Abstract
Gene expression is often controlled by transcriptional repressors during development. Many transcription factors lack intrinsic repressive activity but recruit co-factors that inhibit productive transcription. Here we discuss new insights and models for repression mediated by the Groucho/Transducin-Like Enhancer of split (Gro/TLE) family of co-repressor proteins.
Keywords: Groucho/TLE, repressor, transcriptional repression, RNA polymerase pausing, transcription factor, Drosophila
Abbreviations
- Drosophila
Drosophila melanogaster, Gro, Groucho
- Gro/TLE
Groucho/Transducin-like enhancer of split
- ChIP-seq
chromatin immunoprecipitation followed by high throughput sequencing, qPCR, quantitative PCR
- kb
kilobase
- TSS
transcription start site
- RNAP II
RNA polymerase II
- E(spl)
enhancer of split
- bHLH
basic helix-loop-helix
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeat
- TALENs
Transcription Activator-Like Effector Nucleases
- GAF
GAGA Factor; NELF, Negative Elongation Factor
- P-TEFb
Positive Elongation Factor b
Drosophila Groucho was the original member of the Groucho/Transducin-Like Enhancer of split (Gro/TLE) family of co-repressor proteins. This family is conserved across metazoa and includes four human (TLE1-4) and mouse Gro-related gene (Grg1-4) orthologues.1,2 Gro/TLE family proteins do not bind DNA directly, but act as repressive co-factors for a numerous and diverse range of sequence-specific DNA-binding transcription factors, including members of the Hes, Runx, Nkx, LEF1/Tcf, Pax, Six, and Fox families. Several transcription factors that recruit Gro/TLE proteins are effectors of cell fate specification pathways, including Notch, Wnt, NFKB and Estrogen Receptor signalling. Thus, Gro/TLE proteins participate in a diverse range of biological processes, including neurogenesis, somitogenesis, osteogenesis, haematopoiesis, stem cell maintenance, and proliferation. Human Gro family members have been linked to cancer pathogenesis. For example, epigenetic inactivation of the human TLE1 gene contributes to the development of hematologic malignancies,3 and Estrogen Receptor binding and transcriptional activity is regulated by TLE1 in breast cancer cells.4
Gro/TLE family members are characterized by highly conserved N-terminal and C-terminal domains.2 The conserved N-terminal domain is glutamine rich and so is known as the Q-domain. The Q-domain has been shown to mediate oligomerization of Gro/TLE proteins into tetramers and to facilitate binding of a subset of recruiting transcription factors. The highly conserved C-terminal domain contains WD-repeats and folds into a seven-blade β-propeller that binds many transcription factors.5
Although it is now over 20 years since it was first established that Drosophila Gro acts as a co-repressor,6 the precise molecular mechanisms underlying Gro/TLE-mediated repression have remained enigmatic. To gain new insight into Gro function, we took advantage of recent advances in genome-wide profiling technology and used chromatin immunoprecipitation (ChIP) followed by high-throughput sequencing (ChIP-seq) to determine how Gro is recruited to target genes and to identify factors associated with its binding.7 We opted to use cell lines (Kc167 and S2) to avoid the complications of interpreting data from mixed cell populations (e.g., embryos) where peaks obtained by ChIP may represent binding to overlapping or adjacent regulatory elements used in different cell types. In addition, use of Kc167 cells facilitated direct comparison of our data with the numerous published genome-wide datasets for transcriptional regulator binding and chromatin modifications in Kc167 cells from the modENCODE project and from the van Steensel group.8,9
Drosophila Gro was first described as mediating "long-range" repression (acting over distances of >1 kb) in 1997 in two independent studies that considered Gro acting as a co-repressor for Hairy10 and Dorsal.11 Gro was subsequently also shown to act locally over “short” distances.12 However, its reputation as a “long-range” repressor stuck, and models for Gro function have generally assumed that it acts at a distance from target promoters. One long-standing model for Gro long-range repression was that once recruited to the genome by a transcription factor, Gro oligomerized along the DNA (via its Q-domain) over several kilobases to silence gene expression (Fig. 1A).13 This model was based largely on overexpression studies comparing the ability of wild-type and non-oligomerizing Gro variants to repress specific target genes. One study by Martinez et al.14 used ChIP followed by quantitative PCR (qPCR) and detected Gro at specific sites up to 4 kb away from the site of recruitment. However, these sites were spaced 1-2 kb apart, so it was not certain that the Gro binding represented a single continuous peak from the site of recruitment.
