SUMMARY
Human rhinovirus (HRV) contains a 7.2 Kb messenger-sense RNA genome which is the template for reproducing progeny viruses after it enters the cytoplasm of a host cell. Reverse genetics refers to the regeneration of progeny viruses from an artificial cDNA copy of the RNA genome of an RNA virus. It has been a powerful molecular genetic tool for studying HRV and other RNA viruses because the artificial DNA stage makes it practical to introduce specific mutations into the viral RNA genome. This chapter uses HRV-16 as the model virus to illustrate the strategy and the methods for constructing and cloning the artificial cDNA copy of a full-length HRV genome, identifying the infectious cDNA clone isolates, and selecting the most vigorous cDNA clone isolate to serve as the standard parental clone for future molecular genetic study of the virus.
Keywords: positive single-stranded RNA virus, full-length infectious cDNA clone, in vitro transcription, transfection
1. INTRODUCTION
HRV is a small single-stranded RNA virus. Its naked positive-sense RNA genome is infectious, that is, it can initiate a complete viral reproduction cycle that produces infectious progeny viruses when it is transfected into a permissive cell (1–3).
After its invention in the early 1970s, recombinant DNA technology/genetic engineering quickly became a powerful tool for studying DNA viruses. Unfortunately, genetic engineering technique is not easily applied to RNA molecules. In hope of being able to use genetic engineering as a routine technique for studying RNA viruses, researchers explored new ways to regenerate RNA viruses from the cDNA copies of their genomes. In 1978, Taniguchi and coworkers reported the regeneration of RNA bacteriophage Qbeta from a plasmid that carried the full-length cDNA copy of the viral RNA genome (4). However, regeneration of RNA virus directly from a cloned cDNA copy of its genome is very inefficient and only works for some RNA viruses. For example, 1 μg of plasmid DNA carrying the cDNA copy of the poliovirus 1 genome produced only 7 infectious units of progeny virus and the same amount of HRV14 plasmid generated no virus when they were transfected into HeLa cells (2, 5). These problems were soon overcome by the use of in vitro T7 RNA transcripts derived from the cloned cDNA copy of a viral RNA genome. In 1984, Paul Ahlquist and coworkers reported the regeneration of the bromo mosaic virus by transfecting the T7 RNA transcripts of the cDNA clone of its genome into the host cells (6). T7 RNA transcripts can be as efficient as the viral RNA genome in regenerating progeny virus and the method works for every positive, single-stranded RNA virus. For example, 1 μg of T7 RNA transcripts from the cDNA clones of poliovirus 1 and HRV14 produced 1.5 million and 2 million infectious units of virus, respectively (2).
Making an infectious full-length cDNA clone of the large linear single-stranded RNA genome of HRV is a complex project that requires the use of multiple methods. Here, we use the HRV-16 laboratory strain as the model virus to illustrate the overall strategy and methods for cloning the cDNA copy of a full-length HRV genome (viral RNA extraction, cDNA synthesis, PCR amplification of the HRV genome, cloning of the PCR fragments, assembly of the PCR fragments into a full-length cDNA clone), identifying the infectious cDNA clones (making in vitro RNA transcripts, transfecting the RNA transcripts into HeLa cells and testing viral infectivity), and selecting one of the infectious clones to serve as the standard parental clone for future molecular genetic work (determining the growth kinetics of their progeny viruses and the infectivity of their RNA transcripts).
2. Materials
2.1 Construction of the full-length cDNA clone
Cloning vector: pMJ3 plasmid DNA (Note 1).
-
Equipment:
Incubator shaker.
37°C incubator for bacterial plates.
Eppendorf thermomixer.
PCR machine.
PCR primers: they are typically ordered from Eurofins-MWG-Operon or Sigma-Proligo. The basic “desalt” grade is sufficient for PCR.
Restriction enzymes: they are typically ordered from New England Biolab (NEB), and come with 10x reaction buffer and BSA.
-
Sterile disposables for molecular cloning
1.5-mL DNA LoBind (LB) eppi tube (Eppendorf 022431021).
0.5-mL DNA LoBind (LB) eppi tube (Eppendorf 022431005).
20- to 200-μl ART barrier pipette tips.
100- to 1000-μl ART barrier pipette tips.
0.2-ml thin-wall polypropylene 8-tube strips for PCR.
Domed 8-cap strips for 0.2-ml PCR tubes.
50-ml Falcon blue cap tubes.
14-ml polypropylene tubes (BD Falcon 352059).
5-ml polystyrene tubes (BD Falcon 352054).
-
Reagents for viral RNA extraction:
Phenol-UltraPure (Invitrogen 15509-037).
Chloroform (Fisher BP1145-1).
2-Propanol (Fisher A416P-4).
Ethanol-Absolute (Fisher BP2818-500).
Glycoblue, 15 μg/μl ((Invitrogen AM9515).
DNase/RNase-free distilled water (Invitrogen 10977-023).
Distilled water (Invitrogen 15230147).
-
7. 5x Nucleic Acids (NA) extraction buffer:
To 220 ml distilled water, add the following ingredients:
125 ml 2M Tris, pH 7.5 (Invitrogen 15567-027).
150 ml 5M NaCl (Invitrogen AM9759).
5 ml 0.5M EDTA (Invitrogen 15575020).
-
Buffered phenol:
To prepare buffered phenol, follow the steps below.
Add 300 ml distilled water into 500 g phenol crystals in a brown bottle.
Incubate the bottle in a 65°C waterbath until all phenol crystals become liquid.
Aliquot 30 ml liquid phenol (lower layer) and 5 ml water into a 50-ml blue-cap tube.
Store the phenol at 4°C (water-saturated phenol is stable at 4°C for years).
-
To buffer phenol from acidic to neutral pH.
Remove the upper water layer from one phenol tube.
Add 15 ml 2x NA extraction buffer and mix well.
Centrifuge (2000 xg, 10 min) to separate the phenol and aqueous phases.
Remove the upper aqueous phase.
-
Phenol/chloroform:
To 20 ml buffered phenol, add 20 ml chloroform, mix well, spin at 2000 xg for 10 min, then remove the upper aqueous phase.
