Abstract
Recombinant human erythropoietin (rhEPO) has been extensively used as pharmaceutical product for treatment of anemia. Glycosylation of rhEPO affects the biological activity, immunogenicity, pharmacokinetics and in vivo clearance rate of rhEPO. Characterization of the glycosylation status of rhEPO is of great importance for its quality control. In this study, we established a fast and comprehensive approach for reliable characterization and relative quantitation of rhEPO glycosylation, which combines multiple enzyme digestion, hydrophilic interaction chromatography (HILIC) enrichment of glycopeptides, and tandem mass spectrometry (MS) analysis. The N-linked and O-linked intact glycopeptides were analyzed with high resolution and high accuracy (HR/AM) mass spectrometry on an Orbitrap. Totally, 74 intact glycopeptides from four glycosylation sites at N24, N38, N83 and O126 were identified with the simultaneous determination of peptide sequences and glycoform compositions. The extracted ion chromatograms based on the HR/AM data allowed relative quantification of glycoforms. Our results could be extended to the quality control of rhEPO or help to establish detection approaches for glycosylation of other proteins.
Keywords: site specific glycosylation, recombinant human erythropoietin, tandem mass spectrometry
1 Introduction
The glycoprotein human erythropoietin (EPO) is a hormone mainly synthesized by kidney and liver[1], which regulates erythrocyte (red blood cell) production through controlling the proliferation, differentiation and survival of the erythroid progenitors[2] and plays an important role in other biological processes[3,4] such as wound healing and response to neuronal injury. Because of its significant biological functions, recombinant human erythropoietin (rhEPO) was first hematopoietic growth factor to be cloned[5] and has been extensively used as pharmaceutical product for the treatment of anemia in chronic kidney disease, cancer chemotherapy and many other diseases[6], ever since it first became available as a drug in 1988[7]. Moreover, it is well-known for its misuse in endurance sports, and has been forbidden by the World Anti-Doping Agency since 1989[8]. Currently, various types of rhEPO are commercially available from different manufacturers.
EPO has three N-linked glycosites at Asn-24, Asn-38, Asn-83 and a single O-linked glycosylation site at Ser-126. Previous studies have demonstrated that glycosylation regulates the pharmacological properties of rhEPO[9]. The glycoform distributions borne on each site cause great heterogeneities on the mature protein, whereas the absence of glycosylation greatly decreases the stability of the intermediate species, and changes the folding kinetics of erythropoietin[10]. The N-Glycan moieties, in particular the sialic acid residues that are generally attached at the non-reducing end of the glycans, have a significant impact on the in vivo bioactivity of rhEPO [11,12].
On the other hand, there are differences in the microheterogeneity of glycosylation between exo- and endogenous EPO[13]. rhEPO therapeutics produced in nonhuman mammalian cell lines are often modified with the nonhuman sialic acid N-glycolylneuraminic acid (Neu5Gc) instead of N-acetylneuraminic acid (Neu5Ac)[14], a feature which has generally been ignored in drug development. However, recent research has indicated that humans can produce Neu5Gc-specific antibodies which can promote drug clearance[15]. Given its significant influence on the function of the protein, a comprehensive analysis of the glycosylation status of EPO is important for the development of biopharmaceuticals from the viewpoints of efficacy, safety, and the manufacturing process[16].
In practice, intact rhEPO glycoprotein isoforms have been mainly separated and analyzed by IEF[14], CZE[17], SDS-PAGE[18] and CIEF[19] at the protein level, without revealing the detailed information on the glycoforms and the existence of special glycans, such as Neu5Gc. In several recent reports, the detection of the glycosylation sites and glycan structures have been accomplished separately, with the glycans being released from the protein by either chemical or enzymatic cleavage, followed by mapping of the glycosylation sites by protease digestion. The pool of released glycans was analyzed by MS after treatment by either an array of different exo-glycosidases or by fluorescent labeling[20]. These methods could not supply information on the site-specific glycoforms. Recently, the site-specific glycan information of rhEPO was obtained by a multidimensional strategy comprising a shallow anion exchange gradient in the first dimension, followed by hydrophilic interaction chromatography (HILIC) in the second dimension[21]. Unfortunately, the complexity of the processes and the time it requirrement constrain application of the method. Therefore, a highly effective method for rapid and extensive characterization of site-specific glycosylation of rhEPO is necessary for verification of the quality of different preparations of biopharmaceutical products.
Intact glycopeptides analysis is an ideal strategy which could provide information on the glycosites and glycan isoforms at the same time. Recently, liquid chromatography electrospray ionization mass spectrometry (LC-ESI-MS) has become a powerful technique for the analysis of intact glycopeptides, which can recognize the peptide sequence and the site-specific glycan structures simultaneously[22,23]. However, analysis of intact glycopeptides is still a challenging task, since glycosylation sites are usually resistant to protease digestion, and produce missed cleavages which could result in large glycopeptides[24] that fall out of the practical working mass range for tandem MS, thus optimized digestion condition should be considered such as the using of different enzyme or the combined use of more than one enzyme[23]. Furthermore, heterogeneity and limited ionization of glycopeptides in peptide mixtures usually result in low response upon mass spectrometry detection[25].
In the present study, we have developed a systematic strategy based on high resolution/accurate mass spectrometry (HR/AM), involving multiple enzyme digestion, HILIC enrichment and MS/MS analysis for detailed elucidation of site-specific N- and O-glycosylation status of rhEPO. The combined use of GluC and trypsin digestion effectively generates rhEPO glycopeptides appropriate for MS detection. HILIC capillary has been proved to be useful for glycopeptides separation[26]. In this study homemade HILIC SPE mirco-tips were used to effectively remove non-glycosylated peptides, which may suppress ionization of glycopeptides in MS analysis[27]. MS information including accurate precursor mass and the characteristic fragmentation patterns of intact glycopeptides, improves confidence in the detection of the glycosylation sites and their microheterogeneities. Then, a relative quantification of the site-specific glycoforms is achieved on the basis of the extracted ion chromatograms (XIC) of identified glycopeptides. In general, our results show that the presented approach is facile and powerful in simultaneous characterization and relative quantification of site-specific N- and O-glycosylation in rhEPO, and may be extended to the analysis of other glycoproteins.
