Abstract
Transcriptional regulation is pivotal for development and differentiation of organisms. Transcription of eukaryotic protein-coding genes by RNA polymerase II (Pol II) initiates at the core promoter. Core promoters, which encompass the transcription start site, may contain functional core promoter elements, such as the TATA box, initiator, TCT and downstream core promoter element. TRF2 (TATA-box-binding protein-related factor 2) does not bind TATA box-containing promoters. Rather, it is recruited to core promoters via sequences other than the TATA box. We review the recent findings implicating TRF2 as a basal transcription factor in the regulation of diverse biological processes and specialized transcriptional programs.
Key words: RNA Pol II transcription, TBP-related factors, TRF2, core promoter elements/motifs, DPE, TCT, histone gene cluster, embryonic development, spermiogenesis, ribosomal protein genes
Abbreviations
- BREd
downstream TFIIB recognition element
- BREu
upstream TFIIB recognition element
- ChIP
Chromatin immunoprecipitation
- DPE
downstream core promoter element
- Inr
initiator
- MTE
motif ten element
- PIC
preinitiation complex
- Pol II
RNA polymerase II
- TAF
TBP-associated factor
- TBP
TATA-box binding protein
- TFIIA (transcription factor
RNA polymerase II A)
- TFIIB (transcription factor
RNA polymerase II B)
- TFIID (transcription factor
RNA polymerase II D)
- TRF
TATA-box-binding protein-related factor
- TSS
transcription start site
Introduction
Transcriptional regulation is pivotal for the proper function of multiple cellular processes and signaling pathways. In eukaryotes, transcription of protein-coding genes by Pol II initiates by formation of the preinitiation complex (PIC) at the core promoter (for a review see ref. 1). The PIC is a multisubunit complex composed of Pol II and several basal transcription factors, also termed general transcription factors (or GTFs), which ensure accurate initiation of transcription. But how general are the general transcription factors? Several studies published in the last few years suggest that genes with specific core promoter composition recruit a specialized basal transcription factor, demonstrating the diversity of transcriptional regulation. This is the story of TRF2, TATA-box-binding protein-related factor 2, which was shown to be essential for transcription of genes having a unique role in multiple biological processes, including early embryonic development and differentiation.
Transcription initiation can occur in a focused manner (at a single nucleotide or within a narrow region of several nucleotides), a dispersed manner (at multiple weak start sites over a broad region of about 50 to 100 nucleotides) or in a manner that combines both focused and dispersed transcription initiation. Focused core promoters encompass the RNA start site and are typically 80 nucleotides in length (−40 to +40 relative to the +1 transcription start site). 2-5 Core promoters may contain one or more functional DNA sequence elements, termed core promoter elements or motifs, such as the TATA box, TFIIB recognition elements (BREu and BREd), initiator (Inr), TCT motif, motif 10 element (MTE), and downstream core promoter element (DPE), which confer specific properties to the core promoter (for a review see 3, 4, 6). There is no universal core promoter composition. Notably, dispersed promoters generally lack TATA, BRE, MTE and DPE motifs. 5, 7, 8 From this point onwards, this review will mainly relate to studies performed with focused core promoters.