Figure 1.
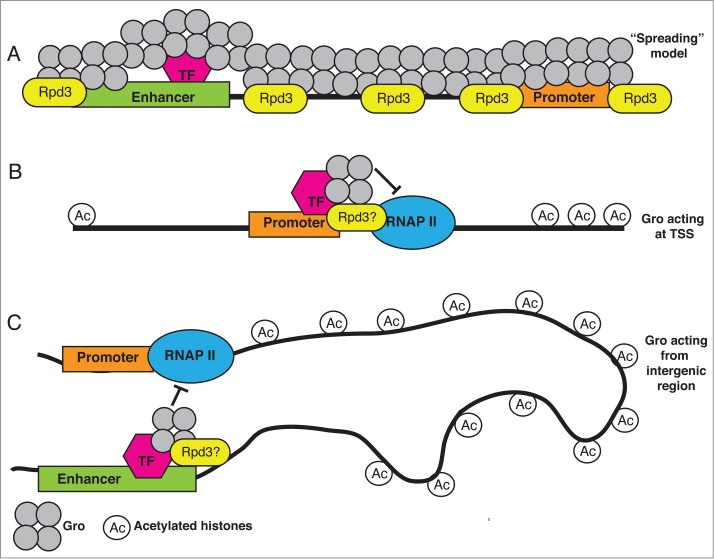
Models for Gro repression. (A) The original "spreading" model (adapted from Chen and Courey13) for long range action of Gro predicted the presence of broad domains (kilobases) of continuous Gro recruitment, which we did not observe in ChIP-seq analysis of Gro. In this model Gro recruits Rpd3 that leads to chromatin that is not acetylated. Speculative and simplified models based on our ChIP-seq analysis7 of how Gro may act both locally (B) and from a distance (C) to repress transcription. In both cases sequence-specific DNA-binding proteins recruit Gro to the chromatin, which somehow interacts (either directly or indirectly) with the RNAP II complex to inhibit transcription when bound at the TSS (B), or via chromatin looping from a distance (C). Rpd3 co-localizes with Gro at a small majority of recruitment sites. Peaks of Gro binding are associated with hypoacetylated histones, however acetylated histones are frequently observed flanking Gro peaks.
Our ChIP-seq analysis generated a series of observations in direct conflict with predictions made by the aforementioned “spreading” model in which Gro oligomerizes along several kilobases of DNA to mediate silencing at distant (>1 kb away) promoters.7 First, Gro typically bound the genome at discrete peaks spanning less than one kilobase; the median peak width was 708 bp in Kc167 cells, and 425 bp in S2 cells and fewer than 3% of peaks were greater than 2 kb. Second, oligomerization via the Q-domain did not contribute to the peak width of Gro recruitment. The average peak width bound by a GFP-tagged variant of Gro unable to oligomerize was actually slightly broader than that of GFP-(wild-type)Gro. Third, Gro was frequently found bound at annotated transcription start sites (TSSs); approximately 40% of Gro peaks overlapped TSSs in Kc167 cells (25% in S2 cells). Finally, genes that had Gro bound in peaks either upstream, at the TSS, or within the gene, were not completely silenced; these were genes that recruited RNA polymerase II (RNAP II) and expressed mRNA at some level.
From these observations, it is clear that Gro does not spread over multi-kilobase regions of chromatin in either Kc167 or S2 cells. Furthermore, meta-analysis of ChIP-seq data for the human Gro orthologue TLE3 in MCF7 cells15 revealed that the average peak width for TLE3 was not significantly different to that of Gro, indicating that it is recruited in a similar manner. However, we did notice that discrete peaks of Gro binding were frequently found in clusters (e.g., peaks within the Enhancer of split complex [E(spl)-C], Fig. 2) that could potentially be mistaken for longer continuous binding of Gro by low resolution mapping techniques (e.g., ChIP-qPCR, DamID). While it is clear that oligomerization has a role to play in Gro-mediated repression, our genome-wide binding data indicates that oligomerization does not influence the breadth of peaks of Groucho recruitment as had been previously proposed.
Figure 2.
