SUMMARY
The colonization of bacteria in complex fluid flow networks, such as those found in host vasculature, remains poorly understood. Recently, it was reported that many bacteria, including Bacillus subtilis [1], Escherichia coli [2], and Pseudomonas aeruginosa [3, 4], can move in the opposite direction of fluid flow. Upstream movement results from the interplay between fluid shear stress and bacterial motility structures and such rheotactic-like behavior is predicted to occur for a wide range of conditions [1]. Given the potential ubiquity of upstream movement, its impact on population-level behaviors within hosts could be significant. Here, we find that P. aeruginosa communities use a diverse set of motility strategies, including a novel surface motility mechanism characterized by counter-advection and transverse diffusion, to rapidly disperse throughout vasculature-like flow networks. These motility modalities give P. aeruginosa a selective growth advantage, enabling it to self-segregate from other human pathogens such as Proteus mirabilis and Staphylococcus aureus that outcompete P. aeruginosa in well-mixed non-flow environments. We develop a quantitative model of bacterial colonization in flow networks, confirm our model in vivo in plant vasculature, and validate a key prediction that colonization and dispersal can be inhibited by modifying surface chemistry. Our results show that the interaction between flow mechanics and motility structures shapes the formation of mixed-species communities and suggest a general mechanism by which bacteria could colonize hosts. Furthermore, our results suggest novel strategies for tuning the composition of multi-species bacterial communities in hosts, preventing inappropriate colonization in medical devices, and combatting bacterial infections.
P. aeruginosa is an opportunistic pathogen that infects a broad range of hosts including plants and animals. In humans, it is a major cause of vascular-related illnesses including lung infections, urinary tract infections, bacteremia and sepsis [5–7]. In fluid flow environments, P. aeruginosa cells attach to surfaces using type IV pili (TFP). These TFP are localized to the bacterial cell poles [8] such that upon attaching to the surface, flow causes the bacteria to orient with the TFP pole pointed in the opposite direction of the flow (Fig. 1A) [3, 4]. The repeated extension and retraction of TFP in this position drives P. aeruginosa to move upstream along the surface (Fig. 1A). The upstream movement is a direct response to surface shear stress and is not due to chemotaxis [4]. P. aeruginosa cells also swim through fluid environments using flagella but upstream movement occurs without flagella [4].
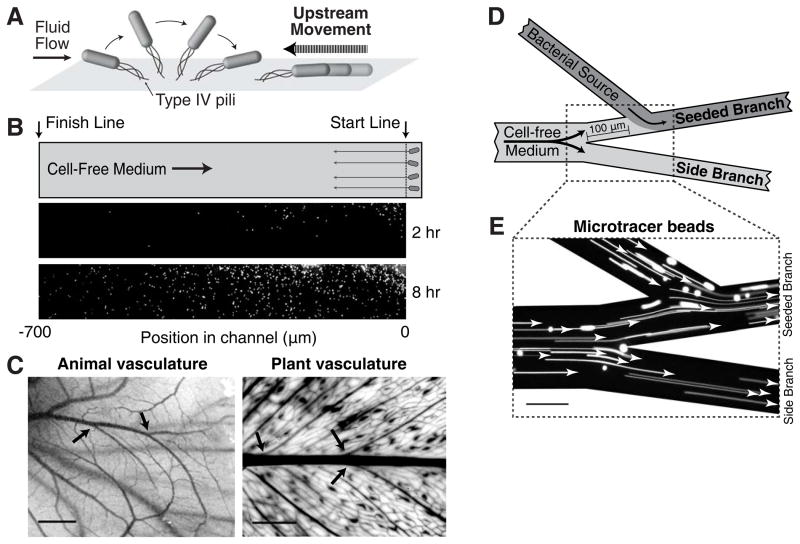
Figure 1. Upstream dispersal of P. aeruginosa populations in flow networks.