-
Reagents for cDNA synthesis and PCR:
AMV-RTase, 10U/μl and 5x reaction buffer (Promega M510F).
Random hexa-primers (Promega C1181).
RNasin, 40U/μl (Promega N2615).
10 mM dNTP (Promega U151B).
Platinum PCR SuperMix HF (Invitrogen 12532-016).
-
Cloning reagents:
CIP, alkaline phosphatase, calf intestinal (NEB M0290L).
T4 kinase with reaction buffer (NEB ML0201L).
PCR-terminator enzyme kit with reaction buffer (Lucigen 40037-2).
T4 DNA ligase with reaction buffer (NEB M0202L).
10 mM ATP (Invitrogen 18330-019).
-
Reagents for DNA fragment isolation and analysis:
10x TBE (Invitrogen AM9863)
Low melting point (LMP) agarose (Invitrogen 16520100)
Agarose (Invitrogen 16500500)
10x BlueJuice gel loading buffer (Invitrogen 10816-015)
1Kb DNA ladder (Invitrogen 15615-024)
Geneclean kit (MP Biomedicals 111001400)
-
TE buffer:
To 494 ml distilled water, add the following ingredients, and then filter-sterilize.
5 ml 1 M Tris-HCl buffer, pH 8.0 (Invitrogen 15567025).
1 ml 0.5M EDTA (Invitrogen 15575020).
Ammonium acetate solution, 7.5 M (Sigma A2706).
-
Ampicillin stock (100 mg/ml, 1000x) solution (Note 2):
Dissolve 1 g ampicillin sodium salt (Sigma A9518) in 10 ml sterilized water.
-
2x YT bacterial liquid medium for suspension culture:
In 1000 ml distilled water, dissolve the following ingredients, autoclave, cool to 70°C, and then add 1 ml ampicillin stock.
10 g Bacto yeast extract (BD #212750).
16 g Bacto tryptone (BD #211705).
5 g NaCl.
-
2x YT bacterial solid medium for plating:
In 1000 ml distilled water, dissolve the following ingredients, autoclave, cool to 70°C, add 1 ml ampicillin stock and then aliquot 25 ml into each petri dish (Falcon #1001).
10 g Bacto yeast extract (BD #212750).
16 g Bacto tryptone (BD #211705).
5 g NaCl. (d) 20 g Bacto agar (BD #214010).
-
Other supplies for the cloning work:
E. coli DH5 alpha competent cells (Bioline BIO-85026) (Note 3).
SOC medium (Invitrogen #15544-034).
Colony Fast-Screen (Restriction Screen) kit (Epicentre #FS0472H).
Qiaprep Spin Miniprep kit (Qiagen #27106).
2.2. Identification of infectious full-length cDNA clones
H1-HeLa cells (see Chapter X1).
MRC-5 cells (see Chapter X2).
Incubator shaker (New Brunswick G24).
CO2 incubator.
High-speed centrifuge (Sorvall RC6).
−80°C freezer.
-
Sterile disposables for cell culture works:
60-mm dishes (BD Falcon 353002).
48-well plate (BD Falcon 353078).
5-ml sterile pipettes.
10-ml sterile pipettes.
Cell lifter (Corning 3008).
1000-ml 0.20μm filter unit (Nalgene 158-0020).
-
Medium A-NCS
To 500 ml MEM (Invitrogen 11090081), add the following ingredients:
5 ml nonessential amino acids (Invitrogen 1114050).
5 ml L-glutamine (Invitrogen 2503081).
5 ml penicillin, streptomycin (Invitrogen 15140163).
50 ml newborn calf serum (Invitrogen 16010-159).
-
Medium A-FBS:
To 500 ml MEM (Invitrogen 11090081), add the following ingredients:
5 ml nonessential amino acids (Invitrogen 1114050).
5 ml L-glutamine (Invitrogen 2503081).
5 ml penicillin, streptomycin (Invitrogen 15140163).
50 ml fetal bovine serum (Invitrogen 10437-028).
-
Reagents for in vitro transcription:
T7 RNA polymerase, 50 U/μl (Invitrogen 18033019), comes with 5x reaction buffer and 0.1 M DTT.
RNasin, 40U/μl (Promega N2615).
10 mM dNTP (Promega U151B).
-
Reagents for transfection:
Lipofectamine 2000 transfection reagent (Invitrogen 11668-019).
OptiMEM reduced-serum medium (Invitrogen 31985070).
PBSI: PBS with calcium and magnesium (Lonza/BioWhittaker 17-513F).
-
PBSA1 (0.1% BSA):
To 500 ml PBSI, add 6.7 ml 7.5% bovine albumin fraction V (Invitrogen 15260-037)
1M HEPES buffer, pH 7.2–7.5 (Invitrogen 15630-080).
2.3. Identification of the most vigorous infectious cDNA clone isolate for future molecular genetic work
-
HBS (1x):
To make 1 liter HBS, dissolve the following ingredients in 900 ml distilled water. Then adjust pH to 7.2 with 1N NaOH (Sigma S2770), bring the final volume to 1000 ml with distilled water, and sterilize the buffer with a 0.20μm filter unit (Nalgene 158-0020).
5 g HEPES (Sigma H4034).
8g Sodium chloride (Sigma S5886).
0.37 g Potassium chloride (Sigma P5405).
0.1 g Sodium phosphate dibasic, Na2HPO4 (Sigma 71639).
1 g Dextrose (Sigma G7021).
0.15 g Calcium chloride dihydrate, CaCl2.2H2O (Sigma C2536).
DEAE-dextran (Sigma 30461-25G).
3. Methods
3.1. Construction of the full-length cDNA clone
3.1.1. Determination of the complete sequence of your target HRV serotype/strain for PCR primer design
The genome sequences of the prototypes and lab strains of all 100 HRV-A and -B serotypes and 17 HRV-C types have been published (2, 8–19). These sequences are available at GenBank. For example, the GenBank accession number of the genome sequence of HRV-16 lab strain is L24917. If your target HRV has not yet been sequenced, you should be able to determine the sequence promptly using a randomly primed PCR method and high-throughput sequencing (20).