The TATA box is the first eukaryotic element discovered.9 Its consensus sequence is TATAWAAR, where the upstream T is located about −31/−30 relative to the A+1 of the transcription start site (TSS). The TATA box is conserved from archaeabacteria to humans. 10 Although the TATA box is well-known and extensively studied, it is only present in 10%-15% of mammalian core promoters, and in about 20% of Drosophila genes. 7, 11-15 The BRE motifs function together with the TATA box. The BREu is located immediately upstream of the TATA box (with a SSRCGCC consensus sequence), and the BREd is located immediately downstream of the TATA box (with a RTDKKKK consensus sequence). 16-18 The initiator (Inr) is the most commonly occurring motif among core promoter elements. 12-15, 19 The Inr encompasses the TSS 20 and its consensus is YYANWYY in humans and TCAKTY in Drosophila. Focused transcription generally starts at the A nucleotide of the Inr consensus that is referred to as “+1” of the TSS (whether transcription starts at this nucleotide or nearby). The TCT motif is a polypyrimidine initiator located from −2 to +6 relative to the TSS, which has been identified in genes encoding ribosomal proteins as well as other proteins involved in translation. 21 Both the MTE and DPE motifs are located at a specific position relative to the Inr (with strict spacing requirements from the A+1 of the Inr) and their function depends on the Inr. The MTE is located from +18 to +27 relative to the A+1 of the Inr, and its consensus sequence is CSARCSSAAC. 19, 22 It is conserved from Drosophila to humans and functions cooperatively with the Inr. 22, 23 The DPE, which has been identified in Drosophila as a TFIID-bound downstream core promoter element in promoters lacking a TATA box, is precisely located from +28 to +33 relative to the A+1 nucleotide of the Inr and has a functional range set of DSWYVY. 24-26 The DPE is conserved from Drosophila to humans. 25 The DPE has been shown to be prevalent among developmentally regulated genes, such as the Hox gene network and Dorsal target genes, 15, 27 suggesting that the composition of the core promoter is a major contributor to transcriptional regulation.
Pol II is a 12-subunit molecular machine that catalyzes the synthesis of RNA from the template DNA. It is, however, unable to discriminate between the core promoter region and other DNA regions. Pol II is recruited to the core promoter by the basal transcription machinery (TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH; for a review see ref. 1). TFIID, a protein complex composed of the TBP (TATA-box binding protein) and TBP-associated factors (TAFs), is the first protein complex that recognizes the core promoter: TBP binds the TATA-box motif, TAF1 and TAF2 bind the Inr motif and TAF6 and TAF9 bind the DPE and MTE motifs. 23, 25, 28-31 Several tissue-specific variants of TAFs have been discovered: TAF4 variants are important for ovarian development and spermatogenesis in mice, while Drosophila and human TAF5 and TAF7 paralogs are implicated in male gametogenesis, thus providing the TFIID with unique functions in a tissue-specific transcription environment. 32-37 Following the binding of TFIID, TFIIA binds and stabilizes the association of TFIID with the TATA-box. The formation of the TFIID-TFIIA-DNA complex is followed by the sequential binding of TFIIB, Pol II/TFIIF, TFIIE and TFIIH, resulting in the assembly of the PIC. The hierarchical recruitment of the basal transcription factors to the core promoter was discovered using the TATA box-containing adenovirus major late promoter. Remarkably, these basal transcription factors, which are necessary for TATA box-dependent transcription, are insufficient to transcribe DPE-dependent promoters. 38, 39 Moreover, whereas TBP binds and activates TATA-dependent transcription, it down regulates DPE-dependent transcription. 40 NC2 and MOT1, which are positive regulators of DPE-dependent transcription, counteract TBP and relieve its inhibition of DPE transcription. 40-42 Hence, the known basal transcription factors are not “general” and additional basal transcription factors that support DPE-dependent transcription exist.
TBP-related factors
The C-terminal core domain of TBP contains two structural repeats that fold into a saddle-like structure that is essential for the interaction with the TATA box. 29-31 Using low-stringency hybridization, two distinct TBPs were identified in Arabidopsis thaliana and then in maize.43, 44 Since then, three TBP-related factors (TRFs; TRF1, TRF2 and TRF3) have been discovered in the animal kingdom based on their homology to the C-terminal core domain of TBP (for a review see refs. 45-49). The existence of the variety of TRFs is another manifestation of the diversity and complexity of transcriptional regulation.
