1455–1464. doi: 10.1002/hep.23831. [DOI] [PubMed] [Google Scholar]
- 23.Cai SY, Gautam S, Nguyen T, Soroka CJ, Rahner C, Boyer JL. ATP8B1 deficiency disrupts the bile canalicular membrane bilayer structure in hepatocytes, but FXR expression and activity are maintained. Gastroenterology. 2009;136:1060–1069. doi: 10.1053/j.gastro.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cariou B, Bouchaert E, Abdelkarim M, Dumont J, Caron S, Fruchart JC, Burcelin R, Kuipers F, Staels B. FXR-deficiency confers increased susceptibility to torpor. FEBS Lett. 2007;581:5191–5198. doi: 10.1016/j.febslet.2007.09.064. [DOI] [PubMed] [Google Scholar]
- 26.Cariou B, van Harmelen K, Duran-Sandoval D, van Dijk TH, Grefhorst A, Abdelkarim M, Caron S, Torpier G, Fruchart JC, Gonzalez FJ, Kuipers F, Staels B. The farnesoid X receptor modulates adiposity and peripheral insulin sensitivity in mice. J Biol Chem. 2006;281:11039–11049. doi: 10.1074/jbc.M510258200. [DOI] [PubMed] [Google Scholar]
- 27.Carlton VE, Harris BZ, Puffenberger EG, Batta AK, Knisely AS, Robinson DL, Strauss KA, Shneider BL, Lim WA, Salen G, Morton DH, Bull LN. Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat Genet. 2003;34:91–96. doi: 10.1038/ng1147. [DOI] [PubMed] [Google Scholar]
- 28.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: Opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 29.Chen F, Ananthanarayanan M, Emre S, Neimark E, Bull LN, Knisely AS, Strautnieks SS, Thompson RJ, Magid MS, Gordon R, Balasubramanian N, Suchy FJ, Shneider BL. Progressive familial intrahepatic cholestasis, type 1, is associated with decreased farnesoid X receptor activity. Gastroenterology. 2004;126:756–764. doi: 10.1053/j.gastro.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 30.Chen G, Liang G, Ou J, Goldstein JL, Brown MS. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci U S A. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang JY. Bile Acid regulation of gene expression: Roles of nuclear hormone receptors. Endocr Rev. 2002;23:443–463. doi: 10.1210/er.2000-0035. [DOI] [PubMed] [Google Scholar]
- 32.Chiang JY. Regulation of bile acid synthesis: Pathways, nuclear receptors, and mechanisms. J Hepatol. 2004;40:539–551. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 33.Chiang JY. Bile acids: Regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiang JYL. Regulation of bile acid synthesis. Front Biosci. 1998;3:D176–D193. doi: 10.2741/a273. [DOI] [PubMed] [Google Scholar]
- 35.Chong CP, Mills PB, McClean P, Gissen P, Bruce C, Stahlschmidt J, Knisely AS, Clayton PT. Bile acid-CoA ligase deficiency–a new inborn error of bile acid metabolism. J Inherit Metab Dis. 2012;35:521–530. doi: 10.1007/s10545-011-9416-3. [DOI] [PubMed] [Google Scholar]
- 36.Cipriani S, Mencarelli A, Chini MG, Distrutti E, Renga B, Bifulco G, Baldelli F, Donini A, Fiorucci S. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS One. 2011;6:e25637. doi: 10.1371/journal.pone.0025637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cipriani S, Mencarelli A, Palladino G, Fiorucci S. FXR activation reverses insulin resistance and lipid abnormalities and protects against liver steatosis in Zucker (fa/fa) obese rats. J Lipid Res. 2010;51:771–784. doi: 10.1194/jlr.M001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Claudel T, Staels B, Kuipers F. The Farnesoid X receptor: A molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol. 2005;25:2020–2030. doi: 10.1161/01.ATV.0000178994.21828.a7. [DOI] [PubMed] [Google Scholar]
- 39.Clayton PT. Disorders of bile acid synthesis. J Inherit Metab Dis. 2011;34:593–604. doi: 10.1007/s10545-010-9259-3. [DOI] [PubMed] [Google Scholar]
- 40.Clayton PT, Leonard JV, Lawson AM, Setchell KD, Andersson S, Egestad B, Sjovall J. Familial giant cell hepatitis associated with synthesis of 3 beta, 7 alpha-dihydroxy-and 3 beta,7 alpha, 12 alpha-trihydroxy-5-cholenoic acids. J Clin Invest. 1987;79:1031–1038. doi: 10.1172/JCI112915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: Old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cordenonsi M, Montagner M, Adorno M, Zacchigna L, Martello G, Mamidi A, Soligo S, Dupont S, Piccolo S. Integration of TGF-beta and Ras/MAPK signaling through p53 phosphorylation. Science. 2007;315:840–843. doi: 10.1126/science.1135961. [DOI] [PubMed] [Google Scholar]
- 43.Davidson MH, Dillon MA, Gordon B, Jones P, Samuels J, Weiss S, Isaacsohn J, Toth P, Burke SK. Colesevelam hydrochloride (cholestagel): A new, potent bile acid sequestrant associated with a low incidence of gastrointestinal side effects. Arch Intern Med. 1999;159:1893–1900. doi: 10.1001/archinte.159.16.1893. [DOI] [PubMed] [Google Scholar]
- 44.Dawson PA, Hubbert M, Haywood J, Craddock AL, Zerangue N, Christian WV, Ballatori N. The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J Biol Chem. 2005;280:6960–6968. doi: 10.1074/jbc.M412752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Day CP, James OF. Steatohepatitis: A tale of two “hits”? Gastroenterology. 1998;114:842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 46.De Fabiani E, Mitro N, Gilardi F, Caruso D, Galli G, Crestani M. Coordinated control of cholesterol catabolism to bile acids and of gluconeogenesis via a novel mechanism of transcription regulation linked to the fasted-to-fed cycle. J Biol Chem. 2003;278:39124–39132. doi: 10.1074/jbc.M305079200. [DOI] [PubMed] [Google Scholar]
- 47.DeBose-Boyd RA, Ou J, Goldstein JL, Brown MS. Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc Natl Acad Sci U S A. 2001;98:1477–1482. doi: 10.1073/pnas.98.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Del Castillo-Olivares A, Campos JA, Pandak WM, Gil G. Role of FTF/LRH-1 on bile acid biosynthesis. A known nuclear receptor activator that can Act as a suppressor of bile acid biosynthesis. J Biol Chem. 2004;279:16813–16821. doi: 10.1074/jbc.M400646200. [DOI] [PubMed] [Google Scholar]
