Abstract
Purpose
Exosomes are cell derived extracellular nanovesicles that relay molecular signals pertinent to both normal physiologic and disease processes. The ability to modify and track exosomes in vivo is essential to understanding exosome pathogenesis, and for utilizing exosomes as effective diagnostic and therapeutic nanocarriers to treat diseases.
Methods
We recently reported a new electroporation method that allow exosomes to be loaded with superparamagnetic iron oxide nanoparticles for magnetic resonance tracking.
Results
Building on this approach, we now demonstrate for the first time using a C57BL/6 mouse model that melanoma exosomes can be imaged in vitro, and within lymph nodes in vivo with the use of standard MRI approaches.
Conclusion
These findings demonstrate proof of principle that exosome biology can be followed in vivo and pave the way for the development of future diagnostic and therapeutic applications.
INTRODUCTION
Exosomes are nanovesicles typically smaller than 100 nm (1) that are released by cells into the extracellular environment (2). Exosome biology has emerged as one of the most active fields in cancer research over the last five years in light of the wealth of information available for biomarker studies, and with respect to the intriguing and fundamental mechanistic aspects of cellular communication under both physiological and pathological conditions (2). We have previously reported the use of dynamic light scattering and fluorescence imaging to rapidly isolate and label exosomes for in vitro and in vivo tracking and experimentation (3,4). With the use of these techniques, we showed that melanoma exosomes facilitate angiogenesis (3) and pre-metastatic niche formation in lymph nodes that promotes local population and growth of tumor cells in a "prepared turf" (4).
More recently, electroporation has been used to load exosomes with drug (5) or RNA cargo (6–8) or 5 nm superparamagnetic iron oxide nanoparticles (SPION5) (9). Building on these previous findings, we now demonstrate that SPION5 loaded melanoma exosomes can be detected by MRI, and they retain the same lymph node homing properties previously demonstrated for fluorescently labeled exosomes (4).
METHODS
Exosome Isolation
Mouse B16-F10 (CRL 6475) melanoma cells and media were purchased from American Type Culture Collection (August 2008), MAP (mitogen-activated protein), and mycoplasma tested for purity and kept frozen at − 80ºC under liquid nitrogen until resuscitated for use. B16-F10 melanoma cells were maintained in normal culture media containing 90% DMEM (Dulbecco's modified Eagle's medium) and 10% heat inactivated FBS at 37ºC and 5% CO2. Cultures were grown to 70% confluence in three 300 cm2 flask. Culture media was removed and cells washed in PBS. Cells were cultured for 48 hours in the presence of conditioned media. Conditioned culture media was prepared by subjecting normal culture media to overnight ultracentrifugation at 110,000 x g to remove bovine exosomes (10). B16 melanoma exosomes were collected from 48 hour culture in conditioned media through standard differential centrifugation steps using a 70 Ti rotor as previously described (10). Briefly, culture media was diluted 1:1 in 50 mM trehalose PBS (9), spun and supernatants collected from 300 x g for 10 min, 2000 x g for 10 min, to remove residual cells and debris, 10,000 x g for 30 min to remove microparticles (11), and 100,000 x g for 2 h in the presence of 1.0 μM DiI (InVitrogen, CA) to fluorescently label the exosome membranes (3). Exosome pellets were resuspended in 1 ml of 50 mM trehalose PBS and stored at −80ºC until use (9). Protein content was measured by bicinchoninic acid assay (Thermo Fisher Scientific Inc., IL).
SPION5 Loading of Exosomes
Exosomes (50 μg total protein) were re-suspended in 0.75 ml of 50 mM trehalose PBS containing 0.25 μg/ml SPION5 iron (Ocean Nanotech, SHP05 water soluble iron oxide nanoparticles (4.5 nm in size by manufacturer TEM) with a carboxylic acid coating) at 4ºC as described previously (9). Suspended exosomes were electroporated (EP) in 4 mm path length electroporation cuvettes using a BTX Harvard Apparatus ECM 399 system (Holliston MA) (9). A single pulse was applied to each exosome sample under the high voltage setting and at an electric field of 0.75 kV/cm. Following electroporation, SPION5 loaded exosomes were washed and re-isolated by ultracentrifugation at 100,000 x g for 2 hours to remove extravesicular SPION5 (9).