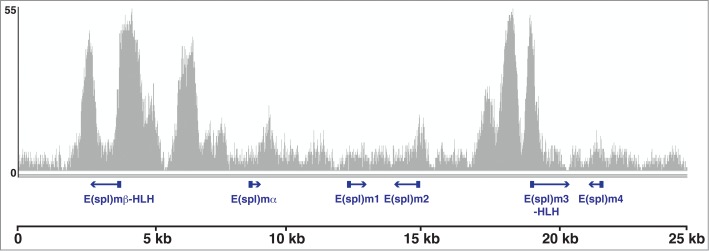
Gro binding in the Enhancer of split complex [E(spl)-C] illustrates some of the key features of typical Gro binding in Drosophila Kc167 cells. Groucho binds in distinct peaks located at transcription start sites (TSSs) as well as in intergenic regions. Gro peaks frequently occur in clusters.
Another long-standing issue we attempted to address was the relationship between Gro and histone acetylation status. Gro has been shown to physically and genetically interact with the histone deacetylase HDAC1, referred to as Rpd3 in Drosophila.16 These observations led to the model that Gro recruits Rpd3 to deacetylate histones, generating chromatin that is not permissive for transcription. Support for this model came from a series of published reports studying ectopic Gro recruitment,14,16-18 though it had long been acknowledged that Gro repression could, at most, be only partially dependent on Rpd3 in vivo.19,20 Embryos with reduced maternal Rpd3 have a segmentation phenotype that may reflect failure of repression by the products of segmentation genes that recruit Gro (e.g., Hairy, Eve, and Runt). However, embryos lacking maternal Rpd3 do not exhibit the characteristic proneural phenotype found in embryos depleted of maternal Gro, resulting from failure of Gro-mediated repression by E(spl)-HLH proteins.6
Our ChIP-seq analysis revealed that a small majority (59%) of Gro peaks in Kc167 cells overlapped peaks of Rpd3 binding, largely consistent with the previous supposition that Gro repression involves Rpd3 in some, but not all, contexts. Gro peaks were generally associated with genomic regions where histones are not highly acetylated. However, depleting cells of Gro by RNAi did not lead to detectable changes in the pattern of histone acetylation at the sites where Gro bound in wild-type cells. This result held true at the E(spl)mβ-HLH locus where transcript production was significantly upregulated in gro-RNAi cells, indicating that increased histone acetylation at Gro targets is not absolutely required for de-repression. However, we could not formally rule out the possibility that the residual levels of Gro left after RNAi treatment were sufficient to maintain histone hypoacetylation and Gro-mediated repression at most loci. Indeed, we saw relatively few significant changes in gene expression in gro-RNAi treated cells. Recent advances in genome engineering techniques including CRISPR and TALENS21 should, in the future, expedite the construction of gronull cells, allowing us to resolve the effect of Gro on Rpd3 recruitment and histone acetylation status at target loci.
ChIP-seq analysis of Gro recruitment in Kc167 cells also allowed us to look for additional factors, including histone modifications associated with Gro binding. Even though Gro acts as a repressor, we found that Gro peaks were most commonly associated with binding of factors and chromatin modifications found in “active chromatin” that is permissive for transcription. This observation was entirely consistent with results from Filion et al., who used the DamID technique to profile the genome-wide binding of 53 transcriptional regulators, including Gro.9 Moreover, Gro peaks were found at sites associated with DNase I hypersensitivity, a further indication that Gro binds in open, active chromatin.
Peaks of Gro at transcription start sites (TSSs) co-localize with RNAP II; therefore, the presence of Gro does not block RNAP II recruitment. However, we found evidence that Gro can alter RNAP II activity. The transition from transcription initiation to elongation involves a checkpoint that is a key point of control of the expression of many developmentally regulated genes (recently reviewed in refs.22-24). When this checkpoint is rate-limiting, the amount of RNAP II observed around the TSS is higher than that through the gene body; this phenomenon is often described as promoter proximal RNAP II pausing. The “pause ratio” of a transcript is the ratio of total RNAP II at the TSS to that within the gene body. In Kc167 cells, almost half of transcripts where Gro is bound at the TSS have a very high pause ratio (in the top 10% of all transcripts). Furthermore, a majority of Gro peaks overlap peaks of GAGA Factor (GAF) binding, which has previously been linked to promoter proximal pausing at many genes in Drosophila.25,26 Although we did not detect any global effects on RNAP II pausing in gro-RNAi treated cells, loss of Gro led to decreased RNAP II pausing at the E(spl)mβ-HLH locus, a validated target of Gro repression in Kc167 cells.