TFR1 exists in Drosophila and Anopheles, but not in yeast or humans. 50, 51 TRF1 has 63% identity to the C-terminal core domain repeats of TBP. In vitro experiments showed that TRF1 can bind to TATA-box together with TFIIA and TFIIB, and can efficiently replace TBP in transcription of TATA-dependent promoters. In addition, whole genome ChIP analysis demonstrated that TRF1 mainly activates transcription through Pol III, and a minor group of genes transcribed by Pol II. Drosophila TRF1 associates with Brf1, a TFIIB-related Pol III transcription factor to form a 300 kDa complex. 51
TRF3 (also known as TBP2 (TATA-binding protein 2) and TBPL2 (TBP-like protein 2)), which is the TRF that is most closely related to TBP, is unique to vertebrates. 52-55 Similarly to TBP and TRF1, TRF3 can associate with TFIIA and TFIIB. 52, 54 TRF3 binds TATA-box containing promoters and activates them. TRF3 has been shown to be important for initiation of hematopoiesis during zebrafish embryogenesis. 53, 56 TRF3 has also been shown to form a complex with TAF3 and play a role in the ex-vivo differentiation of mouse myoblast to myotubes. 57, 58. As skeletal muscle differentiation is unaffected in TRF3 knockout mice 59, the involvement of TRF3 in terminal differentiation in different species awaits further investigation. Importantly, TRF3 of Xenopus and zebrafish is mainly expressed in oocytes and is essential for embryogenesis. 52, 54 Mouse TRF3, which is exclusively expressed in oocytes, is essential for the differentiation of female germ cells but not for embryonic development.59
TRF2 is the TRF protein with the least similarity to TBP. 60-65 In 1999-2000, when different research groups have cloned TRF2 from multiple species, different names were coined. The name TRF2 was used for Drosophila 64 and humans65, as it followed the discovery of TRF1 in Drosophila. It was also named TLP (TATA-like protein; identified in mouse and humans) 62, 63, TLF (TBP-like factor; identified in C. elegans)60, 66, TRP (TBP-related protein; identified in humans)61 and TBPL1 (TBP-like 1; L is used by the Human Gene Nomenclature Committee to denote paralogs of named genes), based on its homology to TBP and perhaps, taking into account that there is no vertebrate TRF1. TRF2 is involved in Pol II transcription. 65 Similarly to TRF1 and TRF3, TRF2 can directly interact with TFIIA 61, 65, 67 and TFIIB. 61 Interestingly, Drosophila trf2 encodes two protein products - a short (632 aa) protein, and a protein of 1715aa (which will henceforth be referred to as long TRF2), in which the same short amino acid sequence is preceded by an N-terminal domain composed of coil-coiled motifs. 68 Translation of the short isoform probably results from an internal translation initiation by an IRES mechanism.68 Both proteins show similarity to the C-terminal core domain of TBP. The long TRF2 has only been identified in Drosophila. The short TRF2, which is highly conserved in evolution, 45, 55, 61, 64, 69, 70 has been extensively studied and will henceforth be referred to as TRF2. The TRF2 core domain shows 40% identity and 60% similarity to the TBP core domain. 64 Despite the homology between the TRF2 and TBP core domains, the TATA-interacting Phe residues of TBP are not conserved in TRF2, and indeed TRF2 cannot bind the TATA-box. 61, 64, 70 Hence, the variety of TBP-related factors adds another level of complexity to the regulation of transcription. As discussed below, recent evidence suggests that TRF2 can direct Pol II to subsets of TATA-less promoters and mediate diverse biological processes and transcriptional programs (Figure 1).
Figure 1.
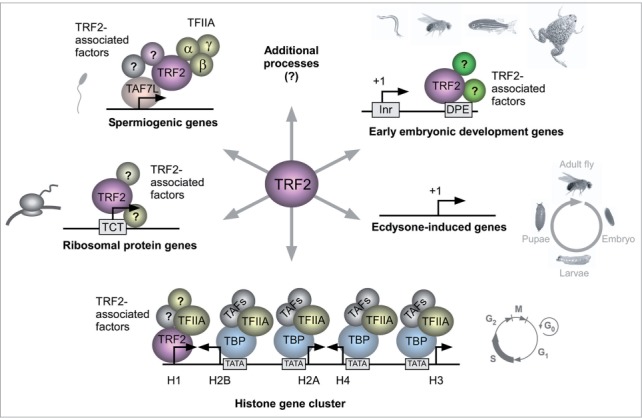
Schematic model depicting the regulation of diverse biological processes and transcriptional programs by TRF2.
