Abstract
Although the concept of programmed cell death (PCD) in bacteria has been met with skepticism, a growing body of evidence suggests that it can no longer be ignored. Several recent studies indicate that the phenotypic manifestations of apoptosis, processes associated with ordered cellular disassembly, are conserved in bacteria. In this Opinion article, I propose a model for the coordinated control of potential bacterial PCD effectors, and argue that the processes involved are functionally analogous to eukaryotic PCD systems.
Introduction
Perhaps the biggest impediment to recognizing the existence of bacterial programmed cell death (PCD) is the long-held belief that bacteria live solely as unicellular organisms and as such, cellular suicide seems counterintuitive to our understanding of evolutionary processes and the driving forces of natural selection. After all, what possible benefit could there be to maintaining genes whose functions are to mediate the self-destruction of a free-living individual? Of course, there is no direct advantage to that individual. However, as argued previously1 the species as a whole could benefit if an individual's demise results in an advantage to its siblings. In many ways, multicellular biofilm communities provide an ideal context for understanding bacterial PCD. For example, studies of biofilm development have demonstrated the importance of cell death and lysis for the release of genomic DNA (referred to as eDNA), which becomes incorporated into the biofilm matrix and serves as an adherence molecule2-11. Furthermore, as an interdependent assembly of cells with differentiated structures that serve specialized functions, bacterial biofilms are similar to complex multicellular eukaryotic organisms, in which PCD has a prominent role in development12.
As a starting point, it is important to define what is meant by “PCD”. First and foremost, the phrase “programmed cell death” is reserved for all genetically-encoded processes that lead to cellular suicide. Although the process of apoptosis is most commonly associated with eukaryotic PCD, other PCD mechanisms also exist, including autophagic death and programmed necrosis13,14 (Box 1). All of these mechanisms require metabolic energy and are typically induced in response to physiological or developmental signals. However, apoptosis is the best-characterized mechanism and was first described in 197215. In this original article, the morphological manifestations associated with apoptosis, including chromatin condensation, chromosomal DNA fragmentation, membrane blebbing, cell shrinkage and disassembly of the cell into membrane-enclosed vesicles were described. These processes were later found to be a consequence of the activation of cysteine proteases, known as caspases, which orchestrate apoptosis by inducing a range of cellular activities that result in the dismantling of the cell16. Now it is known that apoptosis proceeds through one of two major signaling pathways: the intrinsic pathway, which involves mitochondrial outer membrane permeabilization (MOMP) and is induced primarily as a result of a cellular insult (for example, DNA damage or oxidative stress)17; and the extrinsic or “death receptor-mediated” pathway, which is typically induced by developmental signals initiated by receptor-ligand interactions at the cell surface and is MOMP independent18. In fact, cell death induced by the extrinsic pathway is largely independent of the mitochondria and is triggered via the direct activation of caspases, which leads to cellular destruction. In contrast, the intrinsic pathway is widely thought to be initiated by dysfunctional mitochondria, resulting from cellular stress (e.g. DNA damage or oxidative stress), which then leads to caspase activation. Thus, both pathways involve caspase activation; the differences lie primarily in how caspase activity is induced.
In this Opinion article, I will go beyond a discussion of why bacterial PCD exists, to focus specifically on the growing number of studies describing PCD-like activities in bacteria, and propose a model pathway to clarify how the processes involved might be coordinated. I argue that the intrinsic pathway to apoptosis in eukaryotic organisms, including some of their molecular control strategies, is conserved in bacteria, where it provides essential functions in response to stress. Furthermore, I speculate that other bacterial processes commonly associated with death, namely toxin-antitoxin (TA) systems and peptidoglycan hydrolase activity, function in analogous roles comparable to autophagic death and programmed necrosis, respectively.
A prelude to death
For several years now, the potential involvement of TA systems in PCD has generated a great deal of interest. These systems comprise a stable toxin and a labile antitoxin that counteracts toxin activity19. They were originally described as plasmid “addiction modules”, by virtue of the fact that the plasmid-encoded toxin components of these systems are more stable (protease resistant) relative to their antitoxin counterparts, thus, causing a bacterial cell to become “addicted” to the plasmid and its capacity to renew the supply of antitoxin. However, a broader role for these systems seemed likely following the observation that most bacterial genomes encode multiple types of TA systems. The best characterized of the chromosomal TA systems is MazEF of Escherichia coli, in which MazF is the toxin and MazE is the antitoxin20, which are induced in response to a variety of stress-inducing compounds20. As a site-specific endoribonuclease, MazF cleaves ACA sequences in mRNA molecules, thereby inhibiting their translation into protein products21. Although this inhibition of translation causes growth arrest, the precise mechanism by which this system leads to death remains to be determined. It has been proposed that the action of MazF results in the selective elimination of transcripts encoding inhibitors of cell death, whereas the transcripts encoding death effectors remain intact22. This model suggests that MazEF (and probably other TA systems) might promote a bacteriostatic and reversible stage in the pathway to death (Fig. 1). Consistent with this are studies indicating that cell death induced by activation of TA systems can be reversed by expression of the antitoxin23. Thus, growth inhibition caused by the action of TA systems may function to reduce metabolic activity, or even fuel the stress response, in an attempt to “ride out the storm” until a fresh supply of nutrients is available, or the offending substance is eliminated and the damage to the cell is repaired.