2 Materials and methods
2.1 Chemicals
rhEPO produced in a Chinese hamster ovary cell line was obtained from Harbin Pharmaceutical Group (Harbin, CHN). Trypsin (Sequencing grade modified) and endoproteinase Glu-C (Glu-C) were supplied by Promega (Madison, WI, USA). PNGase F was obtained from New England Biolabs (Beverly, MA); Formic acid (FA), ammonium hydrogen carbonate (NH4HCO3) and HPLC grade acetonitrile (ACN) were purchased from Sigma-Aldrich (Sigma, USA). Dithiothreitol (DTT) and iodoacetamide (IAA) were obtained from ACROS (Belgium). A Milli-Q water purification system was used to obtain deionized water and 3-kDa Microcon centrifugal filter devices were purchased from Millipore (Bedford, MA, USA). ZIC-HILIC material was bought from Sequant (Umea, Sweden); the 3M Empore C8 disk was bought from 3M Bioanalytical Technologies (Minnesota, USA); 10 μL extended length pipet tips were obtained from Axygen (California, USA). All chemicals used in the preparation of buffers and solutions were of analytical reagent grade or better.
2.2 Sample treatment
Excipients in the rhEPO sample were removed by passage through a 3-kDa Microcon centrifugal filter device and buffer-exchanged into 50 mM NH4HCO3 (pH 7.8) to obtain a final protein concentration of about 1 μg/μL. The above solution was incubated with 10 mM DTT at 56 °C for 1 h, and then the glycoprotein was alkylated with 50 mM IAA at room temperature in the dark for 30 min. Endoproteinase Glu-C was added in an enzyme-to-sample weight ratio of 1:50, and the mixture was vortexed and incubated at 37 °C in a water bath for 10 h. The Glu-C digestion was stopped by heating the reaction mixture in boiling water for 5 min. Trypsin was added with an enzyme-to-sample weight ratio of 1:50 and the solution was incubated in a water bath overnight at 37 °C. The resulting mixture of peptides and glycopeptides was heated in boiling water for 5 min to stop digestion and stored at -20 °C until further use.
De-glycosylation of N-linked peptides was performed at 37 °C with PNGase F at a concentration of 0.1 U/μL in 50 NH4HCO3 (pH 7.8) for 24 h. The reaction was stopped by heating the product mixture for 5 min in boiling water.
For enrichment of the intact glycopeptides, homemade HILIC SPE mirco-tips were used. A small piece of C8 membrane was taken from a 3M Empore C8 disk and pushed into the end of a 10 μL extended length pipet tip using a blunt needle, and then 500 μg ZIC-HILIC material was packed as previously described [28]. The HILIC SPE micro-tip was washed three times with 50 μL of 80 % ACN/1 % FA. For sample application to the HILIC SPE micro-tip, 40 μL of 80 % ACN/1 % FA was added to 10 μL of a tryptic rhEPO digest, and the mixture was loaded on to the micro-tip 3 times to allow binding of glycopeptides. The micro-tip was washed with 80% of ACN/1% FA to remove the non-glycopeptides, and the glycopeptides were eluted with 5% of ACN and 1% FA. Then, the flow-through was dried and stored at -20 °C until further use.
2.3 Liquid chromatography and electrospray ionization mass spectrometry
Peptide mixtures or enriched glycopeptides were analyzed using an online nanoflow liquid chromatography and electrospray ionization mass spectrometry system (EASY-nLC Q Exactive). The sample was loaded by an auto-sample loading system and then separated by a 10 cm long, 75 μm inner-diameter analytical column packed with reversed-phase 3 μm C18 material. Mobile phase A consisted of 0.1 % formic acid and 2 % ACN in water, and mobile phase B consisted of 0.1 % formic acid and 2 % water in ACN; a linear mobile phase B gradient from 5 % to 30 % with a flow rate of 450 nL/min was applied for 40 min. The Q-Exactive ion source was operated at 2.0 kV. For full MS survey scans, the AGC target was 3 × e6, the scan range was from m/z 400 to 2000 with the resolution of 70,000. The 20 most intense peaks with charge state ≥ 2 were selected for sequencing and fragmented in the HCD cell with the normalized collision energy of 25%. For data dependent MS/MS scans, the AGC target was 1 × e5, the maximum injection time was 120 ms with the resolution of 17,500. The exclude isotope option was on and the dynamic exclusion time was set as 9 s.
Raw files from the Q-Exactive were searched by Proteome Discovery version 1.3 using the SEQUEST search engine with the Percolator function. The mass tolerance was set to be 20 ppm for precursor ions, and 20 mmu for product ions. Oxidation (Met), carbamidomethyl (N-terminus), deamidated (Asn) were chosen as dynamic modifications; carbamidomethyl (Cys) was chosen as a fixed modification. A user-defined enzyme was used with a rule that digests proteins after amino acid K (Lys)/R (Arg)/E (Glu) sites, excluding KP (Pro)/RP, and three missed cleavages were allowed. A false discovery rate (FDR) of 1% was estimated, and applied at the peptide level.
3 Results and discussion
3.1 Optimization of the analytical strategy
The aim of the study is to provide a scheme for analysis of intact glycopeptides. However, glycosylation frequently brings the tryptic N-glycopeptides beyond the range of transmission through the mass spectrometer due to the added mass from glycans; in addition, the glycans tend to shield the proximal protease cleavage sites. Specifically, for the EPO analysis, there is no cleavage site between N24 and N38, and the resulting peptide EAENITTGCAEHCSLNENITVPDTK is too large for MS analysis if bearing glycans, as shown in Table 1. Therefore, endoproteinase Glu-C was chosen as a substitute for trypsin since it can separate the two glycosylation sites effectively[29]. While Glu-C digestion results in a large peptide for site N83. Therefore, in this study, rhEPO was processed with serial digestion by Glu-C and trypsin in NH4HCO3, since the combined proteases cleave the protein at E/K/R. In this way, rhEPO glycopeptides with suitable molecular weights and charge states are generated for mass spectrometry detection.
Table 1.