In vitro characterization of exosomes
Exosome size was determined with dynamic light scattering as described previously (3). SPION5 concentration of exosome pellets was measured in units of mg/ml of iron according to manufacturer’s instructions (Ocean Nanotech). The iron content of electroporated exosomes was calculated using the equation: , where OD is the optical density at a particular wavelength of light and 4.3 is the extinction coefficient for SPION5. Using this equation, exosomes loaded with SPION5 by EP were found to contain 0.549 μg iron per μg exosomal protein, which is consistent with the total iron content previously reported by inductively coupled plasma optical emission spectroscopy in purified exosome peak fractions (9).
In vitro MRI
To provide a standard curve that could be used to quantify exosome content in vivo by MRI, T2*-weighted images of prepared sample phantoms made by serial dilution of known concentrations of exosomes were acquired at 4.7 T with a Varian small animal scanner using a custom-built solenoid radio frequency (RF) coil and multi-echo gradient echo sequence. The exosome concentration in each phantom was 2.8, 0.56, 0.11, 0.022 and 0.0044×108 exosomes per ml of 50 mM trehalose PBS solution. Exosome concentrations were estimated using a standard curve relating known concentrations of synthetic liposomal exosome mimetics to dynamic light scattering counts per second as described previously (4). The imaging parameters for the sample phantoms included a repetition time (TR) of 1000 ms, an echo time (TE) of 5 to 40 ms, a volume element (voxel) size of 0.2 mm×0.2 mm×1 mm, and a flip angle (FA) of 10 degrees.
Animal preparation
All procedures, including animal treatments and MRI conformed to the guidelines and approvals by the Animal Studies Committee at Washington University in St. Louis. C57BL/6 mice (n=20) were divided into two equal groups. The first group underwent in vivo MRI for dynamic tracking of free SPION5 or SPION5 loaded exosomes for detection of homing to popliteal lymph nodes (PLN). The other group was used for ex vivo imaging with the use of a Xenogen in vivo imaging system (IVIS) spectrum workstation (Caliper Life Science, USA) followed by histological analysis with fluorescence microscopy. In each group, animals received a bolus administration of 50 μl of either 27.45 μg of free SPION5 (n =5) iron or a bolus administration of exosomes (n =5, 50 μg of exosome protein content) containing a total of 27.45 μg of SPION5 iron in 50 mM trehalose PBS into the left food pad to facilitate tracking to local lymph nodes as described previously (4). Ipsilateral and contralateral PLNs were visualized with both MRI and in vivo fluorescent imaging. Animals were sacrificed 48-hour after treatment and both left and right PLNs were dissected. PLNs were weighed on an analytical balance (Mettler Toledo, USA) and embedded and frozen in OCT media (Tissue-Trek, Japan) for node fluorescent imaging ex vivo, and for histology and fluorescence microscopic analysis.
In vivo MRI
In vivo MRI was performed on a 4.7T Varian small animal scanner. A custom-built, actively decoupled, transmit and receive coil pair was employed to achieve optimized detection sensitivity. The transmit coil (5 cm by 10 cm) was built to a mouse with superior field homogeneity and power transmission efficiency. Mice were imaged in supine position and the surface receive coil (2 cm by 2.5 cm) was placed around the knees and adjusted to match body curvature. Based on our previous investigations (4), three time points were selected for in vivo MRI including: baseline image prior to treatment, 1-hour post treatment, and 48-hour post treatment. During in vivo imaging, mice were anesthetized with 1% isoflurane mixed with oxygen at a flow rate of 1 L/min. For each imaging time point, a multi-slice T1-weighted and T2*-weighted gradient echo image was acquired in the transverse plane to visualize both ipsilateral and contralateral PLNs. A fat-saturation pre-pulse was employed to improve visualization of lymph nodes within surrounding fat tissue. Detailed imaging parameters included: TR = 100 ms, TE= 5 ms (for T1-weighed images) or 15 ms (for T2*-weighted images), voxel size = 0.2 mm × 0.2 mm ×1 mm and flip angle = 10°. Quantitative evaluation of the voxel-wise T2* value was derived from the T2*-weighted and T1-weighted images using two-point mono-exponential fitting.