- 49.Denson LA, Sturm E, Echevarria W, Zimmerman TL, Makishima M, Mangelsdorf DJ, Karpen SJ. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology. 2001;121:140–147. doi: 10.1053/gast.2001.25503. [DOI] [PubMed] [Google Scholar]
- 50.Dent P, Fang Y, Gupta S, Studer E, Mitchell C, Spiegel S, Hylemon PB. Conjugated bile acids promote ERK1/2 and AKT activation via a pertussis toxin-sensitive mechanism in murine and human hepatocytes. Hepatology. 2005;42:1291–1299. doi: 10.1002/hep.20942. [DOI] [PubMed] [Google Scholar]
- 51.Drury JE, Mindnich R, Penning TM. Characterization of disease-related 5beta-reductase (AKR1D1) mutations reveals their potential to cause bile acid deficiency. J Biol Chem. 2010;285:24529–24537. doi: 10.1074/jbc.M110.127779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duane WC, Javitt NB. 27-hydroxycholesterol. Production rates in normal human subjects. J Lipid Res. 1999;40:1194–1199. [PubMed] [Google Scholar]
- 53.Duran-Sandoval D, Mautino G, Martin G, Percevault F, Barbier O, Fruchart JC, Kuipers F, Staels B. Glucose regulates the expression of the farnesoid X receptor in liver. Diabetes. 2004;53:890–898. doi: 10.2337/diabetes.53.4.890. [DOI] [PubMed] [Google Scholar]
- 54.Edwards PA, Tabor D, Kast HR, Venkateswaran A. Regulation of gene expression by SREBP and SCAP. Biochim Biophys Acta. 2000;1529:103–113. doi: 10.1016/s1388-1981(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 55.Eloranta JJ, Kullak-Ublick GA. The role of FXR in disorders of bile acid homeostasis. Physiology (Bethesda) 2008;23:286–295. doi: 10.1152/physiol.00020.2008. [DOI] [PubMed] [Google Scholar]
- 56.Engelking LJ, Kuriyama H, Hammer RE, Horton JD, Brown MS, Goldstein JL, Liang G. Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J Clin Invest. 2004;113:1168–1175. doi: 10.1172/JCI20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fang Y, Han SI, Mitchell C, Gupta S, Studer E, Grant S, Hylemon PB, Dent P. Bile acids induce mitochondrial ROS, which promote activation of receptor tyrosine kinases and signaling pathways in rat hepatocytes. Hepatology. 2004;40:961–971. doi: 10.1002/hep.20385. [DOI] [PubMed] [Google Scholar]
- 58.Fang Y, Studer E, Mitchell C, Grant S, Pandak WM, Hylemon PB, Dent P. Conjugated bile acids regulate hepatocyte glycogen synthase activity in vitro and in vivo via Galphai signaling. Mol Pharmacol. 2007;71:1122–1128. doi: 10.1124/mol.106.032060. [DOI] [PubMed] [Google Scholar]
- 59.Farrell GC, Larter CZ. Nonalcoholic fatty liver disease: From steatosis to cirrhosis. Hepatology. 2006;43:S99–S112. doi: 10.1002/hep.20973. [DOI] [PubMed] [Google Scholar]
- 60.Ferdinandusse S, Denis S, Faust PL, Wanders RJ. Bile acids: Role of peroxisomes. J Lipid Res. 2009;50:2139–2147. doi: 10.1194/jlr.R900009-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferdinandusse S, Overmars H, Denis S, Waterham HR, Wanders RJ, Vreken P. Plasma analysis of di- and trihydroxycholestanoic acid di-astereoisomers in peroxisomal alpha-methylacyl-CoA racemase deficiency. J Lipid Res. 2001;42:137–141. [PubMed] [Google Scholar]
- 62.Fiorucci S, Baldelli F. Farnesoid X receptor agonists in biliary tract disease. Curr Opin Gastroenterol. 2009;25:252–259. doi: 10.1097/MOG.0b013e328324f87e. [DOI] [PubMed] [Google Scholar]
- 63.Fiorucci S, Cipriani S, Mencarelli A, Renga B, Distrutti E, Baldelli F. Counter-regulatory role of bile acid activated receptors in immunity and inflammation. Curr Mol Med. 2010;10:579–595. doi: 10.2174/1566524011009060579. [DOI] [PubMed] [Google Scholar]
- 64.Foretz M, Pacot C, Dugail I, Lemarchand P, Guichard C, Le Liepvre X, Berthelier-Lubrano C, Spiegelman B, Kim JB, Ferre P, Foufelle F. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol Cell Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Francis GA, Fayard E, Picard F, Auwerx J. Nuclear receptors and the control of metabolism. Annu Rev Physiol. 2003;65:261–311. doi: 10.1146/annurev.physiol.65.092101.142528. [DOI] [PubMed] [Google Scholar]
- 66.Frankenberg T, Rao A, Chen F, Haywood J, Shneider BL, Dawson PA. Regulation of the mouse organic solute transporter alpha-beta, Ostalpha-Ostbeta, by bile acids. Am J Physiol Gastrointest Liver Physiol. 2006;290:G912–G922. doi: 10.1152/ajpgi.00479.2005. [DOI] [PubMed] [Google Scholar]
- 67.Fu L, John LM, Adams SH, Yu XX, Tomlinson E, Renz M, Williams PM, Soriano R, Corpuz R, Moffat B, Vandlen R, Simmons L, Foster J, Stephan JP, Tsai SP, Stewart TA. Fibroblast growth factor 19 increases metabolic rate and reverses dietary and leptin-deficient diabetes. Endocrinology. 2004;145:2594–2603. doi: 10.1210/en.2003-1671. [DOI] [PubMed] [Google Scholar]
- 68.Gadaleta RM, Oldenburg B, Willemsen EC, Spit M, Murzilli S, Salvatore L, Klomp LW, Siersema PD, van Erpecum KJ, van Mil SW. Activation of bile salt nuclear receptor FXR is repressed by pro-inflammatory cytokines activating NF-kappa B signaling in the intestine. Biochim Biophys Acta. 2011;1812:851–858. doi: 10.1016/j.bbadis.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 69.Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen EC, Renooij W, Murzilli S, Klomp LW, Siersema PD, Schipper ME, Danese S, Penna G, Laverny G, Adorini L, Moschetta A, van Mil SW. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–472. doi: 10.1136/gut.2010.212159. [DOI] [PubMed] [Google Scholar]
- 70.Ginsberg HN, Zhang YL, Hernandez-Ono A. Metabolic syndrome: Focus on dyslipidemia. Obesity (Silver Spring) 2006;14(Suppl 1):41S–49S. doi: 10.1038/oby.2006.281. [DOI] [PubMed] [Google Scholar]
- 71.Goldberg RB, Fonseca VA, Truitt KE, Jones MR. Efficacy and safety of colesevelam in patients with type 2 diabetes mellitus and inadequate glycemic control receiving insulin-based therapy. Arch Intern Med. 2008;168:1531–1540. doi: 10.1001/archinte.168.14.1531. [DOI] [PubMed] [Google Scholar]
