Abstract
Transcription factors (TFs) play an important role in the regulation of plant growth and development. The study of the structure and function of TFs represents a research frontier in plant molecular biology. The findings of these studies will provide significant information regarding genetic improvement traits in crops. Currently, a large number of TFs have been cloned, and their function has been verified. However, relatively few studies that genetically map TFs in cotton are available. To genetically map TFs in cotton in this study, specific primers were designed for TF genes that were published in the Plant Transcription Factor Database. A total of 977 TF primers were obtained, and 31 TF polymorphic loci were mapped on 15 cotton chromosomes. These polymorphic loci were clearly preferentially distributed on chromosomes 5, 11, 19 and 20; and TFs from the same family mapped to homologous cotton chromosomes. In-silico mapping verified that many mapped TFs were mapped on their corresponding chromosomes or their homologous chromosomes’ corresponding chromosomes in the diploid genomes. QTL mapping for fiber quality revealed that TF-Ghi005602-2 mapped on Chr19 was associated with fiber length. Eighty-five TF genes were selected for RT-PCR analysis, and 4 TFs were selected for qRT-PCR analysis, revealing unique expression patterns across different stages of fiber development between the mapping parents. Our data offer an overview of the chromosomal distribution of TFs in cotton, and the comparative expression analysis between Gossypium hirsutum and G. barbadense provides a rough understanding of the regulation of TFs during cotton fiber development.
Introduction
Transcription factors (TFs) are a class of the most widely studied and important trans-acting factors, regulating gene expression at the transcriptional level. A typical TF from a higher-order plant typically contains a DNA-binding domain, a transcription regulation domain, an oligomerization site, and a nuclear localization domain [1]. TFs play an important role in the regulation of plant growth and development, organ morphogenesis, secondary metabolism, hormonal signal transduction, and plant responses to various environmental stresses [2–7]. Therefore, it is likely that investigating and exploiting the function of TFs could provide an alternative approach for improving cotton fiber quality and production.
To date, a large number of TFs have been reported, and their functions have been successively investigated and verified in cotton. Among them, the majority of research on TFs has focused on the ethylene-response factor (ERF) family, myeloblastosis (MYB) family, WKRY family, and basic helix-loop-helix (bHLH) family. A large amount of research has confirmed that the MYB family plays a functional role in cotton fiber differentiation and development [8–13]. ERF genes are induced by biotic and abiotic stresses in cotton and are involved in regulating plant disease resistance pathways [14,15]. The bHLH family plays an important role in regulating plant secondary metabolism [16,17] and morphogenesis [6,7,18]. Plant WKRY TFs have important functions in the transcriptional regulation of a variety of biological processes that are related to growth and development [19–21], various environmental stimuli [22–24], and disease resistance pathways [25–27].
As of April 2012, approximately 1,116 TFs from 50 families were annotated in Gossypium hirsutum (http://planttfdb.cbi.edu.cn/index.php?sp=Ghi). However, previous research has focused mainly on identifying and verifying the biological functions of these TFs rather than their genomic distributions. In this study, the TFs were mapped to reveal their genomic distribution in cotton using specific primers designed based on transcription factor sequences available in the Plant Transcription Factor Database (http://planttfdb.cbi.edu.cn/). We also confirmed their chromosomal location by in-silico mapping the experimental mapped TFs in two sequenced diploid genomes, A2 genome of G. arboretum and D5 genome of G. raimondii. QTL mapping was conducted to identify TFs related to fiber quality. RT-PCR and qRT-PCR analysis were also conducted to detect differences in expression during fiber development between G. hirsutum and G. barbadense in selected TFs from each family.
Materials and Methods
Plant materials
Polymorphisms of the designed TF primers were detected using G. hirsutum cv. Emian22 and G. barbadense acc. 3–79, which are the parents of the BC1 mapping population [(Emian22 × 3–79) × Emian22] [28,29], using single-strand conformation polymorphism (SSCP) with minor modifications [30]. The BC1 population, which consisted of 141 plants, was used as the mapping population for all of the polymorphic TF markers.
Marker development
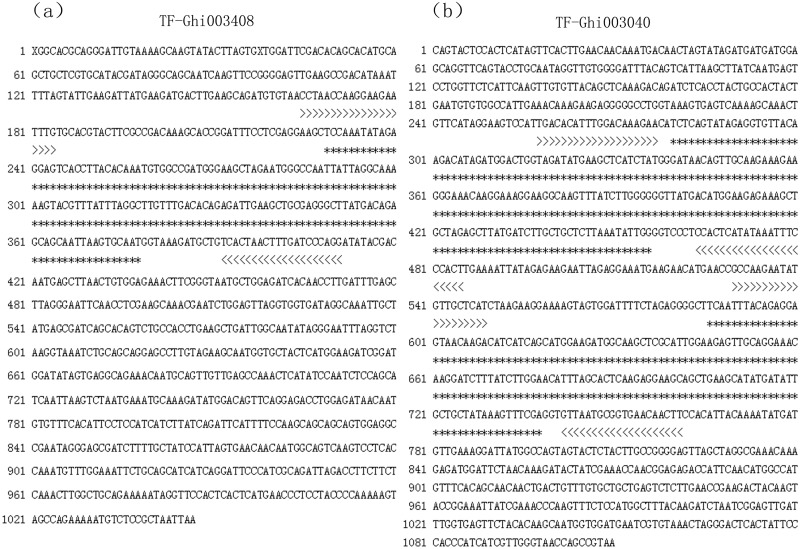
Cotton TFs were obtained from the Plant Transcription Factor Database V2.0 (http://planttfdb.cbi.edu.cn/), which contains 1,116 G. hirsutum TFs classified into 50 families (Table 1). The primers were designed based on the sequences surrounding specific motifs of the TF genes using Primer 3.0 (http://frodo.wi.mit.edu/primer3/). For those TFs with multiple motifs, if the sequence interval between motifs was too long, individual primers were designed for each motif (examples are presented in Fig 1). The criteria for the primer design were as follows: a primer length of 18 to 25 bp (20 bp is optimal), a GC content of 35 to 70% (50% is optimal), an annealing temperature of 50 to 65°C (55°C is optimal), and a PCR product size ranging from 100 to 1,000 bp. The primers were named “TF-Ghi ××××××”. For TFs with multiple motifs, the primers were named “TF-Ghi ××××××-1” and “TF-Ghi ××××××-2”.