TRF2 is involved in transcription of specific pathways
In Drosophila, TRF2 was shown to bind polytene chromosomes at sites distinct from those of TBP, suggesting that TRF2 regulates a subset of genes that differ from TBP-regulated genes. 64, 71 Specifically, TRF2 has been shown to regulate the TATA-less Histone H1 gene, whereas TBP regulates the core histone genes. 71 Recent single cell imaging analysis of the endogenous histone gene cluster in Drosophila cells, showed differential transcription kinetics of TRF2-directed histone H1 gene expression (transcribed throughout S phase) versus TBP-directed core histones gene expression (only transcribed in a short pulse during early S phase). 72
Furthermore, ChIP-chip analysis revealed that Drosophila TRF2, but not TBP, is associated with a large group of TATA-less core promoters, including core promoters of ribosomal protein genes. 71 The core promoters of most ribosomal protein genes in Drosophila and humans contain a functional TCT motif, which is not recognized by the TBP/TFIID complex. 21 Remarkably, TRF2, but not TBP, has recently been shown to mediate the transcription of ribosomal protein genes that lack a TATA box and have functional TCT motifs. 73 In vitro transcription analysis using Drosophila TRF2-depleted embryonic nuclear extracts demonstrated that transcription of four TCT-dependent ribosomal genes (RpL30, RpLP1, RpS12 and RpS15) was reduced, while no effect on transcription of TATA-dependent genes was observed. 73 Moreover, the addition of recombinant TRF2 to the TRF2-depleted extracts restored the transcriptional activity, further supporting the hypothesis that TRF2 regulates transcription of specific pathways.
ChIP-seq analysis of TRF2 on early (2 to 4 h) Drosophila embryos showed a peak of TRF2 in the TSS region. 73 Interestingly, MEME analysis performed on the top TRF2-bound genes revealed enrichment of the DPE motif, while no enriched motif was identified in the least TRF2-bound genes. 74 Drosophila short TRF2 was recently identified by a biochemical complementation analysis as an enriched factor in fractions that support DPE-dependent transcription. 74 Microarray analysis of Drosophila S2R+ cells that over-express inducible short TRF2 identified multiple DPE-dependent targets. In vitro transcription followed by primer extension analysis of four TRF2 target genes (pvf2, sodh-2, bgm and inx3) indicated that their transcription was highly dependent on an intact DPE motif. 74 Furthermore, using microfluidics affinity analysis, protein extracts from S2R+ cells that over-express inducible TRF2, were shown to bind these DPE-containing promoters in a sequence-specific manner. Taken together, these findings suggest existence of specialized transcriptional systems that do not involve TBP and regulate diverse biological pathways.
TRF2 is involved in embryonic development, differentiation and morphogenesis
TRF2 has been shown to be essential for embryonic development of C. elegans, Drosophila, zebrafish and Xenopus. 60, 66, 68, 75, 76 Interestingly, Xenopus TRF2 was shown to play a role in the expression of catabolic genes during embryonic development. 77 Recent analysis of Drosophila genes with high TRF2 occupancy revealed a strong relationship between TRF2 and processes such as cell differentiation and development. 70, 73 An evolutionary conservation analysis indicated that TRF2 evolved by duplication of the TBP gene. 70 TRF2 is highly conserved in evolution and is present in all bilaterian organisms containing three germ layers: endoderm, mesoderm and ectoderm. TRF2 was not found in any of the non-bilaterian genomes that are currently available. 70 As more ancient animals only contain two germ layers (endoderm and ectoderm) and the emergence of the bilateria was at the same evolutionary point as that of trf2, one can speculate that TRF2 is important for mesoderm formation. Notably, analysis of core promoter composition of Drosophila genes involved in embryonic development and differentiation reveals the prevalence of the DPE motif. 15, 78 Moreover, Drosophila embryos with reduced TRF2 have previously shown homeotic and segmentation defects, as well as dorsal-ventral abnormalities. 68 Unlike TRF2 in Drosophila, zebrafish and Xenopus, mouse TRF2 is not required for embryonic development. 79, 80
TRF2 is widely expressed in the adult animal. 61-63, 65 Mouse TRF2 is essential for spermiogenesis. 79-81 It was recently demonstrated that TRF2 works in concert with the tissue-specific TAF7L, which is associated with testis-specific promoters, to regulate a subset of postmeiotic genes directing spermiogenesis. 82 Notably, Drosophila TRF2 has been shown to be involved in differentiation of germ cells of both male and female. 68 Interestingly, it was recently demonstrated that Taspase1-mediated proteolytic cleavage of the TFIIAα-β precursor (into the α and β subunits of TFIIA) is necessary for the activation TRF2-specified transcriptional spermiogenic program in the juvenile and adult mouse testes. 83 Hence, TRF2 may form tissue-specific preinitiation complexes that regulate transcription of specific subsets of genes necessary for germ cell differentiation.