- 72.Goldfisher S, Moore CL, Johnson AB, Spiro AI, Valsamis MP, Wisniewski HK, Ritch RH, Norton WT, Rapin I, Gartner M. Peroxisomal and mitochondrial defects in the cerebro-hepato-renal syndrome. Science. 1997;182:62–64. doi: 10.1126/science.182.4107.62. [DOI] [PubMed] [Google Scholar]
- 73.Gonzales E, Cresteil D, Baussan C, Dabadie A, Gerhardt MF, Jacquemin E. SRD5B1 (AKR1D1) gene analysis in delta(4)-3-oxosteroid 5beta-reductase deficiency: Evidence for primary genetic defect. J Hepatol. 2004;40:716–718. doi: 10.1016/j.jhep.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 74.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 75.Groen AK, Oude Elferink RP, Verkade HJ, Kuipers F. The ins and outs of reverse cholesterol transport. Ann Med. 2004;36:135–145. doi: 10.1080/07853890310020635. [DOI] [PubMed] [Google Scholar]
- 76.Guo GL, Lambert G, Negishi M, Ward JM, Brewer HB, Jr, Kliewer SA, Gonzalez FJ, Sinal CJ. Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J Biol Chem. 2003;278:45062–45071. doi: 10.1074/jbc.M307145200. [DOI] [PubMed] [Google Scholar]
- 77.Hageman J, Herrema H, Groen AK, Kuipers F. A role of the bile salt receptor FXR in atherosclerosis. Arterioscler Thromb Vasc Biol. 2010;30:1519–1528. doi: 10.1161/ATVBAHA.109.197897. [DOI] [PubMed] [Google Scholar]
- 78.Han S, Chiang JY. Mechanism of vitamin D receptor inhibition of cholesterol 7alpha-hydroxylase gene transcription in human hepatocytes. Drug Metab Dispos. 2009;37:469–478. doi: 10.1124/dmd.108.025155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hansen M, Sonne DP, Mikkelsen KH, Gluud LL, Vilsboll T, Knop FK. Effect of bile acid sequestrants on glycaemic control: Protocol for a systematic review with meta-analysis of randomised controlled trials. BMJ open. 2012;2 doi: 10.1136/bmjopen-2012-001803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hanson RF, Szczepanik-Van Leeuwen P, Williams GC, Grabowski G, Sharp HL. Defects of bile acid synthesis in Zellweger’s syndrome. Science. 1979;203:1107–1108. doi: 10.1126/science.424737. [DOI] [PubMed] [Google Scholar]
- 81.He J, Nishida S, Xu M, Makishima M, Xie W. PXR prevents cholesterol gallstone disease by regulating biosynthesis and transport of bile salts. Gastroenterology. 2011;140:2095–2106. doi: 10.1053/j.gastro.2011.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hofmann AF. Detoxification of lithocholic acid, a toxic bile acid: Relevance to drug hepatotoxicity. Drug Metab Rev. 2004;36:703–722. doi: 10.1081/dmr-200033475. [DOI] [PubMed] [Google Scholar]
- 83.Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, Donahee M, Wang da Y, Mansfield TA, Kliewer SA, Goodwin B, Jones SA. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hong C, Bradley MN, Rong X, Wang X, Wagner A, Grijalva V, Castellani LW, Salazar J, Realegeno S, Boyadjian R, Fogelman AM, Van Lenten BJ, Reddy ST, Lusis AJ, Tangirala RK, Tontonoz P. LXRalpha is uniquely required for maximal reverse cholesterol transport and atheroprotection in ApoE-deficient mice. J Lipid Res. 2012;53:1126–1133. doi: 10.1194/jlr.M022061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. Embo J. 2006;25:1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 89.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, Mangelsdorf DJ, Kliewer SA. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ito S, Fujimori T, Furuya A, Satoh J, Nabeshima Y. Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J Clin Invest. 2005;115:2202–2208. doi: 10.1172/JCI23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jacquemin E. Role of multidrug resistance 3 deficiency in pediatric and adult liver disease: one gene for three diseases. Semin Liver Dis. 2001;21:551–562. doi: 10.1055/s-2001-19033. [DOI] [PubMed] [Google Scholar]
- 92.Jahan A, Chiang JY. Cytokine regulation of human sterol 12{alpha}-hydroxylase (CYP8B1) gene. Am J Physiol Gastrointest Liver Physiol. 2005;288:G685–G695. doi: 10.1152/ajpgi.00207.2004. [DOI] [PubMed] [Google Scholar]
- 93.Jansen PL, Sturm E. Genetic cholestasis, causes and consequences for hepatobiliary transport. Liver Int. 2003;23:315–322. doi: 10.1034/j.1478-3231.2003.00856.x. [DOI] [PubMed] [Google Scholar]
- 94.Kast HR, Nguyen CM, Sinal CJ, Jones SA, Laffitte BA, Reue K, Gonzalez FJ, Willson TM, Edwards PA. Farnesoid x-activated receptor induces apolipoprotein c-ii transcription: A molecular mechanism linking plasma triglyceride levels to bile acids. Mol Endocrinol. 2001;15:1720–1728. doi: 10.1210/mend.15.10.0712. [DOI] [PubMed] [Google Scholar]
- 95.Katsuma S, Hirasawa A, Tsujimoto G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun. 2005;329:386–390. doi: 10.1016/j.bbrc.2005.01.139. [DOI] [PubMed] [Google Scholar]
- 96.Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 97.Keitel V, Cupisti K, Ullmer C, Knoefel WT, Kubitz R, Haussinger D. The membrane-bound bile acid receptor TGR5 is localized in the epithelium of human gallbladders. Hepatology. 2009;50:861–870. doi: 10.1002/hep.23032. [DOI] [PubMed] [Google Scholar]
- 98.Keitel V, Donner M, Winandy S, Kubitz R, Haussinger D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun. 2008;372:78–84. doi: 10.1016/j.bbrc.2008.04.171. [DOI] [PubMed] [Google Scholar]
- 99.Keitel V, Nies AT, Brom M, Hummel-Eisenbeiss J, Spring H, Keppler D. A common Dubin-Johnson syndrome mutation impairs protein maturation and transport activity of MRP2 (ABCC2) Am J Physiol Gastrointest Liver Physiol. 2003;284:G165–G174. doi: 10.1152/ajpgi.00362.2002. [DOI] [PubMed] [Google Scholar]
- 100.Keitel V, Reinehr R, Gatsios P, Rupprecht C, Gorg B, Selbach O, Haussinger D, Kubitz R. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology. 2007;45:695–704. doi: 10.1002/hep.21458. [DOI] [PubMed] [Google Scholar]
- 101.Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell DW, Schwarz M. Loss of nbuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim I, Ahn SH, Inagaki T, Choi M, Ito S, Guo GL, Kliewer SA, Gonzalez FJ. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48:2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 103.Kullak-Ublick GA, Meier PJ. Mechanisms of cholestasis. Clin Liver Dis. 2000;4:357–385. doi: 10.1016/s1089-3261(05)70114-8. [DOI] [PubMed] [Google Scholar]