Once the complete genome sequence is known, PCR primers can be made to amplify the viral cDNA for cloning. The PCR fragments can be cloned and then cut and pasted together to form the full-length clone with standard protocol and commercially available reagents.
3.1.2. The plan of the construction of full-length cDNA clone of HRV-16
-
Overall strategy:
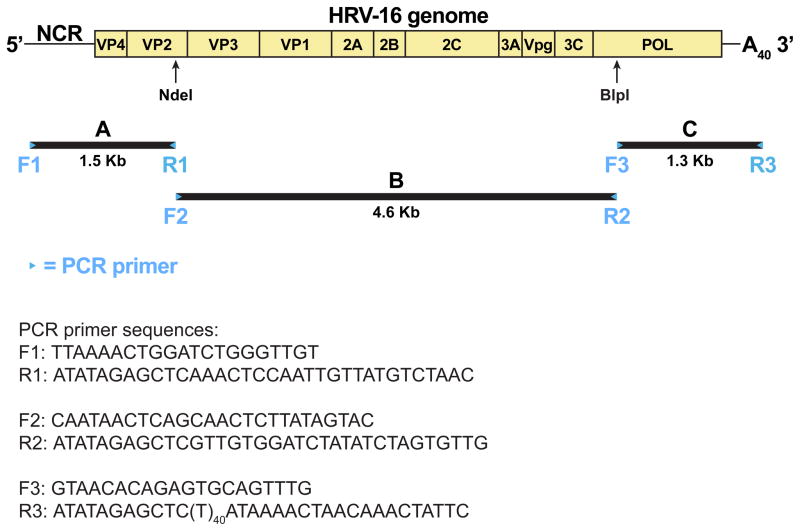
The cDNA of the HRV-16 genome is cloned as three overlapping fragments (A [5′], B [middle] and C [3′] as described below). The three fragments are then assembled into the full-length clone (Figure 1).
The efficiency of PCR amplification of the 5′ and 3′ ends of the viral genome are limited by the sequence of the primers that locate at the genome’s termini. These two primers need to match the exact 5′ and 3′ terminal sequences, which may make suboptimal PCR primers. Therefore, smaller amplicons (about 1.5 Kbps) are designed for the 5′ (A) and 3′ (C) end fragments. Fortunately, better PCR primers can be made for more efficient amplification of the middle fragment because the locations of these primers are flexible. Therefore, a longer amplicon (about 5 Kbps) is designed for the middle (B) fragment.
Fragment A (1498 bp): base 1 to 1487, including the NdeI unique site for full-length cDNA assembly.
Fragment B (4582 bp): base 1407 to 5977, including the NdeI and BlpI sites (Note 4) for full-length cDNA assembly.
Fragment C (1281 bp): base 5888 to 7117 and poly A40 tail, including the BlpI sites for full-length cDNA assembly.
-
PCR primer design:
-
Fragment A primers (F1 and R1):
Primer F1 (TTAAAACTGGATCTGGGTTGT) is composed of the first 21 bases of HRV-16 genome.
Primer R1 (ATATAGAGCTCAAACTCCAATTGTTATGTCTAAC) includes the reverse complement sequence of base# 1465-1487, an artificial SacI site (GAGCTC) for cloning and terminal ATATA for effective SacI-digestion of the PCR fragment.
-
Fragment B primers (F2 and R2):
Primer F2 (CAATAACTCAGCAACTCTTATAGTAC) is composed of base# 1407-1432.
Primer R2 (ATATAGAGCTCGTTGTGGATCTATATCTAGTGTTG) includes the reverse complement sequence of base# 5954-5977, an artificial SacI site (GAGCTC) for cloning and terminal ATATA for effective SacI-digestion of the PCR fragment.
-
Fragment C primers:
Primer F3 (GTAACACAGAGTGCAGTTTG) is composed of base# 5888-5907.
Primer R3 (ATATAGAGCTCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTATAA AACTAACAAACTATTC) includes the reverse complement sequence of the last 20 bases of HRV-16 genome and a poly A tail of 40 bases. It also carries an artificial SacI site (GAGCTC) for cloning and subsequent linearization of the full-length cDNA for in vitro transcription, and terminal ATATA for effective SacI-digestion of the PCR fragment.
-
-
PCR amplification and cloning of DNA fragments A, B and C:
The three DNA fragments are amplified using the same PCR conditions, but with different elongation times (2 minutes for A, 5 minutes for B and 1.5 minutes for C) according to their sizes.
The PCR products of all three fragments are kinase-treated, digested with SacI and then ligated with StuI-SacI double-digested and CIP-treated pMJ3 DNA. The ligation mixture is transformed into competent E. coli. Colonies are screened for the presence of the correct insert by restriction analysis and also by sequencing in the case of fragment A. Four good colonies for each fragment are isolated. Four independent plasmid isolates are needed for each fragment because some of the cDNA fragments may have lethal or disabling mutations that were created by the error-prone viral RNA polymerase and PCR DNA polymerase, and the cloning process.
The clones for each PCR fragment:
Clone A (4673 bp) contains fragment A with complete 5′ HRV-16 sequence fused correctly with the T7 promoter.
Clone B (7757 bp) contains fragment B.
Clone C (4456 bp) contains fragment C with a polyA tail of 40 bases.
-
Assembly of the three fragments into a full-length clone:
For each clone, equal amounts of plasmid DNA of the four good isolates are pooled. For clone A, DNA is digested with NdeI and SacI and followed by CIP-treatment; for clone B, DNA is digested with NdeI and BlpI; and for clone C, DNA is digested with BlpI and SacI.
The desired restriction (R) fragments are purified with LMP agarose gel method. They are:
R fragment A: 4615 bp containing pMJ3 vector and viral bases 1-1436.
R fragment B: 4511 bp containing viral bases 1435-5948.
R fragment C: 1217 bp containing viral bases 5946-7117, poly A40 and the SacI site.
Figure 1.