Drosophila trf2 has been shown to be required for transcriptional and developmental responses to ecdysone during Drosophila metamorphosis. 84 Hypomorphic trf2 mutations display defects in major ecdysone-triggered biological responses, including puparium formation, anterior spiracle eversion, gas bubble translocation, adult head eversion, and larval salivary gland cell death. Drosophila trf2 appears to be required for the proper timing and levels of ecdysone-regulated gene expression required for entry into metamorphosis. 84 Additional support for the involvement of TRF2 in Drosophila metamorphosis comes from RNAi depletion of TRF2 in larval salivary glands that results in a significant reduction in the sizes of the cells and the glands. 71 Although these mutant embryos develop to the third instar larval stage, a majority of them fail to pupate or die during pupal stages. Hence, even though a detailed mechanism has not been demonstrated, TRF2 is involved in the regulation of Drosophila metamorphosis.
Conclusions and future perspectives
Recent findings have highlighted the involvement of TRF2 in the regulation of diverse biological processes and specialized transcription programs (Figure 1). It is possible that TRF2 regulates additional pathways and systems that remain to be discovered.
Historically, promoters have been classified as TATA box-containing or TATA-less. It is now clear that it takes much more than the presence of a TATA box to define the characteristics of a core promoter. Likewise, it is clear that there is no universal transcription machinery and the term general transcription machinery should be replaced by basal transcription machinery. Since its discovery, it has been known that TRF2 does not bind TATA-containing DNA. In fact, based on multiple studies in the last 15 years implying that TRF2 regulates specialized transcription systems, the name TBP-related factor 2, rather than TBP-like factor (TLF) or TBP-like protein (TLP), seems more appropriate. Hence, based on functionality, rather than homology to TBP, we suggest adopting the TRF terminology for the TBP family of proteins.
It remains to be determined whether TRF2 binds sequence-specific DNA directly. The Kadonaga Lab did not observe sequence-specific DNA binding under an extensive range of conditions with many different template DNAs and methodologies in the absence or presence of different combinations of purified TFIIA and TFIIB. 73 ChIP-seq data indicated that TRF2 binds in the vicinity of the TSS. 73 Microfluidic affinity analysis has demonstrated DNA binding of TRF2-containing complexes to DPE-containing promoters. 74 It is likely that there are TRF2-associated factors (like TBP-associated factors), which assist TRF2 in binding to its target promoters. Such TRF2-associated factors remain to be discovered.
Acknowledgments
We thank Dr. Itai Tkacz for graphic design assistance. We thank Jim Kadonaga, Yehuda Danino, Sascha Duttke, Anna Sloutskin and Hila Shir-Shapira for critical reading of the manuscript. TRF2-related research in the Juven-Gershon lab was supported by grants from the Israel Science Foundation (no. 798/10) and the European Union Seventh Framework Programme (Marie Curie International Reintegration Grant; no. 256491).