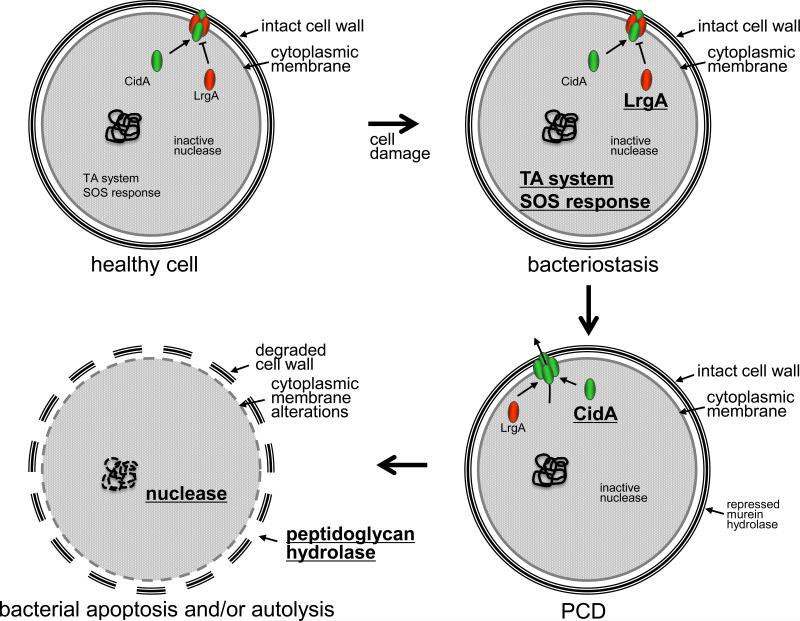
Figure 1. Model pathway of bacterial PCD.
In this model, a healthy cell that has endured some type of damage (e.g. UV-induced mutations) will initially, and likely simultaneously, respond by inducing toxin-antitoxin (TA) systems (e.g. MazEF) and repair mechanisms (the SOS response). The Cid and Lrg proteins are present but are kept inactive at this point because of the inhibitory effect of the Lrg proteins on the Cid PCD effector proteins. The activity of the TA system, by virtue of its inhibitory effect on translation, results in the transition of the cell to a quiescent state (bacteriostasis) that maximizes the energy and resources needed to repair the damage. If the damage is beyond repair, a “point of no return” is reached and the inhibitory effect of the Lrg proteins on Cid is released by an unknown mechanism and cell death ensues. Finally, post-mortem signaling results in the activation of apoptosis-like processes and/or autolysis, including nuclease activation (such as BapE) and DNA fragmentation, membrane alterations, and peptidoglycan hydrolase-mediated cell wall degradation.
Executioners of death
The eukaryotic process
The intrinsic pathway of apoptosis is initiated by a variety of signals, including those associated with stressful conditions, such as DNA damage, oxidative stress, nutrient deprivation and infection. At the heart of this process is the B cell lymphoma 2 (Bcl-2) family of proteins, which are divided into three groups based on the Bcl-2 homology (BH) domains they possess. Two groups are pro-apoptotic: the effectors Bax, Bak, and Bok, which contain BH1, BH2, and BH3 domains; and the “BH3-only proteins”, Bid, Bim, Bad, Bik, Bmf, Bnip3, Hrk, Noxa, and Puma. The third group is comprised of anti-apoptotic proteins (Bcl-2, Bcl-XL, Bcl-W, A1, and Mcl-1) that contain BH1, BH2, BH3, and BH4 domains24. Although the molecular details are incompletely understood, the activities of these proteins are associated with oligomerization of the effector Bcl-2 proteins in the mitochondrial outer membrane, leading to MOMP and the release of cytochrome C, two hallmarks of apoptosis25. Once released, cytochrome C initiates the caspase cascade and the downstream morphological changes associated with cellular disassembly. As might be expected, the entire process is subject to complex regulatory control, primarily via the BH3-only proteins, which function by either blocking the inhibitory effect of the anti-apoptotic Bcl-2 proteins, or by directly stimulating the functions of the pro-apoptotic effector Bcl-2 proteins24.
The bacterial process
It has been hypothesized that CidA and LrgA proteins, which were originally identified in Staphylococcus aureus but are widely conserved in bacteria, are holin-like molecules that possess functions analogous to the pro-apoptotic effector and anti-apoptotic members of the Bcl-2 protein family1,26. Although holins were originally characterized as bacteriophage-encoded proteins that coordinate the death and lysis of infected bacteria, their hypothesized role in bacterial PCD is well established, as well as the similarities to eukaryotic PCD proteins1,26-28. The cidABC and lrgAB operons have been shown to modulate death and lysis via an ill-defined process involving the posttranslational regulation of peptidoglycan hydrolase activity29,30. The cidA gene encodes a putative holin, with a positive effect on death and lysis, while lrgA encodes an antiholin, with an inhibitory effect on these processes (Fig. 1). Furthermore, studies indicate that these proteins oligomerize within the bacterial membrane31, suggesting that they fulfill an analogous role to that of the Bcl-2 proteins as effectors of cell death.
Parallels between eukaryotic and bacterial processes
Upon insertion into the membrane, both holins and Bcl-2 family proteins cause membrane disruption, leading to depolarization of the bacterial cytoplasmic membrane and MOMP (eventually causing depolarization of the inner membrane), respectively, and both are regulated by homologous proteins that oppose their functions. Additionally, both protein families lead to the activation of downstream activities (caspases in eukaryotes and peptidoglycan hydrolases in prokaryotes) that cause cellular disassembly. Remarkably, experimental evidence supporting a relationship between these proteins was obtained by demonstrating that members of the Bcl-2 family are functional in an E. coli system designed to detect holin-like activity (in which cell lysis is dependent on the presence of a functional holin)32. In this study, two pro-apoptotic effectors of apoptosis in eukaryotes, Bax and Bak, were found to oligomerize in the E. coli cytoplasmic membrane and induce cell death and lysis32. In a striking functional correlation, this study also demonstrated that Bax mutants defective in the induction of apoptosis were also unable to induce cell death and lysis in E. coli. Moreover, co-expression of Bax in E. coli with the anti-apoptotic protein, Bcl-XL, resulted in the inhibition of Bax-mediated death and lysis. By contrast, co-expression of Bax with the pro-apoptotic BH3-only protein, tBid (a constitutively active derivative of Bid), stimulated death and lysis. Finally, replacement of the native holin in the lambda bacteriophage genome with the gene encoding Bax resulted in fully functional, infectious bacteriophage particles32. Together, these data provide compelling evidence that key members of the Bcl-2 family function using a holin-like mechanism, and may even point to a bacterial origin for these important apoptosis regulatory proteins.