Strategy for cloning the full-length cDNA of the HRV-16 genome. The genome structure of HRV-16 is shown on the top. The NdeI and BlpI restriction enzyme sites were used for cloning. Viral RNA was extracted from infected cell lysate and converted into cDNA. Three pairs of PCR primers (F1/R1, F2/R2 and F3/R3) were used to generate three overlapping PCR fragments (A, B and C). Each PCR fragment was cloned and the plasmid isolates containing the correct sequences were identified by restriction analysis and sequencing. Then the fragments A, B and C were assembled into a full-length clone with NdeI and BlpI restriction sites.
To assemble the full-length clone, the three R fragments are ligated at a molecular ratio of 1:1:1 and the ligation mixture is transformed into competent E coli. Colonies are screened for the presence of full-length insert by restriction analysis. Twenty independent good colonies should be isolated for the first round of infectivity screening. Five infectious clone isolates will be selected for further characterization. The infectivity of their in vitro transcripts and growth kinetics of their progeny virus are determined for selecting the most vigorous cDNA clone isolate to become the standard parental clone for future molecular genetic work.
3.1.3. Extraction of viral RNA
-
Add the following material into a 1.5-ml LB eppi tube.
320 μl of infected cell lysate of HRV-16 lab strain.
80 μl 5x NA extraction buffer.
2 μl glycoblue (15 μg/μl) (Note 5).
300 μl buffered phenol.
Vortex vigorously in an Eppendorf thermomixer for 2–3 min.
Microfuge for 5 min at room temperature (RT).
Transfer the upper aqueous layer into a 1.5-ml LB eppi tube containing 200 μl phenol/choloform.
Repeat steps 2 and 3.
Transfer the upper aqueous layer into a 1.5-ml LB eppi tube containing 350 μl isopropanol.
Mix well and then incubate at RT for > 1 hr for RNA to precipitate.
Microfuge (10 min, RT) to pellet the RNA precipitant (Note 6).
Remove supernatant and add 0.6 ml 75% ethanol.
Vortex vigorously for 15 sec and then microfuge for 2 min at RT.
Remove all supernatant.
Repeat steps 9 to 11 and then air-dry the RNA pellet.
Dissolve each RNA pellet in 40 μl DNase/RNase-free water.
3.1.4. cDNA synthesis
-
Add the following material to a 0.2-ml PCR tube:
12.25 μl DNase/RNase-free distilled water.
10 μl 5x AMV-RTase buffer.
5 μl 10 mM dNTP.
1.25 μl random primers.
0.75 μl RNasin.
20 μl RNA.
0.75 μl AMV-RTase.
-
Run the following reverse transcription program in a PCR machine:
25°C for 5 min,
42°C for 10 min,
50°C for 20 min,
85°C for 5 min.
Transfer the reaction mixture to 0.5-ml LB eppi tube and store at −20°C.
3.1.5. PCR amplification of PCR fragments A, B and C
-
Add the following material into a 0.2-ml PCR tube:
90 μl Platinum PCR SuperMix HF.
2 μl 25 μM Forward primer.
2 μl 25 μM Reverse primer.
10 μl cDNA.
-
Run the following PCR program:
94°C for 2 min,
94°C for 20 sec,
52°C for 30 sec,
68°C for 4 min (1 min for 1 Kb fragment),
Repeat steps (b) to (d) 27 times,
68°C for 10 min,
4°C forever.
Transfer the reaction mixture to 0.5-ml LB eppi tube and store at −20°C.
3.1.6. Preparation of PCR DNA fragments for cloning
3.1.6.1. Remove proteins and nucleotides from the PCR fragment
-
Add the following reagents into each PCR product (~100 μl) in a 1.5-ml LB eppi tube:
110 μl TE.
130 μl 6 M ammonium acetate.
2 μl Glycoblue (15 μg/μl).
100 μl phenol/chloroform.
Vortex for 10 sec and then microfuge at RT for 3 min.
Transfer the upper aqueous phase into a 1.5-ml LB eppi tube containing 900 μl 100% EtOH.
Mix well, and then incubate at RT for >1 hr.
Microfuge (10 min, RT) to pellet DNA.
Remove supernatant and add 0.6 ml 75% ethonol.
Vortex vigorously for 10 sec and then microfuge for 2 min at RT.
Remove all supernatant.
Repeat steps 6 to 8 and then air-dry the RNA pellet.
Dissolve the DNA pellet in 20 μl DNase/RNase-free water.
3.1.6.2. Kinase treatment and end-repair the PCR DNA fragments (Note 7)
-
Add the following material into 20 μl PCR DNA fragment:
6 μl 5x PCR-terminator buffer.
3 μl dd-water.
1 μl PCR-terminator enzyme (Lucigen 40037-2).
Mix gently and incubate the mixture at RT (25°C) for 15 min (Note 8).
-
Add the following material to each reaction mixture:
180 μl TE.
130 μl 6 M ammonium acetate.
1 μl Glycoblue (15 μg/μl).
100 μl phenol/chloroform.
Perform extraction, ethanol precipitation, micro-centrifugation, washing and air-drying as described in steps 2 to 9 of section 3.1.6.1 above.
Dissolve the DNA pellet in 20 μl DNase/RNase-free water.
3.1.6. 3. Restriction digestion and gel purification of the PCR DNA fragments
-
1
Complete digestion of each PCR DNA fragment with SacI restriction enzyme.
-
2
Prepare a 1% LMP agarose gel in 1x TBE buffer in a UV-transmissible gel box (Note 9).
-
2
Add 10 μl 10x loading dye to 100 μl PCR product and then load the mixture into a well of the gel (Note 10).
-
3
Run and then stain the gel with EtBr in the UV-transmissible gel box (Note 11).
-
4
Visualize the DNA bands with a long-wave UV lamp (Note 12).
-
5
Excise the agarose fragment containing the desired band with a razor blade and spatula into a 1.5-ml LB eppi tube.
-
6
Purify the PCR DNA fragment from agarose with Geneclean kit according to manufacturer’s instruction.
-
7
Elute the DNA fragment with 20 μl DNase/RNase-free water.
