Abstract
Transplant arteriosclerosis, the major manifestation of chronic rejection, develops after allogeneic (Lewis to F344) but not syngeneic (Lewis to Lewis) rat cardiac transplantation. To identify transcriptionally regulated mediators associated with chronic cardiac rejection, we adapted the differential mRNA display technique for in vivo transplant specimens. Gene transcript patterns in four allogeneic hearts showing early signs of chronic rejection were compared with those in two syngeneic hearts exposed to the same surgical procedure but histologically normal. Twelve differentially expressed cDNA bands were identified. We improved the probability of isolating one or more allograft-specific cDNAs from a single display band by first using recovered and reamplified PCR products as probes in RNA blot analysis. cDNA fragments cloned from individual bands were then used in a second RNA blot analysis, which allowed for the correlation of specific mRNA transcripts with cDNA clones. Five cDNA clones produced time-dependent, allograft-specific hybridization. Sequence analysis demonstrated that two of these cDNAs corresponded to unknown genes, whereas the other three represented known genes not previously associated with chronic rejection. The latter group included the macrophage lectin specific for galactose/N-acetylgalactosamine (a cell-surface receptor), the nuclear P1 gene (a homologue of a yeast replication protein), and a ubiquitin-like gene. Our application of the differential display technique allowed the direct identification of potential mediators under in vivo conditions that preserve the environment of the disease process--including infiltrating cell populations critical to the inflammatory response.
Full text
PDF
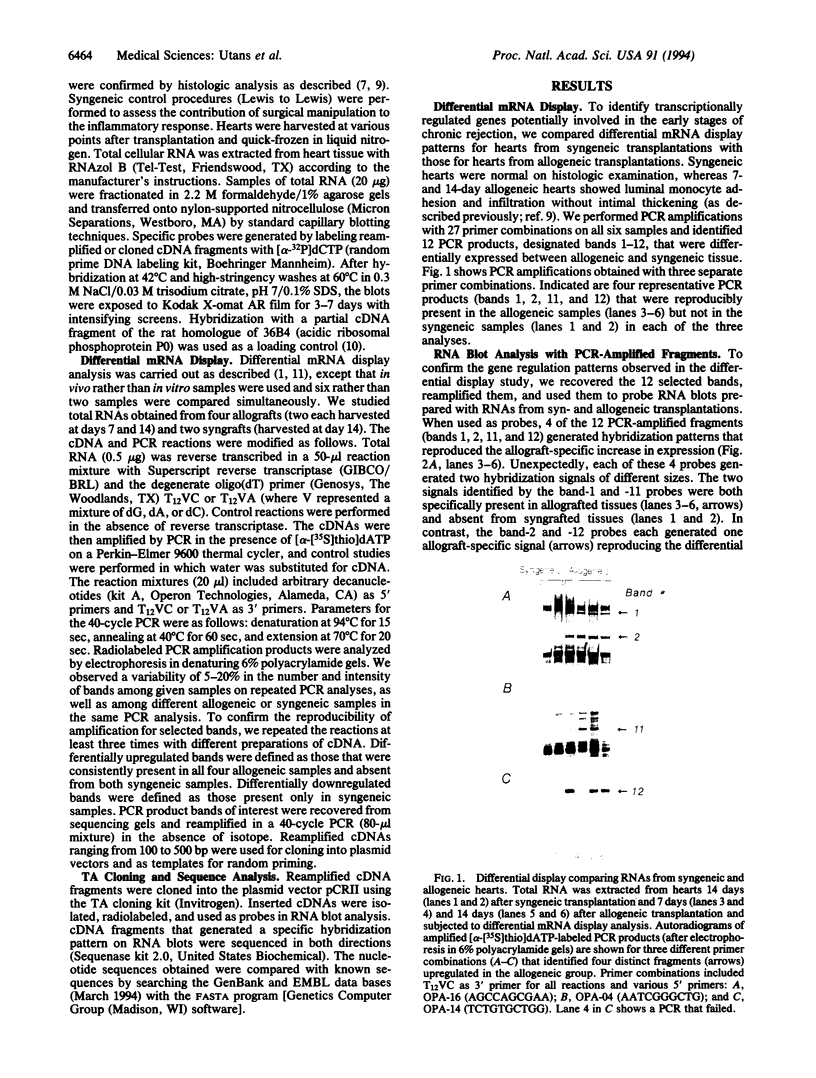
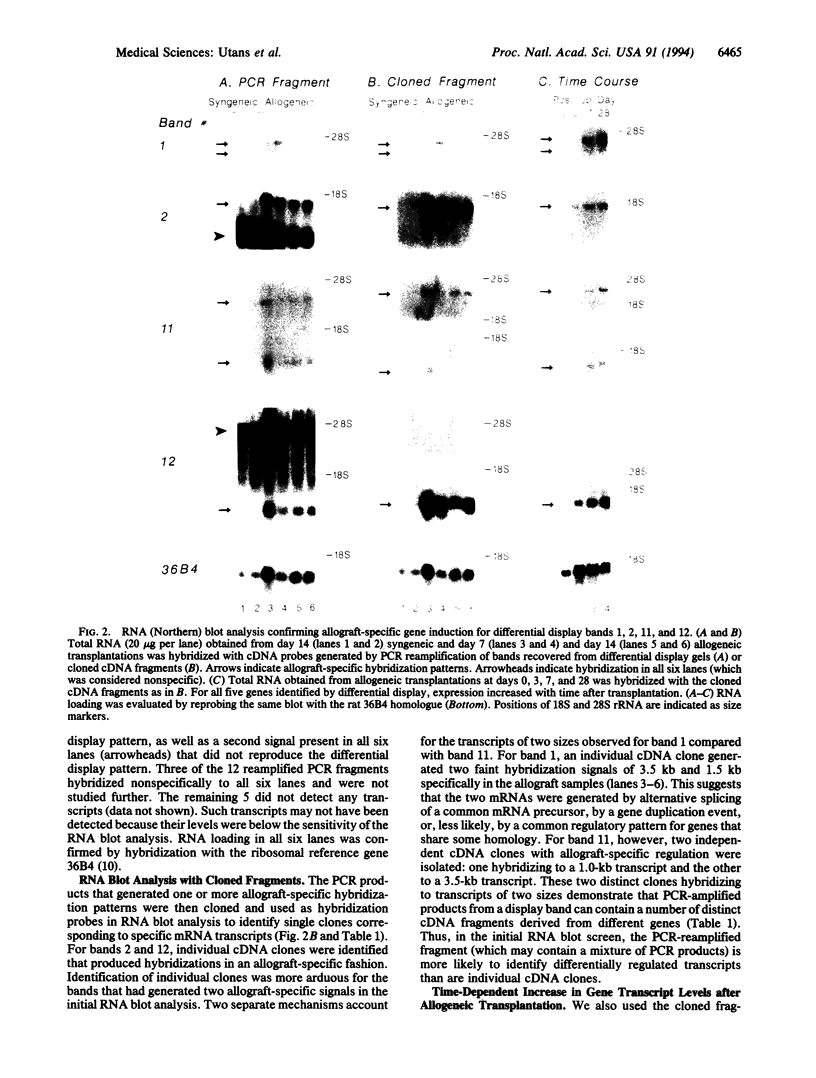


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams D. H., Tilney N. L., Collins J. J., Jr, Karnovsky M. J. Experimental graft arteriosclerosis. I. The Lewis-to-F-344 allograft model. Transplantation. 1992 May;53(5):1115–1119. [PubMed] [Google Scholar]