Despite the paucity of information related to the mechanism of action of the CidA/LrgA and Bcl-2 family of proteins, the experimental evidence that has accumulated supports their function as holins and suggests a common mechanism underlying the control of PCD in bacteria and eukaryotes. It has been proposed that the Cid/Lrg family of proteins was transferred to the eukaryotic cell during the endosymbiotic relationship between a bacterium and a host cell, which led to what we now know as the mitochondria33,34. It is envisioned that both the molecular and physiological processes controlling bacterial autolysis remain key aspects of apoptosis control in mammalian cells, including the physical disruption (or lysis) of the mitochondrial outer membrane, which leads to the induction of the caspase cascade26. Interestingly, recent studies have revealed the presence of an Arabidopsis thaliana lrgAB-like gene involved in the control of plant PCD35,36, suggesting that holin activity may also be the foundation of PCD in plants. The product of this gene, AtLrgB, was shown to localize to the chloroplast and its absence caused interveinal chlorosis and prematurely necrotic leaves, suggesting that is has anti-apoptotic activity. Similar to the mitochondria, the proposed bacterial origin of the chloroplast (which is an important organelle in plant PCD), suggests a common origin, or possibly the convergent evolution of the molecular machinery controlling PCD37. Overall, despite the fact that there is no sequence conservation between the Cid/Lrg proteins (in plants or bacteria) and the Bcl-2 family, the conservation of holin-like activity suggests the existence of a universal mechanism underlying the control of PCD38.
Post-mortem events
As outlined above, one of the best-characterized PCD mechanisms in eukaryotic organisms is apoptosis, which in the intrinsic pathway is initiated by a well-studied process that leads to MOMP and the release of cytochrome C39,40. In the intrinsic pathway of apoptosis (as opposed to the extrinsic pathway) the released cytochrome C has an important role in the next phase, which involves the activation of caspase enzymes that induce cellular disassembly41. This “degradation” phase is essentially a post-mortem process39 and the release of cytochrome C appears to have no greater role than simply to signal that the mitochondria have been compromised (cell death has occurred) and that the post-mortem events should be initiated. The caspases are considered the effectors that stimulate hydrolytic enzyme-mediated degradation, trigger chromosomal DNA fragmentation, chromatin condensation, membrane blebbing, cell shrinkage and disassembly of the cell into membrane-enclosed vesicles16. Several of these processes form the basis of a variety of assays that are useful for the identification of cells undergoing apoptosis (Box 2).
Apoptosis-like processes in bacteria
Although there have been sporadic references to bacterial PCD in the past two decades42-45, evidence for apoptosis-like processes in bacteria has grown in recent years. The first described example of bacterial apoptosis-like processes was that associated with “rapid cell death” (RCD) in Xanthomonas campestris, which is induced by growth in protein-rich medium46,47. During RCD, X. campestris cells exhibit the hallmarks of apoptosis, including positive Annexin V and TUNEL staining (Box 2) and the production of a protein with caspase activity. Growth in the presence of starch inhibited RCD and the apoptosis-like processes that were associated with cell death. In follow-up studies, the protein-rich media-dependent induction of RCD was shown to be a function of the presence of pyruvate-generating amino acids, such as glycine and alanine, in the growth medium48. The authors demonstrated that this caused activation of the TCA cycle, which increased the reducing potential of the cell and stimulated electron transport chain (ETC) activity49. Consistent with this, X. campestris cells undergoing RCD exhibited increased production of ROS, probably as a result of increased ETC activity. Moreover, the addition of ROS scavengers to the growth medium resulted in the inhibition of RCD, suggesting that these molecules are involved in the induction of cell death49.
In 2004, a study reported that the marine cyanobacterium Trichodesmium spp. undergoes “autocatalytic” PCD in response to phosphorus and nitrogen starvation50. Dramatic population reductions are well-documented in eukaryotic planktonic species51, challenging the dogma that PCD is restricted to multicellular organisms. What makes the Trichodesmium spp. study especially interesting is that an apoptosis-like process was observed. Similar to RCD in X. campestris, nutrient deprivation triggered cell death, which was accompanied by DNA degradation, as well as the production of a caspase-like protein (referred to as a metacaspase) that reacted with human caspase-3 polyclonal antisera and was inhibited by a caspase inhibitor. This study also revealed that the dead cells exhibited the degradation of subcellular organelles (including thylakoids, carboxysomes and gas vesicles) and cell shrinkage, again, hallmarks of apoptosis.