Imaging and histology of PLNs ex vivo
Fluorescence images of excised exosome treated PLNs were captured using IVIS. Frozen tissue blocks were sectioned into 8 μm slices for histology and fluorescence microscopy. Prussian blue staining and fluorescent imaging were performed on both left (ipsilateral) and right (contralateral) PLNs to visualize iron delivery and detect fluorescent exosomes. Both histological and fluorescence images were acquired with an Olympus BX61 fluorescent microscope. DAPI Vectashield mounting media (Vector Laboratories, Inc., USA) was used to visualize cell nuclei. For calculating changes in cross sectional area by MRI, the slice with the largest cross sectional nodal area was used for defining a region of interest (ROI). The area of the ROI was calculated using the fat saturated T1 weighted MRI image of the slice given that it allows clear delineation of node boundaries.
Data processing and statistics
Image processing was performed with a custom-designed program written in Matlab (MathWorks, USA), including T2* fitting and image co-registration. All statistics were performed in Origin (OriginLab, USA), in which paired-t tests and ANOVA of repeated measurements were used for hypothesis testing.
RESULTS
MRI of SPION5 loaded exosomes
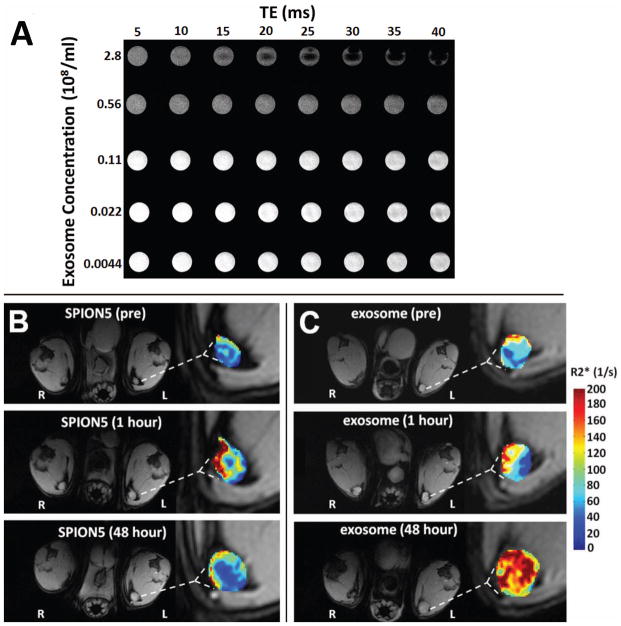
Previously we determined by TEM that exosomes loaded with 5 nm SPIONs exhibit a diameter of approximately 100 nm, which does not differ from that of unmodified exosomes (9). The iron concentration of SPION5 exosomes was determined by optical density to be ~ 0.5 μg iron per μg exosome protein. In vitro MRI revealed that as the concentration of SPION5 exosomes in solution increased, T2*-weight images exhibited stronger negative contrast, thereby confirming the expected field inhomogeneity effect generated by SPION5-loaded exosomes (Fig. 1A).
Figure 1.
MRI of SPION5 loaded melanoma exosomes in vitro and in vivo. (A) T2*-weighted multi-echo gradient echo images of SPION5 loaded exosomes in 50 mM trehalose PBS at different concentrations in vitro. (B) Representative free SPION5 and (C) SPION5 loaded exosome trafficking to ipsilateral (left) and contralateral (right) PLNs as visualized by T1-weighted images with R2* mapping of PLNs ipsilateral to the injection site.