3.1.7. Ligation of the PCR DNA fragments with StuI-SacI digested and CIP treated pMJ3 DNA
-
Prepare the following ligation mixture for each PCR DNA fragment.
14.4 μl water.
3 μl 10x ligase buffer.
10 μl purified PCR fragment.
0.1 μl linearized/CIP treated pMJ3 vector DNA (~50 ng) (Note 13).
2.5 μl T4 DNA ligase.
Mix gently and incubate at 16°C in a PCR machine overnight.
3.1.8. Transformation of the ligation mixture into competent E. coli
Thaw a tube of competent E. coli in ice-water.
Chill four labeled 5-ml Falcon tubes in ice-water for 2 min.
Add 40 μl competent E. coli gently into the bottom of each chilled Falcon tube.
Add 5 μl ligation mixture (Note 14) to the competent cells and swirl the pipet tip gently to mix.
Incubate the Falcon tubes in ice-water for 30 min.
Heat shock the transformation mixtures at 41°C for 50 sec.
Incubate the transformation tubes in ice-water for 2 min.
Add 0.4 ml 37°C SOC into each transformation tube.
Incubate the transformation tubes in a 37°C shaker (100 rpm) for 1 hr.
Plate the transformation mixture on a 2x YT agar plate containing 100μg/ml ampicillin.
Incubate plates in a 37°C bacterial incubator overnight.
3.1.9. Screen for the clones containing the correct HRV inserts
Pick 6 to 12 colonies for each PCR fragment.
Inoculate each colony into 0.5 ml 2x YT/ampicillin medium in a well of a 48-well plate.
Incubate plates in a 37°C shaker (100 rpm) overnight.
Transfer 200 μl culture from each well into a 1.5-ml LB eppi tube.
Microfuge the tubes at low-speed to pellet the bacteria and then remove the supernatant.
Perform Fast Restriction Screen (Epicentre) of the plasmids according to the instruction.
For each PCR fragment, identify six colonies containing the correct insert by restriction analysis.
3.1.10. Preparation of plasmid DNA for the assembly of full-length cDNA clone
Grow a 6 ml culture of each clone in a 14-ml tube.
Purify plasmid DNA with a Qiagen spin column according to the instructions.
-
Verify the presence of cloning restriction sites in each isolate by restriction analysis:
Clone A: NdeI and SacI.
Clone B: NdeI and BlpI.
Clone C: BlpI and SacI.
-
Identify 4 good isolates for each clone by restriction pattern analysis and additional analysis for Clone A and C.
Clone A: correct fusion of the T7-promoter and 5′ end of HRV-16 cDNA by sequencing the junction.
Clone C: polyA tail of 40 bases by restriction analysis (Note 15).
For each clone, equal amounts of plasmid DNA of the 4 good isolates is pooled.
-
Double restriction digestion of plasmid DNA in NEB buffer 4:
Clone A: NdeI and SacI.
Clone B: NdeI and BlpI.
Clone C: BlpI and SacI.
Treat digested DNA of Clone A with 50 units CIP at 37°C overnight.
-
Purify the desired restriction (R) fragments with the LMP agarose gel method.
R fragment A: 4615 bp containing pMJ3 vector and viral bases 1-1436.
R fragment B: 4511 bp containing viral bases 1435-5948.
R fragment C: 1217 bp containing viral bases 5946-7117, polyA40.
3.1.11. Construction of full-length cDNA clone
Ligate the three R fragments (at a molecular ratio of 1:1:1) as described in section 3.1.7 above.
Transform the ligation mixture into competent E. coli as described in section 3.1.8 above.
Screen the colonies for the presence of the full-length insert by restriction analysis.
Select 20 independent full-length isolates for infectivity screening (Note 16).
Grow a 12 ml culture of each isolate (in two 14-ml tubes).
Isolate plasmid DNA with a Qiagen miniprep spin column according to the instruction and elute DNA with 200 μl water.
To each DNA sample, add 200 μl of 2x NA extraction buffer and 300 μl of phenol/chloroform.
Perform extraction, ethanol precipitation, micro-centrifugation, washing and air-drying as described in steps 2 to 9 of section 3.1.6.1 above.
Dissolve plasmid DNA of each isolate in 50 μl DNase/RNase-free water and then measure the DNA concentration by spectrophotometer at OD260nm.
3.2. Identification of infectious full-length cDNA clones
Some of the full-length cDNA clones may be noninfectious because they have lethal mutations that were created by the error-prone viral RNA polymerase and PCR DNA polymerase, and the cloning process.
3.2.1. Generation of in vitro transcripts of full-length HRV-16 cDNA
-
Completely linearize 3 μg of plasmid DNA for each full-length isolate with SacI by incubating the following digestion mixture at 37°C for 4 hours.
12.8 μl DNase/RNase-free water.
5 μl 10x NEB buffer.
0.2 μl 100 μg/ml BSA.
30 μl of DNA (0.1 μg/μl).
2 μl Sac I (20 units/ul).
Run 2 μl of each digest in a 0.8% agarose gel to verify the completion of digestion.
Store the digested DNA at −20°C until in vitro transcription.
-
Prepare the following in vitro transcription mixture:
13.5 μl water.
10 μl 5x T7/T3 buffer.
2.5 μl 10 mM NTP.
5 μl 0.1 M DTT.
17 μl DNA digest (~1 μg DNA).
1 μl RNasin (40 U/ul).
1 μl T7 RNA pol (50 U/ul, Invitrogen 18033019).
Incubate the mixture at 37°C for 45 min.
Remove 2 μl of the transcription mixture for agarose gel analysis and store the rest at −80°C until transfection.