(A) Schematic summarizing upstream movement by P. aeruginosa. Fluid flow orients polar-attached cells such that the attachment pole is positioned upstream and pilus retraction moves cells upstream. (B) P. aeruginosa surface colonization in a linear microfluidic channel. Cells are loaded behind a “start line” in which cell-free medium flows from the left to right. Within 2 hours, cells at the leading edge of the population reach the “finish line”. (C) Vascular flow networks in chicken embryos and E. aureum plant leaves contain many branching intersections (arrows). (D) Schematic of a branched flow network microfluidic device used to track colonization, competition, and dispersal. Cell-free medium flowing through the main branch diverges into two branches. Cells are inoculated into the “seeded branch”. Arrows indicate the direction of flow. (E) Tracking of fluorescent micro-tracer beads indicates a flow pattern that is laminar, stable, and uni-directional. Scale bars represent 2 mm in (C) and 100 μm in (E).
While individual bacteria move upstream over short distances and small timescales [1, 2, 4], it is unknown whether populations can migrate physiological distances relevant for infection or colonize surfaces for extended periods in fluid flow. In particular, P. aeruginosa cells are dislodged from the surface by the fluid flow force and are subsequently pushed downstream [4]. This finding raises a paradox: if cells are ejected from the surface and move backwards along a flow streamline, the same streamline would carry cells back downstream upon detachment from the surface, nullifying any effect of the upstream movement. Furthermore, while individual cells move upstream, it is unknown whether this mechanism could drive the expansion of a multicellular population.
Upstream dispersal involves multiple phenotypically diverse single-cell motility modes
To investigate P. aeruginosa colonization dynamics in flow, we first imaged the leading edge of a population in a linear microfluidic channel. Cell-free medium was flowed steadily through the device (200×50 μm, WxH) at wall shear stresses between 0.2 to 2 Pa (1–10 μL/min, vfluid = 2–20 mm/s), which correspond to the dimensions, shear stresses, and flow speeds typically observed in the vasculature of plants and animals [9–12]. Note that these flow rates are significantly higher than typical bacterial swimming rates. Cells were initially seeded at one side of the channel behind a “start line” (Fig. 1B). In the presence of flow, cells exhibited three distinct motility behaviors: (i) movement in the opposite direction of the flow towards the “finish line” (700 μm upstream), (ii) detachment from the surface (and subsequent downstream movement with the flow), or (iii) no motility. After 2 hours, the fastest cells at the leading edge reached the upstream finish line (Fig. 1B) while slower cells continued upstream migration. Cells were also found at the start line and downstream. The dispersal of the population to upstream and downstream regions in the device, which we refer to collectively as “upstream dispersal”, is due to the phenotypic diversity of motility modes within a single population. As the behavioral heterogeneity at the single-cell level benefits the community as a whole, this could represent a form of bet-hedging by P. aeruginosa that enables the species to explore upstream and downstream environments while maintaining colonization of the initial environment.
P. aeruginosa gains a selective growth advantage by dispersing upstream
We next sought to determine if upstream dispersal provides bacteria with a selective growth advantage over bacteria that lack this motility mode. For these studies we modified the microfluidic device to incorporate a branched network geometry, which enabled us to quantify colonization, competition, and dispersal in the same device. Branched networks are a defining feature of animal and plant vasculatures (Fig. 1C), with bifurcation of vasculature promoting the uniform distribution of nutrients [13, 14]. To establish the flow pattern in this complex network (Fig. 1D), we imaged micro-tracer beads that verified that the flow was laminar, stable, and unidirectional (Fig. 1E, Movie S1).