- 104.Lang C, Meier Y, Stieger B, Beuers U, Lang T, Kerb R, Kullak-Ublick GA, Meier PJ, Pauli-Magnus C. Mutations and polymorphisms in the bile salt export pump and the multidrug resistance protein 3 associated with drug-induced liver injury. Pharmacogenet Genomics. 2007;17:47–60. doi: 10.1097/01.fpc.0000230418.28091.76. [DOI] [PubMed] [Google Scholar]
- 105.Lang T, Haberl M, Jung D, Drescher A, Schlagenhaufer R, Keil A, Mornhinweg E, Stieger B, Kullak-Ublick GA, Kerb R. Genetic variability, haplotype structures, and ethnic diversity of hepatic transporters MDR3 (ABCB4) and bile salt export pump (ABCB11) Drug Metab Dispos. 2006;34:1582–1599. doi: 10.1124/dmd.105.008854. [DOI] [PubMed] [Google Scholar]
- 106.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 107.Lemonde HA, Custard EJ, Bouquet J, Duran M, Overmars H, Scambler PJ, Clayton PT. Mutations in SRD5B1 (AKR1D1), the gene encoding delta(4)-3-oxosteroid 5beta-reductase, in hepatitis and liver failure in infancy. Gut. 2003;52:1494–1499. doi: 10.1136/gut.52.10.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lewis GF, Rader DJ. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ Res. 2005;96:1221–1232. doi: 10.1161/01.RES.0000170946.56981.5c. [DOI] [PubMed] [Google Scholar]
- 109.Li T, Chanda D, Zhang Y, Choi HS, Chiang JYL. Glucose stimulates cholesterol 7alpha-hydroxylase gene transcription in human hepatocytes. J Lipid Res. 2010;51:832–842. doi: 10.1194/jlr.M002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li T, Chiang JY. Mechanism of rifampicin and pregnane X receptor (PXR) inhibition of human cholesterol 7{alpha}-hydroxylase gene (CYP7A1) transcription. Am J Physiol Gastrointest Liver Physiol. 2005;288:G74–G84. doi: 10.1152/ajpgi.00258.2004. [DOI] [PubMed] [Google Scholar]
- 111.Li T, Chiang JY. A novel role of transforming growth factor beta1 in transcriptional repression of human cholesterol 7alpha-hydroxylase gene. Gastroenterology. 2007;133:1660–1669. doi: 10.1053/j.gastro.2007.08.042. [DOI] [PubMed] [Google Scholar]
- 112.Li T, Francl JM, Boehme S, Ochoa A, Zhang Y, Klaassen CD, Erickson SK, Chiang JY. Glucose and insulin induction of bile acid synthesis: Mechanisms and implication in diabetes and obesity. J Biol Chem. 2012;287:1861–1873. doi: 10.1074/jbc.M111.305789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li T, Holmstrom SR, Kir S, Umetani M, Schmidt DR, Kliewer SA, Mangelsdorf DJ. The G protein-coupled bile acid receptor, TGR5, stimulates gallbladder filling. Mol Endocrinol. 2011;25:1066–1071. doi: 10.1210/me.2010-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li T, Jahan A, Chiang JY. Bile acids and cytokines inhibit the human cholesterol 7alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 2006;43:1202–1210. doi: 10.1002/hep.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li T, Kong X, Owsley E, Ellis E, Strom S, Chiang JY. Insulin regulation of cholesterol 7{alpha}-hydroxylase expression in human hepatocytes: Roles of forkhead box o1 and sterol regulatory element-binding protein 1c. J Biol Chem. 2006;281:28745–28754. doi: 10.1074/jbc.M605815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li T, Matozel M, Boehme S, Kong B, Nilsson LM, Guo G, Ellis E, Chiang JY. Overexpression of cholesterol 7alpha-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology. 2011;53:996–1006. doi: 10.1002/hep.24107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li T, Owsley E, Matozel M, Hsu P, Novak CM, Chiang JY. Transgenic expression of cholesterol 7alpha-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology. 2010;52:678–690. doi: 10.1002/hep.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li YC, Wang DP, Chiang JY. Regulation of cholesterol 7 alpha-hydroxylase in the liver. Cloning, sequencing, and regulation of cholesterol 7 alpha-hydroxylase mRNA. J Biol Chem. 1990;265:12012–12019. [PubMed] [Google Scholar]
- 119.Li YT, Swales KE, Thomas GJ, Warner TD, Bishop-Bailey D. Farnesoid x receptor ligands inhibit vascular smooth muscle cell inflammation and migration. Arterioscler Thromb Vasc Biol. 2007;27:2606–2611. doi: 10.1161/ATVBAHA.107.152694. [DOI] [PubMed] [Google Scholar]
- 120.Lin BC, Wang M, Blackmore C, Desnoyers LR. Liver specific activities of FGF19 require KLOTHO beta. J Biol Chem. 2007;282:27277–27284. doi: 10.1074/jbc.M704244200. [DOI] [PubMed] [Google Scholar]
- 121.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 122.Lundasen T, Galman C, Angelin B, Rudling M. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J Intern Med. 2006;260:530–536. doi: 10.1111/j.1365-2796.2006.01731.x. [DOI] [PubMed] [Google Scholar]
- 123.Lundasen T, Liao W, Angelin B, Rudling M. Leptin induces the hepatic high density lipoprotein receptor scavenger receptor B type I (SR-BI) but not cholesterol 7alpha-hydroxylase (Cyp7a1) in leptin-deficient (ob/ob) mice. J Biol Chem. 2003;278:43224–43228. doi: 10.1074/jbc.M302645200. [DOI] [PubMed] [Google Scholar]
- 124.Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest. 2006;116:1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maeda Y, Seidel SD, Wei G, Liu X, Sladek FM. Repression of hepatocyte nuclear factor 4alpha tumor suppressor p53: Involvement of the ligand-binding domain and histone deacetylase activity. Mol Endocrinol. 2002;16:402–410. doi: 10.1210/mend.16.2.0769. [DOI] [PubMed] [Google Scholar]
- 126.Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 127.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 128.Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, Itadani H, Tanaka K. Identification of membrane-type receptor for bile acids (M-BAR) Biochem Biophys Res Commun. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 129.Maruyama T, Tanaka K, Suzuki J, Miyoshi H, Harada N, Nakamura T, Miyamoto Y, Kanatani A, Tamai Y. Targeted disruption of G protein-coupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J Endocrinol. 2006;191:197–205. doi: 10.1677/joe.1.06546. [DOI] [PubMed] [Google Scholar]