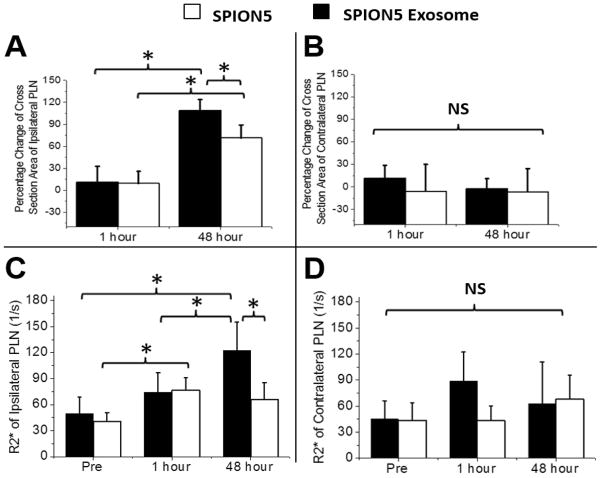
Having successfully imaged SPION5 exosomes in vitro we sought to determine whether they would travel to and be detected in mouse PLNs in vivo, in analogy to our previous reports for fluorescently labeled exosomes (4). T1-weighted images revealed that both free SPION5 (Fig. 1B) and SPION5 exosomes (Fig. 1C) induced visually appreciable nodal enhancement and apparent enlargement 48 hours after initial food pad injection. Moreover, animals receiving SPION5 loaded exosomes displayed a significantly larger (p < 0.05) increase in the cross sectional area (+110 %) of ipsilateral PLNs over pre-treatment baseline as compared to free SPION5 (+80%) (Fig. 2A). This observation is likely a consequence of either SPION5 exosome trapping or induction of inflammatory signaling pathways within lymph nodes (4) beyond that of free SPION5. The cross sectional area of contralateral PLNs however, was not affected by either treatment (Fig. 2B). No ipsilateral or contralateral nodal enlargement was observed at 1-hour post injection, suggesting that exosomal trapping was less likely the cause of nodal enlargement as compared with inflammatory responses.
Figure 2.
MRI analysis of lymph nodal cross sectional area and SPION5 uptake. (A) Ipsilateral and (B) contralateral PLN cross sectional area following treatment with free SPION5 and SPION5 melanoma exosomes. The percentage increment was calculated based on the difference between post-treatment and pre-treatment cross sectional area. * = p < 0.05. R2* values indicative of SPION5 accumulation in both ipsilateral (C) and contralateral (D) PLNs . Accumulation of iron in lymph nodes was directly correlated with the enhancement of nodal R2* signal. * = p < 0.05.
Further analysis of the data revealed that despite injecting the same quantify of SPION5 material, the deposition of SPION5 in the ipsilateral node was significantly greater for SPION5 carried by exosomes than for free SPION5 at 48 hours (Fig. 2C). Additionally, we observed no difference in the MRI signal from the nodes treated with SPION5 exosomes when comparing pre-injection and 1 hour post injection images. In contrast, the signal from nodes treated with free SPION5 was significantly higher after 1 hour than at pre-injection. However, between 1 hour and 48 hours post injection, free SPION5 signal did not increase, whereas SPION5 exosome signal increased significantly. No difference was observed between free SPION5 and exosome carried SPION5 in the lymph node contralateral to the injection site at any time point (Fig. 2D).
Histology and fluorescence microscopy of lymph nodes
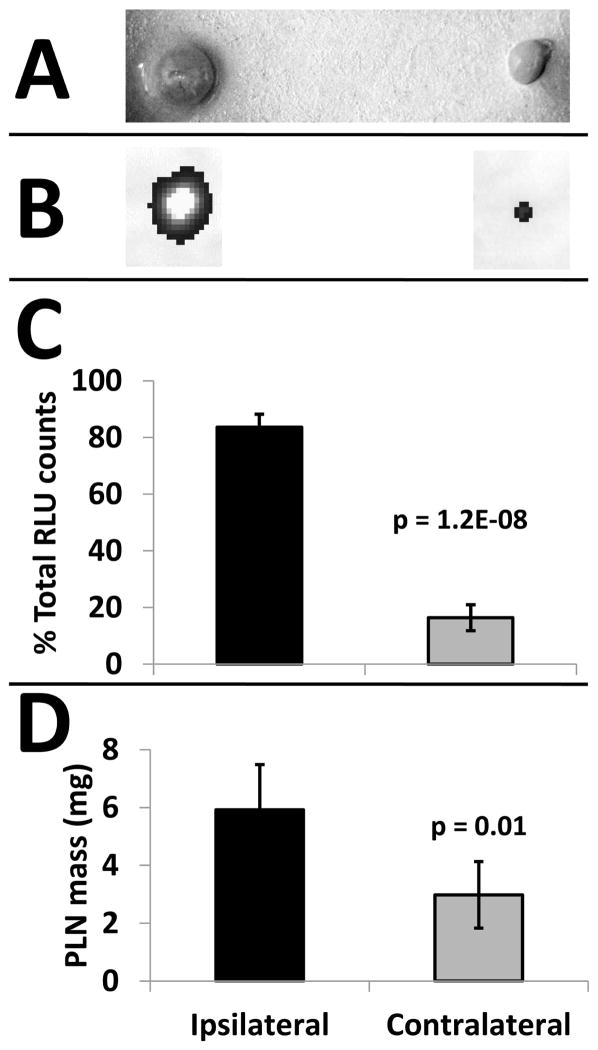