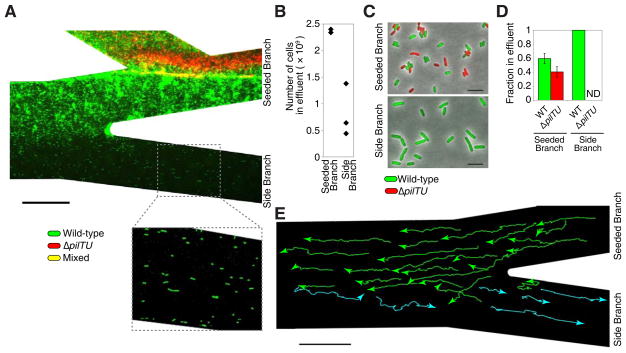
We inoculated one branch, the “seeded branch” (Fig. 1D), with an equal number of wild-type P. aeruginosa (expressing GFP) and ΔpilTU mutants (expressing mCherry) that are defective in surface motility because they lack the TFP PilT and PilU retraction motors [8] but retain swimming motility (Fig. S1A in Supplemental Information). After 15 hours, wild-type cells colonized both the seeded and side branches while the ΔpilTU cells colonized only the seeded branch (Fig. 2A, Movie S2A). We collected the fluid effluent from each branch, which contains both planktonic (swimming) cells and cells that detached from the surface, and cultured the effluent for 3.5 hours at 37°C in order to characterize cell physiology and determined the relative proportion of wild-type and ΔpilTU cells using single-cell fluorescence microscopy. The effluent from the seeded branch contained ~2.4 × 109 cells, of which 60% were wildtype and 40% were ΔpilTU cells (Fig. 2B–D). In contrast, the side branch contained only wild-type cells (Fig. 2A–D). The total number of wild-type cells in both branches outnumbered ΔpilTU cells by more than two-fold (~1.3 × 109 more cells), indicating that upstream migration results in a selective growth advantage. The greater number of wild-type cells is not due to a difference in growth rates as both strains are represented comparably in planktonic co-culture competitions (Fig. S1B). Furthermore, wild-type cells from the side branch were 40% larger than those from the seeded branch (Figs. 2C and S1C), indicating the increased availability of nutrients in the side branch [15]. Thus, by entering the side branch, the upstream-migrating population escapes nutrient limitation imposed by the competing ΔpilTU cells.
Figure 2. Upstream dispersal provides P. aeruginosa with a selective growth advantage in flow.
(A) Surface colonization of wild-type (green) and pilus-defective ΔpilTU (red) P. aeruginosa cells in a branched network. An equal number of each strain was initially co-inoculated into the seeded branch (not shown). After 15 hours, wild-type cells colonized all areas of the device while ΔpilTU cells remained in the seeded branch. The effluent (containing planktonic and surface-detached cells) from each branch was collected and cultured for an additional 3.5 hours. (B) The total number of cells in each effluent was measured through optical density and the (C–D) effluent population composition was determined using fluorescence microscopy (see Experimental Procedures in Supplemental Information). ΔpilTU cells were not detected (ND) in the side branch. Error bars indicate standard deviation. (E) Trajectories of individual wild-type cells on surfaces, which move upstream (green) or downstream (cyan) in “zig-zag” paths that cross laterally into different streamlines. Each trajectory was acquired at 30 second intervals for 30 minutes. Scale bars indicate 100 μm for (A) and (E) and 5 μm for (C). All experiments were performed in triplicate.
Dispersal through zig-zag paths on surfaces
Given that cells that move upstream should simply return downstream along the same streamline when they are released from the surface, our findings raised the question of how P. aeruginosa dispersed to the side branches. By tracking the movements of individual cells (Fig. 2E), we found that upstream motility has a component whose direction is perpendicular to the flow. P. aeruginosa cells always migrated in a zig-zag path, with some trajectories crossing over into streamlines that flow into the side branch. Cells that entered these streamlines were frequently detached from the surface, carried into the side branch, and subsequently re-attached to the surface of the side branch (Movie S2A). This zig-zag motion was observed in upstream movement in linear channels [4] and in a network geometry in which branches converge at the intersection instead of diverging (Fig. S2A, Movie S2B). The motion can be attributed to the radial organization of TFP at the cell pole (Fig. S2B) and their non-synchronous activity [16]. Zig-zag trajectories are also observed through flagellar-mediated swimming in sperm rheotaxis [17], indicating that similar upstream motions can be achieved using distinct motility mechanisms.