Table 1. Distribution of TFs in various families.
| Family | No. | Family | No. | Family | No. | Family | No. | Family | No. |
|---|---|---|---|---|---|---|---|---|---|
| AP2 | 8 | ARF | 7 | ARR-B | 2 | B3 | 23 | BBR/BPC | 6 |
| BES1 | 7 | C2H2 | 56 | C3H | 44 | CAMTA | 2 | CO-like | 4 |
| CPP | 2 | DBB | 12 | Dof | 22 | E2F/DP | 3 | EIL | 10 |
| ERF | 112 | FAR1 | 7 | G2-like | 31 | GATA | 24 | GRAS | 46 |
| GRF | 13 | GeBP | 5 | HB-other | 15 | HD-ZIP | 40 | HSF | 17 |
| LBD | 14 | M-type | 2 | MIKC | 21 | MYB | 75 | MYB_related | 59 |
| NAC | 50 | NF-YA | 5 | NF-YB | 10 | NF-YC | 11 | Nin-like | 7 |
| RAV | 1 | S1Fa-like | 2 | SBP | 13 | SRS | 4 | TALE | 14 |
| TCP | 23 | Trihelix | 38 | VOZ | 5 | WOX | 6 | WRKY | 55 |
| Whirly | 1 | YABBY | 4 | ZF-HD | 15 | bHLH | 92 | bZIP | 71 |
Fig 1. Primer design strategies for TFs.
(a) TFs with one motif (GenBank acc No. Ghi003408); (b) TFs with two motifs (GenBank acc No. Ghi003040). *: Target sequence; >>>>>: Primer region.
PCR amplification and electrophoresis
Polymerase chain reaction (PCR) of the TF markers was performed in 10 μL of solution containing 25 ng DNA template, 1 × buffer, 2.0 mmol L-1 MgCl2, 0.25 mmol L-1 dNTPs, 0.16 μmol L-1 forward primer, 0.16 μmol L-1 reverse primer, 0.8 units Taq DNA polymerase, and ddH2O to a final volume of 10 μl. PCR was performed using the following parameters: 95°C for 5 min; 34 cycles of 94°C for 50 sec, 56°C for 45 sec, and 72°C for 60 sec; and a final extension of 5 min at 72°C. The PCR products were then separated in an 8% non-denaturing polyacrylamide gel at a constant voltage of 15 W for approximately 4 h at room temperature. After electrophoresis, all of the DNA fragments were visualized by silver staining.
Experimental mapping and identification of TFs related to fiber quality
The polymorphic loci were integrated into the interspecific BC1 genetic linkage map [29] using JoinMap V3.0 [31]. The logarithm of odds (LOD) threshold was 5.0. Map distances were measured in centi-Morgans (cM), which were calculated using the Kosambi mapping function [32]. The linkage map was generated using MapChart V2.2 software [33]. QTL mapping of TFs related to fiber quality was performed using the genetic linkage map integrated with the TF markers. The phenotype data of fiber quality and QTL mapping methods were as same as Li et al. [30].
In-silico mapping TFs in two diploid genomes
To verify our genetic mapping results, we blasted the mapped TF sequences against the diploid A2 genome of G. arboretum and D5 genome of G. raimondii (http://www.phytozome.net/cotton.php) with an E-value cut-off 1e-10. Then, the best match for each TF sequence was retained; one TF was only mapped to A2 or D5 genome rather than both.