References
- 1.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Critical Reviews in Biochemistry and Molecular Biology 2006; 41:105–78. [DOI] [PubMed] [Google Scholar]
- 2.Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Current opinion in cell biology 2008; 20:253–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 2010; 339:225–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Wiley interdisciplinary reviews Developmental biology 2012; 1:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet 2012; 13:233–45. [DOI] [PubMed] [Google Scholar]
- 6.Muller F, Tora L. Chromatin and DNA sequences in defining promoters for transcription initiation. Biochimica et biophysica acta 2014; 1839:118–28. [DOI] [PubMed] [Google Scholar]
- 7.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al.. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 2006; 38:626–35. [DOI] [PubMed] [Google Scholar]
- 8.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet 2007; 8:424–36. [DOI] [PubMed] [Google Scholar]
- 9.Ph.D. thesis.. Stanford University; 1979. [Google Scholar]
- 10.Reeve JN. Archaeal chromatin and transcription. Molecular microbiology 2003; 48:587–98. [DOI] [PubMed] [Google Scholar]
- 11.Cooper SJ, Trinklein ND, Anton ED, Nguyen L, Myers RM. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome research 2006; 16:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.FitzGerald PC, Sturgill D, Shyakhtenko A, Oliver B, Vinson C. Comparative genomics of Drosophila and human core promoters. Genome biology 2006; 7:R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gershenzon NI, Trifonov EN, Ioshikhes IP. The features of Drosophila core promoters revealed by statistical analysis. BMC genomics 2006; 7:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature 2005; 436:876–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zehavi Y, Kuznetsov O, Ovadia-Shochat A, Juven-Gershon T. Core promoter functions in the regulation of gene expression of Drosophila dorsal target genes. The Journal of biological chemistry 2014; 289:11993–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng W, Roberts SG. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes & development 2005; 19:2418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng W, Roberts SG. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma 2007; 116:417–29. [DOI] [PubMed] [Google Scholar]
- 18.Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes & development 1998; 12:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohler U, Liao GC, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome biology 2002; 3:RESEARCH0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell 1989; 57:103–13. [DOI] [PubMed] [Google Scholar]
- 21.Parry TJ, Theisen JW, Hsu JY, Wang YL, Corcoran DL, Eustice M, Ohler U, Kadonaga JT. The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes & development 2010; 24:2013–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim CY, Santoso B, Boulay T, Dong E, Ohler U, Kadonaga JT. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes & development 2004; 18:1606–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theisen JW, Lim CY, Kadonaga JT. Three key subregions contribute to the function of the downstream RNA polymerase II core promoter. Molecular and cellular biology 2010; 30:3471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & development 1996; 10:711–24. [DOI] [PubMed] [Google Scholar]
- 25.Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes & development 1997; 11:3020–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutach AK, Kadonaga JT. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Molecular and cellular biology 2000; 20:4754–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juven-Gershon T, Hsu JY, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes & development 2008; 22:2823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chalkley GE, Verrijzer CP. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. The EMBO journal 1999; 18:4835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JL, Burley SK. 1.9 A resolution refined structure of TBP recognizing the minor groove of TATAAAAG. Nature structural biology 1994; 1:638–53. [DOI] [PubMed] [Google Scholar]
- 30.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 1993; 365:520–7. [DOI] [PubMed] [Google Scholar]
- 31.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature 1993; 365:512–20. [DOI] [PubMed] [Google Scholar]
- 32.Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, Morris PL, Tjian R, Richards JS. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes & development 2005; 19:794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falender AE, Shimada M, Lo YK, Richards JS. TAF4b, a TBP associated factor, is required for oocyte development and function. Dev Biol 2005; 288:405–19. [DOI] [PubMed] [Google Scholar]