An alternative explanation for the presence of bacteria in the side channels is through flagella-mediated upstream movement. However, ΔpilTU mutants retain swimming motility (Fig. S1A) but do not reach the side channel (Fig. 2A–D), supporting the conclusion that migration to the side channel is driven by surface motility. We also tested whether B. subtilis or E. coli migrates toward the side-channel, as both have previously been shown to move upstream through flagella-mediated rheotactic motion [1, 2]. Following 15 hours of inoculation in which either species was inoculated into the seeded channel, we found no cells on the surface or in effluent from the side channel (Fig. S2C–D), suggesting that the flagella-mediated rheotactic behavior does not result in dispersal. Together, our data show that colonization and dispersal is due to surface-mediated upstream migration.
Selective advantage enables competition against faster-growing pathogens
In natural settings, bacteria compete against other species for shared resources [18]. To determine whether upstream dispersal confers P. aeruginosa with a competitive advantage against other species, we co-inoculated the seeded branch with equal numbers of P. aeruginosa and Proteus mirabilis, a Gram-negative bacterium that grows at a faster rate than P. aeruginosa (Fig. S3A–B) and moves on surfaces at significantly higher velocities (Fig. S3C, Movie S3). Both pathogens cause nosocomial infections, colonize the urinary tract and gut, and may be considered natural competitors [5, 19–23]. In addition, P. mirabilis swims using a flagella-mediated mechanism similar to E. coli [24, 25], which suggests that P. mirabilis can move upstream.
We flowed media continuously for 15 hours (Fig. 3A), collected effluent containing planktonic and surface-detached cells from each branch, and performed the analysis described above (Fig. 3B–D). As predicted from its higher growth rate, P. mirabilis outgrew P. aeruginosa in the seeded branch and accounted for 85% of the bacteria in the branch effluent (Fig. 3D). In contrast, only P. aeruginosa cells colonized the side branch (Fig. 3A,C–D and Movie S4A). Examination of individual cell trajectories showed that P. mirabilis cells did not disperse upstream (Movie S4A), thereby explaining their restriction to the seeded branch. Furthermore, P. aeruginosa cells in the seeded branch were 29% smaller than those in the side branch (Figs. 3C, S3D), indicating that P. aeruginosa cells were nutrient-limited in the co-culture with P. mirabilis. These results demonstrate that upstream migration by a surface-motility mechanism enables P. aeruginosa to self-segregate from the co-culture to colonize a separate niche in flow. P. aeruginosa thus gains a competitive advantage over a bacterium that would otherwise outgrow P. aeruginosa in the absence of flow. We observed similar findings when P. aeruginosa was co-cultured with Staphylococcus aureus (Fig. S3E, Movie S4B), which co-colonizes the lungs of cystic fibrosis patients alongside P. aeruginosa [26], and with the pathogen Salmonella enterica serovar Typhimurium (Fig. S3F). These results demonstrate that in a flow environment, bacterial species can self-organize to co-exist in separate micro-environments, despite one species having an apparent growth advantage over the other.
Figure 3. Pathogens self-segregate to co-exist in a branched flow network.
(A) Surface colonization of P. aeruginosa (green) and P. mirabilis (red) cells in a branched network. Both strains were initially co-inoculated in equal numbers into the seeded branch. After 15 hours of continuous flow, P. aeruginosa colonized the upper, seeded, and side branches while P. mirabilis colonized only the seeded branch. Dashed box (top) shows a region that is 100 μm upstream from the indicated section. The effluent (containing planktonic and surface-detached cells) from each branch was analyzed for (B) total cell number using optical density and (C–D) population composition using fluorescence microscopy. P. mirabilis was the dominant species in the seeded branch while P. aeruginosa was the only species that colonized the side branch. ND indicates cells were not detected. Error bars in (D) indicate standard deviation. Scale bars in (A) and (C) indicate 100 μm and 5 μm, respectively. Experiments were performed in triplicate.