In Streptococcus pneumoniae apoptosis-like events were observed after treatment with a human milk lipid-protein complex known as HAMLET52. This complex of alpha-lactalbumin and oleic acid was originally studied because of its ability to induce apoptosis in Jurkat leukemia cells53,54. However, exposure of S. pneumoniae to HAMLET revealed that it is bactericidal, producing a greater than six order of magnitude loss of viability in one hour, which was accompanied by lysis of the cells. In addition, the dead cells displayed several phenotypic traits of apoptosis, including cell shrinkage, DNA condensation, DNA fragmentation and calcium-dependent membrane depolarization. The latter was particularly striking as the kinetics of S. pneumoniae membrane depolarization were similar to that of the mitochondria and both were inhibited by the Ca2+-transport inhibitor, Ruthenium Red, a compound known to inhibit mitochondrial membrane depolarization in eukaryotic cells. Finally, HAMLET was found to induce cell death in other streptococcal species and Haemophilus influenzae, suggesting a conservation of mechanism in both Gram-positive and Gram-negative bacteria.
One of the more recent reports of bacterial apoptosis revealed that Escherichia coli cells treated with DNA damaging agents and antibiotics have several characteristics typically associated with apoptosis, including DNA fragmentation, chromosome condensation, extracellular exposure of phosphatidylserine (PS) and membrane depolarization55. Furthermore, the authors demonstrated that deletion of the recA gene resulted in a reduction in the apoptosis-like processes that occur in response to antibiotic treatment. Of particular interest was the finding that treatment with DNA damaging agents and antibiotics resulted in the production of a protein that interacts with the fluorescent caspase substrate peptide, FITC-Z-VD-FMK. Subsequent analysis identified this protein as RecA and suggested the involvement of the ClpXP protease complex in the regulation of this activity.
It was also recently demonstrated that DNA damage induces an apoptosis-like process in Caulobacter crescentus that included chromosome condensation (referred to as “perturbations in chromosomal organization”), DNA fragmentation and membrane depolarization56. All of these processes were found to be dependent on a previously uncharacterized endonuclease (BapE) that was shown to be specific for supercoiled DNA. Interestingly, BapE was previously found to be a member of the SOS regulon57 and is expressed at a late stage after treatment with the DNA damaging agent, mitomycin C56. These results are consistent with the proposed role of BapE in an apoptosis-like process (rather than promoting repair) and suggest that PCD and cellular disassembly are a last resort in response to DNA damage in bacteria (Fig. 1).
Finally, another recent study proposed that there are two PCD pathways in E. coli, one mediated by the MazEF system and another distinct pathway that exhibits the characteristics of apoptosis58. In wild-type cells, DNA damage induced by the antibiotics nalidixic acid or norfloxacin results in what appears to be MazEF-mediated cell death; however, in a mazEF deletion mutant, cell death is still induced by these agents but by a process described as apoptosis-like death. The latter pathway was induced in a recA- and lexA-dependent manner, consistent with the demonstrated role of RecA in PCD55, as well as the finding that the BapE gene was under SOS control57. Furthermore, the cell death induced by the latter pathway was associated with membrane depolarization and DNA fragmentation, again, processes commonly associated with apoptosis. The authors propose that the induction of MazEF-mediated cell death inhibits “apoptosis-like death”, thus masking this process in wild-type cells. Overall, these recent reports not only demonstrate the existence of apoptosis-like events in bacteria, but also suggest that these processes are linked to the SOS response.
A conservation of PCD pathways
The discovery of apoptosis-like events in bacteria highlights the possibility that widely divergent organisms possess remarkably conserved and previously unrecognized PCD pathways. A closer inspection of these pathways suggests that their regulation might also be conserved (Fig. 2). Consider the role that RecA has in the control of apoptosis-like processes in bacteria, suggesting that PCD is part of the SOS response55,56,58. As proposed several years ago, the induction of bacterial PCD in response to damage might be a last resort option that occurs only if the cost of repair exceeds the cost of building a new cell1. The results of the E. coli55 and C. cresentus56 studies described above support this notion, considering that PCD in these organisms appears to occur only after attempts to repair the damage have failed. Based on these findings, along with the observation that the bacteriostatic effect of the MazEF system is only a temporary metabolic state that is reversible upon addition of nutrients or repair of the cellular damage23, it seems reasonable to propose that the activation of the MazEF system might redirect the energy normally utilized for growth to fuel DNA repair mechanisms. In this model, the MazEF system is envisioned to delay the induction of the primary PCD pathway. Thus, rather than the “apoptosis-like pathway” being a backup system for mazEF-associated functions as was previously suggested58, it is proposed here that the apoptosis-like events observed in a mazEF deletion mutant are actually a manifestation of the primary PCD pathway that is prematurely induced in the absence of MazEF. This model could explain the “point of no return” that has been observed in which the action of MazF can no longer be counteracted by MazE59. Furthermore, it is consistent with the hypothesis that “MazF is a regulator of cell death rather than the cell executioner”22.
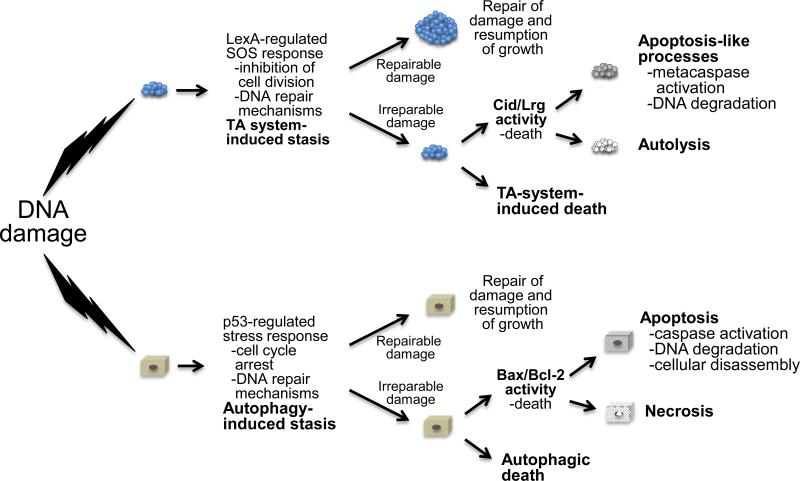
Figure 2. A conservation of responses to cellular stress.