Glycopeptides of rhEPO generated by three digestion strategies (Trypsin (K/R), GluC (D/E), GluC (E)+ Trypsin (K/R)).
| Enzyme | N24 | N38 | N83 | O126 |
|---|---|---|---|---|
| Trypsin (K/R) | 21EAEN*ITTGCAEHCSLNEN*ITVPDTK45 | 77GQALLVN*SSQPWEPLQLHVDK97 | 117EAISPPDAAS*AAPLR131 | |
| GluC (D/E) | 22AEN*ITTGCAE31 | 32HCSLNEN*ITVPD43 | 73AVLRGQALLVN*SSQPWEPLQLHVD96 | 124AAS*AAPLRTITAD136 |
| GluC (E)+ Trypsin (K/R) | 22AEN*ITTGCAE31 | 32HCSLNEN*ITVPDTK45 | 77GQALLVN*SSQPWEPLQLHVDK97 | 118AISPPDAAS*AAPLR131 |
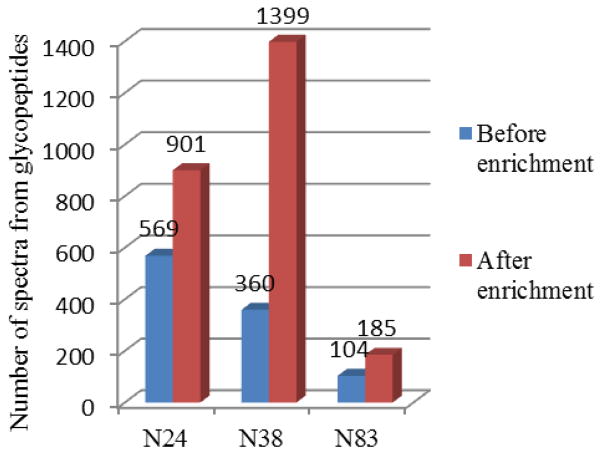
Furthermore, microheterogeneity and limited ionization of glycopeptides in peptide mixtures usually result in low response during mass spectral analysis. For a thorough investigation of all glycopeptide structures by mass spectrometry, enrichment or purification steps are often crucial to remove non-glycosylated peptides and improve the ionization and detection efficiency. Hydrophilic interaction chromatography (HILIC) provides retention of highly polar hydrophilic compounds that might be difficult to retain on reversed phase (RP) material, and is thus very suitable for the separation and enrichment of glycopeptides[30]. As can be seen clearly from Fig 1, the number of spectra from glycopeptides obtained after HILIC enrichment was about 1.5 to 3 times as that from total digest mixture. Before HILIC enrichment, we could only detect 569 spectra containing Y1 (peptide+HexNAc) product ions from N24, 360 spectra from N38, and 104 spectra from N83, while the numbers improved to 901, 1399, 185 respectively after HILIC enrichment.
Fig. 1.
Number of spectra from glycopeptides assigned to three N-glycosylation sites (N24, N38, and N83) from rhEPO digest mixture, before and after HILIC enrichment.
3.2 Sorting and assignment of glycopeptide spectra in tandem MS
The rhEPO digestion mixture or HILIC-enriched glycopeptide fraction was separated by reversed-phase liquid chromatography and then analyzed by mass spectrometry. The parameters for high energy collisional dissociation (HCD) to identify intact glycopeptides had been optimized in our previous study[31]. Owning to the relatively low abundance of glycopeptides, we adjusted the tandem MS method by setting longer injection times and an increased resolution to improve the quality of glycopeptide spectra.
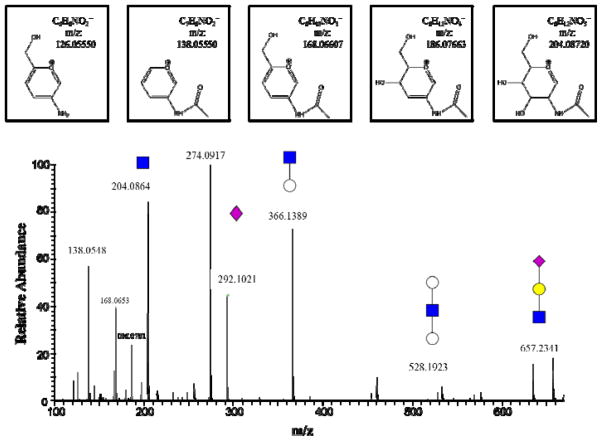
Glycopeptide precursor ions generated a series of specific products, resulting in the observation of mono-, di- or trisaccharide oxonium ions in glycopeptide spectra, as shown in Fig 2, such as the oxonium ions containing HexNAc (m/z 204.0867), and its fragments (m/z 126.0550, 138.0550, 168.0655 and 186.0761), HexNAc+Hex (m/z 366.1395), HexNAc+2Hex (m/z 528.1923), sialic acid Neu5Ac (m/z 292.1027), Neu5Ac-H2O (m/z 274.0921), Neu5Gc (m/z 308.0976), and HexNAc+Hex+Neu5Ac (m/z 657.2349). The ions coming from glycan fragmentation were specific diagnostic ions for sorting the tandem mass spectra of glycopeptides. With the HR/AM performance of the Orbitrap, highly specific extraction of spectra from glycopeptides could be realized. Glycan fragmentation diagnostic ions described above are used to extract glycopeptide spectra from raw data with an error tolerance of 20 ppm. The logarithmic intensities of these diagnostic ions in the glycopeptide spectra are shown in Fig 3.
Fig. 2.
The enlargement of low mass range in the MS/MS spectrum of a glycopeptide from rhEPO digest. The precursor ion is a complex type N-glycopeptide with a sialic acid terminated glycan. The low mass range in the HCD spectrum shows the mono-, di- or trisaccharide oxonium ion peaks. Symbols: blue squares, N-acetyl hexosamine (HexNAc); white circles, Hexose (Hex); yellow circles, Galactose (Gal); cyan rectangles, N-acetyl neuraminic acid (Neu5Ac). And chemic structures above the spectrum show five characteristic oxonium ions of all N-glycopeptides which contain HexNAc (m/z 204.0867), and its fragments (m/z 126.0550, 138.0550, 168.0655 and 186.0761).
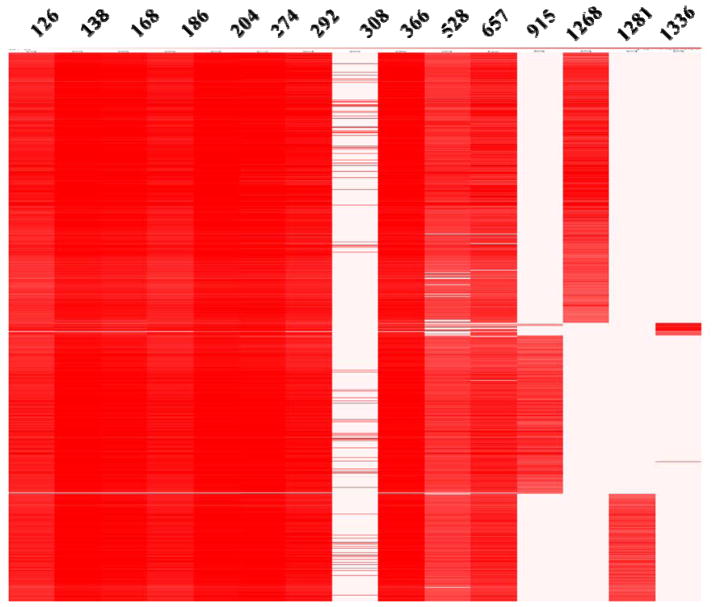
Fig. 3.