Gross (Fig. 3A) and fluorescent (Fig. 3B) visualization of ipsilateral and contralateral lymph nodes 48 hours after treatment with fluorescent SPION5 exosomes further confirmed the MRI findings in that more SPION5 exosomes accumulate in the ipsilateral lymph node (Fig. 3C), and ipsilateral lymph node mass was higher (Fig. 3D). The ratio of ipsilateral (84 +/− 5%) to contralateral (16+/− 5%) in vivo signal retention at 48 hours (Fig. 3C) is consistent with our previous findings demonstrating unilateral homing of exosomes in interconnected lymph nodes networks with increased sentinel node mass (4) and significant ipsilateral retention of exosome associated SPION5 versus free SPION5 (Fig. 2B). The unilateral retention pattern of exosome associated fluorescent or SPION5 cargo contrast with that of control fluorescent liposomal exosome mimetics (4). In a previous study we demonstrated that synthetic exosome mimetic particles exhibited a 50:50 bilateral distribution in lymph node networks compared to exosomes (4). As observed in the current study, free SPION5 exhibits the same 50:50 bilateral distribution pattern (Fig. 2C, D) as do the control exosomal mimetics. These data demonstrate that exosomes are retaining their iron cargo for at least 48 hours or we would have observed a shift in the bilateral distribution pattern of SPION5 that differs from the distribution of the fluorescent label which is not known to exhibit free dye transfer in the absence of a membrane carrier (12). Collectively, these results demonstrate that more SPION5 accumulates in the ipsilateral lymph node if delivered by exosomes, and that the time required for exosome delivery of SPION5 to the lymph nodes is greater than the trafficking time of free SPION5. This finding implicates exosome homing and retention in sentinel lymph nodes, in contrast to unrestricted diffusion of free SPION5 throughout the lymph node network similar to what we previously observed for inert liposomes (4).
Figure 3.
Fluorescent SPION5 exosomes home to the PLN ipsilateral to the injection site. (A) Photograph of representative excised PLNs 48-hours after exosome treatment. (B) Representative IVIS fluorescence images (grayscale) of PLNs 48-hours after exosome treatment. (C) Fluorescence quantification and (D) ex vivo mass measurement of PLNs 48-hour after exosome treatment.
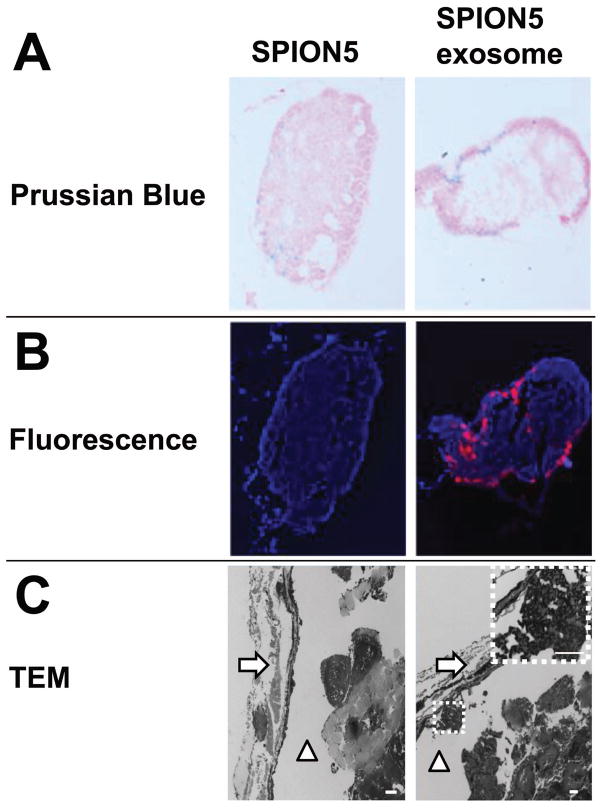
Histological examination of extracted free SPION5 and SPION5 exosome treated ipsilateral PLNs at 48 hours revealed differential Prussian blue iron staining. SPION5 signal was distributed homogeneously throughout the entire lymph node for free SPION5 in contrast to the predominant subcapsular distribution of exosome associated SPION5 (Fig. 4A). Fluorescence microscopy (Fig. 4B) and TEM (Fig. 4C) of lymph node cross sections further confirmed the subcapsular distribution pattern of exosomes. Moreover, clusters of exosomes were observed by TEM lying adjacent to the subcapsular lymph node membrane (Fig. 4C).