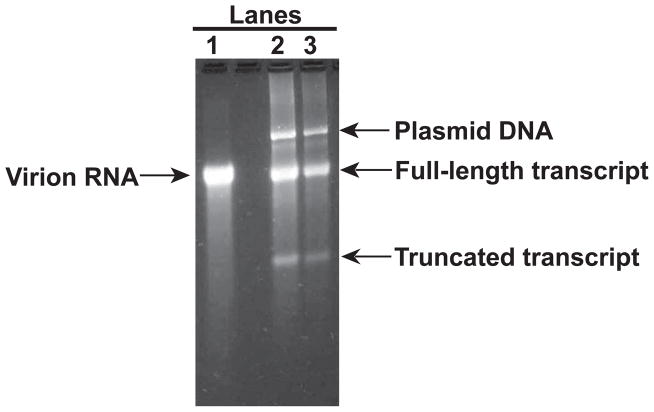
Run 2 and 4 μl RNA transcripts along with 1 μg of virion RNA in a 0.8% agarose/1x TBE gel to check the quality and quantity of the transcripts. The above reaction condition typically yields 0.1 μg of full-length transcript per μl (Figure 2).
Figure 2.
T7 in vitro transcripts of HRV16 full-length cDNA. T7 in vitro transcripts were made as described in the Method section 3.2.1. Virion RNA was extracted from purified virions as described in the Method section 3.1.3. The concentration of virion RNA was measured with OD260nm (1 OD = 40 μg/ml). The RNAs were electrophoresed in a 0.8% agarose/1x TBE gel and the gel was stained with EtBr. Lane 1: 1 μg virion RNA, lane 2: 4 μl T7 transcripts, and Lane 3: 2 μl T7 transcripts.
3.2.2. Transfection of in vitro transcripts into H1-HeLa cell monolayers (Note 17)
Prepare 80–90% confluent HeLa cell monolayers in 60-mm dishes as described in section of 3.2.1 of Chapter X2.
-
Prepare transfection mixtures at RT:
Add 250 μl OptiMEM into the bottom of a 5-ml Falcon tube.
Add 10 μl Lipofectamine 2000 directly into the OptiMEM, and then tap the tube gently to mix.
Incubate the Lipofectamine/OptiMEM mixture at RT for 5 min.
Add 250 μl OptiMEM into the bottom of another 5-ml Falcon tube.
Add 20 μl transcripts (typically about 2 μg) directly into the medium, and then tap the tube gently to mix.
Use a P1000 pipetman to transfer all 270 μl of the transcript/OptiMEM directly into the Lipofectamine/OptiMEM, and then tap the tube gently to mix well.
Incubate the resulting transfection mixture at RT for about 20 min.
-
Prepare the HeLa cell monolayer for transfection at about 15 min after step 2f:
Remove the growth medium.
Wash each monolayer with 4 ml OptiMEM, and then completely remove the media.
Add 1 ml OptiMEM to each monolayer.
Pour the transfection mixture from the 5-ml tube onto the monolayer.
Swirl each dish gently 2–3 times to disperse the transfection mixture.
Wash the remaining transfection mixture from the Falcon tube into the monolayer with 1 ml OptiMEM.
Swirl each dish gently 2–3 times again to disperse the transfection mixture.
Incubate the dishes at 35°C for 3 hr.
Replace transfection medium with 4 ml medium A/FBS.
Continue to incubate the dishes at 35°C for another 20 hr.
3.2.3. Preparation of cell lysate of the transfected HeLa cells
Freeze the dishes at −80°C for >10 min to lyse the cells.
Thaw the cell/medium at RT and then add 40 μl 1M HEPES solution.
Scrape each monolayer with a cell-lifter and then pipet the cell/medium mixture into a 14-ml Falcon tube.
Freeze (−80°C)-thaw cell/medium mixture one more time and then vortex it vigorously for 10 sec.
Remove cell debris by spinning in a high-speed centrifuge (4°C and 10,000 rpm) for 10 min.
Transfer the clarified supernatant into a 5-ml Falcon tube and then store at −80°C until the infectivity assay.
3.2.4. Identification of infectious clones (Note 18)
Prepare one 48-well plate of 90% confluent MRC-5 cells as described in section of 3.1.4 of Chapter X2.
Remove media from each well of MRC-5 cells by aspiration.
Add 10 μl of cell lysate into a well, two wells for each isolate.
Incubate plates for 1 hr at RT to allow virus attachment.
Add 0.5 ml Medium A-FBS.
Incubate the plates at 35°C.
Check and record the appearance of cytopathic effect (CPE) in each well: first check at 3 days after infection and then at days 5 and 7.
Pick five isolates that induce the quickest CPE appearance for further charaterization.
3.3. Identification of the most vigorous infectious cDNA clone isolate for future molecular genetic work
Some of the infectious full-length cDNA clones may produce transcripts with suboptimal infectivity or progeny viruses with sub-optimal growth rates due to a disabling mutation already existing in the original viral genomic RNA or created during the PCR and cloning process. To identify the best infectious cDNA clone isolate for future molecular genetic work, five candidates are screened by measuring the growth kinetics of their progeny viruses and the infectivity of their in vitro transcripts in H1-Hela cells (Note 17).
3.3.1. Production of high titer stock of the infectious isolates for growth kinetics measurement
Prepare 90% confluent HeLa cell monolayers in 100-mm dishes (two dishes for each isolate) by seeding 6×106 cells (in 8 ml medium A-NCS) per dish, and then incubating at 37°C for 12 hrs.
Remove medium and then wash monolayer once with PBSI.
Add 1 ml of virus stock from a transfection dish.
Incubate at RT for 1 hr to allow virus attachment.
Add 4 ml medium A-FBS per dish and then incubate the dishes at 35°C.
Check cell monolayers at 24, 36, 48, 60 and 72 hrs after infection. Transfer the dish to an −80°C freezer when >90% of the cells display CPE. If no CPE appears, transfer the dish to −80°C at 72 hrs after infection.
Thaw the dishes at RT (~10 min).
Add 50 μl 1M HEPES buffer pH7.2.
Scrape cells with a cell lifter and pipette the cell lysate from 2 dishes into a 14-ml Falcon tube for each isolate.
Perform two more freeze (−80°C, 20 min)-thaw (35°C, 10 min) cycles to further break up the cells.
Pellet cell debris in a high-speed centrifuge (4°C and 10,000 rpm) for 10 min.
Aliquot supernatants into 1.5-ml eppi tubes and then store in an −80°C freezer.