RNAP II pausing has been proposed as a means of attenuating the expression of genes primed for rapid activation in reponse to environmental or developmental cues. It is therefore not surprising that Gro is found to associate with paused genes, as Gro is known to repress signal-responsive genes in the absence of both Notch and Wnt signalling. For example, in the absence of incoming signals via Notch, Gro is recruited by Suppressor of Hairless [Su(H)] to primary Notch target genes [including the E(spl)mβ-HLH locus], to repress transcription. Activation of primary Notch target genes occurs swiftly after Notch signalling is triggered (<5 minutes in Drosophila DmD8 cells),27 demonstrating that Gro-mediated repression can be rapidly reversed. We found that in the absence of induced Notch signalling in Kc167 cells, loss of Gro alone was sufficient to cause reduced RNAP II pausing and increased transcript production at the E(spl)mβ-HLH locus. Thus, one action of Gro at E(spl)mβ-HLH is to promote RNAP II pausing. This raises the possibility that the many different sequence-specific DNA-binding transcription factors that recruit Gro may induce repression, at least in part, by promoting RNAP II pausing.
The current understanding of RNAP II pausing revolves largely around its release; much less is known about its establishment. Briefly, after transcribing approximately 30 nucleotides, RNAP II passes through a checkpoint where positive Transcription Elongation Factor b (P-TEFb) must phosphorylate specific amino acid residues in RNAP II, Spt5 (an RNAP II co-factor) and Negative Elongation Factor (NELF) for further transcription elongation to occur.22-24 A number of factors are known to stimulate P-TEFb activity to overcome RNAP II pausing and maximise transcription rates, including the Super Elongation Complexes (SECs) Brd4 and Myc. The factors involved in establishing paused RNAP II at specific genes are not so well understood. The 7SK snRNP inhibits P-TEFb activity, but it is not known if its actions are gene-specific or more general in Drosophila.28 GAF is associated with a subset of paused genes in Drosophila and is thought to promote RNAP II pausing through NELF recruitment.29 Interestingly, we found that the sequences recovered from Gro peaks were enriched for GAF binding motifs and that a high proportion of Gro peaks overlapped GAF peaks (82% at TSSs in Kc167 cells) raising the possibility that GAF and Gro collaborate to promote pausing. Further studies are required to determine if Gro inhibits P-TEFb activity by blocking activating factors, or if Gro recruits factors that impede P-TEFb activity and stabilise pausing.
Although we found many Gro peaks at TSSs, we also found Gro peaks further away from promoters in intergenic regions. Due to the failure to detect continuous regions of Gro bound from distant elements to promoters, we favour a model where Gro bound at distant regulatory elements is brought into the proximity of the target promoter via chromatin looping (Fig. 1). Interestingly, recent results from the Furlong lab30 revealed that the promoters of many differentially expressed genes found in stable chromatin loops were associated with paused RNAP II in Drosophila embryos. They proposed a model in which transcription factors direct loop formation, while RNAP II remains paused until specific activators come along to alleviate pausing. Paused RNAP II is thought to safeguard against premature activation while allowing genes to be poised for rapid activation.
To our initial surprise, the removal of Gro did not lead to a general up-regulation of target genes. However, on reflection, we realised that loss of repression and activation of gene expression are separable processes. We were examining stable populations of cells in culture that were not subject to changes in environmental conditions or cell-cell signalling that had the potential to induce immediate de novo expression and recruitment of activating factors to Gro target genes. Cells in a developing embryo are constantly bombarded with changing signals from neighbouring cells and are likely to be more vulnerable to a loss of repression causing rapid gene activation.
Genome-wide profiling of Gro recruitment in single cell types has led us to re-evaluate the old "spreading" model for Gro recruitment and function, but we have not been able to rule out the possibility that Gro recruits histone deacetylases to attenuate transcription. In addition, we have found that Gro is associated with paused genes and that it facilitates RNAP II pausing at a bona fide target gene. The molecular mechanisms through which Gro acts as a co-repressor for so many different transcription factors remain to be elucidated. However, this study shows that RNAP II pausing and histone acetylation status are likely to be two important aspects of Gro function. Future studies should address how Gro influences RNAP II pausing and histone acetylation status, and whether these two phenomena are linked. It will also be important to expand the set of cell types studied to understand if the patterns we observed in Kc167 and S2 cells are representative, or if the expression of other factors shifts the binding and function of Gro.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The work described in Kaul et al., 20147 was supported by a Wellcome Trust Project grant awarded to BHJ and MRC Career Development Awards to BHJ and EFS.