- 34.Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 2001; 293:2084–7. [DOI] [PubMed] [Google Scholar]
- 35.Grive KJ, Seymour KA, Mehta R, Freiman RN. TAF4b promotes mouse primordial follicle assembly and oocyte survival. Dev Biol 2014; 392:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes & development 2001; 15:1021–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pointud JC, Mengus G, Brancorsini S, Monaco L, Parvinen M, Sassone-Corsi P, Davidson I. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. Journal of cell science 2003; 116:1847–58. [DOI] [PubMed] [Google Scholar]
- 38.Goodrich JA, Tjian R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet 2010; 11:549–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis BA, Sims RJ, 3rd, Lane WS, Reinberg D. Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Molecular cell 2005; 18:471–81. [DOI] [PubMed] [Google Scholar]
- 40.Hsu J-Y, Juven-Gershon T, Marr MT, II, Wright KJ, Tjian R, Kadonaga JT. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes & development 2008; 22:2353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Werven FJ, van Bakel H, van Teeffelen HA, Altelaar AF, Koerkamp MG, Heck AJ, Holstege FC, Timmers HT. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes & development 2008; 22:2359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willy PJ, Kobayashi R, Kadonaga JT. A basal transcription factor that activates or represses transcription. Science 2000; 290:982–5. [DOI] [PubMed] [Google Scholar]
- 43.Gasch A, Hoffmann A, Horikoshi M, Roeder RG, Chua NH. Arabidopsis thaliana contains two genes for TFIID. Nature 1990; 346:390–4. [DOI] [PubMed] [Google Scholar]
- 44.Haass MM, Feix G. Two different cDNAs encoding TFIID proteins of maize. FEBS letters 1992; 301:294–8. [DOI] [PubMed] [Google Scholar]
- 45.Akhtar W, Veenstra GJ. TBP-related factors: a paradigm of diversity in transcription initiation. Cell & bioscience 2011; 1:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller F, Demeny MA, Tora L. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. The Journal of biological chemistry 2007; 282:14685–9. [DOI] [PubMed] [Google Scholar]
- 47.Muller F, Zaucker A, Tora L. Developmental regulation of transcription initiation: more than just changing the actors. Current opinion in genetics & development 2010; 20:533–40. [DOI] [PubMed] [Google Scholar]
- 48.Reina JH, Hernandez N. On a roll for new TRF targets. Genes & development 2007; 21:2855–60. [DOI] [PubMed] [Google Scholar]
- 49.Torres-Padilla ME, Tora L. TBP homologues in embryo transcription: who does what? EMBO reports 2007; 8:1016–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen SK, Takada S, Jacobson RH, Lis JT, Tjian R. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell 1997; 91:71–83. [DOI] [PubMed] [Google Scholar]
- 51.Takada S, Lis JT, Zhou S, Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell 2000; 101:459–69. [DOI] [PubMed] [Google Scholar]
- 52.Bartfai R, Balduf C, Hilton T, Rathmann Y, Hadzhiev Y, Tora L, Orban L, Muller F. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Current biology : CB 2004; 14:593–8. [DOI] [PubMed] [Google Scholar]
- 53.Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature 2007; 450:1082–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci U S A 2004; 101:13525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Persengiev SP, Zhu X, Dixit BL, Maston GA, Kittler EL, Green MR. TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc Natl Acad Sci U S A 2003; 100:14887–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hart DO, Santra MK, Raha T, Green MR. Selective interaction between Trf3 and Taf3 required for early development and hematopoiesis. Developmental dynamics : an official publication of the American Association of Anatomists 2009; 238:2540–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Molecular cell 2008; 32:96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes & development 2007; 21:2137–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gazdag E, Santenard A, Ziegler-Birling C, Altobelli G, Poch O, Tora L, Torres-Padilla ME. TBP2 is essential for germ cell development by regulating transcription and chromatin condensation in the oocyte. Genes & development 2009; 23:2210–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dantonel JC, Quintin S, Lakatos L, Labouesse M, Tora L. TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Molecular cell 2000; 6:715–22. [DOI] [PubMed] [Google Scholar]
- 61.Moore PA, Ozer J, Salunek M, Jan G, Zerby D, Campbell S, Lieberman PM. A human TATA binding protein-related protein with altered DNA binding specificity inhibits transcription from multiple promoters and activators. Molecular and cellular biology 1999; 19:7610–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohbayashi T, Kishimoto T, Makino Y, Shimada M, Nakadai T, Aoki T, Kawata T, Niwa S, Tamura T. Isolation of cDNA, chromosome mapping, and expression of the human TBP-like protein. Biochemical and biophysical research communications 1999; 255:137–42. [DOI] [PubMed] [Google Scholar]