- Adams D. H., Wyner L. R., Karnovsky M. J. Experimental graft arteriosclerosis. II. Immunocytochemical analysis of lesion development. Transplantation. 1993 Oct;56(4):794–799. doi: 10.1097/00007890-199310000-00004. [DOI] [PubMed] [Google Scholar]
- Cramer D. V., Wu G. D., Chapman F. A., Cajulis E., Wang H. K., Makowka L. Lymphocytic subsets and histopathologic changes associated with the development of heart transplant arteriosclerosis. J Heart Lung Transplant. 1992 May-Jun;11(3 Pt 1):458–466. [PubMed] [Google Scholar]
- Fyfe A., Daly P., Galligan L., Pirc L., Feindel C., Cardella C. Coronary sinus sampling of cytokines after heart transplantation: evidence for macrophage activation and interleukin-4 production within the graft. J Am Coll Cardiol. 1993 Jan;21(1):171–176. doi: 10.1016/0735-1097(93)90733-h. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Ii M., Kurata H., Itoh N., Yamashina I., Kawasaki T. Molecular cloning and sequence analysis of cDNA encoding the macrophage lectin specific for galactose and N-acetylgalactosamine. J Biol Chem. 1990 Jul 5;265(19):11295–11298. [PubMed] [Google Scholar]
- Kumar S., Tomooka Y., Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 1992 Jun 30;185(3):1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- Laborda J. 36B4 cDNA used as an estradiol-independent mRNA control is the cDNA for human acidic ribosomal phosphoprotein PO. Nucleic Acids Res. 1991 Jul 25;19(14):3998–3998. doi: 10.1093/nar/19.14.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Averboukh L., Keyomarsi K., Sager R., Pardee A. B. Differential display and cloning of messenger RNAs from human breast cancer versus mammary epithelial cells. Cancer Res. 1992 Dec 15;52(24):6966–6968. [PubMed] [Google Scholar]