Upstream dispersal is characterized by counter-advection and later diffusion
To improve our ability to understand and ultimately disrupt P. aeruginosa dispersal and colonization, we developed a quantitative model of upstream dispersal (for a detailed derivation, see Supplemental Information). Upstream migration within a population is a phenotypically diverse cycle consisting of: upstream movement on surfaces, detachment from the surface, and downstream re-attachment to the surface (Fig. 4A). As cells move upstream along the flow axis (x-axis), cells also move and repeatedly switch direction along the transverse (y) axis, resulting in a zig-zag path (Fig. 2E). This behavior in single cells is described by the differential equation: , where σ(x,y,t) is the population density of cells at positions x and y and at time t that travel upstream at a velocity v, duplicate at a rate α, and detach from the surface at a rate β; D is the effective diffusion coefficient. Upstream dispersal is thus a novel type of movement that is counter-advective along the flow axis and diffusive along the lateral axis. We solved this equation (see Supplemental Information) for a population of cells with distinct upstream velocities [4] in a branched flow network. Our model (see Supplemental Information for parameters) accurately predicts the population density distribution data in linear channels (Fig. 4B–C) and in branched flow networks (Fig. 4D) and shows that the number of cells that enter the streamlines of the side branch increases exponentially with time (Fig. 4E). As bacterial growth is also exponential with time, the combined effects of exponential influx and growth result in rapid colonization of the side branch.
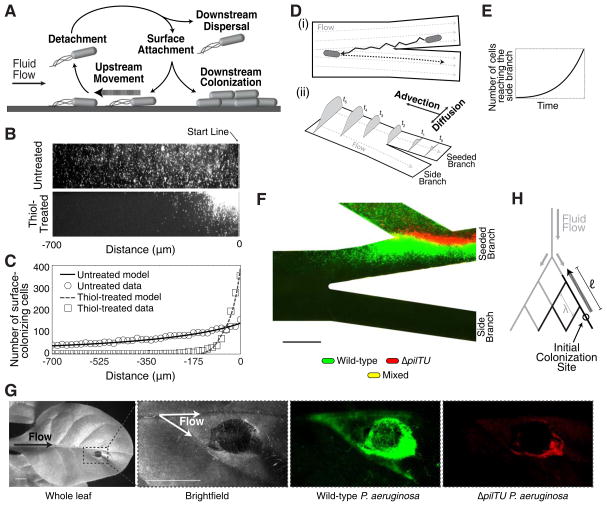
Figure 4. Theoretical and natural host models of upstream dispersal.
(A) Schematic illustrating the diverse motility modes observed during upstream dispersal. (B) Surface colonization of wild-type P. aeruginosa after 15 hours of flow in untreated and thiol-treated linear channels. (C) Corresponding population density data for untreated (circles) or thiol-treated (boxes) devices and population densities predicted by our upstream dispersal model (lines). (D) Schematic depicting the mechanism of upstream dispersal. (i) Cells move upstream in a zig-zag trajectory on surfaces, enter side branch streamlines, detach from the surface, and are carried downstream by the flow. (ii) Time-evolution of a surface population that advances towards a branching intersection through counter-advection and lateral diffusion. (E) Prediction from our model that the number of cells entering side-branch streamlines increases exponentially with time. (F) Surface colonization of wild-type (green) and surface-motility defective ΔpilTU (red) P. aeruginosa cells in a thiol-treated device after 15 hours of continuous flow. No cells were observed in the side channel (see Fig. S4A). (G) Plant colonization assay in which a tobacco plant leaf was inoculated with equal numbers of wild-type and ΔpilTU P. aeruginosa cells. After 7 days, wild-type cells (green) were observed in the upstream vasculature while ΔpilTU cells (red) were found at the inoculation site. (H) Schematic showing a generalized branched flow network (gray) with characteristic pore spacing λ and colonization of the network by bacterial communities (black) that migrate upstream a distance ℓ. Scale bars represent 100 μm in (F) and 5 mm in (G).