Cell stress, such as that elicited by DNA damaging agents, induces a stress response program that includes DNA repair mechanisms and cell death pathways. This response includes mechanisms to inhibit cell division, directing all available resources to repair the damage. If the levels of damage are minimal, the repair mechanisms will be sufficient to restore the cell to working order. Similar to the role of p53 in assessing the extent of damage to the cell, and then coordinating an appropriate response, it is envisioned that the LexA regulator of the SOS response serves a similar role in coordinating the response to DNA damage in bacteria. In both cases, this includes processes that result in the recycling of cytoplasmic components (TA systems and autophagy) in an attempt to fuel DNA repair. If the damage is irreparable, the repair mechanisms will be overwhelmed and PCD, either Cid-/Lrg-induced death (prokaryotes) or Bcl-2 protein family-induced death (eukaryotes), is induced. Alternatively, TA system-induced or autophagic death can also be induced. Finally, post-mortem events, such as those associated with intrinsic apoptosis and necrosis (eukaryotes), and apoptosis-like processes and autolysis (prokaryotes) are activated.
Interestingly, the response to DNA damage in eukaryotic organisms follows a similar pathway that is coordinated by the multifunctional protein p5360. In the presence of DNA damage, p53 coordinates the induction of cell cycle arrest, DNA repair pathways and cell survival genes. Included in this is the induction of autophagy, in which the damaged cell selectively degrades organelles and proteins that are not required for repair (Box 1). Thus, the initial response to stress (such as that induced by DNA damage) in eukaryotes is to reprogram the cell such that non-essential components are degraded and recycled to redirect energy to fuel the stress response (Fig. 2). Indeed, as proposed for the MazEF system, the induction of autophagy has been shown to delay the onset of apoptosis in an attempt to repair the damage61. However, in addition to its pro-survival functions, autophagy in eukaryotes is also known for its ability to induce cell death62,63 in a process referred to as autophagic death13. Although this “Janus role” of autophagy (named after the Roman god who presided over war and peace) appears to be well established14, the observation that the MazEF system produces both pro-survival and pro-death functions22 suggests that this system may also play a Janus role in determining the fate of the bacterial cell, perhaps even having the eventual capacity to directly induce cell death in the absence of apoptotic-like processes as a consequence of the prolonged disruption of the normal cellular transcriptome (Fig. 2).
In response to insurmountable damage in eukaryotes, p53 shifts the cellular response program, to one that is characterized by decreased expression of cell survival genes and that potentiates apoptosis64. As a sequence-specific transcriptional regulator, p53 has the ability to respond to a wide range of signals, including those associated with DNA damage, metabolism, fecundity and nutrient deprivation65. As with autophagy, this incredibly complex “gene network” has a decisive role in determining cell fate66. Using the eukaryotic system as a model, LexA, the master regulator of recA expression and the SOS response in bacteria, could potentially serve a function analogous to that of p53 (Fig. 2). LexA coordinates the transcription of the SOS regulon and manages a scaled response, which results in either repair of the damage (if it is not excessive) or cell death if the damage is extensive and irreparable56. In this sense, LexA appears also to play a decisive role in determining cell fate in bacteria, presiding over the repair of the damage, but inducing cell death as a last resort if the damage is excessive.
As described above, the CidA and LrgA proteins in prokaryotes and the Bcl-2 family in eukaryotes, appear to utilize similar holin-like strategies to disrupt the integrity of membranes and cause cell death. Subsequent to cell death, post-mortem events in eukaryotes can take multiple forms (Fig. 2). As discussed, the induction of MOMP in the intrinsic form of apoptosis is followed by the orderly, caspase-dependent disassembly of cells. In the absence of caspase activity, a less orderly process known as programmed necrosis can occur (Box 1). Outwardly, programmed necrosis shares features in common with bacterial “autolysis” in that they are both characterized by the dissolution of their outer envelopes and release of their cytoplasmic contents into the extracellular milieu. Similarly, compelling evidence indicates that bacterial autolysis is also a post-mortem event, triggered subsequent to loss of viability29,30,67,68. Clearly, differences between these processes exist as a result of the nature of the bacterial cell wall (which requires degradation by peptidoglycan hydrolases). However, as we continue to learn more about the control of autolysis in bacteria, it would not be surprising if more overlap between these systems in prokaryotic and eukaryotic organisms emerge.
Conclusions
The past decade has seen a great deal of progress in our understanding of cell death mechanisms in bacteria. Once thought to be a passive process that occurs as a consequence of a lethal metabolic malfunction – akin to throwing a wrench into a running motor – we now know that bacterial cell death is an active, genetically-encoded and highly coordinated process associated with pre- and post-mortem events that are strikingly similar to PCD processes in eukaryotic cells. Indeed, considering the functional conservation between the processes in bacteria and eukaryotes, it is proposed here that bacterial cell death, like many other metabolic processes, shares many similarities with the eukaryotic systems, possibly even providing the evolutionary nuts and bolts of these systems. As a newly recognized process, we are only beginning to understand how bacterial PCD operates and the purpose of the model described here is to synthesize the observations made to date into a testable hypothesis that will guide further investigations. Many questions about the regulation and functions of PCD-related proteins in bacteria remain. What is the relationship between TA systems and the Cid/Lrg protein family, and what signals might dictate the “decision” to repair cellular damage or proceed through the cell death pathway? Do TA systems have a pro-survival function comparable to that observed for autophagy in eukaryotic organisms? What is the mechanism utilized by the Cid/Lrg proteins to induce cell death and how is this process regulated? What signals lead to the post-mortem events in dead cells? Finally, how do the developmental processes associated with biofilm formation benefit from PCD? Answers to these questions should not only lead to a greater appreciation for developmental processes, physiological responses to antibiotics and novel drug targets in bacteria, but should also enhance our understanding of these complex processes in eukaryotes.