Figure 4.
Fluorescence and Prussian blue staining of PLNs in free SPION5 and exosome treated groups at 48-hour post treatment. (A) Prussian blue iron staining of a central PLN cross section. (B) Fluorescence microscopy of a central PLN cross section; red = DiI-exosomes, blue = DAPI. (C) Transmission electron microscopy of a central PLN cross section; white arrows denote the lymph node capsule, white triangles denote the subcapsular space. The highlighted region denoted by a white dotted line depicts a cluster of exosomes in the subcapsular space. White scale bars = 2 μm.
DISCUSSION
It has been known for some time that free SPIONs (13), cells (14–16) or liposomes (17) loaded with SPIONs can be tracked in vivo by way of MRI for research, diagnostic or therapeutic purposes. For example, SPIONs have served as non-toxic, naturally eliminated theranostic agents in previous investigations to treat tumors by means of magnetic hyperthermia (13). Expanding on the previous applications for SPIONs, the data presented herein demonstrate for the first time that natural exosomes loaded with SPION5 cargo can be imaged and tracked by MRI in vivo. Previously, we demonstrated that melanoma exosomes appear to be trafficking to a specific microanatomical destination in lymph nodes known as the subcapsular sinus (4). TEM now strongly supports this finding for SPION5 loaded exosomes as well (Fig. 4C). These enabling observations contribute to our basic understanding of exosome biology given the ability of melanoma exosomes to modulate production of extracellular matrix factors (4), which are a key component of lymph node scaffolding and organizational structure. The implication is that melanoma exosomes preferentially home to certain resident structural regions of the lymph node. This finding may provide impetus for the development of new diagnostic and therapeutic strategies to inhibit tumor exosome mediated pathogenesis. Given that it is now appreciated that melanoma exosomes can prepare sentinel lymph nodes for metastasis (4), future experiments will be necessary to investigate the exact mechanisms whereby melanoma exosomes participate either directly or indirectly through resident and/or transient lymph node cells to influence subscapular lymph node architecture prior to melanoma metastasis. Moreover, additional investigations will be required to assess exosome retention of SPION5 beyond the 48 hour time point in lymph nodes and may require complex multi-modal imaging approaches to distinguish free SPION5 from free exosomes and exosome associated SPION5. Other potential novel imaging sequences, such as SWIFT, UTE, etc. that can generate positive contrast for SPIO could be exploited to further characterize the heterogeneous distribution of exosomes with high spatial resolution.
Future clinical investigations exploring in depth functional or therapeutic applications of personalized exosomes in vivo might be possible. Recently, it was discovered that exosomes can induce and utilize tunneling nanotubes for intercellular transport (3,18). This highlights a potential advantage exosomal semi-synthetics, expressing natural and conformationally correct targeting receptors, have over pure synthetic nanocarriers. Exosomes have privileged access to transcellular destinations that may not be accessible to pure synthetic nanocarriers using current targeting strategies. Thus, SPION5 loaded exosomes might be specifically tailored using endogenous molecular cell based nanofactories and/or exogenous synthetic exosome modification to simultaneously detect and treat pathogenic microenvironments in vivo (2). One such clinical application might be to detect residual tumor cells in debulked primary tumor and lymph node microenvironments. This would entail limited localized injection of personalized “tracer” exosomal semi-synthetics followed by imaging for residual tumor cells. Further advancements in our understanding of exosome trafficking also may one day allow for intravenous injections of exosomal semi-synthetics and tracking to deep tissue sites harboring melanoma nests for example by MRI.
CONCLUSION
The ability to begin to track natural exosomes in vivo using conventional MRI approaches represents a significant advancement in translational exosome science. This introduces the potential for high resolution and sensitive noninvasive tracking of exosomal nanomedicines.