- Liang P., Averboukh L., Pardee A. B. Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res. 1993 Jul 11;21(14):3269–3275. doi: 10.1093/nar/21.14.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Pardee A. B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992 Aug 14;257(5072):967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Liaw L., Schwartz S. M. Comparison of gene expression in bovine aortic endothelium in vivo versus in vitro. Differences in growth regulatory molecules. Arterioscler Thromb. 1993 Jul;13(7):985–993. doi: 10.1161/01.atv.13.7.985. [DOI] [PubMed] [Google Scholar]
- Mayer R. J., Arnold J., László L., Landon M., Lowe J. Ubiquitin in health and disease. Biochim Biophys Acta. 1991 Jun 13;1089(2):141–157. doi: 10.1016/0167-4781(91)90002-4. [DOI] [PubMed] [Google Scholar]
- Oda S., Sato M., Toyoshima S., Osawa T. Purification and characterization of a lectin-like molecule specific for galactose/N-acetyl-galactosamine from tumoricidal macrophages. J Biochem. 1988 Oct;104(4):600–605. doi: 10.1093/oxfordjournals.jbchem.a122518. [DOI] [PubMed] [Google Scholar]
- Russell M. E., Adams D. H., Wyner L. R., Yamashita Y., Halnon N. J., Karnovsky M. J. Early and persistent induction of monocyte chemoattractant protein 1 in rat cardiac allografts. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6086–6090. doi: 10.1073/pnas.90.13.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Anisowicz A., Neveu M., Liang P., Sotiropoulou G. Identification by differential display of alpha 6 integrin as a candidate tumor suppressor gene. FASEB J. 1993 Jul;7(10):964–970. doi: 10.1096/fasebj.7.10.8344495. [DOI] [PubMed] [Google Scholar]
- Sharon N., Lis H. Lectins as cell recognition molecules. Science. 1989 Oct 13;246(4927):227–234. doi: 10.1126/science.2552581. [DOI] [PubMed] [Google Scholar]
- Thömmes P., Fett R., Schray B., Burkhart R., Barnes M., Kennedy C., Brown N. C., Knippers R. Properties of the nuclear P1 protein, a mammalian homologue of the yeast Mcm3 replication protein. Nucleic Acids Res. 1992 Mar 11;20(5):1069–1074. doi: 10.1093/nar/20.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., Chada K., Dalal S. S., Cheng R., Ralph D., McClelland M. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res. 1992 Oct 11;20(19):4965–4970. doi: 10.1093/nar/20.19.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]