-
If >90% of cells in a dish display CPE within 24 hrs, the cell lysate of this dish will likely have a high enough titer (about 5×108 PFU per ml) for growth kinetics measurement.
If it needs >24 hrs to develop severe CPE, pass the virus one more time by repeating steps 1–12.
Titer the virus stock using plaque assay as described in section 3.2.2 of Chapter X2. Also pay attention to the plaque sizes. Typically, faster growers have larger plaques.
3.3.2. Measurement of viral growth kinetics
Set up an HRV suspension culture by infecting 1×108 HeLa cells with 2×109 PFU of virus as described in section 3.2.1 of Chapter X1. Then culture the infected cells in 20 ml of medium B-FBS at 35°C.
Remove 0.5 ml cells from a culture at 2, 3, 4, 5, 6, 7, 8 and 10 hrs after infection (Note 19). The 0.5 ml sample is added into a 1.5-ml eppi tube containing 5 μl 1M HEPES buffer pH7.2.
Break up the infected cells by three freeze (−80°C, 20 min)-thaw (35°C, 10 min) cycles.
Pellet the cell debris in a microfuge (10,000 rpm, 4°C) for 10 min.
Transfer the supernatant into 1.5-ml eppi tubes and then store at −80°C for plaque assay.
Titer by plaque assay. Pick the clone with the highest titer.
3.3.3. Determination of the infectivity of in vitro transcripts
Prepare 90% confluent HeLa cell monolayers in 60-mm dishes as described in section of 3.2.1 of Chapter X2.
-
Prepare transfection solution D with the following recipe:
1 ml HBS.
200 μg DEAE-dextran.
2 μl RNasin (80 units).
Prepare the transfection mixture by mixing 1 ng, 0.1ng or 0.01ng of transcripts into 0.2 ml tranfection solution D and then incubate the mixture at RT for 5 min.
Wash each monolayer once with 4 ml HBS and then completely remove the HBS.
Add 0.2 ml transfection mixture to each dish.
Incubate the dishes at RT for 1 hr.
. Remove the transfection mixture from each dish and then wash the dish once with 4 ml PBS (Note 20).
Add agar and liquid overlay as described in the plaque assay protocol in steps 8 to 11 of section 3.2.2 of Chapter X2.
Incubate the dishes at 35°C for three days for plaque development as described in steps 12 and 13 of section 3.2.2 of Chapter X2.
Count plaques and calculate infectivity per μg of transcripts.
3.4. Complete sequencing of the chosen infectious cDNA clone
The cDNA clone that is selected to be the parent for future molecular genetic work should be completely sequenced to establish its identity. More importantly, the exact sequence is needed for the planning of future cloning and mutagenesis work.
Footnotes
Vector pMJ3 was constructed by Mike Janda at UW-Madison specifically for the construction of the infectious full-length cDNA clones of positive-stranded RNA viruses (2). It has a modified T7 promoter with a unique blunt StuI site at its 3′ end (TAATACGACTCACTATAGGCCT) for the fusion of the 5′ end of the viral cDNA to the last functional base of the T7 promoter. With this design, the T7 in vitro RNA transcripts of the cDNA clones only have 2 non-viral bases (GG) at its 5′ end. These two G’s have minimal, if any, inhibitory effect on the infectivity of the transcripts. (2, 7) pMJ3 has an ampicillin resistant gene and is available on request.
Ampicillin is very sensitive to heat and light. If you have problems with its degradation, you can use carbenicillin (Sigma C3416) instead. Carbenicillin is a more stable analog of ampicillin. We use it at a concentration of 50 μg/ml.
We also have had good results with E. coli JM109 competent cells (Promega L2005) and XL1-Blue Supercompetent cells (Agilent/Stratagene 200236).
A second BlpI restriction site is located in fragment A. But BlpI-digestion of fragment A is not needed for the assembly of the full-length clone.
Glycoblue has two functions. It helps to precipitate the RNA or DNA as a carrier. Its blue color also helps to visualize the RNA or DNA pellets to avoid discarding the pellets by mistake. However, we have found reverse transcriptase activity is inhibited by high concentration of glycoblue, so use glycoblue according to the recipe.
The RNA precipitant will appear as a small light blue pellet.
Invitrogen’s Platinum PCR SuperMix HF uses Taq polymerase. Taq polymerase does not have proofreading function (3′ exonuclease activity). Therefore, the majority of the PCR product has a single-base A extension at the 3′ termini of the DNA fragment. This single-base A blocks the blunt-end ligation of the PCR fragment and the vector. Lucigen’s PCR-terminator enzyme kit has an exonuclease activity that removes these single-base As and also a kinase activity that adds 5′ phosphates to the DNA fragments for ligation. If polymerases with the proofreading function, such as Vent or Pfu, are used for PCR, the PCR fragments will have blunt-ends. These PCR fragments need only kinase-treatment for ligation.
Avoid over-incubation, which will cause deletion of essential viral bases from the ends of the PCR fragments by the exonuclease activity of the PCR-terminator kit.
LMP agarose gel takes longer to solidify than regular agarose gel.
We use a 1Kb DNA ladder in a control lane to estimate the size of the PCR fragments.
The LMP agarose gel is extremely fragile. Handle it very carefully. Keep the gel in the dark during EtBr staining.
Make sure only long-wave UV is used because short-wave UV induces DNA damage, particularly in the presence of EtBr.
A control ligation without PCR DNA fragments is used to gauge how many colonies might have an empty vector.
Some components of the transformation mixture have an inhibitory effect on transformation of E. coli cells. Avoid using too much ligation mixture in the transformation reaction.
A polyA tail of a minimum of 40 A’s is required for optimal infectivity of the viral RNA transcripts (2). However, some of the clone C isolates may have much shorter polyA tails (<30 A’s) due to polymerase error during PCR. Therefore, the isolates with longer polyA tails will be identified for the construction of the full-length cDNA clone. To roughly determine the length of the polyA tail, plasmid DNA of clone C is digested with restriction enzymes that cut very close to the polyA tail regions to generate a short polyA tail fragment (<300 bps). The digest is then analyzed with a 1.5 to 2% agarose gel.