Box 1. Alternative cell death pathways.
Whereas apoptosis is characterized by very specific and well-studied morphological changes that accompany cell death, programmed necrosis and autophagic death appear to be less orderly forms of PCD. Necrosis has long been considered an uncontrolled process that was brought about primarily by external influences, inducing cell swelling, plasma membrane permeability, the influx of water, and leakage of the cytoplasmic contents into the surrounding environment. The resultant exposure of tissues to these cytoplasmic constituents leads to the characteristic inflammatory response associated with necrosis. However, more recent studies have revealed that this process can be tightly regulated, leading to the designation of “programmed necrosis”. In fact, considerable overlap in the control of programmed necrosis and apoptosis has been demonstrated, although the former is generally considered to be a caspase-independent process. Autophagic death has been proposed to be the acute end stage of autophagy in which a damaged or stressed cell selectively degrades organelles and proteins as a source of energy to fuel costly repair mechanisms needed for cell survival. This process is typified by the formation of “autophagosomes” or double membrane-bound structures that engulf cytoplasmic macromolecules and organelles, such as mitochondria and endoplasmic reticulum, which are destined for recycling. Of note, it has been suggested that autophagy only contributes to cell death by generating energy for apoptosis and necrosis, rather than being a direct effector of cell death. Thus, rather than thinking in terms of autophagic death, it may be more appropriate to consider this form of PCD as “cell death with autophagy”69. For more extensive descriptions of programmed necrosis and autophagic death, the reader is referred to several outstanding reviews of these topics13,14,70,71.
Box 2. Apoptosis assays.
The morphological manifestations of PCD in eukaryotes are referred to as apoptosis and include nuclear and cytoplasmic condensation, as well as the fragmentation of the cell into membrane-bound vesicles (often referred to as “blebbing”). Today, one of the most widely recognized diagnostic indicators of apoptosis is the fragmentation of chromosomal DNA, which is mediated by a specific nuclease, CAD (caspase-activated deoxyribonuclease)72,73. This activity causes the classic DNA laddering that is a common characteristic of apoptotic cells74. The TdT-mediated dUTP-biotin nick end labeling, commonly known as the TUNEL assay, targets these DNA fragments using a specific enzyme called terminal deoxynucleotidyl transferase (TdT) that catalyzes the addition of biotin-labeled dUTPs to the termini of the fragments75. By virtue of the fact that DNA fragmentation is unique to apoptotic cells, this assay specifically identifies cells undergoing apoptosis. Another characteristic of apoptotic cells is the loss of membrane phospholipid asymmetry, resulting in the exposure of phosphotidylserine (which is normally oriented facing the cytoplasm) to the outer face of the cytoplasmic membrane76, which has an important role in the recognition and removal of apoptotic cells by macrophages77. The localization of phosphotidylserine to the outer cell surface provides the basis for the Annexin V stain, which preferentially binds to negatively charged phospholipids such as phosphotidylserine78. Finally, a variety of fluorogenic substrates for the direct detection of activities associated with different caspases induced during apoptosis are available16. These substrates contain amino acid sequences that are specifically recognized and cleaved by different caspases, causing fluorescence as a consequence of the separation of the associated fluorogenic and fluorescence quenching moieties (i.e. Fluorescence Resonance Energy Transfer; FRET). All of these methods allow for the identification of apoptotic cells by epifluorescence microscopy or flow cytometric techniques.
Acknowledgements
The author would like to thank Dr. Xu Luo for lending his expertise on apoptosis, as well as Ms. Kari Nelson for her editorial assistance in the development of this manuscript. Work conducted in my laboratory is supported by grants from the National Institutes of Health (P01-AI83211 and R01-AI038901).
Biography
Kenneth W. Bayles received his training in bacterial genetics with John Iandolo at Kansas State University, Kansas, USA. He has held faculty appointments at the University of Idaho, Idaho, USA and the University of Nebraska Medical Center, Nebraska, USA, where he is a Professor of Microbiology. His laboratory has focused primarily on the role of extracellular DNA as a biofilm matrix molecule, and more recently on the differential control of gene expression during biofilm development.