RT-PCR and qRT-PCR analysis
To evaluate differences in TF expression between G. hirsutum and G. barbadense, RNAs were extracted from developing fibers at 0 days post-anthesis (DPA), 5 DPA, 10 DPA, 15 DPA, 20 DPA, and 25 DPA, RNAs (4 μg) were reverse-transcribed into cDNA using M-MLV-RT Reverse Transcriptase (Invitrogen). For RT-PCR analysis, PCR was performed in 15 μL of solution containing 25 ng cDNA template, 1.7 x buffer, 3.2 mmol L-1 MgCl2, 0.42 mmol L-1 dNTPs, 0.27 μmol L-1 forward primer, 0.27 μmol L-1 reverse primer, 1.3 units Taq DNA polymerase, and ddH2O to a final volume of 15 μl. The PCR program was as follows: denaturation at 95°C for 5 min; 35 cycles of 94°C (50 s), 58°C (45 s), and 72°C (60 s); and a final extension step of 5 min at 72°C. The qRT-PCR analyses were performed according to the methods described by Munis et al. [34] with minor modifications. Ubiquitin (GenBank acc No.: DQ116441; forward primer, 5’GAAGGCATTCCACCTGACCAAC3’; reverse primer, 5’CTTGACCTTCTTCTTCTTGTGCTTG 3’) served as an internal standard to demonstrate equal amounts of first-strand cDNA in each sample.
Results
Primer design and polymorphisms
Primers were designed for the 1,116 TF sequences, and then repeated primers were eliminated by a BLAST analysis. In total, 977 primer pairs were obtained (S1 Table). In the present study, TF primers were screened based on SSCPs. Thirty-four polymorphic primers were obtained, and 37 polymorphic loci were produced with a primer polymorphism rate of 3.48%. These 37 polymorphic loci were from 16 TF families. Seven loci were from the ERF family, and four loci were from the bHLH families. The remaining TF families contained 1–2 polymorphic loci (S1 Table).
Distribution of TFs in the cotton genome and fiber-related QTLs
After linkage analysis, 31 of the 37 TF polymorphic loci were mapped to 15 cotton chromosomes (Chr05, Chr06, Chr07, Chr09, Chr11, Chr12, Chr13, Chr17, Chr19, Chr20, Chr21, Chr22, Chr24, Chr25, and Chr26). Among these chromosomes, 14 TF loci were mapped to 7 chromosomes of the AT genome, and 17 loci were mapped to 8 chromosomes of the DT genome (Fig 2). The 31 TF markers were not equally distributed on the 15 chromosomes. Comparatively, more loci (a total of 15 loci) were mapped to Chr05, Chr11, Chr19, and Chr20. These mapped TF markers belonged to 16 TF families, among which 7 markers were from the ERF family, 4 were from the bHLH family, and 3 were from the WRKY family (S1 Table). As shown in Fig 2, TF-Ghi005868 and TF-Ghi005905 of the ERF family were located on the same chromosome (Chr20) and separated by 0.3 cM. S1 Table indicates that TF-Ghi012239 of the C2H2 family (located on Chr05) and TF-Ghi000600 of the C2H2 family (located on Chr19) were present on homologous chromosomes. In addition, TF-Ghi010629 of the WRKY family has two loci, TF-Ghi010629a and TF-Ghi010629b, which were located on the homologous chromosomes Chr25 and Chr06, respectively. TF-Ghi006349 of the bHLH family and TF-Ghi001350 of the bHLH family were located on the homologous chromosomes, Chr11 and Chr21, respectively.
Fig 2. Locations of polymorphic TF loci on the BC1 genetic linkage map.
TF markers are underlined and bolded. The loci on each chromosome with an average of 10 cM of the original map were selected shown. The top and bottom markers and markers that were closely linked to the TF markers were retained for simplicity.
QTL mapping TF markers associated with fiber quality revealed that only one TF markers, TF-Ghi005602-2 mapped on Chr19, was tightly linked with fiber length with LOD value of 8.70 and explained 12.23% of the phenotypic variance with an additive effect of -0.35. This result may imply that TFs in cotton are involved in developments of many traits rather than fiber development.