Some of the full-length isolates will produce non-infectious transcripts because they have lethal mutations in the cDNA. Some of the viral genomes used for RNA extraction may already have lethal mutations since the viral RNA polymerase has a high error rate. The lethal mutations could also be introduced into the cDNA during PCR and cloning.
H1-HeLa cells are used for the transfection of viral RNA from HeLa-adapted HRV-A and HRV-B serotypes, and HRV-C viruses. For HRV-A and -B clinical isolates that have not been adapted to HeLa cells, MRC-5 or WI-38 cells should be used for transfection. The HeLa transfection procedure is applicable to MRC-5 and WI-38 cells.
Genomic RNA from HRV-C replicates and produces progeny viruses when it is transfected into HeLa cells (8, 21). However, its progeny viruses cannot infect HeLa, MRC-5 or WI-38 cells which do not have the receptor for HRV-C. Therefore, the infectivity of HRV-C progeny viruses should be tested in differentiated airway epithelial cells in air-liquid interface culture or sinus organ culture as described previously (8, 21, 22).
For HRV-A and –B clinical isolates that have not been adapted to HeLa cells, their progeny viruses should be amplified and tested for growth kinetics in MRC-5 or WI-38 cells. For HRV-C, their progeny viruses should be amplified and tested for growth kinetics in differentiated airway epithelial cells in air-liquid interface culture or sinus organ culture (8, 21, 22). Methods for determining the infectivity of in vitro transcripts of HRV-A and –B clinical isolates and HRV-C are not available.
Inject some CO2 from a CO2 tank into the flask every time after sampling. This will help to maintain a proper pH in the medium for viral growth.
DEAE-dextran inhibits the growth of HRV. The excess DEAE-dextran needs to be thoroughly washed away after transfection (2).
References
- 1.Turner RB, Lee W-M. In: Clinical Virology. Richman DD, Whitley RJ, Hayden FG, editors. ASM Press; Washington, D.C: 2009. pp. 1063–82. [Google Scholar]
- 2.Lee WM, Monroe SS, Rueckert RR. J Virol. 1993;67:2110–22. doi: 10.1128/jvi.67.4.2110-2122.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rueckert R. In: Fields Virology. 3. Fields BN, Knipe DM, Howley PM, et al., editors. Lippincott-Raven Publishers; Philadelphia: 1996. pp. 609–54. [Google Scholar]
- 4.Taniguchi T, Palmieri M, Weissmann C. Nature. 1978;274:223–8. doi: 10.1038/274223a0. [DOI] [PubMed] [Google Scholar]
- 5.Racaniello VR, Baltimore D. Proc Natl Acad Sci U S A. 1981;78:4887–91. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahlquist P, French R, Janda M, Loesch-Fries LS. Proc Natl Acad Sci U S A. 1984;81:7066–70. doi: 10.1073/pnas.81.22.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janda M, French R, Ahlquist P. Virology. 1987;158:259–62. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- 8.Bochkov YA, Palmenberg AC, Lee WM, Rathe JA, Amineva SP, Sun X, Pasic TR, Jarjour NN, Liggett SB, Gern JE. Nat Med. 2011;17:627–32. doi: 10.1038/nm.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang T, Wang W, Bessaud M, Ren P, Sheng J, Yan H, Zhang J, Lin X, Wang Y, Delpeyroux F, Deubel V. PLoS One. 2009;4:e6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmenberg AC, Spiro D, Kuzmickas R, Wang S, Djikeng A, Rathe JA, Fraser-Liggett CM, Liggett SB. Science. 2009;324:55–9. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McErlean P, Shackelton LA, Lambert SB, Nissen MD, Sloots TP, Mackay IM. J Clin Virol. 2007;39:67–75. doi: 10.1016/j.jcv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tapparel C, Junier T, Gerlach D, Cordey S, Van Belle S, Perrin L, Zdobnov EM, Kaiser L. BMC Genomics. 2007;8:224. doi: 10.1186/1471-2164-8-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris JR, Racaniello VR. J Virol. 2005;79:5363–73. doi: 10.1128/JVI.79.9.5363-5373.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kistler AL, Webster DR, Rouskin S, Magrini V, Credle JJ, Schnurr DP, Boushey HA, Mardis ER, Li H, DeRisi JL. Virol J. 2007;4:40. doi: 10.1186/1743-422X-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau SK, Yip CC, Tsoi HW, Lee RA, So LY, Lau YL, Chan KH, Woo PC, Yuen KY. J Clin Microbiol. 2007;45:3655–64. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee WM, Wang W, Rueckert RR. Virus Genes. 1995;9:177–81. doi: 10.1007/BF01702661. [DOI] [PubMed] [Google Scholar]
- 17.Hughes PJ, North C, Jellis CH, Minor PD, Stanway G. J Gen Virol. 1988;69 (Pt 1):49–58. doi: 10.1099/0022-1317-69-1-49. [DOI] [PubMed] [Google Scholar]
- 18.Skern T, Sommergruber W, Blaas D, Gruendler P, Fraundorfer F, Pieler C, Fogy I, Kuechler E. Nucleic Acids Res. 1985;13:2111–26. doi: 10.1093/nar/13.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanway G, Hughes PJ, Mountford RC, Minor PD, Almond JW. Nucleic Acids Res. 1984;12:7859–75. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Djikeng A, Halpin R, Kuzmickas R, Depasse J, Feldblyum J, Sengamalay N, Afonso C, Zhang X, Anderson NG, Ghedin E, Spiro DJ. BMC Genomics. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao W, Bernard K, Patel N, Ulbrandt N, Feng H, Svabek C, Wilson S, Stracener C, Wang K, Suzich J, Blair W, Zhu Q. J Virol. 2012;86:13524–32. doi: 10.1128/JVI.02094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashraf S, Brockman-Schneider R, Bochkov YA, Pasic TR, Gern JE. Virology. 2012;436:143–9. doi: 10.1016/j.virol.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]