The heat map of the glycan diagnostic ions and Y1/Y0 ion of N/O-linked glycopeptides from glycopeptide spectrum ranking by retention time. B-type ions including m/z 126, 138, 168 and 186 from HexNAc, m/z 204 (HexNAc), m/z 274 (Neu5Ac-H2O), m/z 292 (Neu5Ac), m/z 308 (Neu5Gc), m/z 366 (HexNAc+Hex), m/z 528 (Hex+HexNAc+Hex), and m/z 657 (HexNAc+Hex+Neu5Ac) are also shown. The Y1-type ions include m/z 915 from N38, m/z 1268 from N24, and m/z 1281 from N83. The Y0-type ion m/z 1336 comes from O126.
It should be noted that N-linked and O-linked glycopeptides each generate specific Y ions. Since N-linked carbohydrates are linked through N-acetylglucosamine to Asn (N) residues with an amido linkage, whereas mucin-type O-linked carbohydrate attachments to proteins involve a linkage between the N-acetylgalactosamine and the amino acids Ser or Thr (S/T) with a glycosidic bond. The different linkage bonds behave differently upon dissociation. At the implementation of proper activation energy in the HCD cell, N-glycopeptides often dissociate between the two innermost GlcNAc residues and generate a typical Y1 (peptide+GlcNAc) ion; upon collision, the O-glycopeptides lose the entire glycan and generate the typical intact peptide ion (Y0). The diagnostic ions, either from glycans, or from the peptides (Y1 and Y0), were used to pick out the spectra from glycopeptides. Here the Y-type ions belonging to four major segments of the glycoprotein, contain m/z 915 (HCSLNENITVPDTK+GlcNAc+H)2+ from N38, m/z 1268 (AENITTGCAE+GlcNAc+H)+ from N24, m/z 1281 (GQALLVNSSQPWEPLQLHVDK+GlcNAc+2H)2+ from N83, m/z 1336 (AISPPDAASAAPLR+H) + from O126. Thus, Fig 3 displays four distinct groups of glycopeptides according to their different peptide sequences in all the glycopeptides sorted by glycan fragmentation diagnostic ions.
3.3 Glycosylation sites characterization
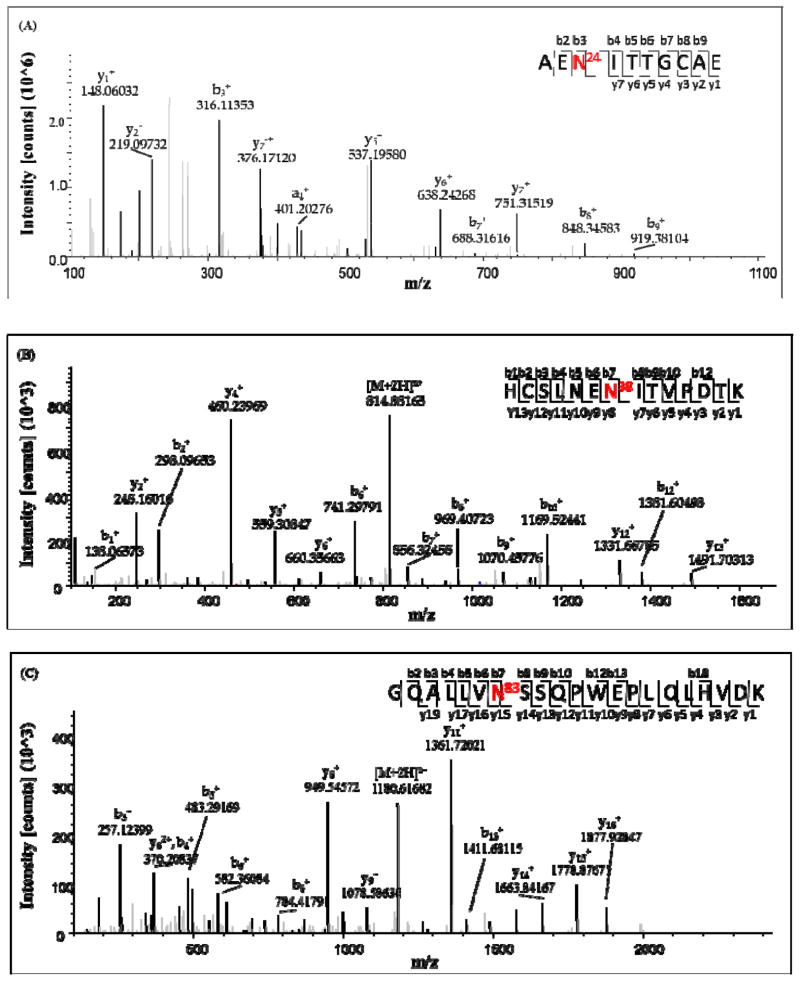
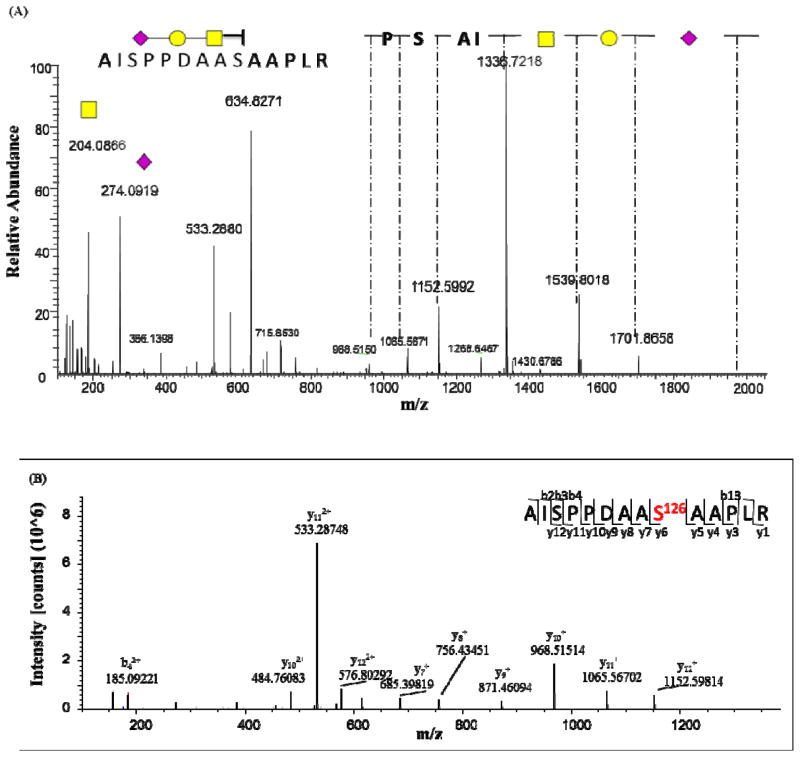
The mixture of rhEPO peptides was deglycosylated by PNGase F and then analyzed by mass spectrometry with the purpose of increasing the coverage of the protein sequence and assignment of the N-glycosylation sites. Fig 4 shows that about 91.57% sequence coverage of rhEPO was obtained in the study. For the N-linked glycopeptides, the asparagine residues were converted to aspartic acid residues during deglycosylation by PNGase F. The MS/MS spectrum of one of the deglycosylated peptides is annotated in Fig 5. Although the O-linked glycans cannot be removed by PNGase F, the tandem mass spectra of glycopeptides provide some limited peptide fragmentation information. The spectrum shown in Fig 6(A) illustrates that the O-linked glycopeptide contains the amino acid sequence AISP from its N-terminus, and thus the whole O-linked peptide sequence (AISPPDAAS126AAPLR) can be deduced; the MS/MS spectrum of the non-glycosylated counterpart peptide is shown in Fig 6(B).