We tested a prediction of our model that decreasing the surface detachment rate (β) and migration velocity restricts population expansion and prevents dispersal to the side branch. We coated channel surfaces with the thiol compound (3-mercaptopropyl)trimethoxysilane to promote sulfide bond formation with proteins on the bacterial cell surface, thereby increasing cell adhesion to the surface (i.e., making the surface more “sticky”). Thiol-treated linear channels inhibited the expansion of the P. aeruginosa population in linear channels (Fig. 4B–C) and prevented cells from entering the side branch (Figs. 4F and S4A), in good agreement with the predictions of our model of upstream dispersal.
Upstream dispersal promotes colonization in plant vasculature
To establish the role of upstream dispersal in the colonization of the vasculature of a natural host of P. aeruginosa, we followed colonization of the tobacco plant Nicotiana tabacum. In these plants, fluid and nutrients flow from the main stem towards the periphery of the leaf vasculature in a branched network (Fig. 4G). We inoculated plant leaves with an equal mixture of wild-type P. aeruginosa (expressing GFP) and ΔpilTU cells (expressing mCherry) at the periphery using syringe infiltration. After 7 days, wild-type cells were found in the vasculature upstream from the inoculation site (towards the main stem) (Fig. 4G and S4B–D). In contrast, ΔpilTU cells were found only at the inoculation site, resulting in a much more localized infection. Our results are thus consistent with upstream motility representing an important factor driving branched vasculature colonization in vivo.
Discussion
Understanding how single-cell behaviors give rise to the population-level dynamics of P. aeruginosa in flow provides a quantitative framework for how P. aeruginosa colonization is established and how infection spreads within a host. Our results suggest that P. aeruginosa responds to fluid flow as a host cue by moving in the opposite direction of the flow. By employing diversified strategies of migrating upstream, downstream, and laterally through counter-advection and diffusion, P. aeruginosa communities gain a selective growth advantage in fluid flow. This combination of behaviors enables P. aeruginosa populations to disperse rapidly throughout a fluid-filled vascular network since the population expands into a new branch each time it reaches an intersection (Fig. 4H). Since the number of cells that reach a branch junction is exponential with respect to time (Fig. 4E), our model suggests that the upstream advancement of P. aeruginosa to a central branch node is catastrophic for the host. Indeed, our experiments demonstrate that only a small number of cells need to reach the branch junction for the establishment of large populations downstream (Figs. 2A–B and 3A–B). To more fully understand the dispersal of bacteria through generalized flow networks, it will be necessary to integrate additional factors into the model, including the network topology, pore spacing between branch intersections (λ), upstream migration distance (ℓ), and migration velocity (Fig. 4H). It will also prove important to study the effects of time-periodic flow patterns such as those observed in the urinary tract.
Importantly, our model highlights strategies for minimizing colonization of flow networks. Specifically, the advantage gained by upstream dispersal can be eliminated by increasing surface adhesion. These findings have immediate implications for the design of medical devices in order to contain P. aeruginosa colonization and suggest alternative approaches to disrupting P. aeruginosa colonization and infection within hosts.
The ability of bacteria to self-segregate into stably co-existing niches has significant implications for pathogens and beneficial commensals. In particular, as fluid flow is prevalent in the digestive tract, understanding how different bacterial species disperse and establish colonization in flow advances our understanding of the forces that shape the composition and structure of microbial communities, as well as our ability to manipulate these populations to improve health. We find that unexpected population-level dynamics emerge from the complex interactions between shear forces, the polar organization of bacterial motility structures, and the architecture of fluidic networks. For example, the polar localization of TFP may have evolved as an adaptation to promote competition in branched flow networks. As many bacteria posses polar-localized TFP [8], upstream dispersal may be a generalized mechanism by which surface-motile bacteria colonize hosts. Thus, while flow has thus far been largely considered in the context of its effects on the motility of individual bacteria, our study shows that flow is a critical factor that drives the colonization, competition, and dispersal of bacterial populations in host environments. Finally, taken together with recent findings [27], the upstream dispersal of P. aeruginosa provides further evidence that the bacterial response to mechanical forces plays an important role in pathogenic processes.