Comparison of the genetically mapped TFs with in-silico mapping in two sequenced diploid genomes
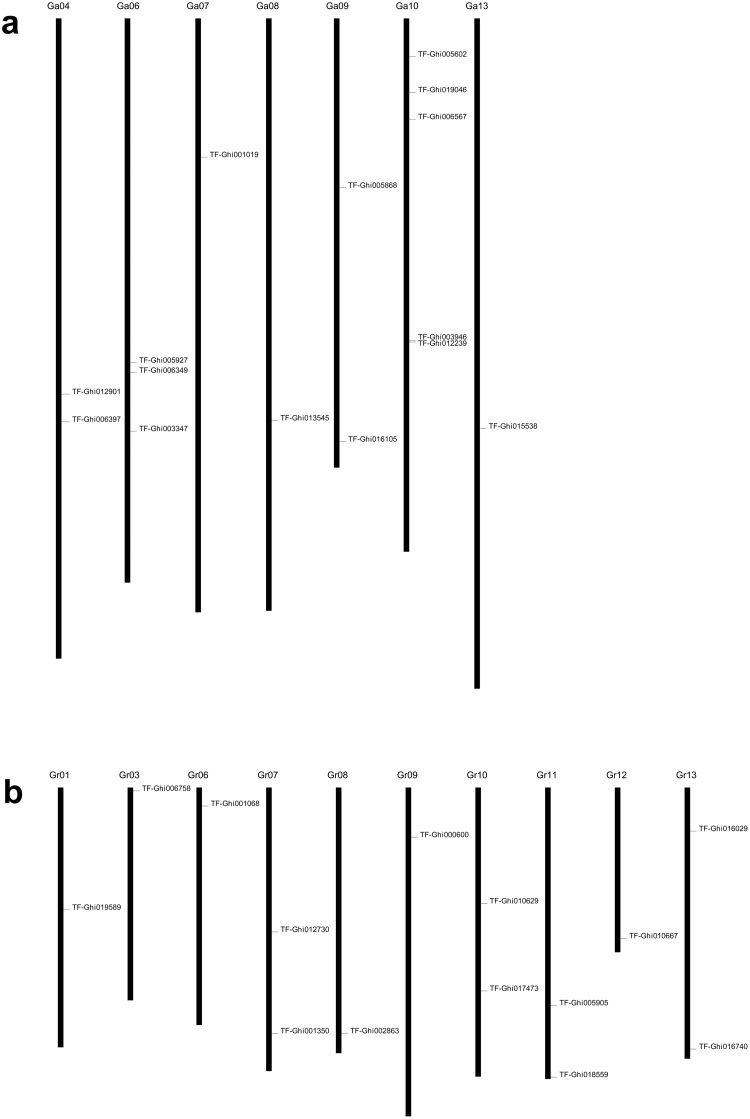
With the availability of cotton genome sequences, we can check the consistency of mapped results with their chromosomal positions by in-silico mapping their sequences to the genome sequence. By unique matching genetically mapped TF sequences to A2 and D5 genomes, 15 TFs were mapped to 7 chromosomes of A2 genome and 14 to 10 chromosomes of D5 genome (Fig 3). The in-silico mapping results was different to genetic mapping results. TF-Ghi018559b, mapped on Chr05, was not mapped on the corresponding Ga10, but on Gr11; Gr10 is the corresponding chromosome of Chr20 which is not the homologous chromosome of Chr19. The two loci on Chr06 were not mapped on the corresponding Ga12 but on Gr10; Gr10 is the corresponding chromosome of Chr25 which is the homologous chromosome of Chr06. TF-Ghi019589 on Chr07 was mapped on Gr01, the corresponding chromosome of Chr16 which is the homologous chromosome of Chr07. TF-Ghi001068 on Chr09 was mapped on Gr06, the corresponding chromosome of Chr23 which is the homologous chromosome of Chr09. TF-Ghi006349 on Chr11 was not mapped on the corresponding Ga04, but on Ga06; TF-Ghi012730 was mapped on Gr07, the corresponding chromosome Chr21 which is the homologous chromosome of Chr11. The two loci on Chr13 were mapped on Gr13, the corresponding chromosome Chr18 which is the homologous chromosome of Chr13.
Fig 3. Chromosomal location of TFs in the A2 genome of G. arboretum (a) and D5 genome of G. raimondii (b).
TF-Ghi015538 on Chr17 was mapped on Ga13, the corresponding chromosome Chr13 which is not the homologous chromosome of Chr17. TF-Ghi005602-2 and TF-Ghi019046 on Chr19 was mapped on Ga10, the corresponding chromosome Chr05 which is the homologous chromosome of Chr19. TF-Ghi005868 and TF-Ghi016105 on Chr20 was mapped on Ga09, the corresponding chromosome Chr10 which is the homologous chromosome of Chr20; TF-Ghi001019 was mapped on Ga07, the corresponding chromosome Chr01 which is not the homologous chromosome of Chr20. TF-Ghi013545 on Chr24 was mapped on Ga08, the corresponding chromosome Chr06 which is not the homologous chromosome of Chr24. TF-Ghi005927 on Chr26 was mapped on Ga06, the corresponding chromosome Chr12 which is the homologous chromosome of Chr26.
RT-PCR and qRT-PCR analysis between mapping parents
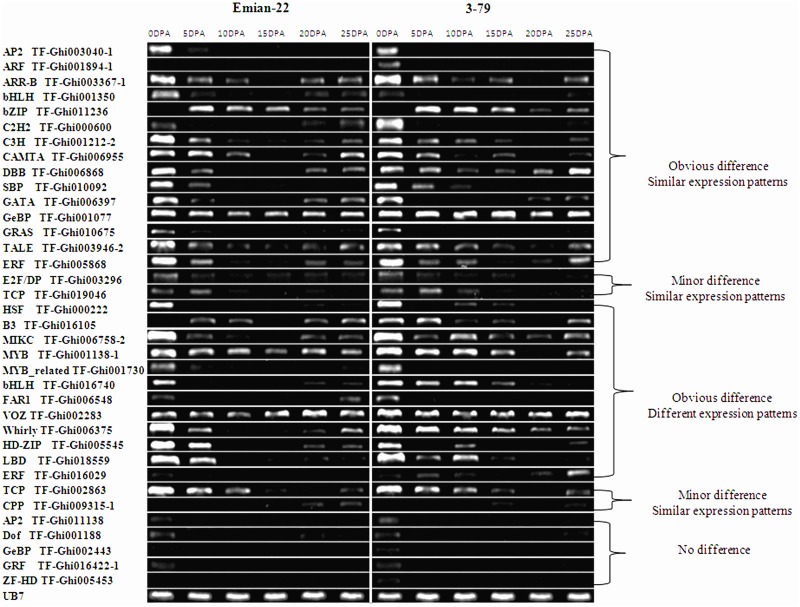
One or two markers were randomly selected from each family, and a total of 85 primer pairs from 45 TF families were used for the RT-PCR analysis. Thirty-six TF primer pairs (42.4%) from 31 families were expressed during the cotton fiber stages (Fig 4, Table 2). Among them, 27 displayed clear differences in expression, 4 exhibited minor differences, and 5 showed no differences. Almost all expressed TFs clearly differed between Emian22 and 3–79 at various stages of fiber development (0, 5, 10, 15, 20, and 25 DPA). Five TFs were weakly expressed or not expressed during any stages in either Emian22 or 3–79, and these TFs were defined as having no differences in expression in this study. Furthermore, 17 TFs displayed similar expression patterns, and 14 had different expression patterns. To further confirm the RT-PCR results, four randomly chosen genes belonging to different categories were analyzed (S1 Fig). Consistent results were observed in both the RT-PCR and the qRT-PCR analyses.