Fig. 4.
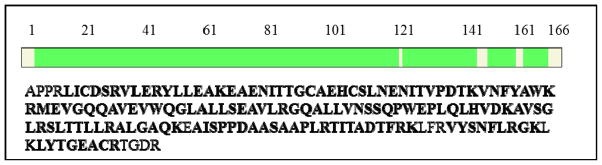
Amino acid sequence and coverage of the rhEPO in this study. Green colored region or bold amino acids are detected by mass spectrometry analysis.
Fig. 5.
Tandem mass spectra of the deglycosylated peptides belonging to three N-linked glycosylation sites. The deglycosylated peptide sequences correspond to rhEPO peptides AEN24ITTGCAE (A), HCSLNEN38ITVPDTK (B), GQALLVN83SSQPWEPLQLHVDK (C), in which the N was converted to D during PNGase F treatment.
Fig. 6.
MS/MS spectrum of the O-linked glycopeptide (AISPPDAAS126AAPLR) with a trisaccharide composed of HexNAc (yellow squares), Gal (yellow circles), Neu5Ac (cyan rectangles) (A) and the peptide without glycan (AISPPDAAS126AAPLR) (B).
3.4 Site-specific characterization of N- and O-glycopeptides
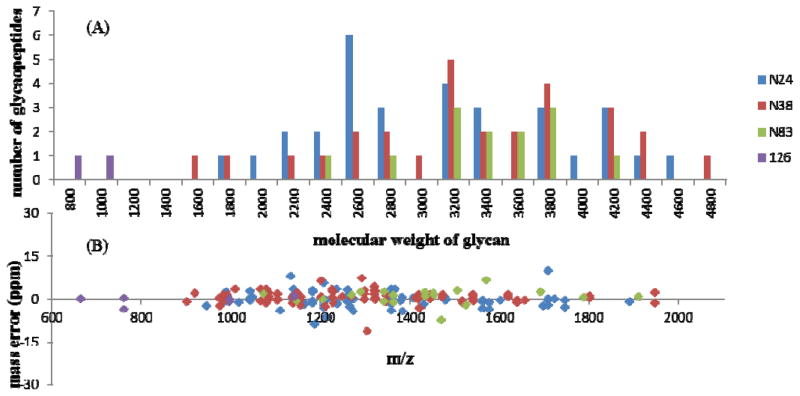
As mentioned above, HCD fragmentation performed with an appropriate energy often results in generation of abundant specific ions, including glycan oxonium ion peaks and Y1 (peptide+GlcNAc) or Y0 (peptide) ions from a glycopeptide precursor ion. The presence of B-type and Y-type ions derived from the glycan fragmentations can assist in determination of the glycan moieties of the glycopeptides. In addition, with the sequence or mass of the peptide known from above, the composition of a glycan attached to the peptide can be calculated by GlycoMod[32]. A list of glycopeptide precursor ions was input as monoisotopic mass, with a mass tolerance of 20 ppm. In total, 74 site-specific glycopeptides isomers were obtained from the tandem mass spectra. Among those, there were 31, 28 and 13 glycopeptides from three N-glycosylation sites, N24, N38, N83, respectively, and two glycopeptides containing O126. The molecular weight distributions of the glycans from those four glycosylation sites is given in Fig 7(A). With the HR/AM performance of the mass spectrometer, the identifications of the glycopeptides have high confidence, since more than 95% of them had mass errors ≤ 5ppm (Fig 7B). A clear difference among the glycoforms on different glycosites can be found from the histogram (Fig 7A). The mass of glycans from the N24 and N38 ranges from 1500 to 4900 Dalton, while those from N83 occur within a narrower region, from 2300 to 4100 Dalton.
Fig. 7.
Molecular weight (A) and mass error (B) distribution of the identified glycopeptides from four glycosylation sites (N24 in blue, N38 in red, N83 in green, and O126 in cyan).
3.5 Site-specific relative abundance of glycopeptide isomers
A well-established method for quantification of the different glycoforms in rhEPO samples is isoelectric focusing gel electrophoresis[14], which separates the intact glycoprotein isomers and allows the intensities of individual glycoprotein spots to be compared across different samples. However, this method has low sensitivity due to the presence of a large number of glycoforms exist. Thus, with their increased sensitivity and mass accuracy, mass spectrometric methods have become increasingly popular for relatively quantitative analysis the of glycan isomers from rhEPO[33]. In this study, the extracted ion chromatograms (XIC) of identified peptides extracted by LFQuant[34] were used to quantify different glycopeptides. The mass-to-charge (m/z) and retention time (RT) of the glycopeptide precursor ions identified above were used to extract the XIC and calculate the peak areas of individual glycopeptides, which are then normalized and shown in Table 2. Although accurate absolute quantitation is not possible in the absence of glycopeptide standards, the relative abundances of various glycoforms could be obtained and the information is meaningful, especially for pharmaceutical quality control.
Table 2.