References
- 1.Bayles KW. Are the molecular strategies that control apoptosis conserved in bacteria? Trends Microbiol. 2003;11:306–311. doi: 10.1016/s0966-842x(03)00144-6. [DOI] [PubMed] [Google Scholar]
- 2.Conover MS, Mishra M, Deora R. Extracellular DNA is essential for maintaining Bordetella biofilm integrity on abiotic surfaces and in the upper respiratory tract of mice. PLoS One. 2011;6:e16861. doi: 10.1371/journal.pone.0016861. doi:10.1371/journal.pone.0016861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harmsen M, Lappann M, Knochel S, Molin S. Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Applied and environmental microbiology. 2010;76:2271–2279. doi: 10.1128/AEM.02361-09. doi:10.1128/AEM.02361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu W, et al. DNA builds and strengthens the extracellular matrix in Myxococcus xanthus biofilms by interacting with exopolysaccharides. PLoS One. 2012;7:e51905. doi: 10.1371/journal.pone.0051905. doi:10.1371/journal.pone.0051905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lappann M, et al. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol Microbiol. 2010;75:1355–1371. doi: 10.1111/j.1365-2958.2010.07054.x. doi:10.1111/j.1365-2958.2010.07054.x. [DOI] [PubMed] [Google Scholar]
- 6.Martins M, et al. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia. 2010;169:323–331. doi: 10.1007/s11046-009-9264-y. doi:10.1007/s11046-009-9264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice KC, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. doi:0610226104 [pii]10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahu PK, Iyer PS, Oak AM, Pardesi KR, Chopade BA. Characterization of eDNA from the clinical strain Acinetobacter baumannii AIIMS 7 and its role in biofilm formation. ScientificWorldJournal. 2012;2012:973436. doi: 10.1100/2012/973436. doi:10.1100/2012/973436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas VC, Thurlow LR, Boyle D, Hancock LE. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. Journal of bacteriology. 2008;190:5690–5698. doi: 10.1128/JB.00314-08. doi:10.1128/JB.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vilain S, Pretorius JM, Theron J, Brozel VS. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Applied and environmental microbiology. 2009;75:2861–2868. doi: 10.1128/AEM.01317-08. doi:10.1128/AEM.01317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 12.Miura M. Apoptotic and nonapoptotic caspase functions in animal development. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a008664. doi:10.1101/cshperspect.a008664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boujrad H, Gubkina O, Robert N, Krantic S, Susin SA. AIF-mediated programmed necrosis: a highly regulated way to die. Cell Cycle. 2007;6:2612–2619. doi: 10.4161/cc.6.21.4842. [DOI] [PubMed] [Google Scholar]
- 14.Ouyang L, et al. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–498. doi: 10.1111/j.1365-2184.2012.00845.x. doi:10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerr JFR, Wylie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 17.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. doi:10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 18.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 19.Rawlings DE. Proteic toxin-antitoxin, bacterial plasmid addiction systems and their evolution with special reference to the pas system of pTF-FC2. FEMS Microbiol Lett. 1999;176:269–277. doi: 10.1111/j.1574-6968.1999.tb13672.x. [DOI] [PubMed] [Google Scholar]
- 20.Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. doi:10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 22.Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genet. 2009;5:e1000390. doi: 10.1371/journal.pgen.1000390. doi:10.1371/journal.pgen.1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen K, Christensen SK, Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 24.Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. doi:10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 25.Walensky LD, Gavathiotis E. BAX unleashed: the biochemical transformation of an inactive cytosolic monomer into a toxic mitochondrial pore. Trends Biochem Sci. 2011;36:642–652. doi: 10.1016/j.tibs.2011.08.009. doi:10.1016/j.tibs.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayles KW. The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 2007;5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 27.Bayles KW. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 2000;8:274–278. doi: 10.1016/s0966-842x(00)01762-5. [DOI] [PubMed] [Google Scholar]
- 28.Rice KC, Bayles KW. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 2008;72:85–109. doi: 10.1128/MMBR.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groicher KH, Firek BA, Fujimoto DF, Bayles KW. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 2000;182:1794–1801. doi: 10.1128/jb.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice KC, et al. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. Journal of bacteriology. 2003;185:2635–2643. doi: 10.1128/JB.185.8.2635-2643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranjit DK, Endres JL, Bayles KW. Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J Bacteriol. 2011;193:2468–2476. doi: 10.1128/JB.01545-10. doi:JB.01545-10 [pii]10.1128/JB.01545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang X, et al. Active Bax and Bak are functional holins. Genes Dev. 2011;25:2278–2290. doi: 10.1101/gad.171645.111. doi:10.1101/gad.171645.111 gad.171645.111 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. doi:10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 34.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. doi:10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, et al. A chloroplast envelope membrane protein containing a putative LrgB domain related to the control of bacterial death and lysis is required for chloroplast development in Arabidopsis thaliana. New Phytol. 2012;193:81–95. doi: 10.1111/j.1469-8137.2011.03867.x. doi:10.1111/j.1469-8137.2011.03867.x. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi M, et al. Loss of the plastid envelope protein AtLrgB causes spontaneous chlorotic cell death in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:125–134. doi: 10.1093/pcp/pcr180. doi:pcr180 [pii]10.1093/pcp/pcr180. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Bayles KW. Programmed cell death in plants: lessons from bacteria? Trends Plant Sci. 2013;18:133–139. doi: 10.1016/j.tplants.2012.09.004. doi:10.1016/j.tplants.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Bayles K. Programmed cell death in plants: lessons from bacteria? Trends in Plant Sciences. 2012 doi: 10.1016/j.tplants.2012.09.004. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 40.Ow YP, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol. 2008;9:532–542. doi: 10.1038/nrm2434. doi:10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 41.Li P, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 42.Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3',5'-bispyrophosphate: a model for programmed bacterial cell death. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen RB, Gerdes K. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 44.Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 45.Yarmolinsky MB. Programmed cell death in bacterial populations. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]