Fig 4. RT-PCR analysis of TFs between mapping parents.
The numbers on the top represent fibers at 0 DPA, 5 DPA, 10 DPA, 15 DPA, 20 DPA, and 25 DPA for Emian22 and 3–79. Similar expression trends between Emian22 and 3–79 were classified as similar expression patterns. Apparent differences in expression between Emian22 and 3–79 were classified as different expression patterns. Obvious differences in expression levels between Emian22 and 3–79 were classified as obvious differences. Minor or no differences in expression levels between Emian22 and 3–79 were classified as no difference. Gene primers and their family names are indicated on the left.
Table 2. Chromosome localization and RT-PCR analysis of TFs.
| c | Primer name | Chromosome | Genome | Results of RT-PCR |
|---|---|---|---|---|
| AP2 | TF-Ghi003040-1 | - | - | 1 |
| TF-Ghi011138 | - | - | 0 | |
| ARF | TF-Ghi001894-1 | - | - | 1 |
| TF-Ghi009171-1 | - | - | - | |
| ARR-B | TF-Ghi001951 | - | - | - |
| TF-Ghi003367-1 | - | - | 1 | |
| B3 | TF-Ghi016105 | Chr 20 | DT | 1 |
| TF-Ghi004897 | - | - | - | |
| BBR/BPC | TF-Ghi013423 | - | - | - |
| bHLH | TF-Ghi001350 | Chr 21 | DT | 1 |
| TF-Ghi016740 | Chr 13 | AT | 1 | |
| bZIP | TF-Ghi011236 | - | - | 1 |
| TF-Ghi000928 | - | - | - | |
| C2H2 | TF-Ghi000600 | Chr 19 | DT | 1 |
| TF-Ghi012239 | Chr 05 | AT | - | |
| C3H | TF-Ghi001212-2 | - | - | 1 |
| TF-Ghi002804-1 | - | - | - | |
| CAMTA | TF-Ghi006955 | - | - | 1 |
| TF-Ghi015073 | - | - | - | |
| CO-like | TF-Ghi011118 | - | - | - |
| TF-Ghi012473-2 | - | - | - | |
| CPP | TF-Ghi005234-2 | - | - | - |
| TF-Ghi009315-1 | - | - | 0 | |
| DBB | TF-Ghi006868 | - | - | 1 |
| TF-Ghi007689 | - | - | - | |
| Dof | TF-Ghi001188 | - | - | 0 |
| TF-Ghi006567 | Chr 05 | AT | - | |
| E2F/DP | TF-Ghi003296 | - | - | 0 |
| TF-Ghi010249 | - | - | - | |
| EIL | TF-Ghi005277 | - | - | - |
| TF-Ghi012794 | - | - | - | |
| ERF | TF-Ghi005868 | Chr 20 | DT | 1 |
| TF-Ghi016029 | Chr 13 | AT | 1 | |
| FAR1 | TF-Ghi001781 | - | - | - |
| TF-Ghi006548 | - | - | 1 | |
| G2-like | TF-Ghi003043 | - | - | - |
| TF-Ghi010667 | Chr 22 | DT | - | |
| GATA | TF-Ghi001019 | Chr 20 | DT | - |
| TF-Ghi006397 | Chr 11 | AT | 1 | |
| GeBP | TF-Ghi001077 | - | - | 1 |
| TF-Ghi002443 | - | - | 0 | |
| GRAS | TF-Ghi002005 | - | - | - |
| TF-Ghi010675 | - | - | 1 | |
| GRF | TF-Ghi005602-2 | Chr 19 | DT | - |
| TF-Ghi016422-1 | - | - | 0 | |
| HB-other | TF-Ghi011486 | - | - | - |
| TF-Ghi015378 | - | - | - | |
| HD-ZIP | TF-Ghi005545 | - | - | 1 |
| TF-Ghi012730 | Chr 11 | AT | - | |
| HSF | TF-Ghi000222 | - | - | 1 |
| TF-Ghi011976 | - | - | - | |
| LBD | TF-Ghi007885 | - | - | - |
| TF-Ghi018559 | Chr 05, Chr 20 | AT, DT | 1 | |
| MIKC | TF-Ghi006758-2 | Chr 17 | DT | 1 |
| TF-Ghi015198-1 | - | - | - | |
| MYB | TF-Ghi001138-1 | - | - | 1 |
| TF-Ghi005417 | - | - | - | |
| MYB_related | TF-Ghi001730 | - | - | 1 |
| TF-Ghi007525 | - | - | - | |
| NAC | TF-Ghi009255 | - | - | - |
| TF-Ghi009258 | - | - | - | |
| NF-YA | TF-Ghi004749 | - | - | - |
| TF-Ghi004977 | - | - | - | |
| NF-YB | TF-Ghi008239 | - | - | - |
| TF-Ghi019211 | - | - | - | |
| NF-YC | TF-Ghi012195 | - | - | - |
| TF-Ghi014911 | - | - | - | |
| Nin-like | TF-Ghi001151 | - | - | - |
| TF-Ghi015538 | Chr 17 | DT | - | |
| RAV | TF-Ghi013999 | - | - | - |
| S1Fa-like | TF-Ghi013563 | - | - | - |
| SBP | TF-Ghi010092 | - | - | 1 |
| SRS | TF-Ghi002427 | - | - | - |
| TALE | TF-Ghi003946-2 | Chr 05 | AT | 1 |
| TF-Ghi012410 | - | - | - | |
| TCP | TF-Ghi002863 | Chr 26 | DT | 0 |
| TF-Ghi019046 | Chr 19 | DT | 0 | |
| Trihelix | TF-Ghi010211 | - | - | - |