A list of highly confident identifications of glycopeptides from the rhEPO with the predicted structures, glycosylation sites and the relative abundance from log2 (peak areas).
| ID | Glycan structure | Relative abundance(%) | |||
|---|---|---|---|---|---|
| N24 | N38 | N83 | O126 | ||
| 1 | (Hex)0(HexNAc)1(NeuAc)1(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 0.4 | |||
| 2 | (Hex)1(HexNAc)1(NeuAc)1(NeuGc)0(Deoxyhex)0 | 53.5 | |||
| 3 | (Hex)1(HexNAc)1(NeuAc)1(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 2.3 | 0.5 | 0.0 | |
| 4 | (Hex)1(HexNAc)1(NeuAc)2(NeuGc)0(Deoxyhex)0 | 46.5 | |||
| 5 | (Hex)1(HexNAc)1(NeuAc)4(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 0.2 | |||
| 6 | (Hex)1(HexNAc)2(NeuAc)1(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 1.2 | |||
| 7 | (Hex)1(HexNAc)2(NeuAc)3(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 0.3 | |||
| 8 | (Hex)2(HexNAc)2(NeuAc)0(NeuGc)1(Deoxyhex)1+(Man)3(GlcNAc)2 | 0.1 | |||
| 9 | (Hex)2(HexNAc)2(NeuAc)1(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 13.1 | 3.1 | ||
| 10 | (Hex)2(HexNAc)2(NeuAc)2(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 28.7 | 6.2 | 1.1 | |
| 11 | (Hex)2(HexNAc)2(NeuAc)3(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 0.2 | |||
| 12 | (Hex)2(HexNAc)2(NeuAc)3(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 0.5 | |||
| 13 | (Hex)2(HexNAc)3(NeuAc)1(NeuGc)1(Deoxyhex)1+(Man)3(GlcNAc)2 | 0.5 | |||
| 14 | (Hex)2(HexNAc)3(NeuAc)2(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 2.3 | 0.3 | ||
| 15 | (Hex)2(HexNAc)3(NeuAc)2(NeuGc)2(Deoxyhex)0+(Man)3(GlcNAc)2 | 0.3 | |||
| 16 | (Hex)3(HexNAc)3(NeuAc)1(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 10.0 | 4.3 | ||
| 17 | (Hex)3(HexNAc)3(NeuAc)1(NeuGc)1(Deoxyhex)1+(Man)3(GlcNAc)2 | 0.2 | |||
| 18 | (Hex)3(HexNAc)3(NeuAc)2(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 11.8 | 11.1 | 4.2 | |
| 19 | (Hex)3(HexNAc)3(NeuAc)3(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 7.2 | 2.7 | 3.0 | |
| 20 | (Hex)3(HexNAc)4(NeuAc)1(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 0.6 | |||
| 21 | (Hex)3(HexNAc)4(NeuAc)2(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 0.4 | |||
| 22 | (Hex)4(HexNAc)4(NeuAc)1(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 2.4 | |||
| 23 | (Hex)4(HexNAc)4(NeuAc)2(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 4.3 | 10.4 | 7.3 | |
| 24 | (Hex)4(HexNAc)4(NeuAc)3(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 0.1 | |||
| 25 | (Hex)4(HexNAc)4(NeuAc)3(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 1.2 | 10.5 | 9.7 | |
| 26 | (Hex)4(HexNAc)4(NeuAc)4(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 1.0 | 2.8 | 0.7 | |
| 27 | (Hex)4(HexNAc)5(NeuAc)3(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 1.1 | |||
| 28 | (Hex)5(HexNAc)5(NeuAc)1(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 0.3 | 2.0 | 36.9 | |
| 29 | (Hex)5(HexNAc)5(NeuAc)1(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 1.9 | 0.6 | ||
| 30 | (Hex)5(HexNAc)5(NeuAc)2(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 2.5 | 9.1 | ||
| 31 | (Hex)5(HexNAc)5(NeuAc)3(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 3.5 | |||
| 32 | (Hex)5(HexNAc)5(NeuAc)4(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 0.7 | 1.9 | ||
| 33 | (Hex)6(HexNAc)6(NeuAc)1(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 1.3 | 15.6 | 6.1 | |
| 34 | (Hex)6(HexNAc)6(NeuAc)1(NeuGc)0Deoxyhex1+(Man)3(GlcNAc)2 | 2.8 | |||
| 35 | (Hex)6(HexNAc)6(NeuAc)2(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 0.6 | 6.3 | 0.6 | |
| 36 | (Hex)6(HexNAc)6(NeuAc)3(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 0.2 | |||
| 37 | (Hex)6(HexNAc)6(NeuAc)4(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 2.6 | 0.8 | ||
| 38 | (Hex)7(HexNAc)7(NeuAc)0(NeuGc)2(Deoxyhex)0+(Man)3(GlcNAc)2 | 0.1 | |||
| 39 | (Hex)7(HexNAc)7(NeuAc)1(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 2.3 | 9.1 | 17.8 | |
| 40 | (Hex)7(HexNAc)7(NeuAc)1(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 0.3 | |||
| 41 | (Hex)7(HexNAc)7(NeuAc)2(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 0.2 | |||
| 42 | (Hex)7(HexNAc)7(NeuAc)4(NeuGc)0(Deoxyhex)1+(Man)3(GlcNAc)2 | 0.1 | |||
| 43 | (Hex)7(HexNAc)8(NeuAc)1(NeuGc)0Deoxyhex1+(Man)3(GlcNAc)2 | 0.6 | |||
| 44 | (Hex)8(HexNAc)8(NeuAc)1(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 0.9 | |||
| 45 | (Hex)8(HexNAc)8(NeuAc)2(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 0.1 | |||
| 46 | (Hex)9(HexNAc)9(NeuAc)1(NeuGc)0(Deoxyhex)0+(Man)3(GlcNAc)2 | 3.8 | |||
| total | 100.0 | 100.0 | 100.0 | 100.0 | |
Moreover, as discussed above, Neu5Gc is known to exhibit significant differences between the rhEPO expressed in nonhuman mammalian cell lines and the endogenous erythropoietin in human serum. Unlike other mammals, humans cannot biosynthesize the Neu5Gc[15]. Therefore, Neu5Gc is recognized as a foreign substance by the immune system and anti-Neu5Gc antibodies may be produced after injection of a Neu5Gc-containing glycoprotein, which can promote the drug clearance. As Neu5Gc is not expressed in humans, but the rhEPO analyzed here was expressed by CHO cells containing both Neu5Ac and minor amounts of Neu5Gc forms, the detection of relative percentage of Neu5Gc (m/z 308) versus Neu5Ac (m/z 292) in glycopeptides could be useful to distinguish between the endogenous and recombinant EPO. It is clear from the data shown in Fig 8, 4.7% of the glycopeptide spectra indicated the presence of Neu5Gc, and 98.5% of the glycopeptide spectra had evidence for Neu5Ac. Further inspection on the spectra showed that most glycopeptides which have Neu5Gc simultaneously exhibit Neu5Ac. Therefore, the content of Neu5Gc glycoforms could be referenced for rhEPO quality control.