- 130.Miao J, Xiao Z, Kanamaluru D, Min G, Yau PM, Veenstra TD, Ellis E, Strom S, Suino-Powell K, Xu HE, Kemper JK. Bile acid signaling pathways increase stability of small heterodimer partner (SHP) by inhibiting ubiquitin-proteasomal degradation. Genes Dev. 2009;23:986–996. doi: 10.1101/gad.1773909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Miyake JH, Duong-Polk XT, Taylor JM, Du EZ, Castellani LW, Lusis AJ, Davis RA. Transgenic expression of cholesterol-7-alpha-hydroxylase prevents atherosclerosis in C57BL/6J mice. Arterioscler Thromb Vasc Biol. 2002;22:121–126. doi: 10.1161/hq0102.102588. [DOI] [PubMed] [Google Scholar]
- 132.Miyake JH, Wang SL, Davis RA. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7alpha-hydroxylase. J Biol Chem. 2000;275:21805–21808. doi: 10.1074/jbc.C000275200. [DOI] [PubMed] [Google Scholar]
- 133.Modica S, Gadaleta RM, Moschetta A. Deciphering the nuclear bile acid receptor FXR paradigm. Nuclear receptor signaling. 2010;8:e005. doi: 10.1621/nrs.08005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Moustafa T, Fickert P, Magnes C, Guelly C, Thueringer A, Frank S, Kratky D, Sattler W, Reicher H, Sinner F, Gumhold J, Silbert D, Fauler G, Hofler G, Lass A, Zechner R, Trauner M. Alterations in lipid metabolism mediate inflammation, fibrosis, and proliferation in a mouse model of chronic cholestatic liver injury. Gastroenterology. 2012;142:140–151. e112. doi: 10.1053/j.gastro.2011.09.051. [DOI] [PubMed] [Google Scholar]
- 135.Mullenbach R, Bennett A, Tetlow N, Patel N, Hamilton G, Cheng F, Chambers J, Howard R, Taylor-Robinson SD, Williamson C. ATP8B1 mutations in British cases with intrahepatic cholestasis of pregnancy. Gut. 2005;54:829–834. doi: 10.1136/gut.2004.058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Myant NB, Mitropoulos KA. Cholesterol 7a-hydroxylase. J Lipid Res. 1977;18:135–153. [PubMed] [Google Scholar]
- 137.Noe J, Kullak-Ublick GA, Jochum W, Stieger B, Kerb R, Haberl M, Mullhaupt B, Meier PJ, Pauli-Magnus C. Impaired expression and function of the bile salt export pump due to three novel ABCB11 mutations in intrahepatic cholestasis. J Hepatol. 2005;43:536–543. doi: 10.1016/j.jhep.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 138.Painter JN, Savander M, Ropponen A, Nupponen N, Riikonen S, Ylikorkala O, Lehesjoki AE, Aittomaki K. Sequence variation in the ATP8B1 gene and intrahepatic cholestasis of pregnancy. Eur J Hum Genet. 2005;13:435–439. doi: 10.1038/sj.ejhg.5201355. [DOI] [PubMed] [Google Scholar]
- 139.Pandak WM, Stravitz RT, Lucas V, Heuman DM, Chiang JY. Hep G2 cells: A model for studies on regulation of human cholesterol 7alpha-hydroxylase at the molecular level. Am J Physiol. 1996;270:G401–G410. doi: 10.1152/ajpgi.1996.270.3.G401. [DOI] [PubMed] [Google Scholar]
- 140.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: Natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 141.Patti ME, Houten SM, Bianco AC, Bernier R, Larsen PR, Holst JJ, Badman MK, Maratos-Flier E, Mun EC, Pihlajamaki J, Auwerx J, Goldfine AB. Serum bile acids are higher in humans with prior gastric bypass: Potential contribution to improved glucose and lipid metabolism. Obesity (Silver Spring) 2009;17:1671–1677. doi: 10.1038/oby.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pauli-Magnus C, Lang T, Meier Y, Zodan-Marin T, Jung D, Breymann C, Zimmermann R, Kenngott S, Beuers U, Reichel C, Kerb R, Penger A, Meier PJ, Kullak-Ublick GA. Sequence analysis of bile salt export pump (ABCB11) and multidrug resistance p-glycoprotein 3 (ABCB4, MDR3) in patients with intrahepatic cholestasis of pregnancy. Pharmacogenetics. 2004;14:91–102. doi: 10.1097/00008571-200402000-00003. [DOI] [PubMed] [Google Scholar]
- 143.Pellicciari R, Costantino G, Camaioni E, Sadeghpour BM, Entrena A, Willson TM, Fiorucci S, Clerici C, Gioiello A. Bile acid derivatives as ligands of the farnesoid X receptor. Synthesis, evaluation, and structure-activity relationship of a series of body and side chain modified analogues of chenodeoxycholic acid. J Med Chem. 2004;47:4559–4569. doi: 10.1021/jm049904b. [DOI] [PubMed] [Google Scholar]
- 144.Pellicciari R, Gioiello A, Macchiarulo A, Thomas C, Rosatelli E, Natalini B, Sardella R, Pruzanski M, Roda A, Pastorini E, Schoonjans K, Auwerx J. Discovery of 6alpha-ethyl-23(S)-methylcholic acid (S-EMCA, INT-777) as a potent and selective agonist for the TGR5 receptor, a novel target for diabesity. J Med Chem. 2009;52:7958–7961. doi: 10.1021/jm901390p. [DOI] [PubMed] [Google Scholar]
- 145.Pellicciari R, Sato H, Gioiello A, Costantino G, Macchiarulo A, Sadeghpour BM, Giorgi G, Schoonjans K, Auwerx J. Nongenomic actions of bile acids. Synthesis and preliminary characterization of 23- and 6,23-alkyl-substituted bile acid derivatives as selective modulators for the G-protein coupled receptor TGR5. J Med Chem. 2007;50:4265–4268. doi: 10.1021/jm070633p. [DOI] [PubMed] [Google Scholar]