| TF-Ghi011360 | - | - | - | |
| VOZ | TF-Ghi002283 | - | - | 1 |
| TF-Ghi007684 | - | - | - | |
| Whirly | TF-Ghi006375 | - | - | 1 |
| WOX | TF-Ghi006162 | - | - | - |
| TF-Ghi011650 | - | - | - | |
| WRKY | TF-Ghi003347 | Chr 12 | AT | - |
| TF-Ghi003889 | - | - | - | |
| YABBY | TF-Ghi001411 | - | - | - |
| TF-Ghi017422 | - | - | - | |
| ZF-HD | TF-Ghi001068 | Chr 09 | AT | - |
| TF-Ghi005453 | - | - | 0 |
-: TF was not mapped on the chromosomes, and expression was not detected.
0: No differences in expression.
1: Obvious difference in expression.
Discussion
Low level of polymorphisms of the TF markers
TFs, as an important type of trans-acting factor, are extensively involved in the development of plants and animals. Various TFs are related to cotton fiber development [3,16,35,36]. A large number of TF functions have been verified. However, few genetic mapping studies of TFs have been performed in cotton [37]. Therefore, we downloaded TF sequences reported in the Plant Transcription Factor Database V2.0 (http://planttfdb.cbi.edu.cn/) (Table 1) and designed specific primers based on the motifs of the TF genes. Thus, the primers developed in the present study were TF-specific. In contrast, the simple sequence repeats (SSRs) primers derived from the TF sequences [37] may not have been TF-specific.
To detect additional polymorphisms in the TF markers, SSCP was applied. We discovered a very low rate of primer polymorphisms in the TF markers (3.48%), which was potentially attributed to the highly conserved nature of TFs. The results also indicated that the genes that were compared between G. hirsutum and G. barbadense are highly conserved. Although the primer polymorphism rate was low, the TF markers examined in this study exhibited more polymorphisms than other TF markers. Li et al. [37] used SSRs designed from 1,116 G. hirsutum TFs in an analysis of polymorphisms and revealed polymorphism ratios of 1.6%, 2.1%, and 2.3% in the (Yumian 1×CCRI35) F2:6, (Yumian 1×T586) F2:7, and (Yumian 1×7235) F2:6 populations, respectively. The higher rate of polymorphisms detected in the present study may have been caused by SSCP, which can detect minor difference between sequences [38]. This result may also be caused by differences between the interspecific and intraspecific populations.
Genetic mapping TFs in cotton
To date, some genetic mapping analyses of TFs have been reported. SNP primers of the MYB family were mapped on the cotton chromosome by An et al. [39]. Myb1Gbmt_238 is located on Chr13, Myb1Gb_500 is located on Chr18, and Myb2Gb_204 is located on Chr8. Guo et al. [40] also mapped TFs of the MYB family on cotton chromosomes; for example, MYB38 is located on Chr16. However, in the present study, TF markers of the MYB family were not mapped on the interspecific linkage map. In 2012, SSR primers designed from the same 1,116 G. hirsutum TFs were mapped on cotton chromosomes by Li et al. [37]. Unfortunately, due to different marker develop strategy, no common TFs were mapped.
In this study, 31 polymorphic TF loci were mapped to 15 chromosomes. Among them, 14 TF loci were mapped to 7 chromosomes of the AT genome and 17 loci on 8 chromosomes of the DT genome (Fig 2). These loci were clearly evenly distributed between the AT and the DT genome. However, Chr05, Chr11, Chr19, and Chr20 contained 15 loci that accounted for 48.4% of the total mapped TF loci. Therefore, genetic mapping revealed preferential distribution of TF loci on cotton chromosomes (Fig 2). Comparison of chromosomal location of TF loci revealed that TFs from the same family mapped to homologous cotton chromosomes.