- 46.Gautam S, Sharma A. Involvement of caspase-3-like protein in rapid cell death of Xanthomonas. Mol. Microbiol. 2002;44:393–401. doi: 10.1046/j.1365-2958.2002.02837.x. [DOI] [PubMed] [Google Scholar]
- 47.Gautam S, Sharma A. Rapid cell death in Xanthomonas campestris pv. glycines. J. Gen. Appl. Microbiol. 2002;48:67–76. doi: 10.2323/jgam.48.67. [DOI] [PubMed] [Google Scholar]
- 48.Raju KK, Gautam S, Sharma A. Molecules involved in the modulation of rapid cell death in Xanthomonas. Journal of bacteriology. 2006;188:5408–5416. doi: 10.1128/JB.00056-06. doi:10.1128/JB.00056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wadhawan S, Gautam S, Sharma A. Metabolic stress-induced programmed cell death in Xanthomonas. FEMS Microbiol Lett. 2010;312:176–183. doi: 10.1111/j.1574-6968.2010.02114.x. doi:10.1111/j.1574-6968.2010.02114.x. [DOI] [PubMed] [Google Scholar]
- 50.Berman-Frank I, Bidle KD, Haramaty L, Falkowsky PG. The demise of the marine cyanobacterium, Trichodesmium spp., via an autocatalyzed cell death pathway. Limnol. Oceanogr. 2004;49:997–1005. [Google Scholar]
- 51.Bidle KD, Falkowski PG. Cell death in planktonic, photosynthetic microorganisms. Nat. Rev. Microbiol. 2004;2:643–655. doi: 10.1038/nrmicro956. [DOI] [PubMed] [Google Scholar]
- 52.Hakansson AP, Roche-Hakansson H, Mossberg AK, Svanborg C. Apoptosis-like death in bacteria induced by HAMLET, a human milk lipid-protein complex. PLoS One. 2011;6:e17717. doi: 10.1371/journal.pone.0017717. doi:10.1371/journal.pone.0017717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hakansson A, Zhivotovsky B, Orrenius S, Sabharwal H, Svanborg C. Apoptosis induced by a human milk protein. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:8064–8068. doi: 10.1073/pnas.92.17.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Svensson M, Hakansson A, Mossberg AK, Linse S, Svanborg C. Conversion of alpha-lactalbumin to a protein inducing apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4221–4226. doi: 10.1073/pnas.97.8.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dwyer DJ, Camacho DM, Kohanski MA, Callura JM, Collins JJ. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol Cell. 2012;46:561–572. doi: 10.1016/j.molcel.2012.04.027. doi:10.1016/j.molcel.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bos J, Yakhnina AA, Gitai Z. BapE DNA endonuclease induces an apoptotic-like response to DNA damage in Caulobacter. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18096–18101. doi: 10.1073/pnas.1213332109. doi:10.1073/pnas.1213332109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.da Rocha RP, Paquola AC, Marques Mdo V, Menck CF, Galhardo RS. Characterization of the SOS regulon of Caulobacter crescentus. Journal of bacteriology. 2008;190:1209–1218. doi: 10.1128/JB.01419-07. doi:10.1128/JB.01419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erental A, Sharon I, Engelberg-Kulka H. Two programmed cell death systems in Escherichia coli: an apoptotic-like death is inhibited by the mazEF-mediated death pathway. PLoS Biol. 2012;10:e1001281. doi: 10.1371/journal.pbio.1001281. doi:10.1371/journal.pbio.1001281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amitai S, Yassin Y, Engelberg-Kulka H. MazF-mediated cell death in Escherichia coli: a point of no return. Journal of bacteriology. 2004;186:8295–8300. doi: 10.1128/JB.186.24.8295-8300.2004. doi:10.1128/JB.186.24.8295-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vousden KH. Outcomes of p53 activation--spoilt for choice. J Cell Sci. 2006;119:5015–5020. doi: 10.1242/jcs.03293. doi:10.1242/jcs.03293. [DOI] [PubMed] [Google Scholar]
- 61.Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. doi:10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 62.Gozuacik D, Kimchi A. Autophagy and cell death. Curr Top Dev Biol. 2007;78:217–245. doi: 10.1016/S0070-2153(06)78006-1. doi:10.1016/S0070-2153(06)78006-1. [DOI] [PubMed] [Google Scholar]
- 63.Yu L, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. doi:10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 64.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. doi:10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 65.Millau JF, Bastien N, Drouin R. P53 transcriptional activities: a general overview and some thoughts. Mutat Res. 2009;681:118–133. doi: 10.1016/j.mrrev.2008.06.002. doi:10.1016/j.mrrev.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 66.Sullivan KD, Gallant-Behm CL, Henry RE, Fraikin JL, Espinosa JM. The p53 circuit board. Biochimica et biophysica acta. 2012;1825:229–244. doi: 10.1016/j.bbcan.2012.01.004. doi:10.1016/j.bbcan.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garrett JM, Young R. Lethal action of bacteriophage lambda S gene. J. Virol. 1982;44:886–892. doi: 10.1128/jvi.44.3.886-892.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moreillon P, Markiewicz Z, Nachman S, Tomasz A. Two bactericidal targets for penicillin in pneumococci: autolysis-dependent and autolysis-independent killing mechanisms. Antimicrob. Agents Chemother. 1990;34:33–39. doi: 10.1128/aac.34.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. doi:10.1038/nrm2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rodriguez-Rocha H, Garcia-Garcia A, Panayiotidis MI, Franco R. DNA damage and autophagy. Mutat Res. 2011;711:158–166. doi: 10.1016/j.mrfmmm.2011.03.007. doi:10.1016/j.mrfmmm.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ryan KM. p53 and autophagy in cancer: guardian of the genome meets guardian of the proteome. Eur J Cancer. 2011;47:44–50. doi: 10.1016/j.ejca.2010.10.020. doi:10.1016/j.ejca.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 72.Enari M, et al. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. doi:10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 73.Liu X, Zou H, Slaughter C, Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 74.Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 75.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. The Journal of cell biology. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Op den Kamp JAF. Lipid asymmetry in membranes. Ann. Rev. Biochem. 1979;48:47–71. doi: 10.1146/annurev.bi.48.070179.000403. [DOI] [PubMed] [Google Scholar]
- 77.Fadok VA, et al. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. Journal of immunology. 1992;148:2207–2216. [PubMed] [Google Scholar]
- 78.Koopman G, et al. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood. 1994;84:1415–1420. [PubMed] [Google Scholar]