- 146.Pols TW, Nomura M, Harach T, Lo Sasso G, Oosterveer MH, Thomas C, Rizzo G, Gioiello A, Adorini L, Pellicciari R, Auwerx J, Schoonjans K. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 2011;14:747–757. doi: 10.1016/j.cmet.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pols TW, Noriega LG, Nomura M, Auwerx J, Schoonjans K. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J Hepatol. 2011;54:1263–1272. doi: 10.1016/j.jhep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Porez G, Prawitt J, Gross B, Staels B. Bile acid receptors as targets for the treatment of dyslipidemia and cardiovascular disease: Thematic review series: New lipid and lipoprotein targets for the treatment of cardiometabolic diseases. J Lipid Res. 2012;53:1723–1737. doi: 10.1194/jlr.R024794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pournaras DJ, Glicksman C, Vincent RP, Kuganolipava S, Alaghband-Zadeh J, Mahon D, Bekker JH, Ghatei MA, Bloom SR, Walters JR, Welbourn R, le Roux CW. The Role of Bile After Roux-en-Y Gastric Bypass in Promoting Weight Loss and Improving Glycaemic Control. Endocrinology. 2012;153:3613–3619. doi: 10.1210/en.2011-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Prawitt J, Abdelkarim M, Stroeve JH, Popescu I, Duez H, Velagapudi VR, Dumont J, Bouchaert E, van Dijk TH, Lucas A, Dorchies E, Daoudi M, Lestavel S, Gonzalez FJ, Oresic M, Cariou B, Kuipers F, Caron S, Staels B. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes. 2011;60:1861–1871. doi: 10.2337/db11-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 152.Pullinger CR, Eng C, Salen G, Shefer S, Batta AK, Erickson SK, Verhagen A, Rivera CR, Mulvihill SJ, Malloy MJ, Kane JP. Human cholesterol 7alpha-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. J Clin Invest. 2002;110:109–117. doi: 10.1172/JCI15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Qiao L, McKinstry R, Gupta S, Gilfor D, Windle JJ, Hylemon PB, Grant S, Fisher PB, Dent P. Cyclin kinase inhibitor p21 potentiates bile acid-induced apoptosis in hepatocytes that is dependent on p53. Hepatology. 2002;36:39–48. doi: 10.1053/jhep.2002.33899. [DOI] [PubMed] [Google Scholar]
- 154.Ratliff EP, Gutierrez A, Davis RA. Transgenic expression of CYP7A1 in LDL receptor-deficient mice blocks diet-induced hypercholesterolemia. J Lipid Res. 2006;47:1513–1520. doi: 10.1194/jlr.M600120-JLR200. [DOI] [PubMed] [Google Scholar]
- 155.Reaven G, Abbasi F, McLaughlin T. Obesity, insulin resistance, and cardiovascular disease. Recent Prog Horm Res. 2004;59:207–223. doi: 10.1210/rp.59.1.207. [DOI] [PubMed] [Google Scholar]
- 156.Reaven GM. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes. 1988;37:1595–1607. doi: 10.2337/diab.37.12.1595. [DOI] [PubMed] [Google Scholar]
- 157.Reinehr R, Becker S, Keitel V, Eberle A, Grether-Beck S, Haussinger D. Bile salt-induced apoptosis involves NADPH oxidase isoform activation. Gastroenterology. 2005;129:2009–2031. doi: 10.1053/j.gastro.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 158.Renga B, Mencarelli A, Vavassori P, Brancaleone V, Fiorucci S. The bile acid sensor FXR regulates insulin transcription and secretion. Biochim Biophys Acta. 2010;1802:363–372. doi: 10.1016/j.bbadis.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 159.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:1370174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 160.Schaap FG, van der Gaag NA, Gouma DJ, Jansen PL. High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology. 2009;49:1228–1235. doi: 10.1002/hep.22771. [DOI] [PubMed] [Google Scholar]
- 161.Schwarz M, Wright AC, Davis DL, Nazer H, Bjorkhem I, Russell DW. The bile acid synthetic gene 3beta-hydroxy-Delta(5)-C(27)-steroid oxidoreductase is mutated in progressive intrahepatic cholestasis. J Clin Invest. 2000;106:1175–1184. doi: 10.1172/JCI10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Setchell KD, Heubi JE, Bove KE, O’Connell NC, Brewsaugh T, Steinberg SJ, Moser A, Squires RH., Jr Liver disease caused by failure to racemize trihydroxycholestanoic acid: Gene mutation and effect of bile acid therapy. Gastroenterology. 2003;124:217–232. doi: 10.1053/gast.2003.50017. [DOI] [PubMed] [Google Scholar]
- 163.Setchell KD, Street JM. Inborn errors of bile acid synthesis. Semin Liver Dis. 1987;7:85–99. doi: 10.1055/s-2008-1040568. [DOI] [PubMed] [Google Scholar]
- 164.Setchell KD, Suchy FJ, Welsh MB, Zimmer-Nechemias L, Heubi J, Balistreri WF. Delta 4-3-oxosteroid 5 beta-reductase deficiency described in identical twins with neonatal hepatitis. A new inborn error in bile acid synthesis. J Clin Invest. 1988;82:2148–2157. doi: 10.1172/JCI113837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Setchell KDR, Schwarz M, O’Connell NC, Lund EG, Davis DL, Lathe R, Thompson HR, Weslie Tyson R, Sokol RJ, Russell DW. Identification of a new inborn error in bile acid synthesis: Mutation of the oxysterol 7α-hydroxylase gene causes severe neonatal liver disease. J Clin Invest. 1998;102:1690–1703. doi: 10.1172/JCI2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shaham O, Wei R, Wang TJ, Ricciardi C, Lewis GD, Vasan RS, Carr SA, Thadhani R, Gerszten RE, Mootha VK. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol. 2008;4:214. doi: 10.1038/msb.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shang Q, Saumoy M, Holst JJ, Salen G, Xu G. Colesevelam improves insulin resistance in a diet-induced obesity (F-DIO) rat model by increasing the release of GLP-1. Am J Physiol Gastrointest Liver Physiol. 2010;298:G419–424. doi: 10.1152/ajpgi.00362.2009. [DOI] [PubMed] [Google Scholar]
- 168.Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem. 1999;274:30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- 169.Shimomura I, Bashmakov Y, Ikemoto S, Horton JD, Brown MS, Goldstein JL. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci U S A. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shin DJ, Campos JA, Gil G, Osborne TF. PGC-1a activates CYP7A1 and bile acid biosynthesis. J Biol Chem. 2003;278:50047–50052. doi: 10.1074/jbc.M309736200. [DOI] [PubMed] [Google Scholar]
- 171.Shneider BL, Setchell KD, Whitington PF, Neilson KA, Suchy FJ. Delta 4-3-oxosteroid 5 beta-reductase deficiency causing neonatal liver failure and hemochromatosis [see comments] J Pediatr. 1994;124:234–238. doi: 10.1016/s0022-3476(94)70310-8. [DOI] [PubMed] [Google Scholar]
- 172.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 173.Sola S, Amaral JD, Aranha MM, Steer CJ, Rodrigues CM. Modulation of hepatocyte apoptosis: cross-talk between bile acids and nuclear steroid receptors. Curr Med Chem. 2006;13:3039–3051. doi: 10.2174/092986706778521823. [DOI] [PubMed] [Google Scholar]