Comparison of the genetically mapped TFs with in-silico mapping results in two sequenced diploid genomes showed that some TFs were not mapped on their corresponding chromosomes in the diploid genome, but on their homologous chromosomes’ corresponding chromosomes in the other diploid genome. It is reasonable that many genes are duplicated on the homologous chromosomes in the tetraploid genome. However, some TFs were not mapped on their corresponding chromosomes or their homologous chromosomes’ corresponding chromosomes in the diploid genomes; the reason may be that these genes translocate after polyploidization.
Differences in the expression of TFs between G. hirsutum and G. barbadense during fiber development
Cotton is an important cash crop. Therefore, improvements in yield, fiber quality, and disease resistance are areas of focus in cotton genetics and breeding. A number of studies have indicated that various TFs are involved in fiber development. Therefore, in the present study, we used RT-PCR analysis to compare TFs between G. hirsutum and G. barbadense during fiber development.
We discovered dynamic expression of TFs during various stages of fiber development in G. hirsutum and G. barbadense. The most expressed TFs (75%) from 25 families exhibited significantly different expression levels during different stages between parents. Further studies of TFs showing different expression patterns between G. hirsutum and G. barbadense may be very helpful for understanding differences in fiber quality between the two species. Unique expression patterns may be associated with a particular function. Additional studies are required to determine the functions and mechanisms of action of these TFs.
Supporting Information
The expression levels of Emian22 and 3–79 are presented. “*” represents P≤0.05, and “**” represents P≤0.01. The following primers were used: A, TF-Ghi001138-1; B, TF-Ghi003946-2; C, TF-Ghi005868; D, TF-Ghi016740.
(TIF)
√: Polymorphic primers. ×: Non-polymorphic primers. -: The primer was not mapped on the chromosome.
(XLS)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by the Special Fund for Agro-scientific Research in the Public Interest (No. 201303008) and the National Science Foundation of China (No. 31371674).
References
- 1. Yang Z, Wang X, Li X, Yang C. Advance on the study of transcription factors in higher plants. Hereditas. 2004, 26: 403–408. [PubMed] [Google Scholar]
- 2. Riechmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors. Curr Opin Plant Biol. 2000, 3(5): 423–434. [DOI] [PubMed] [Google Scholar]
- 3. Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003, 20(5): 735–747. [DOI] [PubMed] [Google Scholar]
- 4. Buck MJ, Atchley WR. Phylogenetic analysis of plant basic helix-loop-helix proteins. J Mol Evol. 2003, 56(6): 742–750. [DOI] [PubMed] [Google Scholar]
- 5. Shen WH, Gigot C. Protein complexes binding to cis elements of the plant histone gene promoters: multiplicity, phosphorylation and cell cycle alteration. Plant Mol Biol. 1997, 33(3): 367–379. [DOI] [PubMed] [Google Scholar]
- 6. Horikawa I, Cable PL, Mazur SJ, Appella E, Afshari CA, Barrett JC. Downstream E-box-mediated regulation of the human telomerase reverse transcriptase (hTERT) gene transcription: Evidence for an endogenous mechanism of transcriptional repression. Mol Biol Cell. 2002, 13(8): 2585–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heisler MGB, Atkinson A, Bylstra YH, Walsh R, Smyth DR. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development. 2001, 128(7): 1089–1098. [DOI] [PubMed] [Google Scholar]
- 8. Kirik V, Schnittger A, Radchuk V, Adler K, Huelskamp M, Bäumlein H. Ectopic expression of the Arabidopsis AtMYB23 gene induces differentiation of trichome cells. Dev Biol. 2001, 235(2): 366–377. [DOI] [PubMed] [Google Scholar]
- 9. Wang S, Wang JW, Yu N, Li CH, Luo B, Gou JY, et al. Control of plant trichome development by a cotton fiber MYB gene. Plant Cell. 2004, 16(9): 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suo JF, Liang XO, Pu L, Zhang YS, Xue YB. Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochim Biophys Acta. 2003, 1630(1): 25–34. [DOI] [PubMed] [Google Scholar]
- 11. Lee JJ, Hassan OS, Gao W, Wei NE, Kohel RJ, Chen XY, et al. Developmental and gene expression analyses of a cotton naked seed mutant. Planta. 2006, 223(3): 418–432. [DOI] [PubMed] [Google Scholar]
- 12. Hsu CY, Jenkins JN, Saha S, Ma DP. Transcriptional regulation of the lipid transfer protein gene LTP3 in cotton fibers by a novel MYB protein. Plant Sci. 2005, 168(1): 167–181. 16159331 [Google Scholar]
- 13. Wu YR, Machado AC, White RG, Llewellyn DJ, Dennis ES. Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant Cell Physiol. 2006, 47(1): 107–127. [DOI] [PubMed] [Google Scholar]
- 14. Huang B, Jin L, Liu J. Molecular cloning and functional characterization of a DREB1/CBF-like gene (GhDREB1L) from cotton. Sci China C Life Sci. 2007, 50(1): 7–14. [DOI] [PubMed] [Google Scholar]
- 15. Meng X, Li F, Liu C, Zhang CJ, Wu Z. Responses of ERF Transcription Factors to Biotic Stresses and its Applications in Improving Cotton Resistance. Mol Plant Breed. 2008, 6: 111–116. [Google Scholar]