EXPERIMENTAL PROCEDURES
Methods summary
Strains were grown in LB Broth to mid-exponential phase. For competition experiments, strains were grown separately and mixed in equal number immediately before loading into devices. Microfluidic devices were constructed using standard soft photolithography techniques. Effluent from each branch of the device was collected into culture tubes and incubated for 3.5 hours at 37°C in order to amplify the total number of cells for single-cell microscopy analysis. The total cell number in the effluent was measured by optical density. P. aeruginosa colonization was tracked in Nicotiana tabacum tobacco plants that were exposed continuously to a fluorescent growth light for 7 days. Details on our model of bacterial dispersal by upstream migration, bacterial strains and culture conditions, microfluidic device construction and experimental conditions, fluorescence microscopy, leaf vasculature imaging, swimming assays, planktonic growth rate measurements and competitions, plant colonization, transmission electron microscopy and surface velocity measurements are described in the Experimental Procedures section of the Supplemental Information.
Supplementary Material
Acknowledgments
We thank A. Binns and N. Ouzounov for plant materials and J. Gleghorn for a chicken embryo vasculature image. This work was supported by the National Institutes of Health (NIH) Director’s New Innovator Award (1DP2OD004389) to Z.G., a National Science Foundation Grant (1330288) to Z.G. and H.A.S., an NIH NRSA postdoctoral fellowship (F32AI095002) to A.S., and a graduate fellowship from the STX Scholarship Foundation to M.K.K.
Footnotes
AUTHOR CONTRIBUTIONS
A.S. and M.K.K. designed and carried out experiments, performed data analysis, and wrote and revised the manuscript. A.S. constructed the quantitative model and H.A.S. revised it. Y.S. carried out experiments and revised the manuscript. H.A.S. and Z.G. designed experiments, provided feedback on results, and revised the manuscript. All authors discussed the results.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Marcos, Fu HC, Powers TR, Stocker R. Bacterial rheotaxis. Proc Natl Acad Sci U S A. 2012;109:4780–4785. doi: 10.1073/pnas.1120955109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill J, Kalkanci O, McMurry JL, Koser H. Hydrodynamic surface interactions enable Escherichia coli to seek efficient routes to swim upstream. Phys Rev Lett. 2007;98:068101. doi: 10.1103/PhysRevLett.98.068101. [DOI] [PubMed] [Google Scholar]
- 3.Meng Y, Li Y, Galvani CD, Hao G, Turner JN, Burr TJ, Hoch HC. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J Bacteriol. 2005;187:5560–5567. doi: 10.1128/JB.187.16.5560-5567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen Y, Siryaporn A, Lecuyer S, Gitai Z, Stone HA. Flow directs surface-attached bacteria to twitch upstream. Biophys J. 2012;103:146–151. doi: 10.1016/j.bpj.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodey GP, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5:279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 6.Fluit AC, Schmitz FJ, Verhoef J, European SPG. Frequency of isolation of pathogens from bloodstream, nosocomial pneumonia, skin and soft tissue, and urinary tract infections occurring in European patients. Eur J Clin Microbiol Infect Dis. 2001;20:188–191. doi: 10.1007/s100960100455. [DOI] [PubMed] [Google Scholar]
- 7.Vincent JL, Bihari DJ, Suter PM, Bruining HA, White J, Nicolas-Chanoin MH, Wolff M, Spencer RC, Hemmer M. The prevalence of nosocomial infection in intensive care units in Europe. Results of the European Prevalence of Infection in Intensive Care (EPIC) Study. EPIC International Advisory Committee. JAMA. 1995;274:639–644. [PubMed] [Google Scholar]