- 174.Song KH, Chiang JY. Glucagon and cAMP inhibit cholesterol 7alpha-hydroxylase (CYP7a1) gene expression in human hepatocytes: Discordant regulation of bile acid synthesis and gluconeogenesis. Hepatology. 2006;43:117–125. doi: 10.1002/hep.20919. [DOI] [PubMed] [Google Scholar]
- 175.Song KH, Ellis E, Strom S, Chiang JY. Hepatocyte growth factor signaling pathway inhibits cholesterol 7alpha-hydroxylase and bile acid synthesis in human hepatocytes. Hepatology. 2007;46:1993–2002. doi: 10.1002/hep.21878. [DOI] [PubMed] [Google Scholar]
- 176.Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49:297–305. doi: 10.1002/hep.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Song MB, Yu XJ, Zhu GX, Chen JF, Zhao G, Huang L. Transfection of HGF gene enhances endothelial progenitor cell (EPC) function and improves EPC transplant efficiency for balloon-induced arterial injury in hypercholesterolemic rats. Vascul Pharmacol. 2009;51:205–213. doi: 10.1016/j.vph.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 178.Sonoda J, Xie W, Rosenfeld JM, Barwick JL, Guzelian PS, Evans RM. Regulation of a xenobiotic sulfonation cascade by nuclear pregnane X receptor (PXR) Proc Natl Acad Sci U S A. 2002;99:13801–13806. doi: 10.1073/pnas.212494599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Staels B. A review of bile acid sequestrants: Potential mechanism(s) for glucose-lowering effects in type 2 diabetes mellitus. Postgrad Med. 2009;121:25–30. doi: 10.3810/pgm.2009.05.suppl53.290. [DOI] [PubMed] [Google Scholar]
- 180.Staels B, Kuipers F. Bile acid sequestrants and the treatment of type 2 diabetes mellitus. Drugs. 2007;67:1383–1392. doi: 10.2165/00003495-200767100-00001. [DOI] [PubMed] [Google Scholar]
- 181.Stayrook KR, Bramlett KS, Savkur RS, Ficorilli J, Cook T, Christe ME, Michael LF, Burris TP. Regulation of carbohydrate metabolism by the farnesoid x receptor. Endocrinology. 2005;146:984–991. doi: 10.1210/en.2004-0965. [DOI] [PubMed] [Google Scholar]
- 182.Strautnieks SS, Bull LN, Knisely AS, Kocoshis SA, Dahl N, Arnell H, Sokal E, Dahan K, Childs S, Ling V, Tanner MS, Kagalwalla AF, Nemeth A, Pawlowska J, Baker A, Mieli-Vergani G, Freimer NB, Gardiner RM, Thompson RJ. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat Genet. 1998;20:233–238. doi: 10.1038/3034. [DOI] [PubMed] [Google Scholar]
- 183.Studer E, Zhou X, Zhao R, Wang Y, Takabe K, Nagahashi M, Pandak WM, Dent P, Spiegel S, Shi R, Xu W, Liu X, Bohdan P, Zhang L, Zhou H, Hylemon PB. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology. 2012;55:267–276. doi: 10.1002/hep.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Subbiah MTR, Yunker RL. Cholesterol 7a-hydroxylase of rat liver: an insulin sensitive enzyme. Biochem Biophys Res Commun. 1984;124:896–902. doi: 10.1016/0006-291x(84)91042-8. [DOI] [PubMed] [Google Scholar]
- 185.Subramaniam P, Clayton PT, Portmann BC, Mieli-Vergani G, Hadzic N. Variable clinical spectrum of the most common inborn error of bile acid metabolism–3beta-hydroxy-Delta 5-C27-steroid dehydrogenase deficiency. J Pediatr Gastroenterol Nutr. 2010;50:61–66. doi: 10.1097/MPG.0b013e3181b47b34. [DOI] [PubMed] [Google Scholar]
- 186.Temel RE, Brown JM. Biliary and nonbiliary contributions to reverse cholesterol transport. Curr Opin Lipidol. 2012;23:85–90. doi: 10.1097/MOL.0b013e3283508c21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Thomas C, Auwerx J, Schoonjans K. Bile acids and the membrane bile acid receptor TGR5–connecting nutrition and metabolism. Thyroid. 2008;18:167–174. doi: 10.1089/thy.2007.0255. [DOI] [PubMed] [Google Scholar]
- 188.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, Pellicciari R, Auwerx J, Schoonjans K. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 190.Tiniakos DG, Vos MB, Brunt EM. Nonalcoholic fatty liver disease: Pathology and pathogenesis. Annu Rev Pathol. 2010;5:145–171. doi: 10.1146/annurev-pathol-121808-102132. [DOI] [PubMed] [Google Scholar]
- 191.Tomlinson E, Fu L, John L, Hultgren B, Huang X, Renz M, Stephan JP, Tsai SP, Powell-Braxton L, French D, Stewart TA. Transgenic mice expressing human fibroblast growth factor-19 display increased metabolic rate and decreased adiposity. Endocrinology. 2002;143:1741–1747. doi: 10.1210/endo.143.5.8850. [DOI] [PubMed] [Google Scholar]
- 192.Trauner M, Meier PJ, Boyer JL. Molecular pathogenesis of cholestasis. N Engl J Med. 1998;339:1217–1227. doi: 10.1056/NEJM199810223391707. [DOI] [PubMed] [Google Scholar]
- 193.Tu H, Okamoto AY, Shan B. FXR, a bile acid receptor and biological sensor. Trends Cardiovasc Med. 2000;10:30–35. doi: 10.1016/s1050-1738(00)00043-8. [DOI] [PubMed] [Google Scholar]
- 194.Uppal H, Toma D, Saini SP, Ren S, Jones TJ, Xie W. Combined loss of orphan receptors PXR and CAR heightens sensitivity to toxic bile acids in mice. Hepatology. 2005;41:168–176. doi: 10.1002/hep.20512. [DOI] [PubMed] [Google Scholar]
- 195.Van Mil SW, Milona A, Dixon PH, Mullenbach R, Geenes VL, Chambers J, Shevchuk V, Moore GE, Lammert F, Glantz AG, Mattsson LA, Whittaker J, Parker MG, White R, Williamson C. Functional variants of the central bile acid sensor FXR identified in intrahepatic cholestasis of pregnancy. Gastroenterology. 2007;133:507–516. doi: 10.1053/j.gastro.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 196.Vassileva G, Golovko A, Markowitz L, Abbondanzo SJ, Zeng M, Yang S, Hoos L, Tetzloff G, Levitan D, Murgolo NJ, Keane K, Davis HR, Jr, Hedrick J, Gustafson EL. Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation. Biochem J. 2006;398:423–430. doi: 10.1042/BJ20060537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol. 2009;183:6251–6261. doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- 198.Wagner M, Zollner G, Trauner M. New molecular insights into the mechanisms of cholestasis. J Hepatol. 2009;51:565–580. doi: 10.1016/j.jhep.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 199.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 200.Wang J, Gafvels M, Rudling M, Murphy C, Bjorkhem I, Einarsson C, Eggertsen G. Critical role of cholic acid for development of hypercholesterolemia and gallstones in diabetic mice. Biochem Biophys Res Commun. 2006;342:1382–1388. doi: 10.1016/j.bbrc.2006.02.108. [DOI] [PubMed] [Google Scholar]