Fig. 8.
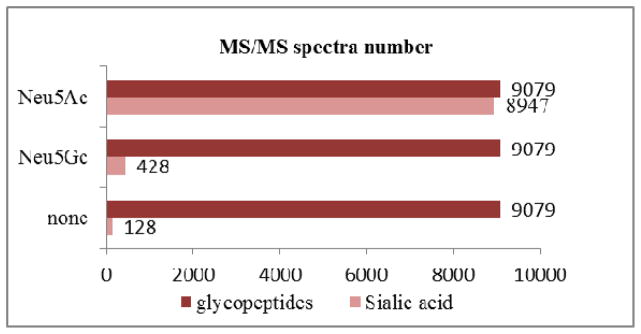
The MS spectrum number of glycopeptides without sialic acid, or with Neu5Ac, Neu5Gc in raw data which contains 9079 glycopeptides spectra in total.
4 Concluding remarks
The approach described here, involving HILIC SPE enrichment, LC-MS detection, glycopeptides grouping and quantification has been successfully applied to the analysis of intact glycopeptides from rhEPO. The coupling of trypsin and GluC digestion allowed a higher sequence coverage and the detection of four glycosylation sites. With high resolution and high mass accuracy, specific ions from the tandem spectra were used as diagnostic ions for the picking and grouping of glycopeptide spectra. Glycosylation site assignments were mainly achieved by accurate mass detection of Y1 or Y0 ions, while the detailed peptide sequences were determined from the MS analysis of deglycosylated peptides. In addition, the MS/MS data provided detailed composition of the oligosaccharide chains, and the extracted ion chromatograms (XICs) from MS data offer glycopeptide relative abundance values. Furthermore, the percentage of Neu5Gc was acquired in a straightforward way, following upon our sorting strategy for glycopeptide spectra. Overall, our strategy enables rapid and sensitive, site-specific glycoform characterization and quantification of N- and O-glycopeptides, which are highly suitable for the differential analysis of rhEPO products from various manufacturers, diverse production cell lines, or different batches.
Acknowledgments
We are grateful for the financial support from the National Key Program for Basic Research of China (2011CB910603, 2013CB911204, 2014CBA02001); National High Technology Research and Development Program of China (2012AA020203); the International Scientific and Technological Cooperation Program (No. 2011DFB30370); National Drug Research and Development Program (2012ZX09301003-010); Beijing Nova Program (Z121107002512014); and the US National Institutes of Health supports the BU Center for Biomedical Mass Spectrometry (P41 GM104603).
References
- 1.Lacombe C, Mayeux P. Biology of erythropoietin. Haematologica. 1998;83 (8):724–732. [PubMed] [Google Scholar]
- 2.Pericoli Ridolfini F, Pagliari M. Erythropoietin in the regulation of erythropoiesis. Policlinico Prat. 1967;74 (14):461–480. [PubMed] [Google Scholar]
- 3.Siren AL, Fratelli M, Brines M, Goemans C, Casagrande S, Lewczuk P, Keenan S, Gleiter C, Pasquali C, Capobianco A, Mennini T, Heumann R, Cerami A, Ehrenreich H, Ghezzi P. Erythropoietin prevents neuronal apoptosis after cerebral ischemia and metabolic stress. Proc Natl Acad Sci U S A. 2001;98 (7):4044–4049. doi: 10.1073/pnas.051606598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haroon ZA, Amin K, Jiang X, Arcasoy MO. A novel role for erythropoietin during fibrin-induced wound-healing response. Am J Pathol. 2003;163 (3):993–1000. doi: 10.1016/S0002-9440(10)63459-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin FK, Suggs S, Lin CH, Browne JK, Smalling R, Egrie JC, Chen KK, Fox GM, Martin F, Stabinsky Z, et al. Cloning and expression of the human erythropoietin gene. Proc Natl Acad Sci U S A. 1985;82 (22):7580–7584. doi: 10.1073/pnas.82.22.7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.John MJ, Jaison V, Jain K, Kakkar N, Jacob JJ. Erythropoietin use and abuse. Indian J Endocrinol Metab. 2012;16 (2):220–227. doi: 10.4103/2230-8210.93739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raddino R, Robba D, Caretta G, Bonadei I, Teli M, Zanini G, Madureri A, Vizzardi E, Dei Cas L. Erythropoietin: a new perspective in cardiovascular therapy. Monaldi Arch Chest Dis. 2008;70 (4):206–213. doi: 10.4081/monaldi.2008.414. [DOI] [PubMed] [Google Scholar]
- 8.Lasne F, de Ceaurriz J. Recombinant erythropoietin in urine. Nature. 2000;405 (6787):635. doi: 10.1038/35015164. [DOI] [PubMed] [Google Scholar]
- 9.Li H, d’Anjou M. Pharmacological significance of glycosylation in therapeutic proteins. Curr Opin Biotechnol. 2009;20 (6):678–684. doi: 10.1016/j.copbio.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Banks DD. The effect of glycosylation on the folding kinetics of erythropoietin. J Mol Biol. 2011;412 (3):536–550. doi: 10.1016/j.jmb.2011.07.061. [DOI] [PubMed] [Google Scholar]
- 11.Jeong YT, Choi O, Lim HR, Son YD, Kim HJ, Kim JH. Enhanced sialylation of recombinant erythropoietin in CHO cells by human glycosyltransferase expression. J Microbiol Biotechnol. 2008;18 (12):1945–1952. [PubMed] [Google Scholar]
- 12.Yanagihara S, Taniguchi Y, Hosono M, Yoshioka E, Ishikawa R, Shimada Y, Kadoya T, Kutsukake K. Measurement of sialic acid content is insufficient to assess bioactivity of recombinant human erythropoietin. Biol Pharm Bull. 2010;33 (9):1596–1599. doi: 10.1248/bpb.33.1596. [DOI] [PubMed] [Google Scholar]
- 13.Skibeli V, Nissen-Lie G, Torjesen P. Sugar profiling proves that human serum erythropoietin differs from recombinant human erythropoietin. Blood. 2001;98 (13):3626–3634. doi: 10.1182/blood.v98.13.3626. [DOI] [PubMed] [Google Scholar]