References
- 1. Cinnamon E, Paroush Z. Context-dependent regulation of Groucho/TLE-mediated repression. Curr Opin Genet Dev 2008; 18:435-40. [DOI] [PubMed] [Google Scholar]
- 2. Jennings BH, Ish-Horowicz D. The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol 2008; 9:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fraga MF, Berdasco M, Ballestar E, Ropero S, Lopez-Nieva P, Lopez-Serra L, Martin-Subero JI, Calasanz MJ, Lopez de Silanes I, Setien F, et al. . Epigenetic inactivation of the Groucho homologue gene TLE1 in hematologic malignancies. Cancer Res 2008; 68:4116-22. [DOI] [PubMed] [Google Scholar]
- 4. Holmes KA, Hurtado A, Brown GD, Launchbury R, Ross-Innes CS, Hadfield J, Odom DT, Carroll JS. Transducin-like enhancer protein 1 mediates estrogen receptor binding and transcriptional activity in breast cancer cells. Proc Natl Acad Sci U S A 2012; 109:2748-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jennings BH, Pickles LM, Wainwright SM, Roe SM, Pearl LH, Ish-Horowicz D. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol Cell 2006; 22:645-55. [DOI] [PubMed] [Google Scholar]
- 6. Paroush Z, Finley RL, Jr., Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 1994; 79:805-15. [DOI] [PubMed] [Google Scholar]
- 7. Kaul A, Schuster E, Jennings B. The Groucho Co-repressor is Primarily Recruited to Local Target Sites in Active Chromatin to Attenuate Transcription. PLoS Genet 2014; 10:e1004595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. . Unlocking the secrets of the genome. Nature 2009; 459:927-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. . Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 2010; 143:212-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barolo S, Levine M. hairy mediates dominant repression in the Drosophila embryo. Embo J 1997; 16:2883-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dubnicoff T, Valentine SA, Chen G, Shi T, Lengyel JA, Paroush Z, Courey AJ. Conversion of Dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev 1997; 11:2952-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nibu Y, Zhang H, Levine M. Local action of long-range repressors in the Drosophila embryo. Embo J 2001; 20:2246-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene 2000; 249:1-16. [DOI] [PubMed] [Google Scholar]
- 14. Martinez CA, Arnosti DN. Spreading of a corepressor linked to action of long-range repressor Hairy. Mol Cell Biol 2008; 28:2792-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mohammed H, D'Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, Rueda OM, Holmes KA, Theodorou V, Robinson JL, et al. . Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep 2013; 3:342-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes Dev 1999; 13:2218-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li LM, Arnosti DN. Long- and short-range transcriptional repressors induce distinct chromatin states on repressed genes. Curr Biol 2011; 21:406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Winkler CJ, Ponce A, Courey AJ. Groucho-mediated repression may result from a histone deacetylase-dependent increase in nucleosome density. PLoS One 2010; 5:e10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mannervik M, Levine M. The Rpd3 histone deacetylase is required for segmentation of the Drosophila embryo. Proc Natl Acad Sci U S A 1999; 96:6797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wheeler JC, VanderZwan C, Xu X, Swantek D, Tracey WD, Gergen JP. Distinct in vivo requirements for establishment versus maintenance of transcriptional repression. Nat Genet 2002; 32:206-10. [DOI] [PubMed] [Google Scholar]
- 21. Beumer KJ, Carroll D. Targeted genome engineering techniques in Drosophila. Methods 2014; 68:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jennings BH. Pausing for thought: Disrupting the early transcription elongation checkpoint leads to developmental defects and tumourigenesis. Bioessays 2013; 35:553-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet 2013; 47:483-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gaertner B, Zeitlinger J. RNA polymerase II pausing during development. Development 2014; 141:1179-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol 2008; 28:3290-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li J, Gilmour DS. Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. EMBO J 2013; 32:1829-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Housden BE, Fu AQ, Krejci A, Bernard F, Fischer B, Tavare S, Russell S, Bray SJ. Transcriptional dynamics elicited by a short pulse of notch activation involves feed-forward regulation by E(spl)/Hes genes. PLoS Genet 2013; 9:e1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nguyen D, Krueger BJ, Sedore SC, Brogie JE, Rogers JT, Rajendra TK, Saunders A, Matera AG, Lis JT, Uguen P, et al. . The Drosophila 7SK snRNP and the essential role of dHEXIM in development. Nucleic Acids Res 2012; 40:5283-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li J, Liu Y, Rhee HS, Ghosh SK, Bai L, Pugh BF, Gilmour DS. Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol Cell 2013; 50:711-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EE. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 2014; 512:96-100. [DOI] [PubMed] [Google Scholar]