- 201.Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, Moore DD. Redundant pathways for negative feedback regulation of bile Acid production. Dev Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 202.Wang L, Liu J, Saha P, Huang J, Chan L, Spiegelman B, Moore DD. The orphan nuclear receptor SHP regulates PGC-1alpha expression and energy production in brown adipocytes. Cell Metab. 2005;2:227–238. doi: 10.1016/j.cmet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 203.Wang YD, Chen WD, Wang M, Yu D, Forman BM, Huang W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology. 2008;48:1632–1643. doi: 10.1002/hep.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-Protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-kappaB) in mice. Hepatology. 2011;54:1421–1432. doi: 10.1002/hep.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wasmuth HE, Glantz A, Keppeler H, Simon E, Bartz C, Rath W, Mattsson LA, Marschall HU, Lammert F. Intrahepatic cholestasis of pregnancy: The severe form is associated with common variants of the hepatobiliary phospholipid transporter ABCB4 gene. Gut. 2007;56:265–270. doi: 10.1136/gut.2006.092742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Watanabe M, Horai Y, Houten SM, Morimoto K, Sugizaki T, Arita E, Mataki C, Sato H, Tanigawara Y, Schoonjans K, Itoh H, Auwerx J. Lowering bile acid pool size with a synthetic farnesoid X receptor (FXR) agonist induces obesity and diabetes through reduced energy expenditure. J Biol Chem. 2011;286:26913–26920. doi: 10.1074/jbc.M111.248203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, Schoonjans K, Bianco AC, Auwerx J. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 208.Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, Auwerx J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Weingartner O, Laufs U, Bohm M, Lutjohann D. An alternative pathway of reverse cholesterol transport: The oxysterol 27-hydroxycholesterol. Atherosclerosis. 2010;209:39–41. doi: 10.1016/j.atherosclerosis.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 210.Wilkinson DS, Ogden SK, Stratton SA, Piechan JL, Nguyen TT, Smulian GA, Barton MC. A direct intersection between p53 and transforming growth factor beta pathways targets chromatin modification and transcription repression of the alpha-fetoprotein gene. Mol Cell Biol. 2005;25:1200–1212. doi: 10.1128/MCB.25.3.1200-1212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wittenburg H, Lyons MA, Li R, Churchill GA, Carey MC, Paigen B. FXR and ABCG5/ABCG8 as determinants of cholesterol gallstone formation from quantitative trait locus mapping in mice. Gastroenterology. 2003;125:868–881. doi: 10.1016/s0016-5085(03)01053-9. [DOI] [PubMed] [Google Scholar]
- 212.Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, Evans RM. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci U S A. 2001;98:3375–3380. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Xie Y, Blanc V, Kerr TA, Kennedy S, Luo J, Newberry EP, Davidson NO. Decreased expression of cholesterol 7{alpha}-hydroxylase and altered bile acid metabolism in apobec-1−/− mice lead to increased gallstone susceptibility. J Biol Chem. 2009;284:16860–16871. doi: 10.1074/jbc.M109.010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci U S A. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yamagata K, Daitoku H, Shimamoto Y, Matsuzaki H, Hirota K, Ishida J, Fukamizu A. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J Biol Chem. 2004;279:23158–23165. doi: 10.1074/jbc.M314322200. [DOI] [PubMed] [Google Scholar]
- 216.Yeh MM, Brunt EM. Pathology of nonalcoholic fatty liver disease. Am J Clin Pathol. 2007;128:837–847. doi: 10.1309/RTPM1PY6YGBL2G2R. [DOI] [PubMed] [Google Scholar]
- 217.Yu C, Wang F, Jin C, Huang X, McKeehan WL. Independent repression of bile acid synthesis and activation of c-Jun N-terminal kinase (JNK) by activated hepatocyte fibroblast growth factor receptor 4 (FGFR4) and bile acids. J Biol Chem. 2005;280:17707–17714. doi: 10.1074/jbc.M411771200. [DOI] [PubMed] [Google Scholar]
- 218.Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, McKeehan WL. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem. 2000;275:15482–15489. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]
- 219.Zhang M, Chiang JY. Transcriptional regulation of the human sterol 12α-hydroxylase gene (CYP8B1): Roles of hepatocyte nuclear factor 4α(HNF4α) in mediating bile acid repression. J Biol Chem. 2001;276:41690–41699. doi: 10.1074/jbc.M105117200. [DOI] [PubMed] [Google Scholar]
- 220.Zhang Y, Breevoort SR, Angdisen J, Fu M, Schmidt DR, Holmstrom SR, Kliewer SA, Mangelsdorf DJ, Schulman IG. Liver LXRalpha expression is crucial for whole body cholesterol homeostasis and reverse cholesterol transport in mice. J Clin Invest. 2012;122:1688–1699. doi: 10.1172/JCI59817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhang Y, Edwards PA. FXR signaling in metabolic disease. FEBS Lett. 2008;582:10–18. doi: 10.1016/j.febslet.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 222.Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A. 2006;103:1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zollner G, Marschall HU, Wagner M, Trauner M. Role of nuclear receptors in the adaptive response to bile acids and cholestasis: Pathogenetic and therapeutic considerations. Mol Pharm. 2006;3:231–251. doi: 10.1021/mp060010s. [DOI] [PubMed] [Google Scholar]
- 224.Zollner G, Trauner M. Nuclear receptors as therapeutic targets in cholestatic liver diseases. Br J Pharmacol. 2009;156:7–27. doi: 10.1111/j.1476-5381.2008.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]