- 16. Endt DV, Kijne JW, Memelink J. Transcription factors controlling plant secondary metabolism: what regulates the regulators? Phytochemistry. 2002, 61(2): 107–114. [DOI] [PubMed] [Google Scholar]
- 17. Ramsay NA, Walker AR, Mooney M, Gray JC. Two basic-helix-loop-helix genes (MYC-146 and GL3) from Arabidopsis can activate anthocyanin biosynthesis in a white-flowered Matthiola incana mutant. Plant Mol Biol. 2003, 52(3): 679–688. [DOI] [PubMed] [Google Scholar]
- 18. Soh MS, Kim YM, Han SJ, Song PS. REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis . Plant Cell. 2000, 12(11): 2061–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xie Z, Zhang ZL, Zou XL, Yang GX, Komatsu S, Shen QJ. Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 2006, 46(2): 231–242. [DOI] [PubMed] [Google Scholar]
- 20. Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis . Plant Mol Biol. 2004, 55(6): 853–867. [DOI] [PubMed] [Google Scholar]
- 21. Lagace M, Matton DP. Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense . Planta. 2004, 219(1): 185–189. [DOI] [PubMed] [Google Scholar]
- 22. Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, et al. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol. 2007, 65(6): 799–815. [DOI] [PubMed] [Google Scholar]
- 23. Marè C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM, Cattivelli L. Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol Biol. 2004, 55(3): 399–416. [DOI] [PubMed] [Google Scholar]
- 24. Oh SK, Yi SY, Yu SH, Moon JS, Park JM, Choi D. CaWRKY2, a chili pepper transcription factor, is rapidly induced by incompatible plant pathogens. Mol Cells. 2006, 22(1): 58–64. [PubMed] [Google Scholar]
- 25. Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002, 415(6875): 977–983. [DOI] [PubMed] [Google Scholar]
- 26. Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010, 15(5): 247–258. 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- 27. Xing DH, Lai ZB, Zheng ZY, Vinod KM, Fan BF, Chen ZX. Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol Plant. 2008, 1(3): 459–470. 10.1093/mp/ssn020 [DOI] [PubMed] [Google Scholar]
- 28. Zhang YX, Lin ZX, Xia QZ, Zhang MJ, Zhang XL. Characteristics and analysis of simple sequence repeats in the cotton genome based on a linkage map constructed from a BC1 population between Gossypium hirsutum and G.barbadense . Genome. 2008, 51(7): 534–546. 10.1139/G08-033 [DOI] [PubMed] [Google Scholar]
- 29. Yu Y, Yuan D, Liang S, Li X, Wang X, Lin Z, et al. (2011) Genome structure of cotton revealed by a genome-wide SSR genetic map constructed from a BC1 population between gossypium hirsutum and G. barbadense . BMC Genomics. 2011, 12(1): 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X, Yuan D, Wang H, Chen X, Wang B, Lin Z, et al. Increasing cotton genome coverage with polymorphic SSRs as revealed by SSCP. Genome. 2012, 55(6): 459–470. 10.1139/g2012-032 [DOI] [PubMed] [Google Scholar]
- 31. Stam P. Construction of integrated genetic linkage maps by means of a new computer package: Join Map. Plant J. 1993, 3(5): 739–744. [Google Scholar]
- 32. Kosambi DD. The estimation of map distance from recombination values. Ann Eugen. 1944, 12(1): 172–175. [Google Scholar]
- 33. Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered. 2002, 93(1): 77–78. [DOI] [PubMed] [Google Scholar]
- 34. Munis MF, Tu L, Deng F, Tan J, Xu L, Xu S, et al. A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco . Biochem Biophys Res Commun. 2010, 393(1): 38–44. 10.1016/j.bbrc.2010.01.069 [DOI] [PubMed] [Google Scholar]
- 35. Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones. 2001, 6(3): 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Singh KB, Foley RC, Onate-Sanchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002, 5(5): 430–436. [DOI] [PubMed] [Google Scholar]
- 37. Li L, Wang S, Liu F, Tang S, Tan Z, Zhang J, et al. Chromosomal Localization of Transcription Factors in Gossypium hirsutum . Acta agronomica Sinica. 2012, 38: 1361–1368. [Google Scholar]
- 38. Liu C, Lin Z, Zhang X. Unbiased genomic distribution of genes related to cell morphogenesis in cotton by chromosome mapping. Plant Cell Tiss Organ Cult. 2012, 108(3): 529–534. [Google Scholar]
- 39. An C, Saha S, Jenkins JN, Ma DP, Scheffler BE, Kohel RJ, et al. Cotton (Gossypium spp.) R2R3-MYB transcription factors SNP identification, phylogenomic characterization, chromosome localization, and linkage mapping. Theor Appl Genet. 2008, 116(7): 1015–1026. 10.1007/s00122-008-0732-4 [DOI] [PubMed] [Google Scholar]
- 40. Guo W, Cai C, Wang C, Han Z, Song X, Wang K, et al. A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium . Genetics. 2007, 176(1): 527–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expression levels of Emian22 and 3–79 are presented. “*” represents P≤0.05, and “**” represents P≤0.01. The following primers were used: A, TF-Ghi001138-1; B, TF-Ghi003946-2; C, TF-Ghi005868; D, TF-Ghi016740.
(TIF)
√: Polymorphic primers. ×: Non-polymorphic primers. -: The primer was not mapped on the chromosome.
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.