- 63.Ohbayashi T, Makino Y, Tamura TA. Identification of a mouse TBP-like protein (TLP) distantly related to the drosophila TBP-related factor. Nucleic acids research 1999; 27:750–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rabenstein MD, Zhou S, Lis JT, Tjian R. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc Natl Acad Sci U S A 1999; 96:4791–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teichmann M, Wang Z, Martinez E, Tjernberg A, Zhang D, Vollmer F, Chait BT, Roeder RG. Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc Natl Acad Sci U S A 1999; 96:13720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaltenbach L, Horner MA, Rothman JH, Mango SE. The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Molecular cell 2000; 6:705–13. [DOI] [PubMed] [Google Scholar]
- 67.Nakadai T, Shimada M, Shima D, Handa H, Tamura TA. Specific interaction with transcription factor IIA and localization of the mammalian TATA-binding protein-like protein (TLP/TRF2/TLF). The Journal of biological chemistry 2004; 279:7447–55. [DOI] [PubMed] [Google Scholar]
- 68.Kopytova DV, Krasnov AN, Kopantceva MR, Nabirochkina EN, Nikolenko JV, Maksimenko O, Kurshakova MM, Lebedeva LA, Yerokhin MM, Simonova OB, et al.. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Molecular and cellular biology 2006; 26:7492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dantonel JC, Wurtz JM, Poch O, Moras D, Tora L. The TBP-like factor: an alternative transcription factor in metazoa? Trends Biochem Sci 1999; 24:335–9. [DOI] [PubMed] [Google Scholar]
- 70.Duttke SH, Doolittle RF, Wang YL, Kadonaga JT. TRF2 and the evolution of the bilateria. Genes & development 2014; 28:2071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Isogai Y, Keles S, Prestel M, Hochheimer A, Tjian R. Transcription of histone gene cluster by differential core-promoter factors. Genes & development 2007; 21:2936–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guglielmi B, La Rochelle N, Tjian R. Gene-specific transcriptional mechanisms at the histone gene cluster revealed by single-cell imaging. Molecular cell 2013; 51:480–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang YL, Duttke SH, Chen K, Johnston J, Kassavetis GA, Zeitlinger J, Kadonaga JT. TRF2, but not TBP, mediates the transcription of ribosomal protein genes. Genes & development 2014; 28:1550–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kedmi A, Zehavi Y, Glick Y, Orenstein Y, Ideses D, Wachtel C, Doniger T, Waldman Ben-Asher H, Muster N, Thompson J, et al.. Drosophila TRF2 is a preferential core promoter regulator. Genes & development 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muller F, Lakatos L, Dantonel J, Strahle U, Tora L. TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Current biology : CB 2001; 11:282–7. [DOI] [PubMed] [Google Scholar]
- 76.Veenstra GJ, Weeks DL, Wolffe AP. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science 2000; 290:2312–5. [DOI] [PubMed] [Google Scholar]
- 77.Jacobi UG, Akkers RC, Pierson ES, Weeks DL, Dagle JM, Veenstra GJ. TBP paralogs accommodate metazoan- and vertebrate-specific developmental gene regulation. The EMBO journal 2007; 26:3900–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Juven-Gershon T, Hsu J-Y, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes & development 2008; 22:2823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martianov I, Fimia GM, Dierich A, Parvinen M, Sassone-Corsi P, Davidson I. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Molecular cell 2001; 7:509–15. [DOI] [PubMed] [Google Scholar]
- 80.Zhang D, Penttila TL, Morris PL, Teichmann M, Roeder RG. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 2001; 292:1153–5. [DOI] [PubMed] [Google Scholar]
- 81.Zhang D, Penttila TL, Morris PL, Roeder RG. Cell- and stage-specific high-level expression of TBP-related factor 2 (TRF2) during mouse spermatogenesis. Mechanisms of development 2001; 106:203–5. [DOI] [PubMed] [Google Scholar]
- 82.Zhou H, Grubisic I, Zheng K, He Y, Wang PJ, Kaplan T, Tjian R. Taf7l cooperates with Trf2 to regulate spermiogenesis. Proc Natl Acad Sci U S A 2013; 110:16886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oyama T, Sasagawa S, Takeda S, Hess RA, Lieberman PM, Cheng EH, Hsieh JJ. Cleavage of TFIIA by Taspase1 activates TRF2-specified mammalian male germ cell programs. Developmental cell 2013; 27:188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bashirullah A, Lam G, Yin VP, Thummel CS. dTrf2 is required for transcriptional and developmental responses to ecdysone during Drosophila metamorphosis. Developmental dynamics : an official publication of the American Association of Anatomists 2007; 236:3173–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