- 14.Llop E, Gutierrez-Gallego R, Segura J, Mallorqui J, Pascual JA. Structural analysis of the glycosylation of gene-activated erythropoietin (epoetin delta, Dynepo) Anal Biochem. 2008;383 (2):243–254. doi: 10.1016/j.ab.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 15.Ghaderi D, Taylor RE, Padler-Karavani V, Diaz S, Varki A. Implications of the presence of N-glycolylneuraminic acid in recombinant therapeutic glycoproteins. Nat Biotechnol. 2010;28 (8):863–867. doi: 10.1038/nbt.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawasaki N, Itoh S, Hashii N, Takakura D, Qin Y, Huang X, Yamaguchi T. The significance of glycosylation analysis in development of biopharmaceuticals. Biol Pharm Bull. 2009;32 (5):796–800. doi: 10.1248/bpb.32.796. [DOI] [PubMed] [Google Scholar]
- 17.Balaguer E, Neususs C. Glycoprotein characterization combining intact protein and glycan analysis by capillary electrophoresis-electrospray ionization-mass spectrometry. Anal Chem. 2006;78 (15):5384–5393. doi: 10.1021/ac060376g. [DOI] [PubMed] [Google Scholar]
- 18.Reichel C, Kulovics R, Jordan V, Watzinger M, Geisendorfer T. SDS-PAGE of recombinant and endogenous erythropoietins: benefits and limitations of the method for application in doping control. Drug Test Anal. 2009;1 (1):43–50. doi: 10.1002/dta.10. [DOI] [PubMed] [Google Scholar]
- 19.Dou P, Liu Z, He J, Xu JJ, Chen HY. Rapid and high-resolution glycoform profiling of recombinant human erythropoietin by capillary isoelectric focusing with whole column imaging detection. J Chromatogr A. 2008;1190 (1–2):372–376. doi: 10.1016/j.chroma.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Doerr A. Glycoproteomics. Nature methods. 2011;9 (1):36–36. [Google Scholar]
- 21.Bones J, McLoughlin N, Hilliard M, Wynne K, Karger BL, Rudd PM. 2D-LC analysis of BRP 3 erythropoietin N-glycosylation using anion exchange fractionation and hydrophilic interaction UPLC reveals long poly-N-acetyl lactosamine extensions. Anal Chem. 2011;83 (11):4154–4162. doi: 10.1021/ac200406z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayampurath AM, Wu Y, Segu ZM, Mechref Y, Tang H. Improving confidence in detection and characterization of protein N-glycosylation sites and microheterogeneity. Rapid Commun Mass Spectrom. 2011;25 (14):2007–2019. doi: 10.1002/rcm.5059. [DOI] [PubMed] [Google Scholar]
- 23.Castilho A, Neumann L, Daskalova S, Mason HS, Steinkellner H, Altmann F, Strasser R. Engineering of sialylated mucin-type O-glycosylation in plants. J Biol Chem. 2012;287 (43):36518–36526. doi: 10.1074/jbc.M112.402685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H. Glycoproteomics Analysis of Human Liver Tissue by Combination of Multiple Enzyme Digestion and Hydrazide Chemistry. Journal of Proteome Research. 2009;8 (2):651–661. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- 25.Kondo A, Thaysen-Andersen M, Hjerno K, Jensen ON. Characterization of sialylated and fucosylated glycopeptides of beta2-glycoprotein I by a combination of HILIC LC and MALDI MS/MS. J Sep Sci. 2010;33 (6–7):891–902. doi: 10.1002/jssc.200900802. [DOI] [PubMed] [Google Scholar]
- 26.Takegawa Y, Ito H, Keira T, Deguchi K, Nakagawa H, Nishimura S. Profiling of N- and O-glycopeptides of erythropoietin by capillary zwitterionic type of hydrophilic interaction chromatography/electrospray ionization mass spectrometry. J Sep Sci. 2008;31 (9):1585–1593. doi: 10.1002/jssc.200700679. [DOI] [PubMed] [Google Scholar]
- 27.Zauner G, Deelder AM, Wuhrer M. Recent advances in hydrophilic interaction liquid chromatography (HILIC) for structural glycomics. Electrophoresis. 2011;32 (24):3456–3466. doi: 10.1002/elps.201100247. [DOI] [PubMed] [Google Scholar]
- 28.Jiang J, Han HH, Ma C, Wang JF, Ying WT, Qian XH. Characterization of N-Glycosylation Status of Micro-Level Glycoprotein Using Solid Phase Extraction Based Techniques. Chinese Journal of Analytical Chemistry. 2012;40 (7):1019–1024. [Google Scholar]
- 29.Gimenez E, Ramos-Hernan R, Benavente F, Barbosa J, Sanz-Nebot V. Analysis of recombinant human erythropoietin glycopeptides by capillary electrophoresis electrospray-time of flight-mass spectrometry. Anal Chim Acta. 2012;709:81–90. doi: 10.1016/j.aca.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 30.Selman MH, Hemayatkar M, Deelder AM, Wuhrer M. Cotton HILIC SPE microtips for microscale purification and enrichment of glycans and glycopeptides. Anal Chem. 2011;83 (7):2492–2499. doi: 10.1021/ac1027116. [DOI] [PubMed] [Google Scholar]
- 31.Jiang J, Ying W, Qian X. Characterization of Intact Glycopeptides by a Combination of Hydrophilic Interaction Liquid Chromatography and Multiple Fragmentation Tandem Mass Spectrometry. Chinese J Anal Chem. 2014;42 (2):159–165. [Google Scholar]
- 32.Cooper CA, Gasteiger E, Packer NH. GlycoMod--a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics. 2001;1 (2):340–349. doi: 10.1002/1615-9861(200102)1:2<340::AID-PROT340>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 33.Giménez E. Capillary electrophoresis time-of-flight mass spectrometry for a confident elucidation of a glycopeptide map of recombinant human erythropoietin. Rapid Commun Mass Spectrom. 2011;25 (16):2307–2316. doi: 10.1002/rcm.5114. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Zhang J, Xu C, Li N, Liu H, Ma J, Zhu Y, Xie H. LFQuant: A label-free fast quantitative analysis tool for high-resolution LC-MS/MS proteomics data. Proteomics. 2012;12 (23–24):3475–3484. doi: 10.1002/pmic.201200017. [DOI] [PubMed] [Google Scholar]