- 8.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 9.Ku DN. Blood flow in arteries. Annual Review of Fluid Mechanics. 1997;29:399–434. [Google Scholar]
- 10.Motomiya M, Karino T. Flow patterns in the human carotid artery bifurcation. Stroke. 1984;15:50–56. doi: 10.1161/01.str.15.1.50. [DOI] [PubMed] [Google Scholar]
- 11.Zimmermann MH. Xylem structure and the ascent of sap. Berlin ; New York: Springer-Verlag; 1983. [DOI] [PubMed] [Google Scholar]
- 12.Dimond AE. Pressure and flow relations in vascular bundles of the tomato plant. Plant Physiol. 1966;41:119–131. doi: 10.1104/pp.41.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zamir M. Distributing and delivering vessels of the human heart. J Gen Physiol. 1988;91:725–735. doi: 10.1085/jgp.91.5.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West GB, Brown JH, Enquist BJ. A general model for the structure and allometry of plant vascular systems. Nature. 1999;400:664–667. [Google Scholar]
- 15.Weart RB, Lee AH, Chien AC, Haeusser DP, Hill NS, Levin PA. A metabolic sensor governing cell size in bacteria. Cell. 2007;130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin F, Conrad JC, Gibiansky ML, Wong GC. Bacteria use type-IV pili to slingshot on surfaces. Proc Natl Acad Sci U S A. 2011;108:12617–12622. doi: 10.1073/pnas.1105073108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kantsler V, Dunkel J, Blayney M, Goldstein RE. Rheotaxis facilitates upstream navigation of mammalian sperm cells. Elife. 2014;3:e02403. doi: 10.7554/eLife.02403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coker C, Poore CA, Li X, Mobley HL. Pathogenesis of Proteus mirabilis urinary tract infection. Microbes Infect. 2000;2:1497–1505. doi: 10.1016/s1286-4579(00)01304-6. [DOI] [PubMed] [Google Scholar]
- 20.Ronald A. The etiology of urinary tract infection: traditional and emerging pathogens. Am J Med. 2002;113(Suppl 1A):14S–19S. doi: 10.1016/s0002-9343(02)01055-0. [DOI] [PubMed] [Google Scholar]
- 21.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerckhoffs AP, Ben-Amor K, Samsom M, van der Rest ME, de Vogel J, Knol J, Akkermans LM. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J Med Microbiol. 2011;60:236–245. doi: 10.1099/jmm.0.022848-0. [DOI] [PubMed] [Google Scholar]
- 23.Finegold SM, Sutter VL, Mathisen GE. Normal indigenous intestinal flora. In: Hentges DJ, editor. Human intestinal microflora in health and disease. New York: Academic Press; 1983. pp. 3–33. [Google Scholar]
- 24.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 25.Tuson HH, Copeland MF, Carey S, Sacotte R, Weibel DB. Flagellum density regulates Proteus mirabilis swarmer cell motility in viscous environments. J Bacteriol. 2013;195:368–377. doi: 10.1128/JB.01537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sagel SD, Gibson RL, Emerson J, McNamara S, Burns JL, Wagener JS, Ramsey BW Inhaled Tobramycin in Young Children Study G, Cystic Fibrosis Foundation Therapeutics Development N. Impact of Pseudomonas and Staphylococcus infection on inflammation and clinical status in young children with cystic fibrosis. J Pediatr. 2009;154:183–188. doi: 10.1016/j.jpeds.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siryaporn A, Kuchma SL, O’Toole GA, Gitai Z. Surface attachment induces Pseudomonas aeruginosa virulence. Proc Natl Acad Sci U S A. 2014;111:16860–16865. doi: 10.1073/pnas.1415712111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.