Acknowledgments
L. Hu designed and performed all imaging experiments, analyzed MRI data, and prepared portions of the manuscript. J. L. Hood and S. A. Wickline conceived the experimental design and concepts, analyzed and reviewed all data, and wrote and edited the manuscript. All authors read and approved the final version of the manuscript. Additionally, we would like to thank Stacy J. Allen who provided expertise in animal care, Michael J. Scott who cultured cells and assisted in exosome isolations, Dr. Junjie Chen Ph.D. for advice on MRI procedures and Marilyn A. Levy for providing technical assistance in obtaining electron microscopy images. Financial support was provided by a combination of mechanisms including an AHA Predoctoral Fellowship Award, NIH-HL073646, NIH-DK095555, NIH-GM107894 and notably the Elsa U. Pardee Foundation.
References
- 1.Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Nolte-'t Hoen EN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH, Hochberg F. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of extracellular vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hood JL, Wickline SA. A systematic approach to exosome-based translational nanomedicine. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2012;4(4):458–467. doi: 10.1002/wnan.1174. [DOI] [PubMed] [Google Scholar]
- 3.Hood JL, Pan H, Lanza GM, Wickline SA. Paracrine induction of endothelium by tumor exosomes. Laboratory investigation; a journal of technical methods and pathology. 2009;89(11):1317–1328. doi: 10.1038/labinvest.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71(11):3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 5.Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, Wei J, Nie G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 6.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature biotechnology. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 7.Wahlgren J, De LKT, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, Valadi H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic acids research. 2012;40(17):e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kooijmans SA, Stremersch S, Braeckmans K, de Smedt SC, Hendrix A, Wood MJ, Schiffelers RM, Raemdonck K, Vader P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. Journal of controlled release : official journal of the Controlled Release Society. 2013;172(1):229–238. doi: 10.1016/j.jconrel.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 9.Hood JL, Scott MJ, Wickline SA. Maximizing exosome colloidal stability following electroporation. Analytical biochemistry. 2014;448:41–49. doi: 10.1016/j.ab.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thery C, Clayton A, Amigorena S, Raposo G. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Current Protocols in Cell Biology. 2006:3.22.21–23.22.29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 11.Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94(11):3791–3799. [PubMed] [Google Scholar]
- 12.Lassailly F, Griessinger E, Bonnet D. “Microenvironmental contaminations” induced by fluorescent lipophilic dyes used for noninvasive in vitro and in vivo cell tracking. Blood. 2010;115(26):5347–5354. doi: 10.1182/blood-2009-05-224030. [DOI] [PubMed] [Google Scholar]
- 13.Santhosh PB, Ulrih NP. Multifunctional superparamagnetic iron oxide nanoparticles: promising tools in cancer theranostics. Cancer letters. 2013;336(1):8–17. doi: 10.1016/j.canlet.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 14.Walczak P, Kedziorek DA, Gilad AA, Lin S, Bulte JW. Instant MR labeling of stem cells using magnetoelectroporation. Magn Reson Med. 2005;54(4):769–774. doi: 10.1002/mrm.20701. [DOI] [PubMed] [Google Scholar]
- 15.Walczak P, Ruiz-Cabello J, Kedziorek DA, Gilad AA, Lin S, Barnett B, Qin L, Levitsky H, Bulte JW. Magnetoelectroporation: improved labeling of neural stem cells and leukocytes for cellular magnetic resonance imaging using a single FDA-approved agent. Nanomedicine. 2006;2(2):89–94. doi: 10.1016/j.nano.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Kedziorek DA, Kraitchman DL. Superparamagnetic iron oxide labeling of stem cells for MRI tracking and delivery in cardiovascular disease. Methods Mol Biol. 2010;660:171–183. doi: 10.1007/978-1-60761-705-1_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De La Pena H, Madrigal JA, Rusakiewicz S, Bencsik M, Cave GW, Selman A, Rees RC, Travers PJ, Dodi IA. Artificial exosomes as tools for basic and clinical immunology. J Immunol Methods. 2009;344(2):121–132. doi: 10.1016/j.jim.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Thayanithy V, Babatunde V, Dickson EL, Wong P, Oh S, Ke X, Barlas A, Fujisawa S, Romin Y, Moreira AL, Downey RJ, Steer CJ, Subramanian S, Manova-Todorova K, Moore MA, Lou E. Tumor exosomes induce tunneling nanotubes in lipid raft-enriched regions of human mesothelioma cells. Experimental cell research. 2014;323(1):178–188. doi: 10.1016/j.yexcr.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]